Stress Modifies the Expression of Glucocorticoid-Responsive Genes by Acting at Epigenetic Levels in the Rat Prefrontal Cortex: Modulatory Activity of Lurasidone
Abstract
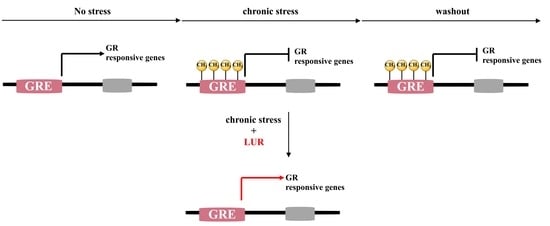
1. Introduction
2. Results
2.1. Effect of Chronic Mild Stress and Lurasidone Treatment on Body Weight and Sucrose Intake Measured in the Sucrose Consumption Test
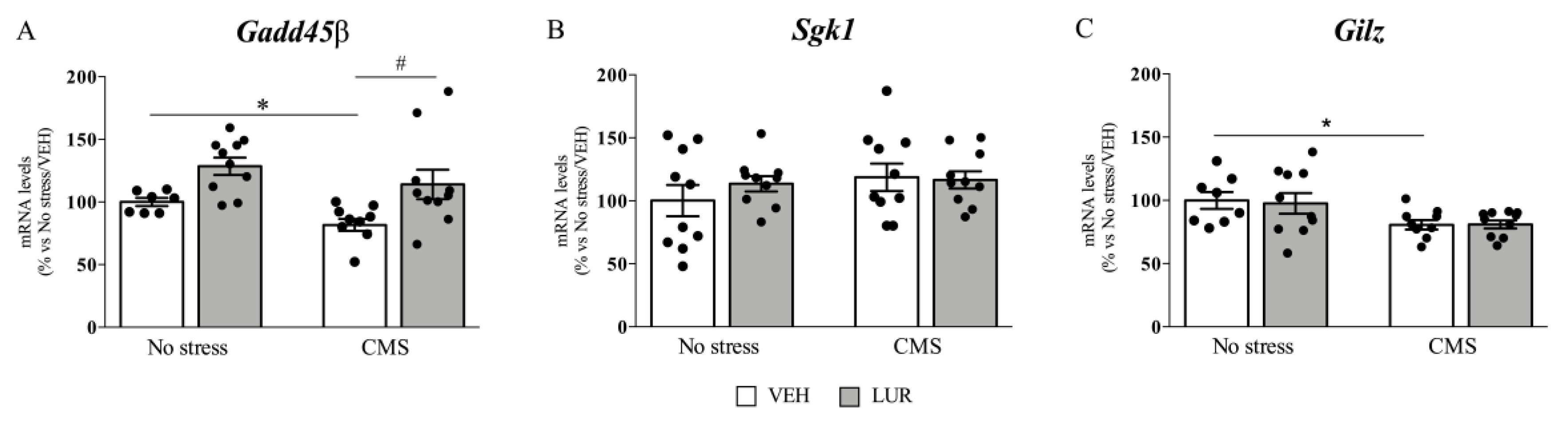
2.2. Modulation of Gadd45β, Sgk1, and Gilz Gene Expression of Rats Exposed to the CMS and Treated with Lurasidone
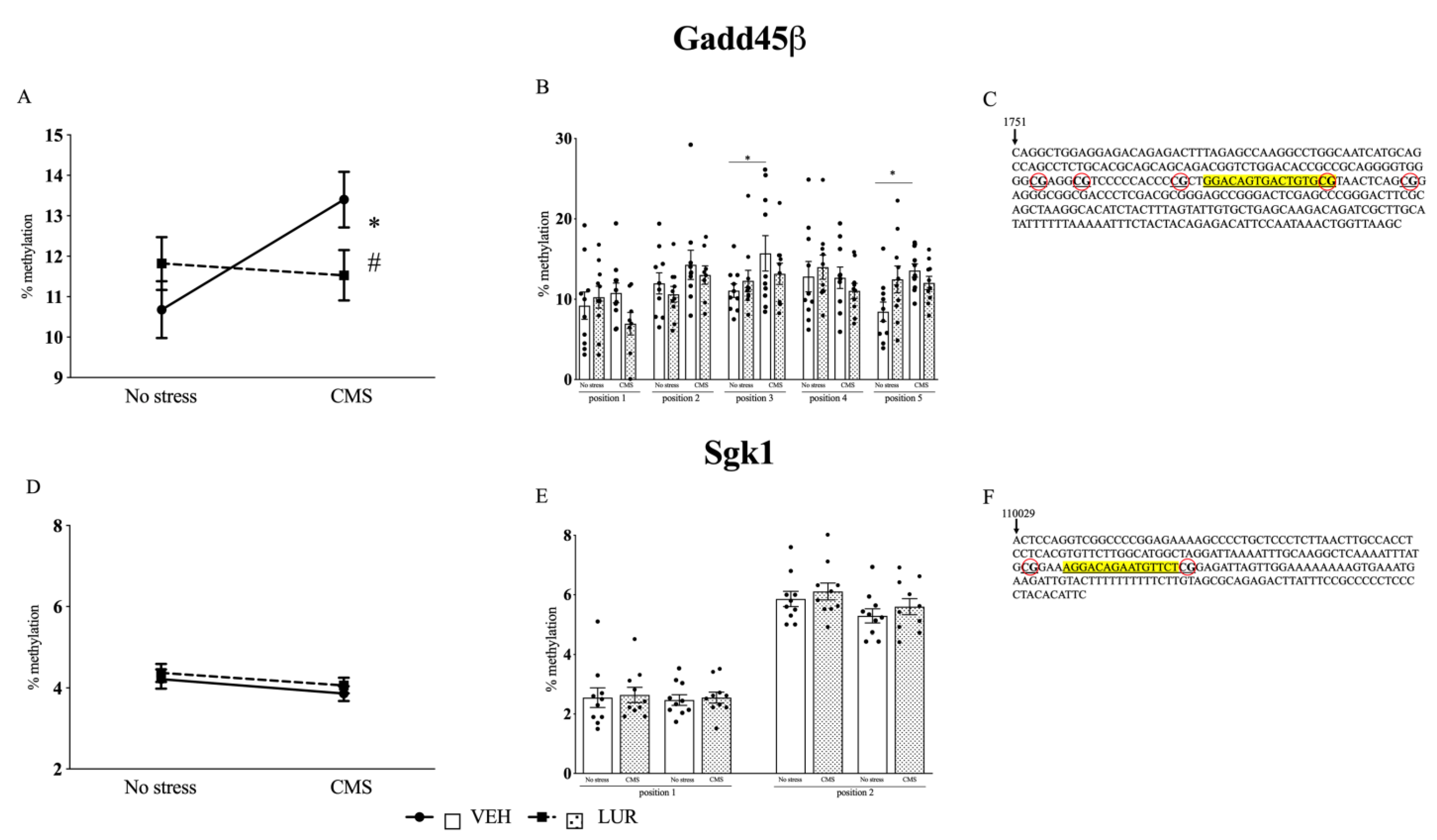
2.3. Modulation of Gadd45β and Sgk1 DNA Methylation Levels in the PFC of Rats Exposed to the CMS and Treated with Lurasidone
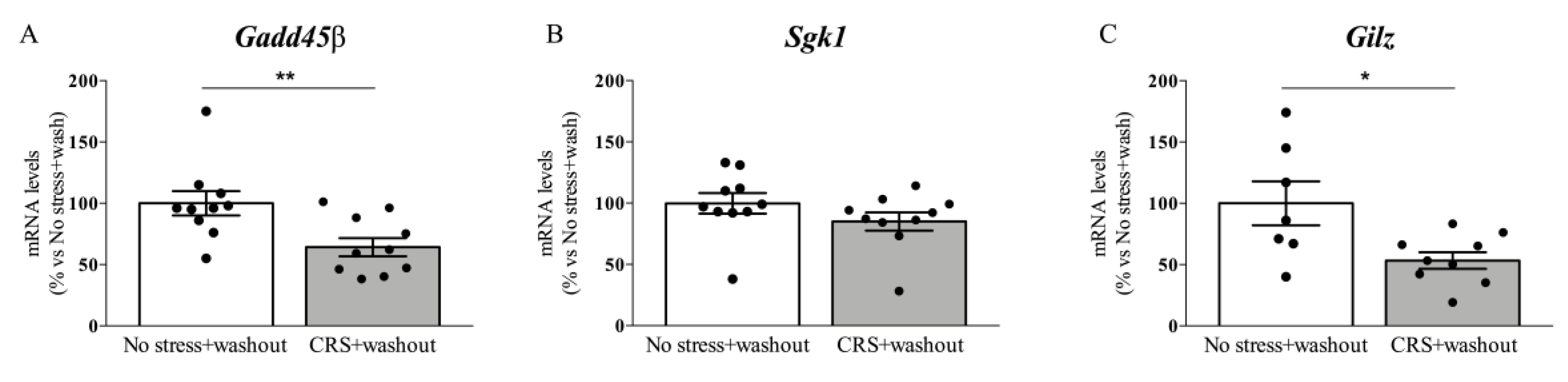
2.4. Analysis of Gadd45β Sgk1, and Gilz Gene Expression in the Prefrontal Cortex of Chronically Stressed (CRS) Rats after a Period of Washout
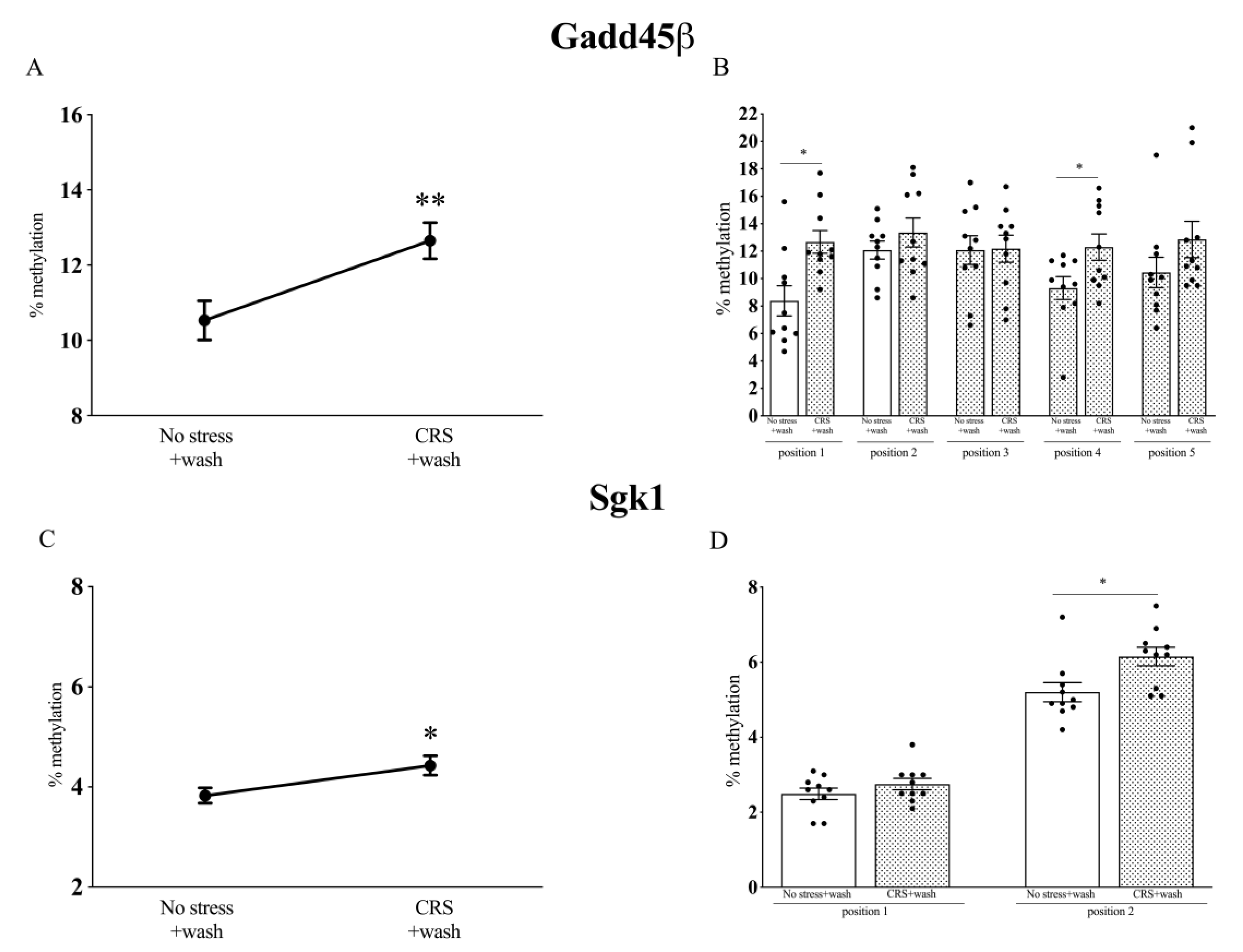
2.5. Analysis of Gadd45β and Sgk1 DNA Methylation Levels in Chronically Stressed (CRS) Rats after a Period of Washout
2.6. Effect of Stress and Lurasidone Treatment on miRNAs Expression
3. Discussion
4. Materials and Methods
4.1. Chronic Stress Procedure
4.2. Drug Treatment
4.3. Sucrose Consumption Test and Body Weight
4.4. Bioinformatic for miRNA/Integrated miRNA/mRNA Network Analysis
4.5. RNA Preparation and Real Time RT-PCR
4.6. DNA Extraction, Bisulfite Treatment, and DNA Methylation Analyses
4.7. Data and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Caspi, A. Influence of Life Stress on Depression: Moderation by a Polymorphism in the 5-HTT Gene. Science 2003, 301, 386–389. [Google Scholar] [CrossRef]
- Fava, M.; Kendler, K.S. Major Depressive Disorder. Neuron 2000, 28, 335–341. [Google Scholar] [CrossRef]
- McEwen, B.S.; Bowles, N.P.; Gray, J.D.; Hill, M.N.; Hunter, R.G.; Karatsoreos, I.N.; Nasca, C. Mechanisms of stress in the brain. Nat. Neurosci. 2015, 18, 1353–1363. [Google Scholar] [CrossRef]
- Holsboer, F. The Corticosteroid Receptor Hypothesis of Depression. Neuropsychopharmacology 2000, 23, 477–501. [Google Scholar] [CrossRef]
- McEwen, B.S. Protective and damaging effects of stress mediators: Central role of the brain. Dialogues Clin. Neurosci. 2006. [Google Scholar] [CrossRef]
- Calabrese, F.; Brivio, P.; Sbrini, G.; Gruca, P.; Lason, M.; Litwa, E.; Niemczyk, M.; Papp, M.; Riva, M.A. Effect of lurasidone treatment on chronic mild stress-induced behavioural deficits in male rats: The potential role for glucocorticoid receptor signalling. J. Psychopharmacol. 2020, 34, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, V.; Nestler, E.J. Linking Molecules to Mood: New Insight into the Biology of Depression. Am. J. Psychiatry 2010, 167, 1305–1320. [Google Scholar] [CrossRef]
- Frieling, H.; Tadić, A. Value of genetic and epigenetic testing as biomarkers of response to antidepressant treatment. Int. Rev. Psychiatry 2013, 25, 572–578. [Google Scholar] [CrossRef]
- Szyf, M.; McGowan, P.O.; Turecki, G.; Meaney, M.J. The Social Environment and the Epigenome. In Formative Experiences; Worthman, C.M., Plotsky, P.M., Schechter, D.S., Cummings, C.A., Eds.; Cambridge University Press: Cambridge, UK, 2010; pp. 53–81. ISBN 9780511711879. [Google Scholar]
- Dwivedi, Y. Evidence demonstrating role of microRNAs in the etiopathology of major depression. J. Chem. Neuroanat. 2011, 42, 142–156. [Google Scholar] [CrossRef]
- Baudry, A.; Mouillet-Richard, S.; Schneider, B.; Launay, J.-M.; Kellermann, O. MiR-16 Targets the Serotonin Transporter: A New Facet for Adaptive Responses to Antidepressants. Science 2010, 329, 1537–1541. [Google Scholar] [CrossRef] [PubMed]
- Farrell, C.; O’Keane, V. Epigenetics and the glucocorticoid receptor: A review of the implications in depression. Psychiatry Res. 2016, 242, 349–356. [Google Scholar] [CrossRef]
- Auger, C.J.; Auger, A.P. Permanent and plastic epigenesis in neuroendocrine systems. Front. Neuroendocrinol. 2013, 34, 190–197. [Google Scholar] [CrossRef]
- Zovkic, L.B.; Meadows, J.P.; Kaas, G.A.; Sweatt, J.D. Interindividual variability in stress susceptibility: A role for epigenetic mechanisms in PTSD. Front. Psychiatry 2013, 4, 60. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, V.; Nestler, E.J. The molecular neurobiology of depression. Nature 2008, 455, 894–902. [Google Scholar] [CrossRef]
- Uchida, S.; Yamagata, H.; Seki, T.; Watanabe, Y. Epigenetic mechanisms of major depression: Targeting neuronal plasticity. Psychiatry Clin. Neurosci. 2018, 72, 212–227. [Google Scholar] [CrossRef] [PubMed]
- Zannas, A.S.; Chrousos, G.P. Epigenetic programming by stress and glucocorticoids along the human lifespan. Mol. Psychiatry 2017, 22, 640–646. [Google Scholar] [CrossRef]
- Vinet, L.; Zhedanov, A. A “missing” family of classical orthogonal polynomials. Neuropsychopharmacology 2010. [Google Scholar] [CrossRef]
- Weaver, I.C.G.; Cervoni, N.; Champagne, F.A.; D’Alessio, A.C.; Sharma, S.; Seckl, J.R.; Dymov, S.; Szyf, M.; Meaney, M.J. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004, 7, 847–854. [Google Scholar] [CrossRef]
- Murgatroyd, C.; Patchev, A.V.; Wu, Y.; Micale, V.; Bockmühl, Y.; Fischer, D.; Holsboer, F.; Wotjak, C.T.; Almeida, O.F.X.; Spengler, D. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat. Neurosci. 2009, 12, 1559–1566. [Google Scholar] [CrossRef]
- Shimizu, S.; Tanaka, T.; Takeda, T.; Tohyama, M.; Miyata, S. The Kampo Medicine Yokukansan Decreases MicroRNA-18 Expression and Recovers Glucocorticoid Receptors Protein Expression in the Hypothalamus of Stressed Mice. Biomed Res. Int. 2015, 2015, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Vreugdenhil, E.; Verissimo, C.S.L.; Mariman, R.; Kamphorst, J.T.; Barbosa, J.S.; Zweers, T.; Champagne, D.L.; Schouten, T.; Meijer, O.C.; Ron de Kloet, E.; et al. MicroRNA 18 and 124a Down-Regulate the Glucocorticoid Receptor: Implications for Glucocorticoid Responsiveness in the Brain. Endocrinology 2009, 150, 2220–2228. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; D’Arcy, C.; Li, X.; Zhang, T.; Joober, R.; Meng, X. What do DNA methylation studies tell us about depression? A systematic review. Transl. Psychiatry 2019, 9, 68. [Google Scholar] [CrossRef]
- Story Jovanova, O.; Nedeljkovic, I.; Spieler, D.; Walker, R.M.; Liu, C.; Luciano, M.; Bressler, J.; Brody, J.; Drake, A.J.; Evans, K.L.; et al. DNA Methylation Signatures of Depressive Symptoms in Middle-aged and Elderly Persons. JAMA Psychiatry 2018, 75, 949. [Google Scholar] [CrossRef] [PubMed]
- Farrell, C.; Doolin, K.; O’ Leary, N.; Jairaj, C.; Roddy, D.; Tozzi, L.; Morris, D.; Harkin, A.; Frodl, T.; Nemoda, Z.; et al. DNA methylation differences at the glucocorticoid receptor gene in depression are related to functional alterations in hypothalamic–pituitary–adrenal axis activity and to early life emotional abuse. Psychiatry Res. 2018, 265, 341–348. [Google Scholar] [CrossRef]
- Anacker, C.; Cattaneo, A.; Musaelyan, K.; Zunszain, P.A.; Horowitz, M.; Molteni, R.; Luoni, A.; Calabrese, F.; Tansey, K.; Gennarelli, M.; et al. Role for the kinase SGK1 in stress, depression, and glucocorticoid effects on hippocampal neurogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 8708–8713. [Google Scholar] [CrossRef]
- Gavin, D.P.; Sharma, R.P.; Chase, K.A.; Matrisciano, F.; Dong, E.; Guidotti, A. Growth Arrest and DNA-Damage-Inducible, Beta (GADD45b)-Mediated DNA Demethylation in Major Psychosis. Neuropsychopharmacology 2012, 37, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Ayroldi, E.; Riccardi, C. Glucocorticoid-induced leucine zipper (GILZ): A new important mediator of glucocorticoid action. FASEB J. 2009, 23, 3649–3658. [Google Scholar] [CrossRef]
- Itani, O.A.; Liu, K.Z.; Cornish, K.L.; Campbell, J.R.; Thomas, C.P. Glucocorticoids stimulate human sgk1 gene expression by activation of a GRE in its 5′-flanking region. Am. J. Physiol. Endocrinol. Metab. 2002. [Google Scholar] [CrossRef]
- Muzikar, K.A.; Nickols, N.G.; Dervan, P.B. Repression of DNA-binding dependent glucocorticoid receptor-mediated gene expression. Proc. Natl. Acad. Sci. 2009, 106, 16598–16603. [Google Scholar] [CrossRef] [PubMed]
- Anacker, C.; Zunszain, P.A.; Cattaneo, A.; Carvalho, L.A.; Garabedian, M.J.; Thuret, S.; Price, J.; Pariante, C.M. Antidepressants increase human hippocampal neurogenesis by activating the glucocorticoid receptor. Mol. Psychiatry 2011, 16, 738–750. [Google Scholar] [CrossRef]
- Asselin-Labat, M.-L.; David, M.; Biola-Vidamment, A.; Lecoeuche, D.; Zennaro, M.-C.; Bertoglio, J.; Pallardy, M. GILZ, a new target for the transcription factor FoxO3, protects T lymphocytes from interleukin-2 withdrawal–induced apoptosis. Blood 2004, 104, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, F.; Savino, E.; Papp, M.; Molteni, R.; Riva, M.A. Chronic mild stress-induced alterations of clock gene expression in rat prefrontal cortex: Modulatory effects of prolonged lurasidone treatment. Pharmacol. Res. 2016, 104, 140–150. [Google Scholar] [CrossRef]
- Calabrese, F.; Brivio, P.; Gruca, P.; Lason-Tyburkiewicz, M.; Papp, M.; Riva, M.A. Chronic Mild Stress-Induced Alterations of Local Protein Synthesis: A Role for Cognitive Impairment. ACS Chem. Neurosci. 2017, 8, 817–825. [Google Scholar] [CrossRef]
- Caputo, V.; Ciolfi, A.; Macri, S.; Pizzuti, A. The Emerging Role of MicroRNA in Schizophrenia. CNS Neurol. Disord. Drug Targets 2015, 14, 208–221. [Google Scholar] [CrossRef]
- Kocerha, J.; Dwivedi, Y.; Brennand, K.J. Noncoding RNAs and neurobehavioral mechanisms in psychiatric disease. Mol. Psychiatry 2015, 20, 677–684. [Google Scholar] [CrossRef]
- Luoni, A.; Riva, M.A. MicroRNAs and psychiatric disorders: From aetiology to treatment. Pharmacol. Ther. 2016, 167, 13–27. [Google Scholar] [CrossRef]
- Grassi, D.; Franz, H.; Vezzali, R.; Bovio, P.; Heidrich, S.; Dehghanian, F.; Lagunas, N.; Belzung, C.; Krieglstein, K.; Vogel, T. Neuronal Activity, TGFβ-Signaling and Unpredictable Chronic Stress Modulate Transcription of Gadd45 Family Members and DNA Methylation in the Hippocampus. Cereb. Cortex 2017, 27, 4166–4181. [Google Scholar] [CrossRef]
- Szyf, M.; Tang, Y.-Y.; Hill, K.G.; Musci, R. The dynamic epigenome and its implications for behavioral interventions: A role for epigenetics to inform disorder prevention and health promotion. Transl. Behav. Med. 2016, 6, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Numata, S.; Ishii, K.; Tajima, A.; Iga, J.; Kinoshita, M.; Watanabe, S.; Umehara, H.; Fuchikami, M.; Okada, S.; Boku, S.; et al. Blood diagnostic biomarkers for major depressive disorder using multiplex DNA methylation profiles: Discovery and validation. Epigenetics 2015, 10, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Pandey, G.N.; Rizavi, H.S.; Ren, X.; Dwivedi, Y.; Palkovits, M. Region-specific alterations in glucocorticoid receptor expression in the postmortem brain of teenage suicide victims. Psychoneuroendocrinology 2013, 38, 2628–2639. [Google Scholar] [CrossRef] [PubMed]
- Frodl, T.; Carballedo, A.; Hughes, M.M.; Saleh, K.; Fagan, A.; Skokauskas, N.; McLoughlin, D.M.; Meaney, J.; O’Keane, V.; Connor, T.J. Reduced expression of glucocorticoid-inducible genes GILZ and SGK-1: High IL-6 levels are associated with reduced hippocampal volumes in major depressive disorder. Transl. Psychiatry 2012, 2, e88. [Google Scholar] [CrossRef] [PubMed]
- Brivio, P.; Sbrini, G.; Corsini, G.; Paladini, M.S.; Racagni, G.; Molteni, R.; Calabrese, F. Chronic Restraint Stress Inhibits the Response to a Second Hit in Adult Male Rats: A Role for BDNF Signaling. Int. J. Mol. Sci. 2020, 21, 6261. [Google Scholar] [CrossRef] [PubMed]
- Luoni, A.; Macchi, F.; Papp, M.; Molteni, R.; Riva, M.A. Lurasidone Exerts Antidepressant Properties in the Chronic Mild Stress Model through the Regulation of Synaptic and Neuroplastic Mechanisms in the Rat Prefrontal Cortex. Int. J. Neuropsychopharmacol. 2015, 18. [Google Scholar] [CrossRef]
- Piechota, M.; Korostynski, M.; Golda, S.; Ficek, J.; Jantas, D.; Barbara, Z.; Przewlocki, R. Transcriptional signatures of steroid hormones in the striatal neurons and astrocytes. BMC Neurosci. 2017. [Google Scholar] [CrossRef]
- McGrath, J.C.; Lilley, E. Implementing guidelines on reporting research using animals (ARRIVE etc.): New requirements for publication in BJP. Br. J. Pharmacol. 2015, 172, 3189–3193. [Google Scholar] [CrossRef]
- Paxinos, G.; Watson, C. The Rat Brain in Stereotaxic Coordinates the New Coronal Set; Elsevier: Amsterdam, The Netherlands, 2004; ISBN 9780120884728. [Google Scholar]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4. [Google Scholar] [CrossRef]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Vlachos, I.S.; Vergoulis, T.; Reczko, M.; Filippidis, C.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-microT web server v5.0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013, 41, W169–W173. [Google Scholar] [CrossRef] [PubMed]
- Reczko, M.; Maragkakis, M.; Alexiou, P.; Grosse, I.; Hatzigeorgiou, A.G. Functional microRNA targets in protein coding sequences. Bioinformatics 2012, 28, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Liu, W.; Wang, X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef]
- Brivio, P.; Sbrini, G.; Riva, M.A.; Calabrese, F. Acute Stress Induces Cognitive Improvement in the Novel Object Recognition Task by Transiently Modulating Bdnf in the Prefrontal Cortex of Male Rats. Cell. Mol. Neurobiol. 2020, 40, 1037–1047. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Bollati, V.; Baccarelli, A.; Hou, L.; Bonzini, M.; Fustinoni, S.; Cavallo, D.; Byun, H.-M.; Jiang, J.; Marinelli, B.; Pesatori, A.C.; et al. Changes in DNA Methylation Patterns in Subjects Exposed to Low-Dose Benzene. Cancer Res. 2007, 67, 876–880. [Google Scholar] [CrossRef] [PubMed]
- Pariante, C.M.; Lightman, S.L. The HPA axis in major depression: Classical theories and new developments. Trends Neurosci. 2008, 31, 464–468. [Google Scholar] [CrossRef] [PubMed]
| (a) | No Stress | CMS | ||
| Vehicle | Lurasidone | Vehicle | Lurasidone | |
| miR-452-3p (U6) | 100 ± 20 | 74 ± 14 | 91 ± 25 | 86 ± 18 |
| miR-19a-3p (U6) | 100 ± 12 | 178 ± 24 * | 116 ± 15 | 231 ± 32 ## |
| miR-19b-3p (U6) | 100 ± 16 | 85 ± 13 | 114 ± 25 | 115 ± 21 |
| miR-143-3p (U6) | 100 ± 13 | 104 ± 17 | 61 ± 11 | 95 ± 13 |
| (b) | No Stress | CRS + Washout | ||
| miR-452-3p (U6) | 100 ± 26 | 62 ± 15 | ||
| miR-19a-3p (U6) | 100 ± 21 | 105 ± 25 | ||
| miR-19b-3p (U6) | 100 ± 20 | 80 ± 16 | ||
| miR-143-3p (U6) | 100 ± 13 | 63 ± 10 * | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brivio, P.; Sbrini, G.; Tarantini, L.; Parravicini, C.; Gruca, P.; Lason, M.; Litwa, E.; Favero, C.; Riva, M.A.; Eberini, I.; et al. Stress Modifies the Expression of Glucocorticoid-Responsive Genes by Acting at Epigenetic Levels in the Rat Prefrontal Cortex: Modulatory Activity of Lurasidone. Int. J. Mol. Sci. 2021, 22, 6197. https://doi.org/10.3390/ijms22126197
Brivio P, Sbrini G, Tarantini L, Parravicini C, Gruca P, Lason M, Litwa E, Favero C, Riva MA, Eberini I, et al. Stress Modifies the Expression of Glucocorticoid-Responsive Genes by Acting at Epigenetic Levels in the Rat Prefrontal Cortex: Modulatory Activity of Lurasidone. International Journal of Molecular Sciences. 2021; 22(12):6197. https://doi.org/10.3390/ijms22126197
Chicago/Turabian StyleBrivio, Paola, Giulia Sbrini, Letizia Tarantini, Chiara Parravicini, Piotr Gruca, Magdalena Lason, Ewa Litwa, Chiara Favero, Marco Andrea Riva, Ivano Eberini, and et al. 2021. "Stress Modifies the Expression of Glucocorticoid-Responsive Genes by Acting at Epigenetic Levels in the Rat Prefrontal Cortex: Modulatory Activity of Lurasidone" International Journal of Molecular Sciences 22, no. 12: 6197. https://doi.org/10.3390/ijms22126197
APA StyleBrivio, P., Sbrini, G., Tarantini, L., Parravicini, C., Gruca, P., Lason, M., Litwa, E., Favero, C., Riva, M. A., Eberini, I., Papp, M., Bollati, V., & Calabrese, F. (2021). Stress Modifies the Expression of Glucocorticoid-Responsive Genes by Acting at Epigenetic Levels in the Rat Prefrontal Cortex: Modulatory Activity of Lurasidone. International Journal of Molecular Sciences, 22(12), 6197. https://doi.org/10.3390/ijms22126197