Gut Serpinome: Emerging Evidence in IBD
Abstract
1. Introduction
2. Impact of Inflammatory Bowel Diseases and Available Treatments
3. Role of the Gut Microbiota in the Pathogenesis of IBD
4. Proteases as Key Targets in Intestinal Inflammation
4.1. Role of Human Proteases in IBD
4.2. Role of Microbial Proteases in IBD
5. Serpins, Natural Inhibitors to Control the Activity of Serine Proteases
5.1. Overview of Serpins
5.2. Non-Gut Microbial Serpins
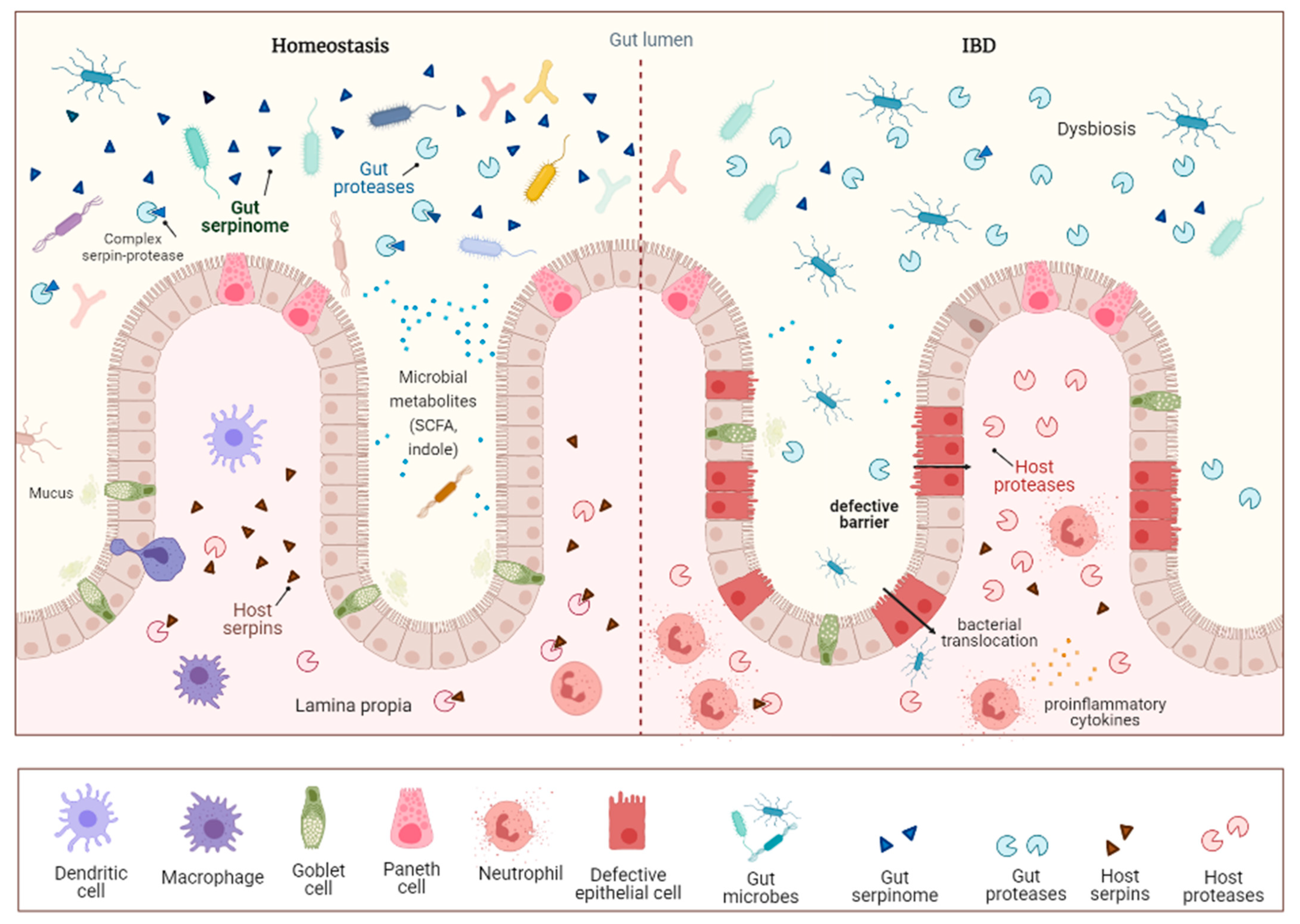
5.3. Gut Serpinome and Inflammatory Bowel Diseases
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Honda, K.; Littman, D.R. The microbiota in adaptive immune homeostasis and disease. Nature 2016, 535, 75–84. [Google Scholar] [CrossRef]
- De Angelis, M.; Ferrocino, I.; Calabrese, F.M.; De Filippis, F.; Cavallo, N.; Siragusa, S.; Rampelli, S.; Di Cagno, R.; Rantsiou, K.; Vannini, L.; et al. Diet influences the functions of the human intestinal microbiome. Sci. Rep. 2020, 10, 4247. [Google Scholar] [CrossRef] [PubMed]
- Dethlefsen, L.; Huse, S.; Sogin, M.L.; Relman, D.A. The pervasive effects of an antibiotic on the human gut microbiota as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008, 6, e280. [Google Scholar] [CrossRef]
- Wlodarska, M.; Kostic, A.D.; Xavier, R.J. An integrative view of microbiome-host interactions in inflammatory bowel diseases. Cell Host Microbe 2015, 17, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Lavelle, A.; Sokol, H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Duboc, H.; Rajca, S.; Rainteau, D.; Benarous, D.; Maubert, M.A.; Quervain, E.; Thomas, G.; Barbu, V.; Humbert, L.; Despras, G.; et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 2013, 62, 531–539. [Google Scholar] [CrossRef]
- Jablaoui, A.; Kriaa, A.; Mkaouar, H.; Akermi, N.; Soussou, S.; Wysocka, M.; Wołoszyn, D.; Amouri, A.; Gargouri, A.; Maguin, E.; et al. Fecal Serine Protease Profiling in Inflammatory Bowel Diseases. Front. Cell. Infect. Microbiol. 2020, 10, 21. [Google Scholar] [CrossRef]
- Kriaa, A.; Jablaoui, A.; Mkaouar, H.; Akermi, N.; Maguin, E.; Rhimi, M. Serine proteases at the cutting edge of IBD: Focus on gastrointestinal inflammation. FASEB J. 2020, 34, 7270–7282. [Google Scholar] [CrossRef]
- Van Spaendonk, H.; Ceuleers, H.; Witters, L.; Patteet, E.; Joossens, J.; Augustyns, K.; Lambeir, A.M.; De Meester, I.; De Man, J.G.; De Winter, B.Y. Regulation of intestinal permeability: The role of proteases. World J. Gastroenterol. 2017, 23, 2106–2123. [Google Scholar] [CrossRef] [PubMed]
- Vergnolle, N. Protease inhibition as new therapeutic strategy for GI diseases. Gut 2016, 65, 1215–1224. [Google Scholar] [CrossRef] [PubMed]
- Mkaouar, H.; Akermi, N.; Mariaule, V.; Boudebbouze, S.; Gaci, N.; Szukala, F.; Pons, N.; Marquez, J.; Gargouri, A.; Maguin, E.; et al. Siropins, novel serine protease inhibitors from gut microbiota acting on human proteases involved in inflammatory bowel diseases. Microb. Cell. Fact. 2016, 15, 201. [Google Scholar] [CrossRef]
- Motta, J.P.; Bermúdez-Humarán, L.G.; Deraison, C.; Martin, L.; Rolland, C.; Rousset, P.; Boue, J.; Dietrich, G.; Chapman, K.; Kharrat, P.; et al. Food-grade bacteria expressing elafin protect against inflammation and restore colon homeostasis. Sci. Transl. Med. 2012, 4, 158ra144. [Google Scholar] [CrossRef]
- Sánchez-Navarro, A.; González-Soria, I.; Caldiño-Bohn, R.; Bobadilla, N.A. An integrative view of serpins in health and disease: The contribution of SerpinA3. Am. J. Physiol. Cell. Physiol. 2021, 320, C106–C118. [Google Scholar] [CrossRef]
- Ng, S.C.; Shi, H.Y.; Hamidi, N.; Underwood, F.E.; Tang, W.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Wu, J.C.Y.; Chan, F.K.L.; et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: A systematic review of population-based studies. Lancet 2017, 390, 2769–2778. [Google Scholar] [CrossRef]
- Burisch, J.; Pedersen, N.; Čuković-Čavka, S.; Brinar, M.; Kaimakliotis, I.; Duricova, D.; Shonová, O.; Vind, I.; Avnstrøm, S.; Thorsgaard, N.; et al. East-West gradient in the incidence of inflammatory bowel disease in Europe: The ECCO-EpiCom inception cohort. Gut 2014, 63, 588–597. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Manne, S.; Treem, W.R.; Bennett, D. Prevalence of Inflammatory Bowel Disease in Pediatric and Adult Populations: Recent Estimates from Large National Databases in the United States, 2007-2016. Inflamm. Bowel. Dis. 2020, 26, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Mak, W.Y.; Zhao, M.; Ng, S.C.; Burisch, J. The epidemiology of inflammatory bowel disease: East meets west. J. Gastroenterol. Hepatol. 2020, 35, 380–389. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.C. Emerging Trends of Inflammatory Bowel Disease in Asia. Gastroenterol. Hepatol. 2016, 12, 193–196. [Google Scholar]
- GBD 2017 Inflammatory Bowel Disease Collaborators. The global regional and national burden of inflammatory bowel disease in 195 countries and territories 1990-2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020, 5, 17–30. [Google Scholar] [CrossRef]
- Zenlea, T.; Peppercorn, M.A. Immunosuppressive therapies for inflammatory bowel disease. World J. Gastroenterol. 2014, 20, 3146–3152. [Google Scholar] [CrossRef] [PubMed]
- Frolkis, A.D.; Dykeman, J.; Negrón, M.E.; Debruyn, J.; Jette, N.; Fiest, K.M.; Frolkis, T.; Barkema, H.W.; Rioux, K.P.; Panaccione, R.; et al. Risk of surgery for inflammatory bowel diseases has decreased over time: A systematic review and meta-analysis of population-based studies. Gastroenterology 2013, 145, 996–1006. [Google Scholar] [CrossRef]
- McLeod, R.S.; Churchill, D.N.; Lock, A.M.; Vanderburgh, S.; Cohen, Z. Quality of life of patients with ulcerative colitis preoperatively and postoperatively. Gastroenterology 1991, 101, 1307–1313. [Google Scholar] [CrossRef]
- Nordin, K.; Påhlman, L.; Larsson, K.; Sundberg-Hjelm, M.; Lööf, L. Health-related quality of life and psychological distress in a population-based sample of Swedish patients with inflammatory bowel disease. Scand. J. Gastroenterol. 2002, 37, 450–457. [Google Scholar] [CrossRef]
- Park, K.T.; Ehrlich, O.G.; Allen, J.I.; Meadows, P.; Szigethy, E.M.; Henrichsen, K.; Kim, S.C.; Lawton, R.C.; Murphy, S.M.; Regueiro, M.; et al. The Cost of Inflammatory Bowel Disease: An Initiative from the Crohn’s & Colitis Foundation. Inflamm. Bowel Dis. 2020, 26, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Human Microbiome Project Consortium. Structure function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age Environment Diet and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Marchesi, J.R. Prokaryotic and eukaryotic diversity of the human gut. Adv. Appl. Microbiol. 2010, 72, 43–62. [Google Scholar] [CrossRef] [PubMed]
- Kho, Z.Y.; Lal, S.K. The Human Gut Microbiome - A Potential Controller of Wellness and Disease. Front. Microbiol. 2018, 9, 1835. [Google Scholar] [CrossRef]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Nageshwar Reddy, D. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787–8803. [Google Scholar] [CrossRef]
- Álvarez-Mercado, A.I.; Navarro-Oliveros, M.; Robles-Sánchez, C.; Plaza-Díaz, J.; Sáez-Lara, M.J.; Muñoz-Quezada, S.; Fontana, L.; Abadía-Molina, F. Microbial Population Changes and Their Relationship with Human Health and Disease. Microorganisms 2019, 7, 68. [Google Scholar] [CrossRef]
- Casén, C.; Vebø, H.C.; Sekelja, M.; Hegge, F.T.; Karlsson, M.K.; Ciemniejewska, E.; Dzankovic, S.; Frøyland, C.; Nestestog, R.; Engstrand, L.; et al. Deviations in human gut microbiota: A novel diagnostic test for determining dysbiosis in patients with IBS or IBD. Aliment. Pharmacol. Ther. 2015, 42, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.W.; Sanderson, J.D.; Churcher, C.; Parkes, G.C.; Hudspith, B.N.; Rayment, N.; Brostoff, J.; Parkhill, J.; Dougan, G.; Petrovska, L. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011, 11, 7. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, B.; Zhang, Y.; Wei, H.; Lei, Y.; Zhao, L. Structural shifts of mucosa-associated lactobacilli and Clostridium leptum subgroup in patients with ulcerative colitis. J. Clin. Microbiol. 2007, 45, 496–500. [Google Scholar] [CrossRef] [PubMed]
- Joossens, M.; Huys, G.; Cnockaert, M.; De Preter, V.; Verbeke, K.; Rutgeerts, P.; Vandamme, P.; Vermeire, S. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut 2011, 60, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Vigsnaes, L.K.; van den Abbeele, P.; Sulek, K.; Frandsen, H.L.; Steenholdt, C.; Brynskov, J.; Vermeiren, J.; van de Wiele, T.; Licht, T.R. Microbiotas from UC patients display altered metabolism and reduced ability of LAB to colonize mucus. Sci. Rep. 2013, 3, 1110. [Google Scholar] [CrossRef] [PubMed]
- Sokol, H.; Seksik, P.; Furet, J.P.; Firmesse, O.; Nion-Larmurier, I.; Beaugerie, L.; Cosnes, J.; Corthier, G.; Marteau, P.; Doré, J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel Dis. 2009, 15, 1183–1189. [Google Scholar] [CrossRef] [PubMed]
- Ott, S.J.; Musfeldt, M.; Wenderoth, D.F.; Hampe, J.; Brant, O.; Fölsch, U.R.; Timmis, K.N.; Schreiber, S. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 2004, 53, 685–693. [Google Scholar] [CrossRef]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humarán, L.G.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef]
- Vich Vila, A.; Imhann, F.; Collij, V.; Jankipersadsing, S.A.; Gurry, T.; Mujagic, Z.; Kurilshikov, A.; Bonder, M.J.; Jiang, X.; Tigchelaar, E.F.; et al. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci. Transl. Med. 2018, 10, eaap8914. [Google Scholar] [CrossRef] [PubMed]
- Machiels, K.; Joossens, M.; Sabino, J.; De Preter, V.; Arijs, I.; Eeckhaut, V.; Ballet, V.; Claes, K.; Van Immerseel, F.; Verbeke, K.; et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014, 63, 1275–1283. [Google Scholar] [CrossRef]
- Breyner, N.M.; Michon, C.; de Sousa, C.S.; Vilas Boas, P.B.; Chain, F.; Azevedo, V.A.; Langella, P.; Chatel, J.M. Microbial Anti-Inflammatory Molecule (MAM) from Faecalibacterium prausnitzii Shows a Protective Effect on DNBS and DSS-Induced Colitis Model in Mice through Inhibition of NF-κB Pathway. Front. Microbiol. 2017, 8, 114. [Google Scholar] [CrossRef] [PubMed]
- Rossi, O.; van Berkel, L.A.; Chain, F.; Tanweer Khan, M.; Taverne, N.; Sokol, H.; Duncan, S.H.; Flint, H.J.; Harmsen, H.J.; Langella, P.; et al. Faecalibacterium prausnitzii A2-165 has a high capacity to induce IL-10 in human and murine dendritic cells and modulates T cell responses. Sci. Rep. 2016, 6, 18507. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, M.; Wang, Y.; Dorfman, R.G.; Liu, H.; Yu, T.; Chen, X.; Tang, D.; Xu, L.; Yin, Y.; et al. Faecalibacterium prausnitzii Produces Butyrate to Maintain Th17/Treg Balance and to Ameliorate Colorectal Colitis by Inhibiting Histone Deacetylase 1. Inflamm. Bowel Dis. 2018, 24, 1926–1940. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, J.R.; Holmes, E.; Khan, F.; Kochhar, S.; Scanlan, P.; Shanahan, F.; Wilson, I.D.; Wang, Y. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J. Proteome Res. 2007, 6, 546–551. [Google Scholar] [CrossRef] [PubMed]
- Bjerrum, J.T.; Wang, Y.; Hao, F.; Coskun, M.; Ludwig, C.; Günther, U.; Nielsen, O.H. Metabonomics of human fecal extracts characterize ulcerative colitis.; Crohn’s disease and healthy individuals. Metabolomics 2015, 11, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Nikolaus, S.; Schulte, B.; Al-Massad, N.; Thieme, F.; Schulte, D.M.; Bethge, J.; Rehman, A.; Tran, F.; Aden, K.; Häsler, R.; et al. Increased Tryptophan Metabolism Is Associated with Activity of Inflammatory Bowel Diseases. Gastroenterology 2017, 153, 1504–1516. [Google Scholar] [CrossRef]
- Zamani, S.; Hesam Shariati, S.; Zali, M.R.; Asadzadeh Aghdaei, H.; Sarabi Asiabar, A.; Bokaie, S.; Nomanpour, B.; Sechi, L.A.; Feizabadi, M.M. Detection of enterotoxigenic Bacteroides fragilis in patients with ulcerative colitis. Gut Pathog. 2017, 9, 53. [Google Scholar] [CrossRef] [PubMed]
- Prindiville, T.P.; Sheikh, R.A.; Cohen, S.H. Bacteroides fragilis enterotoxin gene sequences in patients with inflammatory bowel disease. Emerg. Infect. Dis. 2000, 6, 171–174. [Google Scholar] [CrossRef] [PubMed]
- Sears, C.L. Enterotoxigenic Bacteroides fragilis: A Rogue among Symbiotes. Clin. Microbiol. Rev. 2009, 22, 349–369. [Google Scholar] [CrossRef] [PubMed]
- Chung, L.; Thiele Orberg, E.; Geis, A.L.; Chan, J.L.; Fu, K.; DeStefano Shields, C.E.; Dejea, C.M.; Fathi, P.; Chen, J.; Finard, B.B.; et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe 2018, 23, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Kotlowsky, R.; Bernstein, C.N.; Sepehri, S. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut 2007, 56, 669–675. [Google Scholar] [CrossRef]
- Darfeuille-Michaud, A.; Boudeau, J.; Bulois, P.; Neut, C.; Glasser, A.L.; Barnich, N.; Bringer, M.A.; Swidsinski, A.; Beaugerie, L.; Colombel, J.F. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 2004, 127, 412–421. [Google Scholar] [CrossRef]
- Zamani, S.; Zali, M.R.; Aghdaei, H.A.; Sechi, L.A.; Niegowska, M.; Caggiu, E.; Keshavarz, R.; Mosavari, N.; Feizabadi, M.M. Mycobacterium avium subsp. paratuberculosis and associated risk factors for inflammatory bowel disease in Iranian patients. Gut Pathog. 2017, 9, 1. [Google Scholar] [CrossRef]
- Qin, X.; Singh, K.V.; Weinstick, G.M. Effects of enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect. Immun. 2000, 68, 2579–2586. [Google Scholar] [CrossRef] [PubMed]
- Balish, E.; Warner, T. Enterococcus faecalis induces inflammatory bowel disease in interleukin-10 knockout mice. Am. J. Pathol. 2002, 160, 2253–2257. [Google Scholar] [CrossRef]
- Tripathi, M.K.; Pratap, C.B.; Dixit, V.K.; Singh, T.B.; Shukla, S.K.; Jain, A.K.; Nath, G. Ulcerative Colitis and Its Association with Salmonella Species. Interdiscip. Perspect. Infect. Dis. 2016, 2016, 5854285. [Google Scholar] [CrossRef]
- Di Cera, E. Serine proteases. IUBMB Life 2009, 61, 510–515. [Google Scholar] [CrossRef]
- Wernersson, S.; Pejler, G. Mast cell secretory granules: Armed for battle. Nat. Rev. Immunol. 2014, 14, 478–494. [Google Scholar] [CrossRef]
- Meyer-Hoffert, U.; Wiedow, O. Neutrophil serine proteases: Mediators of innate immune responses. Curr. Opin. Hematol. 2011, 18, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Denadai-Souza, A.; Bonnart, C.; Tapias, N.S.; Marcellin, M.; Gilmore, B.; Alric, L.; Bonnet, D.; Burlet-Schiltz, O.; Hollenberg, M.D.; Vergnolle, N.; et al. Functional Proteomic Profiling of Secreted Serine Proteases in Health and Inflammatory Bowel Disease. Sci. Rep. 2018, 8, 7834. [Google Scholar] [CrossRef] [PubMed]
- Motta, J.P.; Palese, S.; Giorgio, C.; Chapman, K.; Denadai-Souza, A.; Rousset, P.; Sagnat, D.; Guiraud, L.; Edir, A.; Seguy, C.; et al. Increased mucosal thrombin is associated with Crohn’s disease and causes inflammatory damage through Protease-Activated Receptors activation. J. Crohn’s Colitis 2020, jjaa229. [Google Scholar] [CrossRef]
- Liu, B.; Yang, M.Q.; Yu, T.Y.; Yin, Y.Y.; Liu, Y.; Wang, X.D.; He, Z.G.; Yin, L.; Chen, C.Q.; Li, J.Y. Mast Cell Tryptase Promotes Inflammatory Bowel Disease-Induced Intestinal Fibrosis. Inflamm. Bowel Dis. 2021, 27, 242–255. [Google Scholar] [CrossRef] [PubMed]
- Dabek, M.; Ferrier, L.; Annahazi, A.; Bézirard, V.; Polizzi, A.; Cartier, C.; Leveque, M.; Roka, R.; Wittmann, T.; Theodorou, V.; et al. Intracolonic infusion of fecal supernatants from ulcerative colitis patients triggers altered permeability and inflammation in mice: Role of cathepsin G and protease-activated receptor-4. Inflamm. Bowel Dis. 2011, 17, 1409–1414. [Google Scholar] [CrossRef] [PubMed]
- Scudamore, C.L.; Jepson, M.A.; Hirst, B.H.; Miller, H.R. The rat mucosal mast cell chymase RMCP-II alters epithelial cell monolayer permeability in association with altered distribution of the tight junction proteins ZO-1 and occludin. Eur. J. Cell Biol. 1998, 75, 321–330. [Google Scholar] [CrossRef]
- Hermant, B.; Bibert, S.; Concord, E.; Dublet, B.; Weidenhaupt, M.; Vernet, T.; Gulino-Debrac, D. Identification of proteases involved in the proteolysis of vascular endothelium cadherin during neutrophil transmigration. J. Biol. Chem. 2003, 278, 14002–14012. [Google Scholar] [CrossRef] [PubMed]
- Mortier, A.; Loos, T.; Gouwy, M.; Ronsse, I.; Van Damme, J.; Proost, P. Posttranslational modification of the NH2-terminal region of CXCL5 by proteases or peptidylarginine Deiminases (PAD) differently affects its biological activity. J. Biol. Chem. 2010, 285, 29750–29759. [Google Scholar] [CrossRef]
- Chapman, H.A.; Riese, R.J.; Shi, G.P. Emerging roles for cysteine proteases in human biology. Annu. Rev. Physiol. 1997, 59, 63–88. [Google Scholar] [CrossRef]
- Julien, O.; Wells, J.A. Caspases and their substrates. Cell Death. Differ. 2017, 24, 1380–1389. [Google Scholar] [CrossRef]
- Guo, C.; Ahmad, T.; Beckly, J.; Cummings, J.R.; Hancock, L.; Geremia, A.; Cooney, R.; Pathan, S.; Jewell, D.P. Association of caspase-9 and RUNX3 with inflammatory bowel disease. Tissue Antigens 2011, 77, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Flood, B.; Oficjalska, K.; Laukens, D.; Fay, J.; O’Grady, A.; Caiazza, F.; Heetun, Z.; Mills, K.H.; Sheahan, K.; Ryan, E.J.; et al. Altered expression of caspases-4 and -5 during inflammatory bowel disease and colorectal cancer: Diagnostic and therapeutic potential. Clin. Exp. Immunol. 2015, 181, 39–50. [Google Scholar] [CrossRef]
- Turk, V.; Stoka, V.; Vasiljeva, O.; Renko, M.; Sun, T.; Turk, B.; Turk, D. Cysteine cathepsins: From structure, function and regulation to new frontiers. Biochim. Biophys. Acta 2012, 1824, 68–88. [Google Scholar] [CrossRef] [PubMed]
- Menzel, K.; Hausmann, M.; Obermeier, F.; Schreiter, K.; Dunger, N.; Bataille, F.; Falk, W.; Scholmerich, J.; Herfarth, H.; Rogler, G. Cathepsins B, L and D in inflammatory bowel disease macrophages and potential therapeutic effects of cathepsin inhibition in vivo. Clin. Exp. Immunol. 2006, 146, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Culp, E.; Wright, G.D. Bacterial proteases untapped antimicrobial drug targets. J. Antibiot. 2017, 70, 366–377. [Google Scholar] [CrossRef]
- Kim, D.Y.; Kim, K.K. Structure and function of HtrA family proteins the key players in protein quality control. J. Biochem. Mol. Biol. 2005, 38, 266–274. [Google Scholar] [CrossRef]
- Page, M.J.; Di Cera, E. Evolution of peptidase diversity. J. Biol. Chem. 2008, 283, 30010–30014. [Google Scholar] [CrossRef]
- Wilson, R.L.; Brown, L.L.; Kirkwood-Watts, D.; Warren, T.K.; Lund, S.A.; King, D.S.; Jones, K.F.; Hruby, D.E. Listeria monocytogenes 10403S HtrA is necessary for resistance to cellular stress and virulence. Infect. Immun. 2006, 74, 765–768. [Google Scholar] [CrossRef]
- Sela-Abramovich, S.; Chitlaru, T.; Gat, O.; Grosfeld, H.; Cohen, O.; Shafferman, A. Novel and unique diagnostic biomarkers for Bacillus anthracis infection. Appl. Environ. Microbiol. 2009, 75, 6157–6167. [Google Scholar] [CrossRef]
- Russell, T.M.; Delorey, M.J.; Johnson, B.J. Borrelia burgdorferi BbHtrA degrades host ECM proteins and stimulates release of inflammatory cytokines in vitro. Mol. Microbiol. 2013, 90, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Boehm, M.; Haenel, I.; Hoy, B.; Brøndsted, L.; Smith, T.G.; Hoover, T.; Wessler, S.; Tegtmeyer, N. Extracellular secretion of protease HtrA from Campylobacter jejuni is highly efficient and independent of its protease activity and flagellum. Eur. J. Microbiol. Immunol. 2013, 3, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Löwer, M.; Weydig, C.; Metzler, D.; Reuter, A.; Starzinski-Powitz, A.; Wessler, S.; Schneider, G. Prediction of extracellular proteases of the human pathogen Helicobacter pylori reveals proteolytic activity of the Hp1018/19 protein HtrA. PLoS ONE 2008, 3, e3510. [Google Scholar] [CrossRef] [PubMed]
- Wessler, S.; Schneider, G.; Backert, S. Bacterial serine protease HtrA as a promising new target for antimicrobial therapy? Cell Commun. Signal. 2017, 15, 4. [Google Scholar] [CrossRef]
- Boehm, M.; Hoy, B.; Rohde, M.; Tegtmeyer, N.; Bæk, K.T.; Oyarzabal, O.A.; Brøndsted, L.; Wessler, S.; Backert, S. Rapid paracellular transmigration of Campylobacter jejuni across polarized epithelial cells without affecting TER: Role of proteolytic-active HtrA cleaving E-cadherin but not fibronectin. Gut Pathog. 2012, 4, 3. [Google Scholar] [CrossRef]
- Elmi, A.; Nasher, F.; Jagatia, H.; Gundogdu, O.; Bajaj-Elliott, M.; Wren, B.; Dorrell, N. Campylobacter jejuni outer membrane vesicle-associated proteolytic activity promotes bacterial invasion by mediating cleavage of intestinal epithelial cell E-cadherin and occludin. Cell Microbiol. 2016, 18, 561–572. [Google Scholar] [CrossRef]
- Hoy, B.; Geppert, T.; Boehm, M.; Reisen, F.; Plattner, P.; Gadermaier, G.; Sewald, N.; Ferreira, F.; Briza, P.; Schneider, G.; et al. Distinct roles of secreted HtrA proteases from gram-negative pathogens in cleaving the junctional protein and tumor suppressor E-cadherin. J. Biol. Chem. 2012, 287, 10115–10120. [Google Scholar] [CrossRef]
- Abfalter, C.M.; Schubert, M.; Götz, C.; Schmidt, T.P.; Posselt, G.; Wessler, S. HtrA-mediated E-cadherin cleavage is limited to DegP and DegQ homologs expressed by gram-negative pathogens. Cell Commun. Signal. 2016, 14, 30. [Google Scholar] [CrossRef]
- Schmidt, T.P.; Perna, A.M.; Fugmann, T.; Böhm, M.; Jan, H.; Haller, S.; Götz, C.; Tegtmeyer, N.; Hoy, B.; Rau, T.T.; et al. Identification of E-cadherin signature motifs functioning as cleavage sites for Helicobacter pylori HtrA. Sci. Rep. 2016, 6, 23264. [Google Scholar] [CrossRef] [PubMed]
- Gibold, L.; Garenaux, E.; Dalmasso, G.; Gallucci, C.; Cia, D.; Mottet-Auselo, B.; Faïs, T.; Darfeuille-Michaud, A.; Nguyen, H.T.; Barnich, N.; et al. The Vat-AIEC protease promotes crossing of the intestinal mucus layer by Crohn’s disease-associated Escherichia coli. Cell Microbiol. 2016, 18, 617–631. [Google Scholar] [CrossRef]
- Brunder, W.; Schmidt, H.; Karch, H. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol. Microbiol. 1997, 24, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Henderson, I.R.; Czeczulin, J.; Eslava, C.; Noriega, F.; Nataro, J.P. Characterization of pic a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect. Immun. 1999, 67, 5587–5596. [Google Scholar] [CrossRef] [PubMed]
- Pontarollo, G.; Acquasaliente, L.; Peterle, D.; Frasson, R.; Artusi, I.; De Filippis, V. Non-canonical proteolytic activation of human prothrombin by subtilisin from Bacillus subtilis may shift the procoagulant-anticoagulant equilibrium toward thrombosis. J. Biol. Chem. 2017, 292, 15161–15179. [Google Scholar] [CrossRef]
- Giannotta, M.; Tapete, G.; Emmi, G.; Silvestri, E.; Milla, M. Thrombosis in inflammatory bowel diseases: What’s the link? Thromb. J. 2015, 13, 14. [Google Scholar] [CrossRef]
- Potempa, J.; Golonka, E.; Filipek, R.; Shaw, L.N. Fighting an enemy within: Cytoplasmic inhibitors of bacterial cysteine proteases. Mol. Microbiol. 2005, 57, 605–610. [Google Scholar] [CrossRef]
- Monteiro, A.C.; Scovino, A.; Raposo, S.; Gaze, V.M.; Cruz, C.; Svensjö, E.; Narciso, M.S.; Colombo, A.P.; Pesquero, J.B.; Feres-Filho, E.; et al. Kinin danger signals proteolytically released by gingipain induce Fimbriae-specific IFN-gamma- and IL-17-producing T cells in mice infected intramucosally with Porphyromonas gingivalis. J. Immunol. 2009, 183, 3700–3711. [Google Scholar] [CrossRef] [PubMed]
- Carlisle, M.D.; Srikantha, R.N.; Brogden, K.A. Degradation of human alpha- and beta-defensins by culture supernatants of Porphyromonas gingivalis strain 381. J. Innate Immun. 2009, 1, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Pingel, L.C.; Kohlgraf, K.G.; Hansen, C.J.; Eastman, C.G.; Dietrich, D.E.; Burnell, K.K.; Srikantha, R.N.; Xiao, X.; Bélanger, M.; Progulske-Fox, A.; et al. Human beta-defensin 3 binds to hemagglutinin B (rHagB) a non-fimbrial adhesin from Porphyromonas gingivalis.; and attenuates a pro-inflammatory cytokine response. Immunol. Cell Biol. 2008, 86, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Pattamapun, K.; Tiranathanagul, S.; Yongchaitrakul, T.; Kuwatanasuchat, J.; Pavasant, P. Activation of MMP-2 by Porphyromonas gingivalis in human periodontal ligament cells. J. Periodontal. Res. 2003, 38, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Brito, F.; Zaltman, C.; Carvalho, A.T.; Fischer, R.G.; Persson, R.; Gustafsson, A.; Figueredo, C.M. Subgingival microflora in inflammatory bowel disease patients with untreated periodontitis. Eur. J. Gastroenterol. Hepatol. 2013, 25, 239–245. [Google Scholar] [CrossRef]
- Van Dyke, T.E.; Dowell, V.R., Jr.; Offenbacher, S.; Snyder, W.; Hersh, T. Potential role of microorganisms isolated from periodontal lesions in the pathogenesis of inflammatory bowel disease. Infect. Immun. 1986, 53, 671–677. [Google Scholar] [CrossRef]
- Nakajima, M.; Arimatsu, K.; Kato, T.; Matsuda, Y.; Minagawa, T.; Takahashi, N.; Ohno, H.; Yamazaki, K. Oral Administration of P. gingivalis Induces Dysbiosis of Gut Microbiota and Impaired Barrier Function Leading to Dissemination of Enterobacteria to the Liver. PLoS ONE 2015, 10, e0134234. [Google Scholar] [CrossRef]
- Silverman, G.A.; Whisstock, J.C.; Bottomley, S.P.; Huntington, J.A.; Kaiserman, D.; Luke, C.J.; Pak, S.C.; Reichhart, J.M.; Bird, P. Serpins flex their muscle: I. Putting the clamps on proteolysis in diverse biological systems. J. Biol. Chem. 2010, 285, 24299–24305. [Google Scholar] [CrossRef] [PubMed]
- Law, R.H.; Zhang, Q.; McGowan, S.; Buckle, A.M.; Silverman, G.A.; Wong, W.; Rosado, C.J.; Langendorf, C.G.; Pike, R.N.; Bird, P.I.; et al. An overview of the serpin superfamily. Genome Biol. 2006, 7, 216. [Google Scholar] [CrossRef][Green Version]
- Gettins, P.G.W. Serpin structure, mechanism, and function. Chem. Rev. 2002, 102, 4751–4804. [Google Scholar] [CrossRef]
- Zhou, A.; Wei, Z.; Read, R.J.; Carrell, R.W. Structural mechanism for the carriage and release of thyroxine in the blood. Proc. Natl. Acad. Sci. USA 2006, 103, 13321–13326. [Google Scholar] [CrossRef]
- Klieber, M.A.; Underhill, C.; Hammond, G.L.; Muller, Y.A. Corticosteroid-binding globulin, a structural basis for steroid transport and proteinase-triggered release. J. Biol. Chem. 2007, 282, 29594–29603. [Google Scholar] [CrossRef] [PubMed]
- Selbonne, S.; Azibani, F.; Iatmanen, S.; Boulaftali, Y.; Richard, B.; Jandrot-Perrus, M.; Bouton, M.C.; Arocas, V. In vitro and in vivo antiangiogenic properties of the serpin protease nexin-1. Mol. Cell Biol. 2012, 32, 1496–1505. [Google Scholar] [CrossRef]
- Huntington, J.A.; Carrell, R.W. The serpins: Nature’s molecular mousetraps. Sci Prog. 2001, 84, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Irving, J.A.; Pike, R.N.; Lesk, A.M.; Whisstock, J.C. Phylogeny of the serpin superfamily: Implications of patterns of amino acid conservation for structure and function. Genome Res. 2000, 10, 1845–1864. [Google Scholar] [CrossRef]
- Ray, C.A.; Black, R.A.; Kronheim, S.R.; Greenstreet, T.A.; Sleath, P.R.; Salvesen, G.S.; Pickup, D.J. Viral inhibition of inflammation: Cowpox virus encodes an inhibitor of the interleukin-1 beta converting enzyme. Cell 1992, 69, 597–604. [Google Scholar] [CrossRef]
- Schick, C.; Pemberton, P.A.; Shi, G.P.; Kamachi, Y.; Cataltepe, S.; Bartuski, A.J.; Gornstein, E.R.; Brömme, D.; Chapman, H.A.; Silverman, G.A. Cross-class inhibition of the cysteine proteinases cathepsins K, L, and S by the serpin squamous cell carcinoma antigen 1: A kinetic analysis. Biochemistry 1998, 37, 5258–5266. [Google Scholar] [CrossRef]
- Irving, J.A.; Pike, R.N.; Dai, W.; Brömme, D.; Worrall, D.M.; Silverman, G.A.; Coetzer, T.H.; Dennison, C.; Bottomley, S.P.; Whisstock, J.C. Evidence that serpin architecture intrinsically supports papain-like cysteine protease inhibition: Engineering alpha(1)-antitrypsin to inhibit cathepsin proteases. Biochemistry 2002, 41, 4998–5004. [Google Scholar] [CrossRef]
- Sanrattana, W.; Maas, C.; de Maat, S. SERPINs-From Trap to Treatment. Front. Med. 2019, 6, 25. [Google Scholar] [CrossRef]
- Lucas, A.; Yaron, J.R.; Zhang, L.; Ambadapadi, S. Overview of serpins and their roles in biological systems. Methods Mol. Biol. 2018, 1826, 1–7. [Google Scholar] [CrossRef]
- Diebold, I.; Kraicun, D.; Bonello, S.; Görlach, A. The ‘‘PAI-1 paradox’’ in vascular remodeling. Thromb. Haemost. 2008, 100, 984–991. [Google Scholar]
- Gatto, M.; Iaccarino, L.; Ghirardello, A.; Bassi, N.; Pontisso, P.; Punzi, L.; Shoenfeld, Y.; Doria, A. Serpins, immunity and autoimmunity: Old molecules, new functions. Clin. Rev. Allergy. Immunol. 2013, 45, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Irving, J.A.; Cabrita, L.D.; Rossjohn, J.; Pike, R.N.; Bottomley, S.P.; Whisstock, J.C. The 1.5 A crystal structure of a prokaryote serpin: Controlling conformational change in a heated environment. Structure 2003, 11, 387–397. [Google Scholar] [CrossRef]
- Zhang, H.; Fei, R.; Xue, B.; Yu, S.; Zhang, Z.; Zhong, S.; Gao, Y.; Zhou, X. Pnserpin: A Novel Serine Protease Inhibitor from Extremophile Pyrobaculum neutrophilum. Int. J. Mol. Sciv. 2017, 18, 113. [Google Scholar] [CrossRef] [PubMed]
- Gaci, N.; Dobrijevic, D.; Boudebbouze, S.; Moumen, B.; Maguin, E.; Rhimi, M. Patented biotechnological applications of serpin: An update. Recent Pat. DNA Gene Seq. 2013, 7, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Harish, B.S.; Uppuluri, K.B. Microbial serine protease inhibitors and their therapeutic applications. Int. J. Biol. Macromol. 2018, 107, 1373–1387. [Google Scholar] [CrossRef]
- Kantyka, T.; Rawlings, N.D.; Potempa, J. Prokaryote-derived protein inhibitors of peptidases: A sketchy occurrence and mostly unknown function. Biochimie 2010, 92, 1644–1656. [Google Scholar] [CrossRef]
- Irving, J.A.; Steenbakkers, P.J.M.; Lesk, A.M.; Op den Camp, H.J.M.; Pike, R.N.; Whisstock, J.C. Serpins in prokaryotes. Mol. Biol. Evol. 2002, 19, 1881–1890. [Google Scholar] [CrossRef]
- Mkaouar, H.; Akermi, N.; Kriaa, A.; Abraham, A.L.; Jablaoui, A.; Soussou, S.; Mokdad-Gargouri, R.; Maguin, E.; Rhimi, M. Serine protease inhibitors and human wellbeing interplay: New insights for old friends. PeerJ 2019, 7, e7224. [Google Scholar] [CrossRef]
- Roberts, T.H.; Hejgaard, J.; Saunders, N.F.W.; Cavicchioli, R.; Curmi, P.M.G. Serpins in unicellular Eukarya, Archaea, and Bacteria: Sequence analysis and evolution. J. Mol. Evol. 2004, 59, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Goulas, T.; Ksiazek, M.; Garcia-Ferrer, I.; Sochaj-Gregorczyk, A.M.; Waligorska, I.; Wasylewski, M.; Potempa, J.; Gomis-Rüth, F.X. A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome. J. Biol. Chem. 2017, 292, 10883–10898. [Google Scholar] [CrossRef] [PubMed]
- Spence, M.A.; Mortimer, M.D.; Buckle, A.M.; Minh, B.Q.; Jackson, C.J. A comprehensive phylogenetic analysis of the serpin superfamily. Mol. Biol. Evol. 2021, msab081. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Buckle, A.M.; Law, R.H.; Pearce, M.C.; Cabrita, L.D.; Lloyd, G.J.; Irving, J.A.; Smith, A.I.; Ruzyla, K.; Rossjohn, J.; et al. The N terminus of the serpin, tengpin, functions to trap the metastable native state. EMBO Rep. 2007, 8, 658–663. [Google Scholar] [CrossRef]
- Kang, S.; Barak, Y.; Lamed, R.; Bayer, E.A.; Morrison, M. The functional repertoire of prokaryote cellulosomes includes the serpin superfamily of serine proteinase inhibitors. Mol. Microbiol. 2006, 60, 1344–1354. [Google Scholar] [CrossRef]
- Cuív, P.; Gupta, R.; Goswami, H.P.; Morrison, M. Extending the cellulosome paradigm: The modular clostridium thermocellum cellulosomal serpin pinA is a broad-spectrum inhibitor of subtilisin-like proteases. Appl. Environ. Microbiol. 2013, 79, 6173–6175. [Google Scholar] [CrossRef]
- Mushtaq, M.; Asad, M.J.; Hyder, M.Z.; Naqvi, S.M.S.; Malik, S.I.; Mehmood, R.T. Serpins: Purification and characterization of potent protease inhibitors from Clostridium thermocellum. bioRxiv 2020, 1–17. [Google Scholar] [CrossRef]
- Oliveira, J.P.; Salazar, N.; Zani, M.B.; de Souza, L.R.; Passos, S.G.; Sant’Ana, A.M.; de Andrade, R.A.; Marcili, A.; Sperança, M.A.; Puzer, L. Vioserpin, a serine protease inhibitor from Gloeobacter violaceus possibly regulated by heparin. Biochimie 2016, 127, 115–120. [Google Scholar] [CrossRef]
- Hong, T.T.; Dat, T.T.H.; Hoa, N.P.; Dung, T.T.K.; Huyen, V.T.T.; Bui, L.M.; Cuc, N.T.K.; Cuong, P.V. Expression and characterization of a new serine protease inhibitory protein in Escherichia coli. Biomed. Res. Ther. 2020, 7, 3633–3644. [Google Scholar] [CrossRef]
- Ksiazek, M.; Mizgalska, D.; Enghild, J.J.; Scavenius, C.; Thogersen, I.B.; Potempa, J. Miropin, a novel bacterial serpin from the periodontopathogen Tannerella forsythia, inhibits a broad range of proteases by using different peptide bonds within the reactive center loop. J. Biol. Chem. 2015, 290, 658–670. [Google Scholar] [CrossRef] [PubMed]
- Eckert, M.; Mizgalska, D.; Sculean, A.; Potempa, J.; Stavropoulos, A.; Eick, S. In vivo expression of proteases and protease inhibitor, a serpin, by periodontal pathogens at teeth and implants. Mol. Oral Microbiol. 2018, 33, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Sochaj-Gregorczyk, A.; Ksiazek, M.; Waligorska, I.; Straczek, A.; Benedyk, M.; Mizgalska, D.; Thøgersen, I.B.; Enghild, J.J.; Potempa, J. Plasmin inhibition by bacterial serpin: Implications in gum disease. FASEB J. 2020, 34, 619–663. [Google Scholar] [CrossRef] [PubMed]
- Duboux, S.; Golliard, M.; Muller, J.A.; Bergonzelli, G.; Bolten, C.J.; Mercenier, A.; Kleerebezem, M. Carbohydrate-controlled serine protease inhibitor (serpin) production in Bifidobacterium longum subsp. longum. Sci. Rep. 2021, 11, 7236. [Google Scholar] [CrossRef]
- Turroni, F.; Foroni, E.; Motherway, M.O.C.; Bottacini, F.; Giubellini, V.; Zomer, A.; Ferrarini, A.; Delledonne, M.; Zhang, Z.; van Sinderen, D.; et al. Characterization of the serpin-encoding gene of Bifidobacterium breve 210B. Appl. Environ. Microbiol. 2010, 76, 3206–3219. [Google Scholar] [CrossRef]
- Schell, M.A.; Karmirantzou, M.; Snel, B.; Vilanova, D.; Berger, B.; Pessi, G.; Zwahlen, M.C.; Desiere, F.; Bork, P.; Delley, M.; et al. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 2002, 99, 14422–14427. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status taxonomic expansion and functional annotation. Nucleic Acids Res. 2016, 44, 733–745. [Google Scholar] [CrossRef]
- Alvarez-Martin, P.; Motherway, M.O.C.; Turroni, F.; Foroni, E.; Ventura, M.; van Sinderen, D. Two-component regulatory system controls autoregulated serpin expression in Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 2012, 78, 7032–7041. [Google Scholar] [CrossRef]
- Ivanov, D.; Emonet, C.; Foata, F.; Affolter, M.; Delley, M.; Fisseha, M.; Blum-Sperisen, S.; Kochhar, S.; Arigoni, F. A serpin from the gut bacterium Bifidobacterium longum inhibits eukaryotic elastase-like serine proteases. J. Biol. Chem. 2006, 281, 17246–17252. [Google Scholar] [CrossRef]
- Riedel, C.U.; Foata, F.; Philippe, D.; Adolfsson, O.; Eikmanns, B.J.; Blum, S. Anti-inflammatory effects of Bifidobacteria by inhibition of LPS-induced NF-kappaB activation. World J. Gastroenterol. 2006, 12, 3729–3735. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, L.; Delgado, S.; Ruas-Madiedo, P.; Sánchez, B.; Margolles, A. Bifidobacteria and Their Molecular Communication with the Immune System. Front. Microbiol. 2017, 8, 2345. [Google Scholar] [CrossRef] [PubMed]
- Buhner, S.; Hahne, H.; Hartwig, K.; Li, Q.; Vignali, S.; Ostertag, D.; Meng, C.; Hörmannsperger, G.; Braak, B.; Pehl, C.; et al. Protease signaling through protease activated receptor 1 mediate nerve activation by mucosal supernatants from irritable bowel syndrome but not from ulcerative colitis patients. PLoS ONE 2018, 13, e019394. [Google Scholar] [CrossRef] [PubMed]
- Bercik, P.; Park, A.J.; Sinclair, D.; Khoshdel, A.; Lu, J.; Huang, X.; Deng, Y.; Blennerhassett, P.A.; Fahnestock, M.; Moine, D.; et al. The anxiolytic effect of Bifidobacterium longum NCC3001 involves vagal pathways for gut-brain communication. Neurogastroenterol. Motil. 2011, 23, 1132–1139. [Google Scholar] [CrossRef] [PubMed]
- McCarville, J.L.; Dong, J.; Caminero, A.; Bermudez-Brito, M.; Jury, J.; Murray, J.A.; Duboux, S.; Steinmann, M.; Delley, M.; Tangyu, M.; et al. A Commensal Bifidobacterium longum Strain Prevents Gluten-Related Immunopathology in Mice through Expression of a Serine Protease Inhibitor. Appl. Environ. Microbiol. 2017, 83, e01323-17. [Google Scholar] [CrossRef]
- Mukherjee, A.; Lordan, C.; Ross, R.P.; Cotter, P.D. Gut microbes from the phylogenetically diverse genus Eubacterium and their various contributions to gut health. Gut Microbes. 2020, 12, 1802866. [Google Scholar] [CrossRef]
- Akermi, N.; Mkaouar, H.; Kriaa, A.; Jablaoui, A.; Soussou, S.; Gargouri, A.; Coleman, A.W.; Perret, F.; Maguin, E.; Rhimi, M. para-Sulphonato-calix[n]arene capped silver nanoparticles challenge the catalytic efficiency and the stability of a novel human gut serine protease inhibitor. Chem. Commun. 2019, 55, 8935–8938. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Xu, J.; Wu, L.; Yu, P.; Liu, M.; Lu, Y. Effect of two recombinant Trichinella spiralis serine protease inhibitors on TNBS-induced experimental colitis of mice. Clin. Exp. Immunol. 2018, 194, 400–413. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Liu, M.; Yu, P.; Wu, L.; Lu, Y. Effect of recombinant Trichinella spiralis cysteine proteinase inhibitor on TNBS-induced experimental inflammatory bowel disease in mice. Int. Immunopharmacol. 2019, 66, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Beatty, K.; Bieth, J.; Travis, J. Kinetics of association of serine proteinases with native and oxidized alpha-1-proteinase inhibitor and alpha-1-antichymotrypsin. J. Biol. Chem. 1980, 255, 3931–3934. [Google Scholar] [CrossRef]
- Schechter, N.M.; Sprows, J.L.; Schoenberger, O.L.; Lazarus, G.S.; Cooperman, B.S.; Rubin, H. Reaction of human skin chymotrypsin-like proteinase chymase with plasma proteinase inhibitors. J. Biol. Chem. 1989, 264, 21308–21315. [Google Scholar] [CrossRef]
- Zhou, G.X.; Chao, L.; Chao, J. Kallistatin: A novel human tissue kallikrein inhibitor. Purification, characterization, and reactive center sequence. J. Biol. Chem. 1992, 267, 25873–25880. [Google Scholar] [CrossRef]
- Suzuki, K.; Nishioka, J.; Hashimoto, S. Protein C inhibitor. Purification from human plasma and characterization. J. Biol. Chem. 1983, 258, 163–168. [Google Scholar] [CrossRef]
- Han, X.; Fiehler, R.; Broze, G.J., Jr. Characterization of the protein Z-dependent protease inhibitor. Blood 2000, 96, 3049–3055. [Google Scholar] [CrossRef]
- Heiker, J.T.; Klöting, N.; Kovacs, P.; Kuettner, E.B.; Sträter, N.; Schultz, S.; Kern, M.; Stumvoll, M.; Blüher, M.; Beck-Sickinger, A.G. Vaspin inhibits kallikrein 7 by serpin mechanism. Cell. Mol. Life Sci. 2013, 70, 2569–2583. [Google Scholar] [CrossRef] [PubMed]
- Pemberton, P.A.; Stein, P.E.; Pepys, M.B.; Potter, J.M.; Carrell, R.W. Hormone binding globulins undergo serpin conformational change in inflammation. Nature 1988, 336, 257–258. [Google Scholar] [CrossRef]
- Nakagawa, T.; Akaki, J.; Satou, R.; Takaya, M.; Iwata, H.; Katsurada, A.; Nishiuchi, K.; Ohmura, Y.; Suzuki, F.; Nakamura, Y. The His-Pro-Phe motif of angiotensinogen is a crucial determinant of the substrate specificity of renin. Biol. Chem. 2007, 388, 237–246. [Google Scholar] [CrossRef]
- Cooley, J.; Takayama, T.K.; Shapiro, S.D.; Schechter, N.M.; Remold-O’Donnell, E. The serpin MNEI inhibits elastase-like and chymotrypsin-like serine proteases through efficient reactions at two active sites. Biochemistry 2001, 40, 15762–15770. [Google Scholar] [CrossRef] [PubMed]
- Thorsen, S.; Philips, M.; Selmer, J.; Lecander, I.; Astedt, B. Kinetics of inhibition of tissue-type and urokinase-type plasminogen activator by plasminogen-activator inhibitor type 1 and type 2. Eur. J. Biochem. 1988, 175, 33–39. [Google Scholar] [CrossRef]
- Schick, C.; Kamachi, Y.; Bartuski, A.J.; Cataltepe, S.; Schechter, N.M.; Pemberton, P.A.; Silverman, G.A. Squamous cell carcinoma antigen 2 is a novel serpin that inhibits the chymotrypsin-like proteinases cathepsin G and mast cell chymase. J. Biol. Chem. 1997, 272, 1849–1855. [Google Scholar] [CrossRef]
- Dahlen, J.R.; Foster, D.C.; Kisiel, W. The inhibitory specificity of human proteinase inhibitor 8 is expanded through the use of multiple reactive site residues. Biochem. Biophys. Res. Commun. 1998, 244, 172–177. [Google Scholar] [CrossRef]
- Riewald, M.; Schleef, R.R. Molecular cloning of bomapin (protease inhibitor 10), a novel human serpin that is expressed specifically in the bone marrow. J. Biol. Chem. 1995, 270, 26754–26757. [Google Scholar] [CrossRef]
- Rezaie, A.R.; Olson, S.T. Calcium enhances heparin catalysis of the antithrombin-factor Xa reaction by promoting the assembly of an intermediate heparin-antithrombin-factor Xa bridging complex. Demonstration by rapid kinetics studies. Biochemistry 2000, 39, 12083–12090. [Google Scholar] [CrossRef]
- Olson, S.T.; Björk, I. Predominant contribution of surface approximation to the mechanism of heparin acceleration of the antithrombin-thrombin reaction. Elucidation from salt concentration effects. J. Biol. Chem. 1991, 266, 6353–6364. [Google Scholar]
- Pieters, J.; Willems, G.; Hemker, H.C.; Lindhout, T. Inhibition of factor IXa and factor Xa by antithrombin III/heparin during factor X activation. J. Biol. Chem. 1988, 263, 15313–15318. [Google Scholar] [CrossRef]
- Rogers, S.J.; Pratt, C.W.; Whinna, H.C.; Church, F.C. Role of thrombin exosites in inhibition by heparin cofactor II. J. Biol. Chem. 1992, 267, 3613–3617. [Google Scholar] [CrossRef]
- Ellis, V.; Wun, T.C.; Behrendt, N.; Rønne, E.; Danø, K. Inhibition of receptor-bound urokinase by plasminogen-activator inhibitors. J. Biol. Chem. 1990, 265, 9904–9908. [Google Scholar] [CrossRef]
- Knauer, D.J.; Majumdar, D.; Fong, P.C.; Knauer, M.F. SERPIN regulation of factor XIa. The novel observation that protease nexin 1 in the presence of heparin is a more potent inhibitor of factor XIa than C1 inhibitor. J. Biol. Chem. 2000, 275, 37340–37346. [Google Scholar] [CrossRef] [PubMed]
- Wiman, B.; Collen, D. On the mechanism of the reaction between human alpha 2-antiplasmin and plasmin. J. Biol. Chem. 1979, 254, 9291–9297. [Google Scholar] [CrossRef]
- Sim, R.B.; Arlaud, G.J.; Colomb, M.G. Kinetics of reaction of human C1-inhibitor with the human complement system proteases C1r and C1s. Biochim. Biophys. Acta 1980, 612, 433–449. [Google Scholar] [CrossRef]
- Ravindran, S.; Grys, T.E.; Welch, R.A.; Schapira, M.; Patston, P.A. Inhibition of plasma kallikrein by C1-inhibitor: Role of endothelial cells and the amino-terminal domain of C1-inhibitor. Thromb. Haemost. 2004, 92, 1277–1283. [Google Scholar] [CrossRef] [PubMed]
- Widmer, C.; Gebauer, J.M.; Brunstein, E.; Rosenbaum, S.; Zaucke, F.; Drögemüller, C.; Leeb, T.; Baumann, U. Molecular basis for the action of the collagen-specific chaperone Hsp47/SERPINH1 and its structure-specific client recognition. Proc. Natl. Acad. Sci. USA 2012, 109, 13243–13247. [Google Scholar] [CrossRef] [PubMed]
- Krueger, S.R.; Ghisu, G.P.; Cinelli, P.; Gschwend, T.P.; Osterwalder, T.; Wolfer, D.P.; Sonderegger, P. Expression of neuroserpin, an inhibitor of tissue plasminogen activator, in the developing and adult nervous system of the mouse. J. Neurosci. 1997, 17, 8984–8996. [Google Scholar] [CrossRef] [PubMed]
| Clade | Serpin | Biological Functions | References |
|---|---|---|---|
| A | Serpin A1, A3, A4, A5, A10, A12 | Serine protease inhibition | [150,151,152,153,154,155] |
| Serpin A6, A7 and A8 | Hormone transport | [156,157] | |
| B | Serpin B1, B2, B3, B4, B8, B10 | Serine and cysteine protease inhibition | [109,158,159,160,161,162] |
| C | Serpin C1 | Inhibition of thrombin, factor Xa and factor IXa | [163,164,165] |
| D | Serpin D1 | Inhibition of thrombin | [166] |
| E | Serpin E1 and E2 | Serine protease inhibition | [167,168] |
| F | Serpin F2 | Inhibition of plasmin | [169] |
| G | Serpin G1 | Inhibition of C1 proteinase and plasma kallikrein | [170,171] |
| H | Serpin H1 | Chaperone | [172] |
| I | Serpin I1 | Inhibition of plasmin, uPA and tPA | [173] |
| U | Thermopin | Inhibition of chymotrypsin | [115] |
| T | Siropins, Tengpin, Miropin, SerpinBL | Inhibition of eukaryotic proteases | [11,125,131,139] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mkaouar, H.; Mariaule, V.; Rhimi, S.; Hernandez, J.; Kriaa, A.; Jablaoui, A.; Akermi, N.; Maguin, E.; Lesner, A.; Korkmaz, B.; et al. Gut Serpinome: Emerging Evidence in IBD. Int. J. Mol. Sci. 2021, 22, 6088. https://doi.org/10.3390/ijms22116088
Mkaouar H, Mariaule V, Rhimi S, Hernandez J, Kriaa A, Jablaoui A, Akermi N, Maguin E, Lesner A, Korkmaz B, et al. Gut Serpinome: Emerging Evidence in IBD. International Journal of Molecular Sciences. 2021; 22(11):6088. https://doi.org/10.3390/ijms22116088
Chicago/Turabian StyleMkaouar, Héla, Vincent Mariaule, Soufien Rhimi, Juan Hernandez, Aicha Kriaa, Amin Jablaoui, Nizar Akermi, Emmanuelle Maguin, Adam Lesner, Brice Korkmaz, and et al. 2021. "Gut Serpinome: Emerging Evidence in IBD" International Journal of Molecular Sciences 22, no. 11: 6088. https://doi.org/10.3390/ijms22116088
APA StyleMkaouar, H., Mariaule, V., Rhimi, S., Hernandez, J., Kriaa, A., Jablaoui, A., Akermi, N., Maguin, E., Lesner, A., Korkmaz, B., & Rhimi, M. (2021). Gut Serpinome: Emerging Evidence in IBD. International Journal of Molecular Sciences, 22(11), 6088. https://doi.org/10.3390/ijms22116088






