MiR-181a-5p Regulates NIS Expression in Papillary Thyroid Carcinoma
Abstract
1. Introduction
2. Results
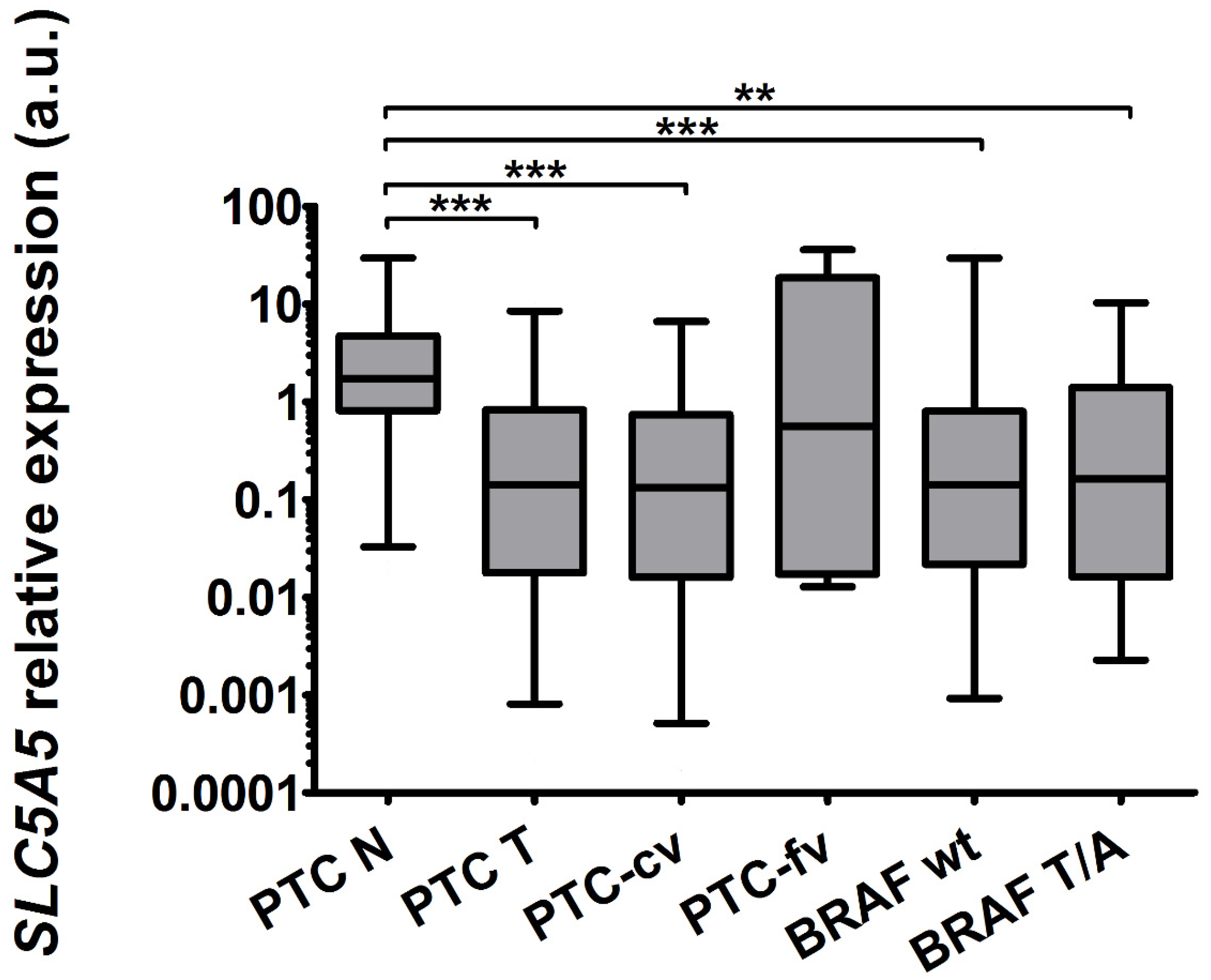
2.1. The Expression of SLC5A5 Is Lowered in PTC
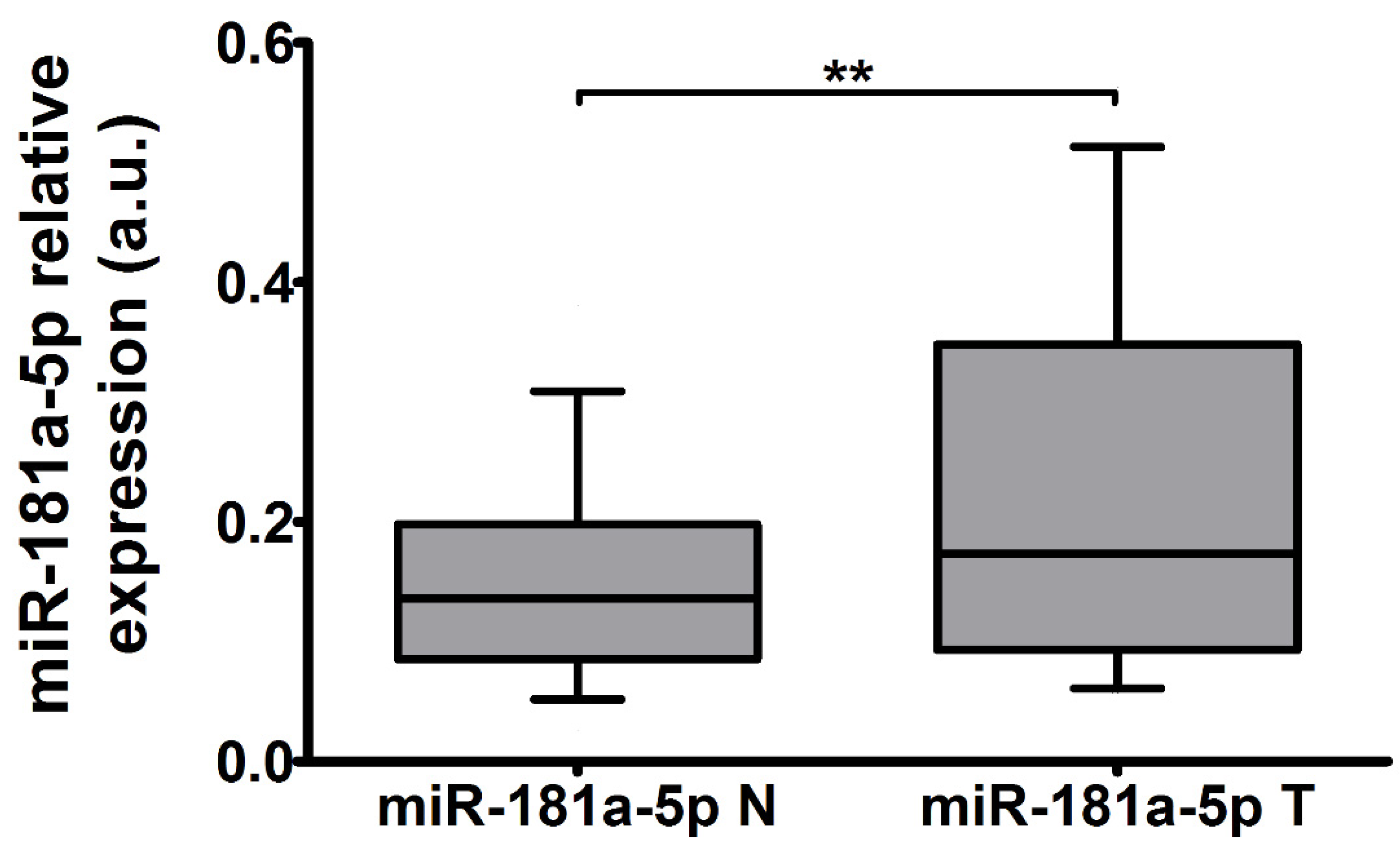
2.2. The Expression of miR-181a-5p Is Upregulated in Tumor Tissue
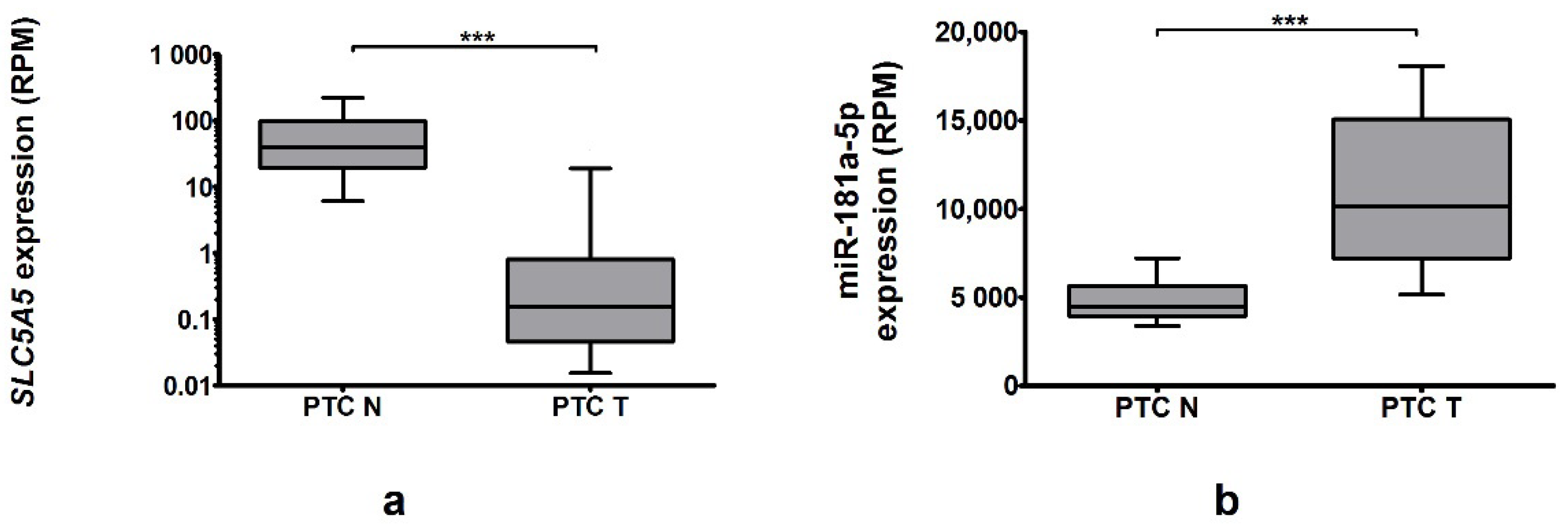
2.3. Genes Expression in TCGA Cohort
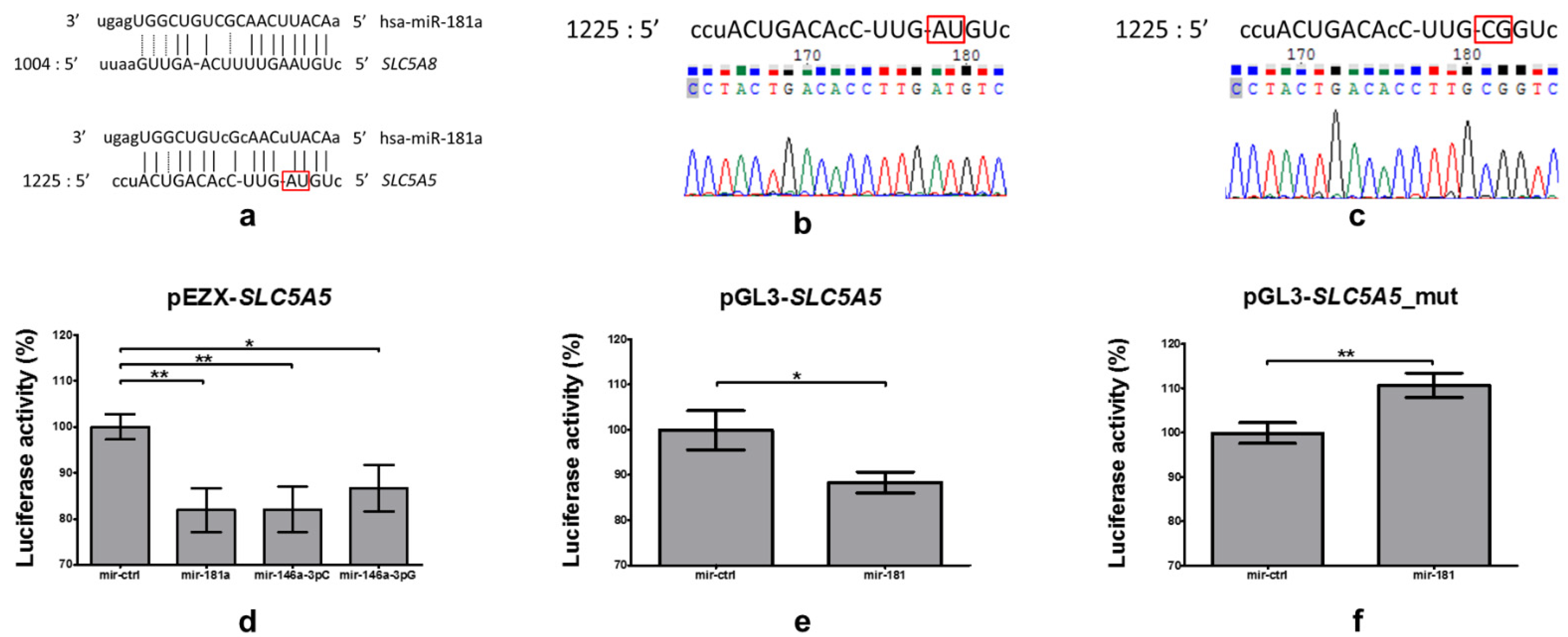
2.4. Investigation of Binding of miR-181a-5p with 3′UTR of SLC5A5
2.4.1. Analysis Employing Broadly Used Algorithms Does Not Reveal Binding of miR-181a-5p with 3′UTR of SLC5A5
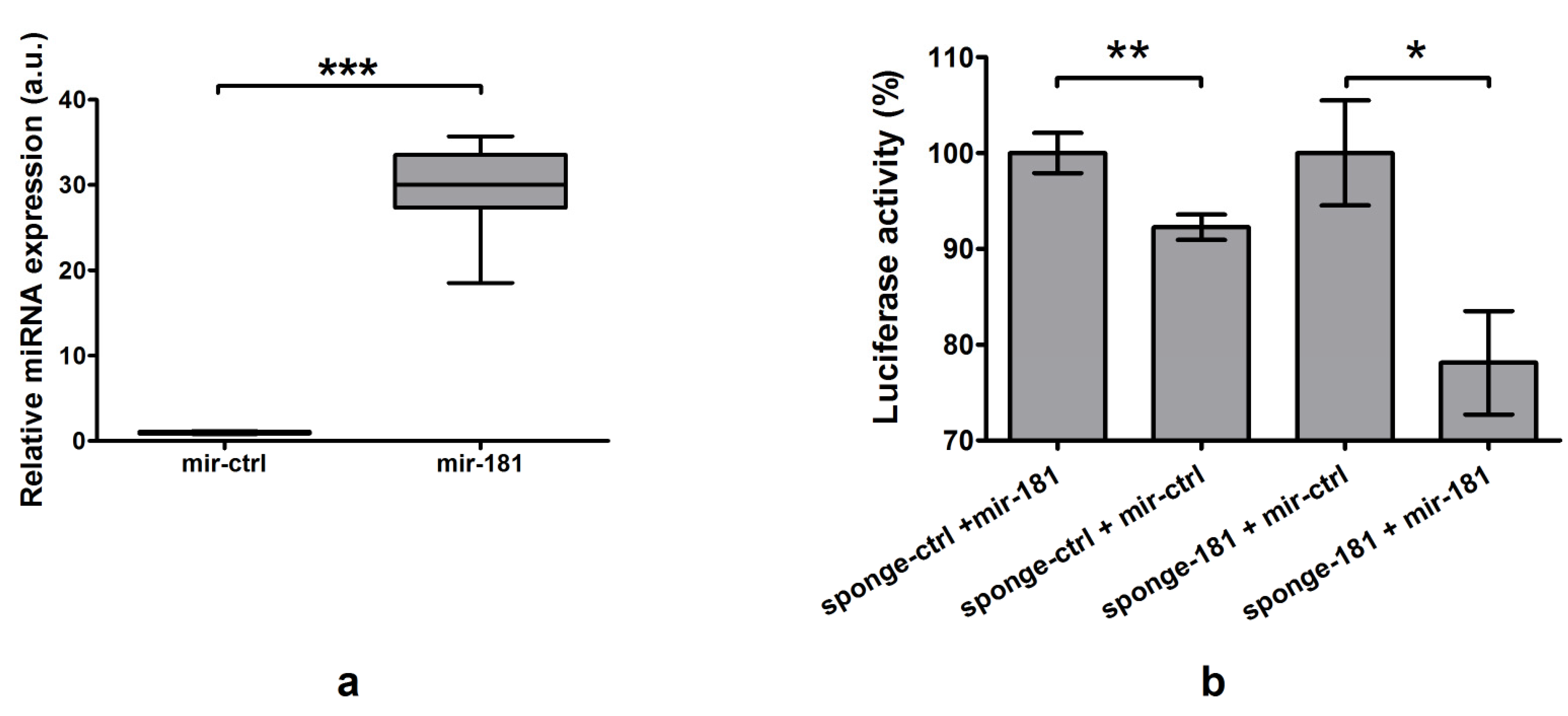
2.4.2. Identification and Confirmation of miR-181a-5p Binding Site
2.5. miR-181a-5p Affects Expression of Endogenous SLC5A5
2.5.1. Plasmid Functionality Verification
2.5.2. Impact of Overexpression and Silencing of miR-181a-5p on NIS Expression and Function
3. Discussion
4. Materials and Methods
4.1. Tissue Samples
4.2. Real-Time PCR
4.3. Comparison of the Results with the TCGA Data
4.4. In Silico Identification of Binding Site of miR-181a-5p in SLC5A5 3′UTR
4.5. MicroRNA Cloning and MicroRNA-Sponges Preparation
4.6. Analysis of miR-181a-5p-Mediated Regulation of SLC5A5
4.7. Protein Quantification
4.8. Radioactive Iodine Uptake Assay
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Baumann, E. Über Das Thyrojodin; Springer: Berlin/Heidelberg, Germany, 1896. [Google Scholar]
- Hertz, S.; Roberts, A.; Means, J.H.; Evans, R.D. Radioactive Iodine as an Indicator in Thyroid Physiology: Iodine Collection by Normal and Hyperplastic Thyroids in Rabbits. Am. J. Physiol. Leg. Content 1940, 128, 565–576. [Google Scholar] [CrossRef]
- Seidlin, S.M.; Marinelli, L.D.; Oshry, E. Radioactive Iodine Therapy; Effect on Functioning Metastases of Adenocarcinoma of the Thyroid. J. Am. Med. Assoc. 1946, 132, 838–847. [Google Scholar] [CrossRef]
- Ravera, S.; Reyna-Neyra, A.; Ferrandino, G.; Amzel, L.G.; Carrasco, N. The Sodium/Iodide Symporter (Nis): Molecular Physiology and Preclinical and Clinical Applications. Annu. Rev. Physiol. 2017, 79, 261–289. [Google Scholar] [CrossRef]
- Wolff, J. Transport of Iodide and Other Anions in the Thyroid Gland. Physiol. Rev. 1964, 44, 45–90. [Google Scholar] [CrossRef]
- Smanik, P.A.; Liu, Q.; Furminger, T.L.; Ryu, K.; Xing, S.; Mazzaferri, E.L.; Jhiang, S.M. Cloning of the Human Sodium Lodide Symporter. Biochem. Biophys. Res. Commun. 1996, 226, 339–345. [Google Scholar] [CrossRef]
- Smanik, P.A.; Ryu, K.Y.; Theil, K.S.; Mazzaferri, E.L.; Jhiang, S.M. Expression, Exon-Intron Organization, and Chromosome Mapping of the Human Sodium Iodide Symporter. Endocrinology 1997, 138, 3555–3558. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Xing, M. Recent Incidences and Differential Trends of Thyroid Cancer in the USA. Endocr. Relat. Cancer 2016, 23, 313–322. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Papillary Thyroid Carcinoma. Cell 2014, 3, 676–690. [Google Scholar]
- Mathur, A.; Moses, W.; Rahbari, R.; Khanafshar, E.; Duh, Q.Y.; Clark, O.; Kebebew, E. Higher Rate of Braf Mutation in Papillary Thyroid Cancer over Time: A Single-Institution Study. Cancer 2011, 117, 4390–4395. [Google Scholar] [CrossRef] [PubMed]
- Riesco-Eizaguirre, G.; Rodríguez, I.; De la Vieja, A.; Costamagna, E.; Carrasco, N.; Nistal, M.; Santisteban, P. The Brafv600e Oncogene Induces Transforming Growth Factor Beta Secretion Leading to Sodium Iodide Symporter Repression and Increased Malignancy in Thyroid Cancer. Cancer Res. 2009, 69, 8317–8325. [Google Scholar] [CrossRef] [PubMed]
- Costamagna, E.; García, B.; Santisteban, P. The Functional Interaction between the Paired Domain Transcription Factor Pax8 and Smad3 Is Involved in Transforming Growth Factor-Beta Repression of the Sodium/Iodide Symporter Gene. J. Biol. Chem. 2004, 279, 3439–3446. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, D.; Murugan, A.K.; Liu, Z.; Xing, M. Histone Deacetylation of Nis Promoter Underlies Braf V600e-Promoted Nis Silencing in Thyroid Cancer. Endocr. Relat. Cancer 2014, 21, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Riesco-Eizaguirre, G.; Gutiérrez-Martínez, P.; García-Cabezas, M.A.; Nistal, M.; Santisteban, P. The Oncogene Braf V600e Is Associated with a High Risk of Recurrence and Less Differentiated Papillary Thyroid Carcinoma Due to the Impairment of Na+/I- Targeting to the Membrane. Endocr. Relat. Cancer 2006, 13, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most Mammalian Mrnas Are Conserved Targets of Micrornas. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Bartel, D.P. Micrornas: Target Recognition and Regulatory Functions. Cell 2009, 2, 215–233. [Google Scholar] [CrossRef]
- Swierniak, M.; Wojcicka, A.; Czetwertynska, M.; Stachlewska, E.; Maciag, M.; Wiechno, W.; Gornicka, B.; Bogdanska, M.; Koperski, L.; de la Chapelle, A.; et al. In-Depth Characterization of the Microrna Transcriptome in Normal Thyroid and Papillary Thyroid Carcinoma. J. Clin. Endocrinol. Metab. 2013, 8, E1401–E1409. [Google Scholar] [CrossRef]
- Wojcicka, A.; Kolanowska, M.; Jazdzewski, K. Mechanisms in Endocrinology: Microrna in Diagnostics and Therapy of Thyroid Cancer. Eur. J. Endocrinol. 2016, 174, R89–R98. [Google Scholar] [CrossRef]
- Lindow, M.; Kauppinen, S. Discovering the First Microrna-Targeted Drug. J. Cell Biol. 2012, 199, 407–412. [Google Scholar] [CrossRef]
- Kotlarek, M.; Kubiak, A.; Czetwertynska, M.; Swierniak, M.; Gierlikowski, W.; Kolanowska, M.; Bakula-Zalewska, E.; Jhiang, S.M.; Jazdzewski, K.; Wojcicka, A. The Rs2910164 Genetic Variant of Mir-146a-3p Is Associated with Increased Overall Mortality in Patients with Follicular Variant Papillary Thyroid Carcinoma. Int. J. Mol. Sci. 2018, 19, 655. [Google Scholar] [CrossRef] [PubMed]
- Riesco-Eizaguirre, G.; Wert-Lamas, L.; Perales-Patón, J.; Sastre-Perona, A.; Fernández, P.L.; Santisteban, P. The Mir-146b-3p/Pax8/Nis Regulatory Circuit Modulates the Differentiation Phenotype and Function of Thyroid Cells During Carcinogenesis. Cancer Res. 2015, 75, 4119–4130. [Google Scholar] [CrossRef]
- Li, L.; Lv, B.; Chen, B.; Guan, M.; Sun, Y.; Li, H.; Zhang, B.; Ding, C.; He, S.; Zeng, Q. Inhibition of Mir-146b Expression Increases Radioiodine-Sensitivity in Poorly Differential Thyroid Carcinoma Via Positively Regulating Nis Expression. Biochem. Biophys. Res. Commun. 2015, 462, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Lakshmanan, A.; Wojcicka, A.; Kotlarek, M.; Zhang, X.; Jazdzewski, K.; Jhiang, S.M. Microrna-339-5p Modulates Na+/I- Symporter-Mediated Radioiodide Uptake. Endocr. Relat. Cancer 2015, 1, 11–21. [Google Scholar] [CrossRef]
- Tang, Y.; Meng, X.; Yu, X.; Shang, H.; Chen, S.; Liao, L.; Dong, J. Inhibition of Microrna-875-5p Promotes Radioiodine Uptake in Poorly Differentiated Thyroid Carcinoma Cells by Upregulating Sodium-Iodide Symporter. J. Endocrinol. Invest. 2020, 43, 439–450. [Google Scholar] [CrossRef] [PubMed]
- Wojcicka, A.; Gierlikowski, W.; Kotlarek, M.; Koperski, L.; Jazdzewski, K. Apical Iodide Transporter (Ait) and Its Microrna -Induced Silencing in Thyroid Malignancies. Endocr. Rev. 2014, 25, P1125. [Google Scholar]
- Jazdzewski, K.; Murray, E.L.; Franssila, K.; Jarzab, B.; Schoenberg, D.R.; de la Chapelle, A. Common Snp in Pre-Mir-146a Decreases Mature Mir Expression and Predisposes to Papillary Thyroid Carcinoma. Proc. Natl. Acad. Sci. USA 2008, 105, 7269–7274. [Google Scholar] [CrossRef]
- Haugen, B.R.; Alexander, E.K.; Bible, K.C.; Doherty, G.M.; Mandel, S.J.; Nikiforov, Y.E.; Pacini, F.; Randolph, G.W.; Sawka, A.M.; Schlumberger, M.; et al. 2015 American Thyroid Association Management Guidelines for Adult Patients with Thyroid Nodules and Differentiated Thyroid Cancer: The American Thyroid Association Guidelines Task Force on Thyroid Nodules and Differentiated Thyroid Cancer. Thyroid 2016, 26, 1–133. [Google Scholar] [CrossRef]
- de Morais, R.M.; Sobrinho, A.B.; Silva, C.M.d.; de Oliveira, J.R.; da Silva, I.C.R.; Nóbrega, O.d. The Role of the Nis (Slc5a5) Gene in Papillary Thyroid Cancer: A Systematic Review. Int. J. Endocrinol. 2018, 2018, 1–11. [Google Scholar] [CrossRef]
- Tavares, C.; Coelho, M.J.; Eloy, C.; Melo, M.; da Rocha, A.G.; Pestana, A.; Batista, R.; Ferreira, L.B.; Rios, E.; Selmi-Ruby, S.; et al. Nis Expression in Thyroid Tumors, Relation with Prognosis Clinicopathological and Molecular Features. Endocr. Connect 2018, 7, 78–90. [Google Scholar] [CrossRef]
- Ge, J.; Wang, J.; Wang, H.; Jiang, X.; Liao, Q.; Gong, Q.; Mo, Y.; Li, G.; Li, H.; Xiong, W.; et al. The Braf V600e Mutation Is a Predictor of the Effect of Radioiodine Therapy in Papillary Thyroid Cancer. J. Cancer 2020, 11, 932–939. [Google Scholar] [CrossRef]
- Daliri, M.; Abbaszadegan, M.R.; Bahar, M.M.; Arabi, A.; Yadollahi, M.; Ghafari, A.; Taghehchian, N.; Zakavi, S.R. The Role of Braf V600e Mutation as a Potential Marker for Prognostic Stratification of Papillary Thyroid Carcinoma: A Long-Term Follow-up Study. Endocr. Res. 2014, 39, 189–193. [Google Scholar] [CrossRef]
- Li, J.; Liang, J.; Zhao, T.; Lin, Y. Noninferior Response in Braf(V600e) Mutant Nonmetastatic Papillary Thyroid Carcinoma to Radioiodine Therapy. Eur. J. Nucl. Med. Mol. Imaging 2016, 43, 1034–1039. [Google Scholar] [CrossRef]
- Buffet, C.; Wassermann, J.; Hecht, F.; Leenhardt, L.; Dupuy, C.; Groussin, L.; Lussey-Lepoutre, C. Redifferentiation of Radioiodine-Refractory Thyroid Cancers. Endocr. Relat. Cancer 2020, 27, R113–R132. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Bo, C.; Guo, L.; Yu, P.; Miao, S.; Gu, X. Bcl2 and Hsa-Mir-181a-5p Are Potential Biomarkers Associated with Papillary Thyroid Cancer Based on Bioinformatics Analysis. World J. Surg. Oncol. 2019, 17, 221. [Google Scholar] [CrossRef]
- Cong, D.; He, M.; Chen, S.; Liu, X.; Liu, X.; Sun, H. Expression Profiles of Pivotal Micrornas and Targets in Thyroid Papillary Carcinoma: An Analysis of the Cancer Genome Atlas. Onco Targets Ther. 2015, 8, 2271–2277. [Google Scholar]
- Celakovsky, P.; Kovarikova, H.; Chrobok, V.; Mejzlik, J.; Laco, J.; Vosmikova, H.; Chmelarova, M.; Ryska, A. Microrna Deregulation in Papillary Thyroid Cancer and Its Relationship with Braf V600e Mutation. In Vivo 2021, 35, 319–323. [Google Scholar] [CrossRef]
- Sun, Y.; Shuang, Y.; Yuanyuan, L.; Fen, W.; Yujie, L.; Haipeng, X. Expression of Mirnas in Papillary Thyroid Carcinomas Is Associated with Braf Mutation and Clinicopathological Features in Chinese Patients. Int. J. Endocrinol. 2013, 5, 675–681. [Google Scholar]
- Chmielik, E.; Rusinek, D.; Oczko-Wojciechowska, M.; Jarzab, M.; Krajewska, J.; Czarniecka, A.; Jarzab, B. Heterogeneity of Thyroid Cancer. Pathobiology 2018, 85, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Zhou, X. Computational Methods for the Identification of Microrna Targets. Open Access Bioinform. 2010, 2, 29–39. [Google Scholar]
- Chen, L.; Heikkinen, L.; Wang, C.; Yang, Y.; Sun, H.; Wong, G. Trends in the Development of Mirna Bioinformatics Tools. Brief. Bioinform. 2019, 20, 1836–1852. [Google Scholar] [CrossRef]
- Lewis, B.P.; Shih, I.H.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of Mammalian Microrna Targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef]
- Chi, S.W.; Hannon, G.J.; Darnell, R.B. An Alternative Mode of Microrna Target Recognition. Nat. Struct. Mol. Biol. 2012, 19, 321–327. [Google Scholar] [CrossRef]
- Chandradoss, S.D.; Schirle, N.T.; Szczepaniak, M.; MacRae, I.J.; Joo, C. A Dynamic Search Process Underlies Microrna Targeting. Cell 2015, 162, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Wee, L.M.; Flores-Jasso, C.F.; Salomon, W.E.; Zamore, D.P. Argonaute Divides Its Rna Guide into Domains with Distinct Functions and Rna-Binding Properties. Cell 2012, 151, 1055–1067. [Google Scholar] [CrossRef]
- Jin, Y.; Chen, Z.; Liu, X.; Zhou, X. Evaluating the Microrna Targeting Sites by Luciferase Reporter Gene Assay. Methods Mol. Biol. 2013, 936, 117–127. [Google Scholar]
- Eichhorn, S.W.; Guo, H.; McGeary, S.E.; Rodriguez-Mias, R.A.; Shin, C.; Baek, D.; Hsu, S.H.; Ghoshal, K.; Villén, J.; Bartel, D.P. Mrna Destabilization Is the Dominant Effect of Mammalian Micrornas by the Time Substantial Repression Ensues. Mol. Cell 2014, 56, 104–115. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan Micrornas. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- Fong, P. Apical Iodide Efflux in Thyroid. Vitam Horm. 2015, 98, 33–62. [Google Scholar]
- Loeb, G.B.; Khan, A.A.; Canner, D.; Hiatt, J.B.; Shendure, J.; Darnell, R.B.; Leslie, C.S.; Rudensky, A.Y. Transcriptome-Wide Mir-155 Binding Map Reveals Widespread Noncanonical Microrna Targeting. Mol. Cell 2012, 48, 760–770. [Google Scholar] [CrossRef] [PubMed]
- Wojcicka, A.; Swierniak, M.; Kornasiewicz, O.; Gierlikowski, W.; Maciag, M.; Kolanowska, M.; Kotlarek, M.; Gornicka, B.; Koperski, L.; Niewinski, G.; et al. Next Generation Sequencing Reveals Microrna Isoforms in Liver Cirrhosis and Hepatocellular Carcinoma. Int. J. Biochem. Cell Biol. 2014, 53, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Scientific &-Educational Software. Clone Manager Professional Suite; Version 8; Scientific &-Educational Software: Denver, CO, USA, 2005. [Google Scholar]
- Wright, M.W.; Bruford, E.A. Naming ‘Junk’: Human Non-Protein Coding Rna (Ncrna) Gene Nomenclature. Hum. Genom. 2011, 5, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Ebert, M.S.; Neilson, J.R.; Shar, P.A.P. Microrna Sponges: Competitive Inhibitors of Small Rnas in Mammalian Cells. Nat. Methods 2007, 4, 721–726. [Google Scholar] [CrossRef] [PubMed]
- Kogai, T.; Schultz, J.J.; Johnson, L.S.; Huang, M.; Brent, G.A. Retinoic Acid Induces Sodium/Iodide Symporter Gene Expression and Radioiodide Uptake in the Mcf-7 Breast Cancer Cell Line. Proc. Natl. Acad. Sci. USA 2000, 97, 8519–8524. [Google Scholar] [CrossRef] [PubMed]
- Unterholzner, S.; Willhauck, M.J.; Cengic, N.; Schütz, M.; Göke, B.; Morris, J.C.; Spitzweg, C. Dexamethasone Stimulation of Retinoic Acid-Induced Sodium Iodide Symporter Expression and Cytotoxicity of 131-I in Breast Cancer Cells. J. Clin. Endocrinol. Metab. 2006, 91, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Lin, X. Regulation of Sodium Iodide Symporter Expression/Function and Tissue-Targeted Gene Transfer of Sodium Iodide Symporter; The Ohio State University: Ohio, OH, USA, 2003. [Google Scholar]
- GraphPad Software. Graphpad Prism; Version 5.03; GraphPad Software: La Jolla, CA, USA, 2009. [Google Scholar]


| Feature | N | (%) | |
|---|---|---|---|
| Sex | Female | 43 | 88% |
| Male | 6 | 12% | |
| Histopathological subtype | PTC cf | 44 | 90% |
| PTC fv | 5 | 10% | |
| No. of foci | Single | 37 | 76% |
| Multiple | 12 | 24% | |
| Tumor diameter | Average | 16.23 | mm |
| Range | 1–73 | mm | |
| pT feature | pT1a | 21 | 43% |
| pT1b | 13 | 27% | |
| pT2 | 6 | 12% | |
| pT3 | 9 | 18% | |
| pT4 | 0 | 0% | |
| pN feature | N0 | 39 | 80% |
| N1a | 5 | 10% | |
| N1b | 5 | 10% | |
| cM feature | M0 | 48 | 98% |
| M1 | 1 | 2% | |
| Vascular invasion | No | 44 | 90% |
| Yes | 5 | 10% | |
| Local invasion | No | 33 | 67% |
| Capsule only | 10 | 20% | |
| Extrathyroidal | 6 | 12% | |
| Stage | I | 39 | 80% |
| II | 2 | 4% | |
| III | 5 | 10% | |
| IVA | 2 | 4% | |
| IVB | 0 | 0% | |
| IVC | 1 | 2% | |
| BRAF status 1 | T (wild type) | 23 | 56% |
| T/A (mutated) | 18 | 44% | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gierlikowski, W.; Broniarek, K.; Cheda, Ł.; Rogulski, Z.; Kotlarek-Łysakowska, M. MiR-181a-5p Regulates NIS Expression in Papillary Thyroid Carcinoma. Int. J. Mol. Sci. 2021, 22, 6067. https://doi.org/10.3390/ijms22116067
Gierlikowski W, Broniarek K, Cheda Ł, Rogulski Z, Kotlarek-Łysakowska M. MiR-181a-5p Regulates NIS Expression in Papillary Thyroid Carcinoma. International Journal of Molecular Sciences. 2021; 22(11):6067. https://doi.org/10.3390/ijms22116067
Chicago/Turabian StyleGierlikowski, Wojciech, Katarzyna Broniarek, Łukasz Cheda, Zbigniew Rogulski, and Marta Kotlarek-Łysakowska. 2021. "MiR-181a-5p Regulates NIS Expression in Papillary Thyroid Carcinoma" International Journal of Molecular Sciences 22, no. 11: 6067. https://doi.org/10.3390/ijms22116067
APA StyleGierlikowski, W., Broniarek, K., Cheda, Ł., Rogulski, Z., & Kotlarek-Łysakowska, M. (2021). MiR-181a-5p Regulates NIS Expression in Papillary Thyroid Carcinoma. International Journal of Molecular Sciences, 22(11), 6067. https://doi.org/10.3390/ijms22116067





