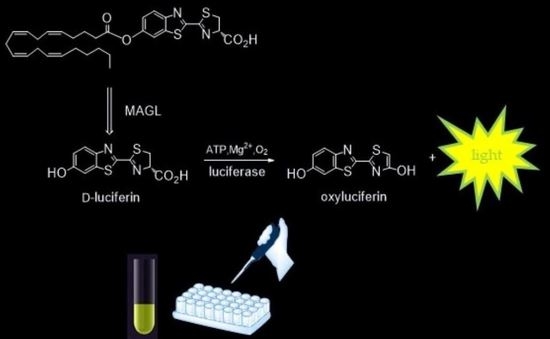
A New Ultrasensitive Bioluminescence-Based Method for Assaying Monoacylglycerol Lipase
Abstract
:1. Introduction
2. Results and Discussion
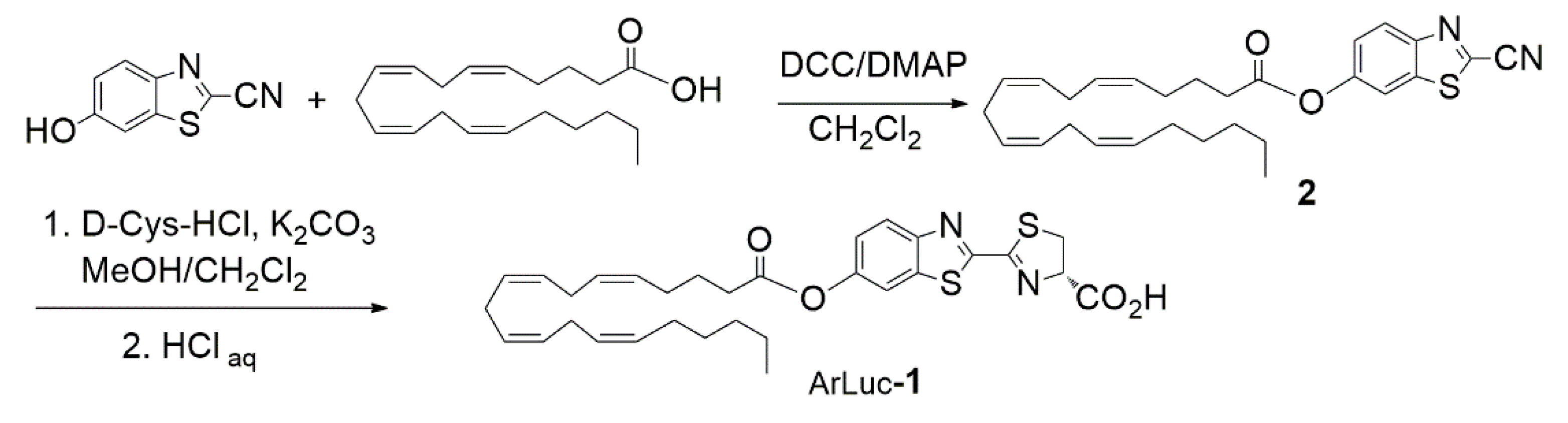
2.1. Design and Synthesis of the Probe
2.2. PLG2 Expression and Purification
2.3. PLG2 Activity: Effects of DMSO and ArLuc-1
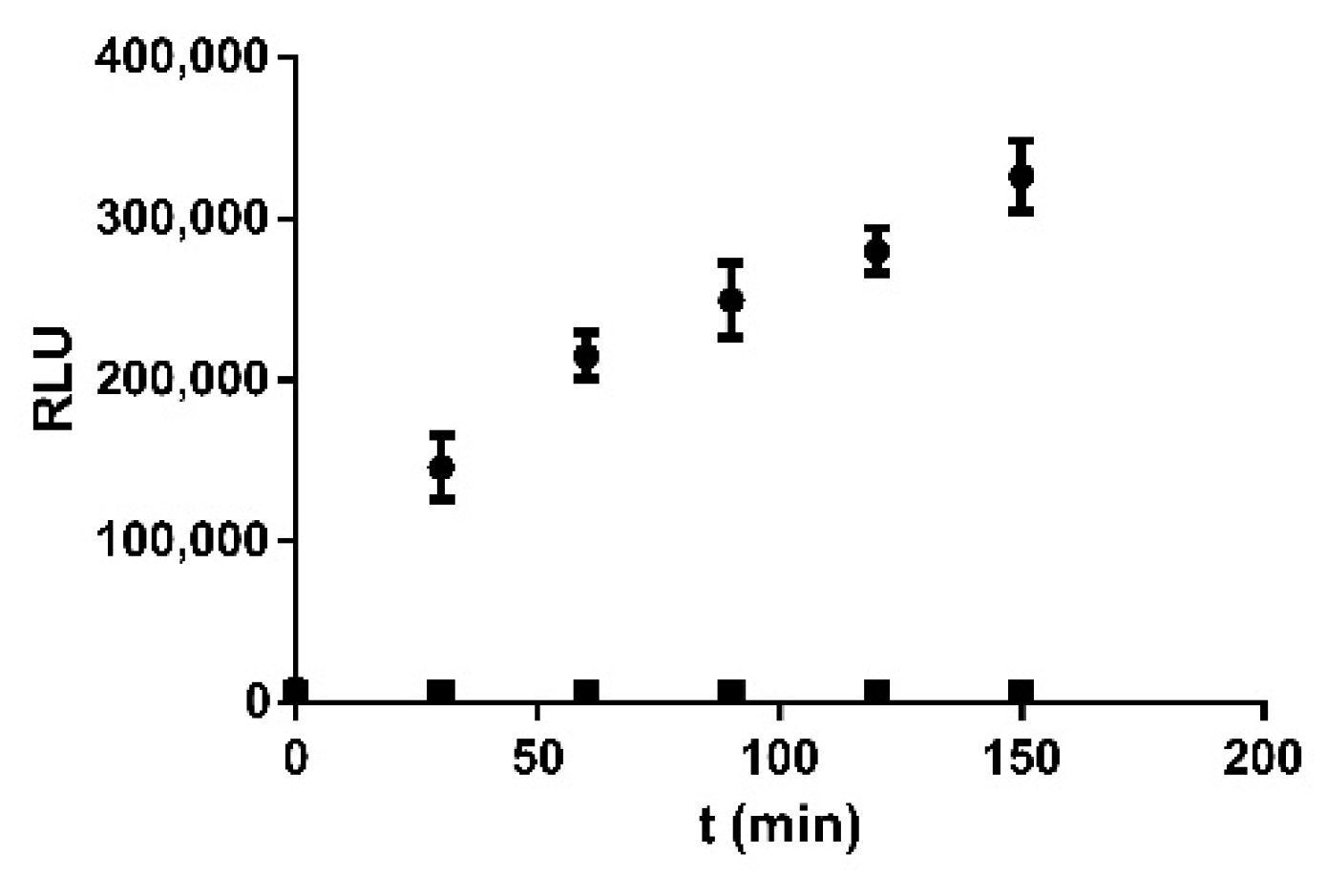
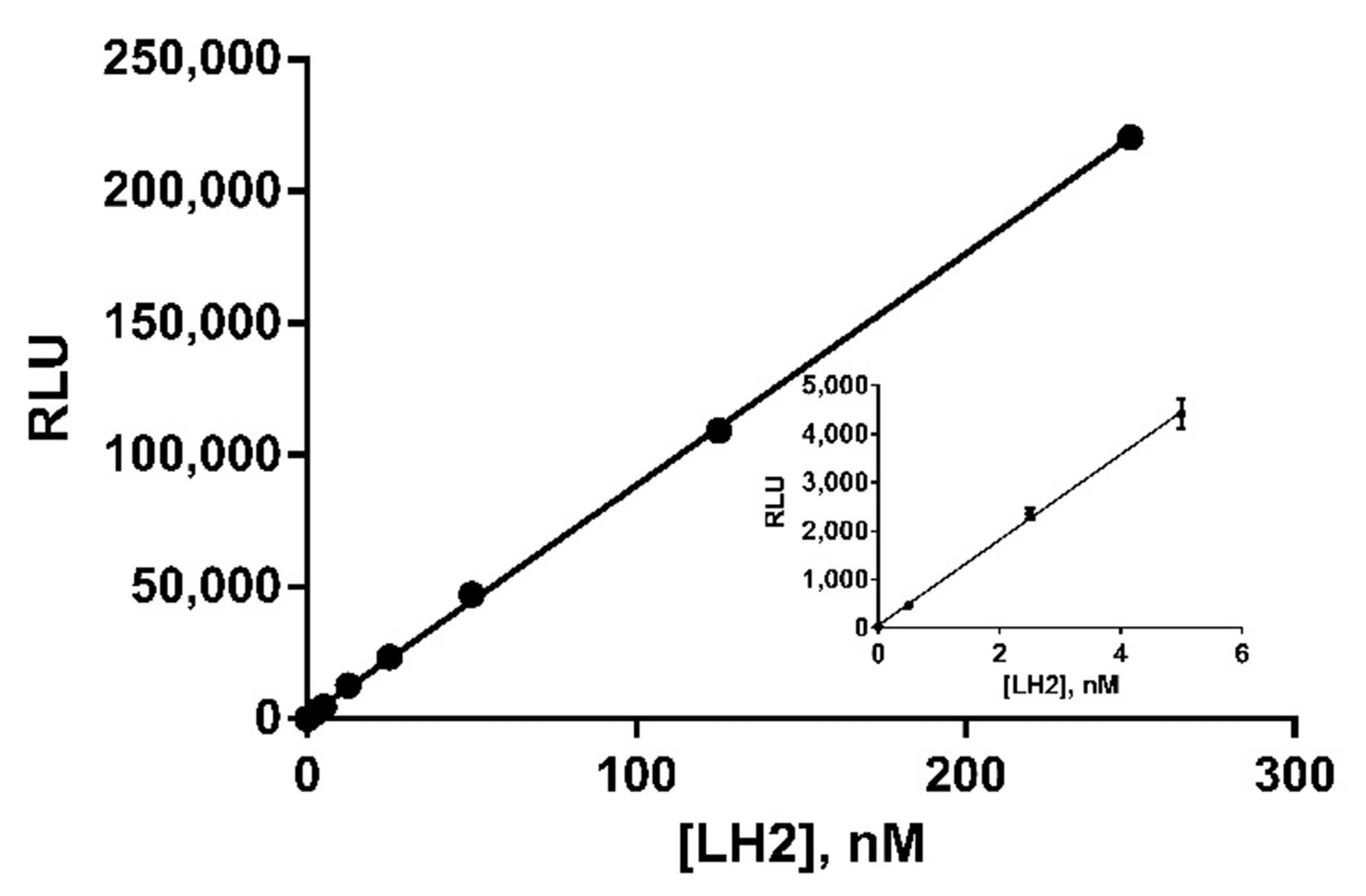
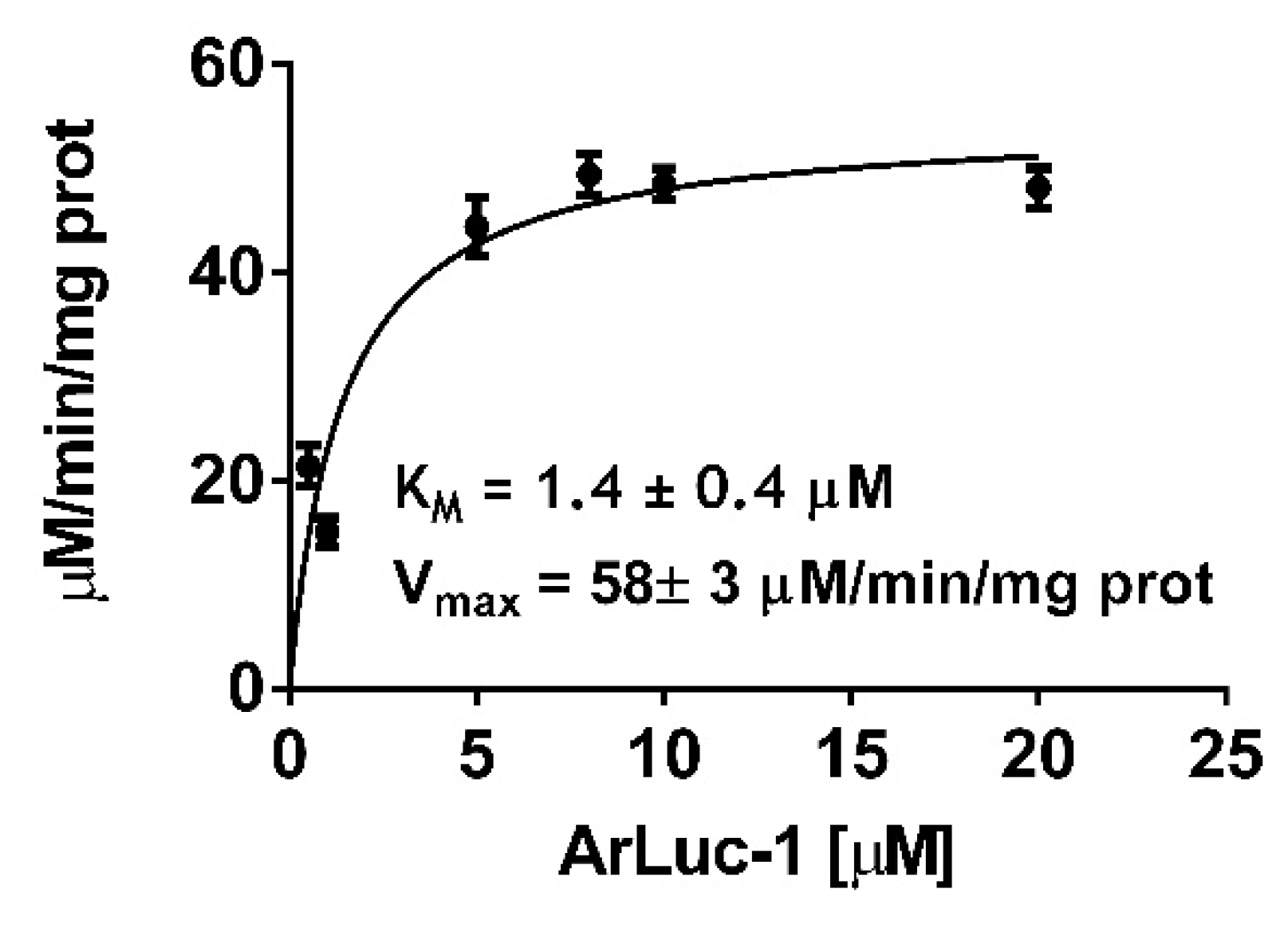
2.4. ArLuc-1 as MAGL Substrate: Assay Development and Validation
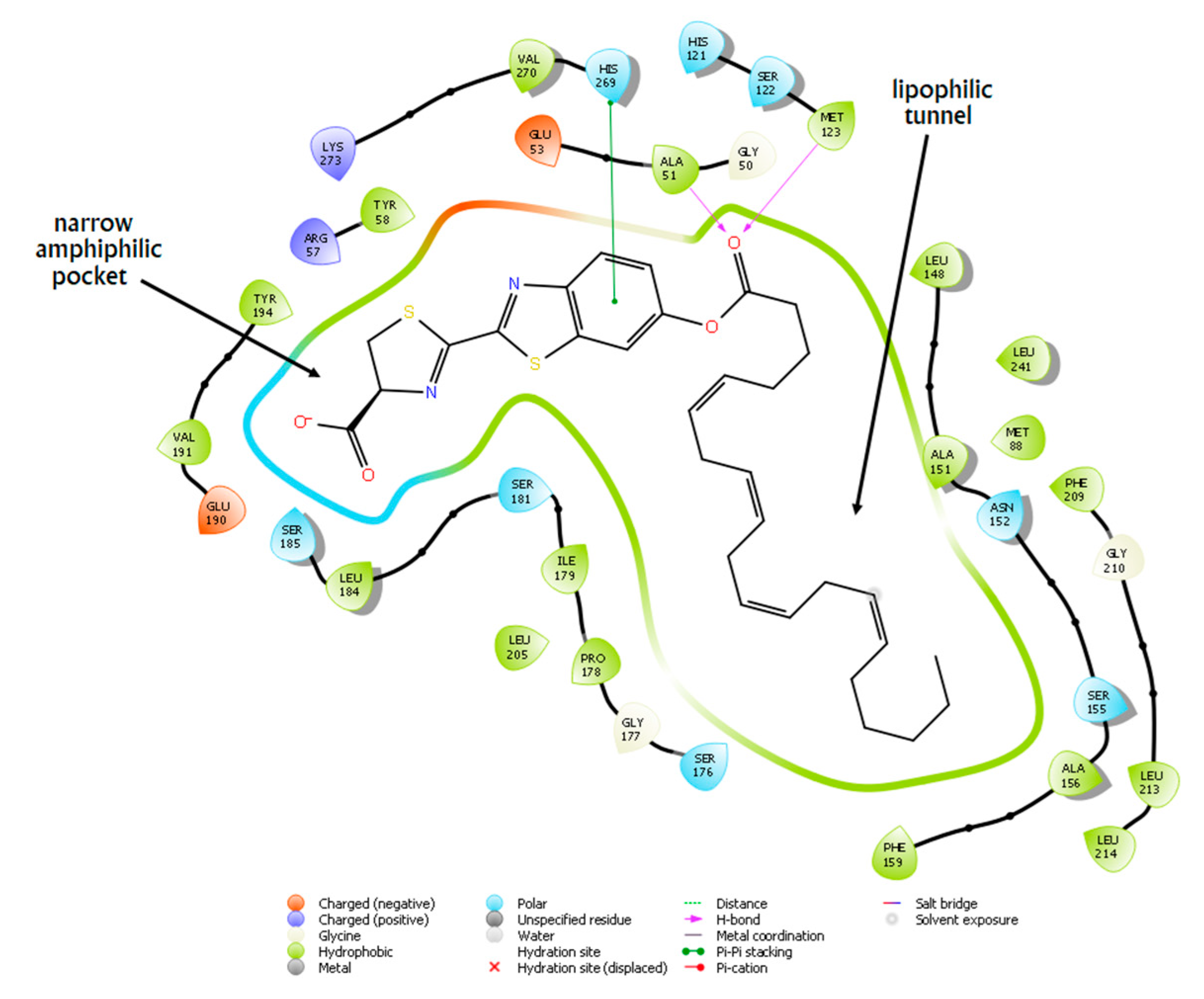
2.5. Docking Studies
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Instruments
3.3. Synthesis of 6-O-Arachidonoylluciferin (ArLuc-1)
3.4. Stability of the Substrate
3.5. PLG2 Expression and Purification
3.6. Stock, Working, and Enzymatic Assay Solutions
3.7. PLG2 DMSO Trials
3.8. PLG2 ArLuc-1 Trials
3.9. Linearity Tests and Sensitivity of the Method
3.10. MAGL Bioluminescent Assays and Kinetic Tests
3.11. MAGL Assay Validation
3.12. In silico Molecular Docking Simulations
3.13. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Joshi, N.; Onaivi, E.S. Endocannabinoid System Components: Overview and Tissue Distribution. Adv. Exp. Med. Biol. 2019, 1162, 1–12. [Google Scholar] [CrossRef]
- Meccariello, R. Endocannabinoid System in Health and Disease: Current Situation and Future Perspectives. Int. J. Mol. Sci. 2019, 1162, 3549. [Google Scholar] [CrossRef]
- Freitas, H.R.; Isaac, A.R.; Malcher-Lopes, R.; Diaz, B.L.; Trevenzoli, I.H.; de Melo Reis, R.A. Polyunsaturated fatty acids and endocannabinoids in health and disease. Nutr. Neurosci. 2018, 21, 695–714. [Google Scholar] [CrossRef]
- Ottria, R.; Cappelletti, L.; Ravelli, A.; Mariotti, M.; Gigli, F.; Romagnoli, S.; Ciuffreda, P.; Banfi, G.; Drago, L. Plasma endocannabinoid behaviour in total knee and hip arthroplasty. J. Biol. Regul. Homeost. Agents 2016, 30, 1147–1152. [Google Scholar] [PubMed]
- Pezzilli, R.; Ciuffreda, P.; Ottria, R.; Ravelli, A.; Melzi d’Eril, G.; Barassi, A. Serum endocannabinoids in assessing pain in patients with chronic pancreatitis and in those with pancreatic ductal adenocarcinoma. Scand. J. Gastroenterol. 2017, 52, 1133–1139. [Google Scholar] [CrossRef] [PubMed]
- Woodhams, S.G.; Sagar, D.R.; Burston, J.J.; Chapman, V. The role of the endocannabinoid system in pain. In Pain Control. Handbook of Experimental Pharmacology; Schaible, H.G., Ed.; Springer: Berlin/Heidelberg, Germany, 2015; Volume 227, pp. 119–143. [Google Scholar] [CrossRef]
- Cristino, L.; Bisogno, T.; Di Marzo, V. Cannabinoids and the expanded endocannabinoid system in neurological disorders. Nat. Rev. Neurol. 2020, 16, 9–29. [Google Scholar] [CrossRef] [PubMed]
- Laezza, C.; Pagano, C.; Navarra, G.; Pastorino, O.; Proto, M.C.; Fiore, D.; Piscopo, C.; Gazzerro, P.; Bifulco, M. The Endocannabinoid System: A Target for Cancer Treatment. Int. J. Mol. Sci. 2020, 21, 747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vago, R.; Ravelli, A.; Bettiga, A.; Casati, S.; Lavorgna, G.; Benigni, F.; Salonia, A.; Montorsi, F.; Orioli, M.; Ciuffreda, P.; et al. Urine Endocannabinoids as Novel Non-Invasive Biomarkers for Bladder Cancer at Early Stage. Cancers 2020, 12, 870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casati, S.; Giannasi, C.; Minoli, M.; Niada, S.; Ravelli, A.; Angeli, I.; Mergenthaler, V.; Ottria, R.; Ciuffreda, P.; Orioli, M.; et al. Quantitative Lipidomic Analysis of Osteosarcoma Cell-Derived Products by UHPLC-MS/MS. Biomolecules 2020, 10, 1302. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Immadi, S.S.; Wu, Z.; Kendall, D.A. Translational potential of allosteric modulators targeting the cannabinoid CB1 receptor. Acta Pharmacol. Sin. 2019, 40, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Morales, P.; Goya, P.; Jagerovic, N. Emerging strategies targeting CB2 cannabinoid receptor: Biased agonism and allosterism. Biochem. Pharmacol. 2018, 157, 8–17. [Google Scholar] [CrossRef]
- Vago, R.; Bettiga, A.; Salonia, A.; Ciuffreda, P.; Ottria, R. Development of new inhibitors for N-acylethanolamine-hydrolyzing acid amidase as promising tool against bladder cancer. Bioorg. Med. Chem. 2017, 25, 1242–1249. [Google Scholar] [CrossRef]
- Malamas, M.S.; Farah, S.I.; Lamani, M.; Pelekoudas, D.N.; Perry, N.T.; Rajarshi, G.; Miyabe, C.Y.; Chandrashekhar, H.; West, J.; Pavlopoulos, S.; et al. Design and synthesis of cyanamides as potent and selective N-acylethanolamine acid amidase inhibitors. Bioorg. Med. Chem. 2020, 28, 115195. [Google Scholar] [CrossRef]
- van Egmond, N.; Straub, V.M.; van der Stelt, M. Targeting Endocannabinoid Signaling: FAAH and MAG Lipase Inhibitors. Annu. Rev. Pharmacol. Toxicol. 2021, 61, 441–463. [Google Scholar] [CrossRef]
- Di Marzo, V. New approaches and challenges to targeting the endocannabinoid system. Nat. Rev. Drug Discov. 2018, 17, 623–639. [Google Scholar] [CrossRef]
- Mulvihill, M.M.; Nomura, D.K. Therapeutic potential of monoacylglycerol lipase inhibitors. Life Sci. 2013, 92, 492–497. [Google Scholar] [CrossRef] [Green Version]
- Saario, S.M.; Savinainen, J.R.; Laitinen, J.T.; Järvinen, T.; Niemi, R. Monoglyceride lipase-like enzymatic activity is responsible for hydrolysis of 2-arachidonoylglycerol in rat cerebellar membranes. Biochem. Pharmacol. 2004, 67, 1381–1387. [Google Scholar] [CrossRef] [PubMed]
- King, A.R.; Lodola, A.; Carmi, C.; Fu, J.; Mor, M.; Piomelli, D. A critical cysteine residue in monoacylglycerol lipase is targeted by a new class of isothiazolinone-based enzyme inhibitors. Br. J. Pharmacol. 2009, 157, 974–983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Chanda, P.; Jones, P.G.; Kennedy, J.D. A fluorescence-based assay for monoacylglycerol lipase compatible with inhibitor screening. Assay Drug Dev. Technol. 2008, 6, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Lauria, S.; Casati, S.; Ciuffreda, P. Synthesis and characterization of a new fluorogenic substrate for monoacylglycerol lipase and application to inhibition studies. Anal. Bioanal. Chem. 2015, 407, 8163–8167. [Google Scholar] [CrossRef]
- Miceli, M.; Casati, S.; Ottria, R.; Di Leo, S.; Eberini, I.; Palazzolo, L.; Parravicini, C.; Ciuffreda, P. Set-Up and Validation of a High Throughput Screening Method for Human Monoacylglycerol Lipase (MAGL) Based on a New Red Fluorescent Probe. Molecules 2019, 24, 2241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCaffrey, A.; Kay, M.A.; Contag, C.H. Advancing molecular therapies through in vivo bioluminescent imaging. Mol. Imaging 2003, 2, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Branchini, B.R.; Southworth, T.L. A Highly Sensitive Biosensor for ATP Using a Chimeric Firefly Luciferase. Methods Enzymol. 2017, 589, 351–364. [Google Scholar] [CrossRef]
- Niwa, K.; Ichino, Y.; Kumata, S.; Nakajima, Y.; Hiraishi, Y.; Kato, D.-I.; Viviani, V.R.; Ohmiya, Y. Quantum yields and kinetics of the firefly bioluminescence reaction of beetle luciferases. Photochem. Photobiol. 2010, 86, 1046–1049. [Google Scholar] [CrossRef]
- Ando, Y.; Niwa, K.; Yamada, N.; Enomoto, T.; Irie, T.; Kubota, H.; Ohmiya, Y.; Akiyama, H. Firefly bioluminescence quantum yield and colour change by pH-sensitive green emission. Nat. Photon 2008, 2, 44–47. [Google Scholar] [CrossRef]
- Luker, K.E.; Smith, M.C.P.; Luker, G.D.; Gammon, S.T.; Piwnica-Worms, H.; Piwnica-Worms, D. Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc. Natl. Acad. Sci. USA 2004, 101, 12288–12293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badr, C.E.; Tannous, B.A. Bioluminescence imaging: Progress and applications. Trends Biotechnol. 2011, 29, 624–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, M.; Ma, X.; Jiang, T.; Zhang, C.; Chen, H.; Gao, Y.; Yang, X.; Du, L.; Li, M. A novel coelenterate luciferin-based luminescent probe for selective and sensitive detection of thiophenols. Org. Biomol. Chem. 2016, 14, 10267–10274. [Google Scholar] [CrossRef]
- van de Bittner, G.C.; Bertozzi, C.R.; Chang, C.J. Strategy for dual-analyte luciferin imaging: In vivo bioluminescence detection of hydrogen peroxide and caspase activity in a murine model of acute inflammation. J. Am. Chem. Soc. 2013, 135, 1783–1795. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Chen, L.; Wu, W.; Zhang, W.; Ma, Z.; Cheng, Y.; Du, L.; Li, M. Discovery of bioluminogenic probes for aminopeptidase N imaging. Anal. Chem. 2014, 86, 2747–2751. [Google Scholar] [CrossRef] [PubMed]
- Feng, P.; Zhang, H.; Deng, Q.; Liu, W.; Yang, L.; Li, G.; Chen, G.; Du, L.; Ke, B.; Li, M. Real-Time Bioluminescence Imaging of Nitroreductase in Mouse Model. Anal. Chem. 2016, 88, 5610–5614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ke, B.; Wu, W.; Liu, W.; Liang, H.; Gong, D.; Hu, X.; Li, M. Bioluminescence Probe for Detecting Hydrogen Sulfide in Vivo. Anal. Chem. 2016, 88, 592–595. [Google Scholar] [CrossRef] [PubMed]
- White, E.H.; Wörther, H.; Seliger, H.H.; McElroy, W.D. Amino Analogs of Firefly Luciferin and Biological Activity Thereof1. J. Am. Chem. Soc. 1966, 88, 2015–2019. [Google Scholar] [CrossRef]
- White, E.H.; McCapra, F.; Field, G.F.; McElroy, W.D. The structure and synthesis of firefly luciferin. J. Am. Chem. Soc. 1961, 83, 2402–2403. [Google Scholar] [CrossRef]
- Bowie, L.J. Synthesis of firefly luciferin and structural analogs. In Bioluminescence and Chemiluminescence; Kaplan, N., Colowick, N., Eds.; Elsevier: Amsterdam, The Netherlands, 1978; Volume 57, pp. 15–28. ISBN 9780121819576. [Google Scholar]
- Toya, Y.; Takagi, M.; Kondo, T.; Nakata, H.; Isobe, M.; Goto, T. Improved Synthetic Methods of Firefly Luciferin Derivatives for Use in Bioluminescent Analysis of Hydrolytic Enzymes; Carboxylic Esterase and Alkaline Phosphatase. BCSJ 1992, 65, 2604–2610. [Google Scholar] [CrossRef]
- Lundin, A.; Rickardsson, A.; Thore, A. Continuous monitoring of ATP-converting reactions by purified firefly luciferase. Anal. Biochem. 1976, 75, 611–620. [Google Scholar] [CrossRef]
- Branchini, B.R.; Southworth, T.L.; Fontaine, D.M.; Kohrt, D.; Talukder, M.; Michelini, E.; Cevenini, L.; Roda, A.; Grossel, M.J. An enhanced chimeric firefly luciferase-inspired enzyme for ATP detection and bioluminescence reporter and imaging applications. Anal. Biochem. 2015, 484, 148–153. [Google Scholar] [CrossRef] [Green Version]
- ICH Q2B Validation of Analytical Procedures: Methodology; International Commission on Harmonisation: London, UK, 1996.
- Shrivastava, A.; Gupta, V. Methods for the determination of limit of detection and limit of quantitation of the analytical methods. Chron Young Sci. 2011, 2, 21. [Google Scholar] [CrossRef]
- Long, J.Z.; Li, W.; Booker, L.; Burston, J.J.; Kinsey, S.G.; Schlosburg, J.E.; Pavón, F.J.; Serrano, A.M.; Selley, D.E.; Parsons, L.H.; et al. Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nat. Chem. Biol. 2009, 5, 37–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savinainen, J.R.; Yoshino, M.; Minkkilä, A.; Nevalainen, T.; Laitinen, J.T. Characterization of binding properties of monoglyceride lipase inhibitors by a versatile fluorescence-based technique. Anal. Biochem. 2010, 399, 132–134. [Google Scholar] [CrossRef]
- Aida, J.; Fushimi, M.; Kusumoto, T.; Sugiyama, H.; Arimura, N.; Ikeda, S.; Sasaki, M.; Sogabe, S.; Aoyama, K.; Koike, T. Design, Synthesis, and Evaluation of Piperazinyl Pyrrolidin-2-ones as a Novel Series of Reversible Monoacylglycerol Lipase Inhibitors. J. Med. Chem. 2018, 61, 9205–9217. [Google Scholar] [CrossRef]
- Labar, G.; Bauvois, C.; Borel, F.; Ferrer, J.-L.; Wouters, J.; Lambert, D.M. Crystal structure of the human monoacylglycerol lipase, a key actor in endocannabinoid signaling. Chembiochem 2010, 11, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Casati, S.; Manzocchi, A.; Ottria, R.; Ciuffreda, P. 1H, 13C and 15N NMR spectral assignments of adenosine derivatives with different amino substituents at C6-position. Magn. Reson. Chem. 2011, 49, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Casati, S.; Manzocchi, A.; Ottria, R.; Ciuffreda, P. 1H, 13C and 15N NMR assignments for N6-isopentenyladenosine/inosine analogues. Magn. Reson. Chem. 2010, 48, 745–748. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miceli, M.; Casati, S.; Allevi, P.; Berra, S.; Ottria, R.; Rota, P.; Branchini, B.R.; Ciuffreda, P. A New Ultrasensitive Bioluminescence-Based Method for Assaying Monoacylglycerol Lipase. Int. J. Mol. Sci. 2021, 22, 6148. https://doi.org/10.3390/ijms22116148
Miceli M, Casati S, Allevi P, Berra S, Ottria R, Rota P, Branchini BR, Ciuffreda P. A New Ultrasensitive Bioluminescence-Based Method for Assaying Monoacylglycerol Lipase. International Journal of Molecular Sciences. 2021; 22(11):6148. https://doi.org/10.3390/ijms22116148
Chicago/Turabian StyleMiceli, Matteo, Silvana Casati, Pietro Allevi, Silvia Berra, Roberta Ottria, Paola Rota, Bruce R. Branchini, and Pierangela Ciuffreda. 2021. "A New Ultrasensitive Bioluminescence-Based Method for Assaying Monoacylglycerol Lipase" International Journal of Molecular Sciences 22, no. 11: 6148. https://doi.org/10.3390/ijms22116148
APA StyleMiceli, M., Casati, S., Allevi, P., Berra, S., Ottria, R., Rota, P., Branchini, B. R., & Ciuffreda, P. (2021). A New Ultrasensitive Bioluminescence-Based Method for Assaying Monoacylglycerol Lipase. International Journal of Molecular Sciences, 22(11), 6148. https://doi.org/10.3390/ijms22116148