Improvement of the Similarity Spectral Unmixing Approach for Multiplexed Two-Photon Imaging by Linear Dimension Reduction of the Mixing Matrix
Abstract
1. Introduction
2. Results
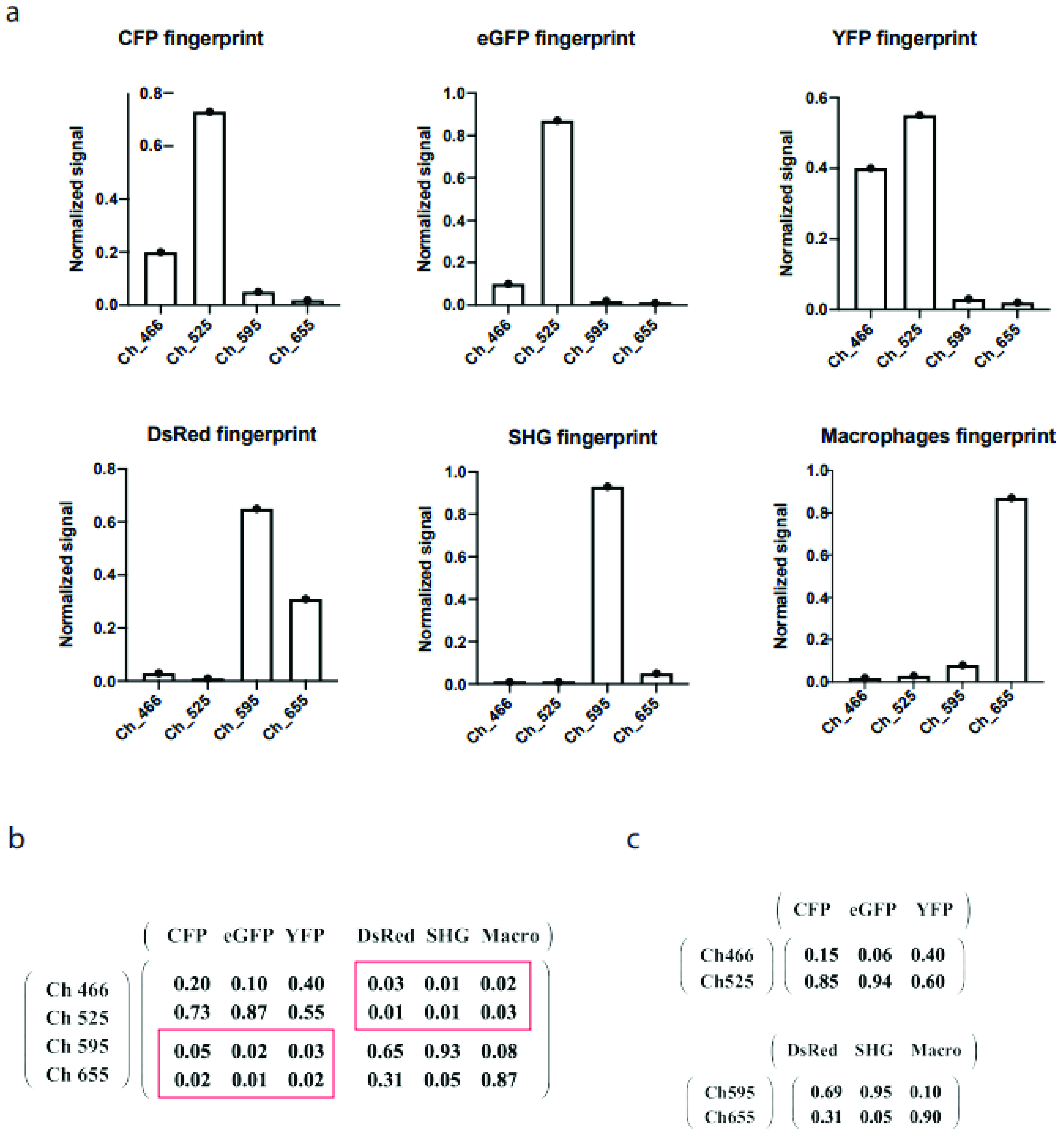
2.1. Dimensionality Reduction of the Spectral Mixing Matrix
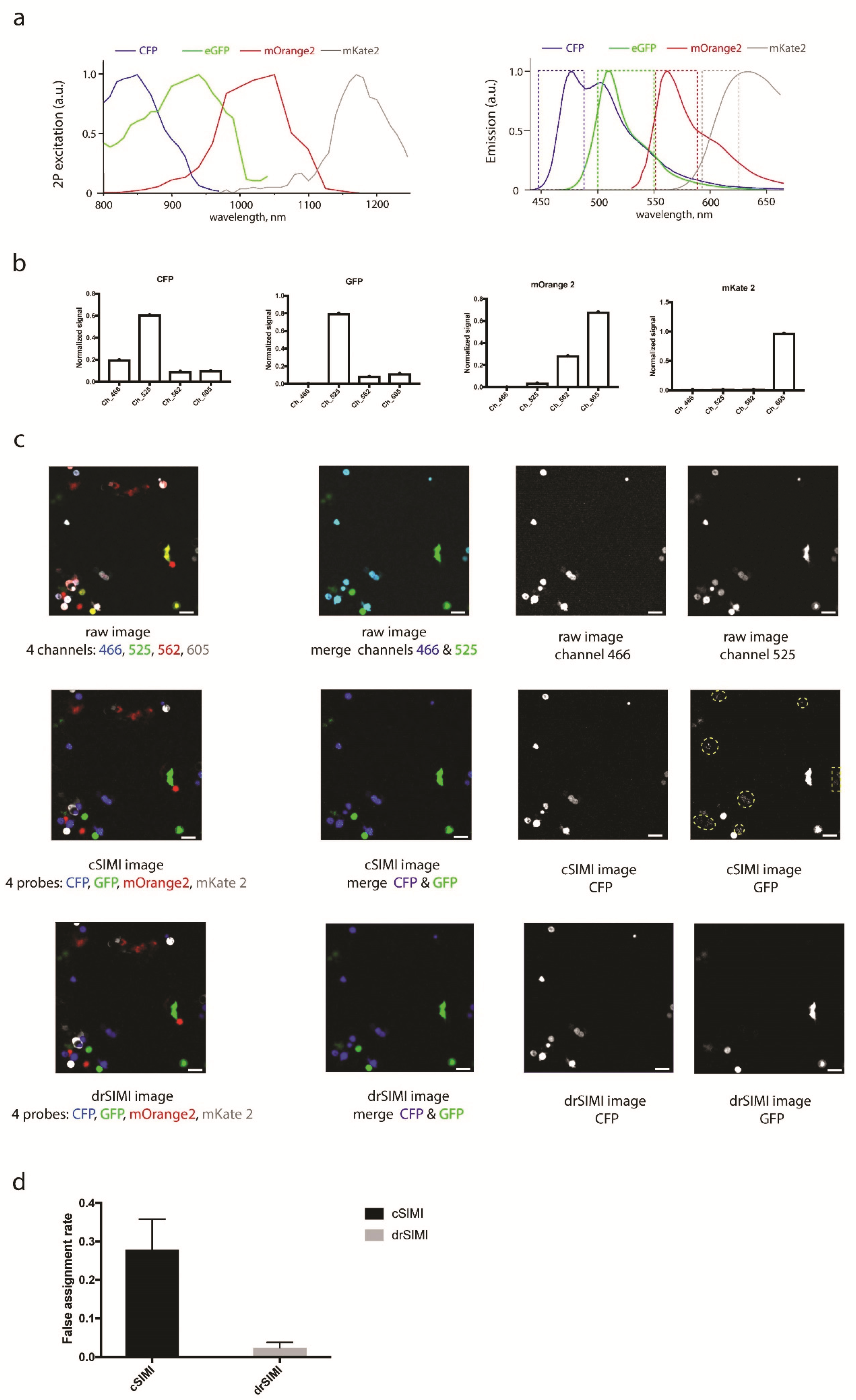
2.2. Benchmarking of drSIMI in Standardized Multiplexed Two-Photon Microscopy Data of Single-Label HEK Cells
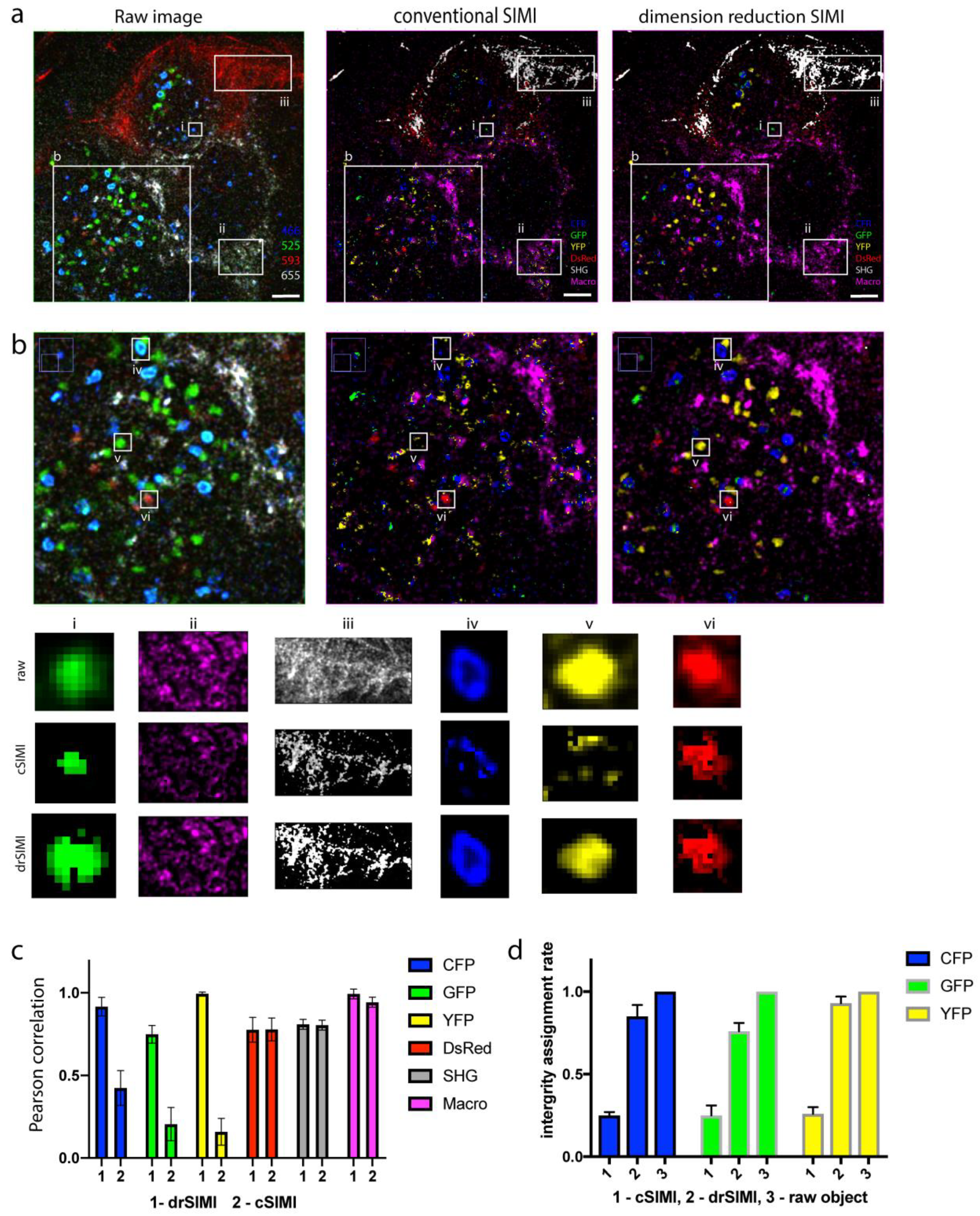
2.3. Superior Performance of drSIMI for Resolving Signals in Multiplexed Deep-Tissue Two-Photon Microscopy Data
3. Discussion
4. Material and Methods
4.1. Multiplexed Two-Photon Laser-Scanning Microscope Setup
4.2. Data Analysis
4.3. HEK Cells Transfection and Imaging
4.4. Mice
4.5. Preparation of Freshly Explanted Popliteal Lymph Node for Imaging
4.6. Recording of Two-Photon Spectra of Fluorescent Proteins, Second-Harmonic Generation, and Autofluorescence
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tang, J.; Van Panhuys, N.; Kastenmüller, W.; Germain, R.N. The future of immunoimaging—Deeper, bigger, more precise, and definitively more colorful. Eur. J. Immunol. 2013, 43, 1407–1412. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.S.; Rodig, S.J. Overview of Tissue Imaging Methods. Methods Mol. Biol. 2019, 2055, 455–465. [Google Scholar] [CrossRef]
- Schubert, W.; Bonnekoh, B.; Pommer, A.J.; Philipsen, L.; Böckelmann, R.; Malykh, Y.; Gollnick, H.; Friedenberger, M.; Bode, M.; Dress, A.W.M. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat. Biotechnol. 2006, 24, 1270–1278. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Reguant, A.; Köhler, R.; Mothes, R.; Bauherr, S.; Hernández, D.C.; Uecker, R.; Holzwarth, K.; Kotsch, K.; Seidl, M.; Philipsen, L.; et al. Multiplexed histology analyses for the phenotypic and spatial characterization of human innate lymphoid cells. Nat. Commun. 2021, 12, 1737. [Google Scholar] [CrossRef]
- Goltsev, Y.; Samusik, N.; Kennedy-Darling, J.; Bhate, S.; Hale, M.; Vazquez, G.; Black, S.; Nolan, G.P. Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell 2018, 174, 968–981.e15. [Google Scholar] [CrossRef]
- Giesen, C.; A O Wang, H.; Schapiro, D.; Zivanovic, N.; Jacobs, A.; Hattendorf, B.; Schüffler, P.J.; Grolimund, D.; Buhmann, J.M.; Brandt, S.; et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 2014, 11, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Rakhymzhan, A.; Leben, R.; Zimmermann, H.; Günther, R.; Mex, P.; Reismann, D.; Ulbricht, C.; Acs, A.; Brandt, A.U.; Lindquist, R.L.; et al. Synergistic Strategy for Multicolor Two-photon Microscopy: Application to the Analysis of Germinal Center Reactions In Vivo. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef]
- Hauser, A.E.; Junt, T.; Mempel, T.R.; Sneddon, M.W.; Kleinstein, S.H.; Henrickson, S.E.; Haberman, A.M. Definition of germinal-center B cell migration in vivo reveals predominant intrazonal circulation patterns. Immunity 2007, 26, 655–667. [Google Scholar] [CrossRef]
- Shcherbakova, D.M.; Verkhusha, V.V. Near-Infrared fluorescent proteins for multicolor in vivo imaging. Nat. Methods 2013, 10, 751–754. [Google Scholar] [CrossRef]
- Shcherbo, D.; Shemiakina, I.I.; Ryabova, A.V.; E Luker, K.; Schmidt, B.T.; A Souslova, E.; Gorodnicheva, T.V.; Strukova, L.; Shidlovskiy, K.M.; Britanova, O.V.; et al. Near-infrared fluorescent proteins. Nat. Methods 2010, 7, 827–829. [Google Scholar] [CrossRef]
- Entenberg, D.; Wyckoff, J.; Gligorijevic, B.; Roussos, E.T.; Verkhusha, V.; Pollard, J.W.; Condeelis, J. Setup and use of a two-laser multiphoton microscope for multichannel intravital fluorescence imaging. Nat. Protoc. 2011, 6, 1500–1520. [Google Scholar] [CrossRef] [PubMed]
- Ricard, C.; Debarbieux, F.C. Six-Color intravital two-photon imaging of brain tumors and their dynamic microenvironment. Front. Cell Neurosci. 2014, 8, 57. [Google Scholar] [CrossRef] [PubMed]
- Herz, J.; Siffrin, V.; Hauser, A.E.; Brandt, A.U.; Leuenberger, T.; Radbruch, H.; Zipp, F.; Niesner, R.A. Expanding Two-Photon Intravital Microscopy to the Infrared by Means of Optical Parametric Oscillator. Biophys. J. 2010, 98, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Mahou, P.; Zimmerley, M.; Loulier, K.; Matho, K.; Labroille, G.; Morin, X.; Supatto, W.; Livet, J.; Débarre, D.; Beaurepaire, E. Multicolor two-photon tissue imaging by wavelength mixing. Nat. Methods 2012, 9, 815–818. [Google Scholar] [CrossRef]
- Tu, H.; Boppart, S.A. Coherent fiber supercontinuum for biophotonics. Laser Photon. Rev. 2013, 7, 628–645. [Google Scholar] [CrossRef]
- Holzapfel, H.Y.; Stern, A.D.; Bouhaddou, M.; Anglin, C.M.; Putur, D.; Comer, S.; Birtwistle, M.R. Fluorescence Multiplexing with Spectral Imaging and Combinatorics. ACS Comb. Sci. 2018, 20, 653–659. [Google Scholar] [CrossRef]
- Seo, J.; Sim, Y.; Kim, J.; Kim, H.; Cho, I.; Yoon, Y.G.; Chang, J.B. PICASSO: Ultra-multiplexed fluorescence imaging of biomolecules through single-round imaging and blind source unmixing. bioRxiv 2021. [Google Scholar] [CrossRef]
- Zimmermann, T. Spectral Imaging and Linear Unmixing in Light Microscopy. Blue Biotechnol. 2005, 95, 245–265. [Google Scholar] [CrossRef]
- Broida, J.G.; Williamson, S.G. A Comprehensive Introduction to Linear Algebra; Addison-Wesley: Boston, MA, USA, 1989. [Google Scholar]
- Livet, J.; Weissman, T.A.; Kang, H.; Draft, R.W.; Lu, J.; Bennis, R.A.; Lichtman, J.W. Trangenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 2007, 450, 56–63. [Google Scholar] [CrossRef]
- Bolte, S.; Cordelières, F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006, 224, 213–232. [Google Scholar] [CrossRef]
- Wang, T.; Ouzounov, D.G.; Wu, C.; Horton, N.G.; Zhang, B.; Wu, C.-H.; Zhang, Y.; Schnitzer, M.J.; Xu, C. Three-photon imaging of mouse brain structure and function through the intact skull. Nat. Methods 2018, 15, 789–792. [Google Scholar] [CrossRef] [PubMed]
- Guesmi, K.; Abdeladim, L.; Tozer, S.; Mahou, P.; Kumamoto, T.; Jurkus, K.; Rigaud, P.; Loulier, K.; Dray, N.; Georges, P.; et al. Dual-color deep-tissue three-photon microscopy with a multiband infrared laser. Light. Sci. Appl. 2018, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Niesner, R.A.; Hauser, A.E.; Entenberg, D. Life through a lense: Technological development and applications in intravital microscopy. Cytometry 2020, 97, 445–447. [Google Scholar] [CrossRef] [PubMed]
- Turcotte, R.; Liang, Y.; Tanimoto, M.; Zhang, Q.; Li, Z.; Koyama, M.; Betzig, E.; Ji, N. Dynamic super-resolution structured illumination imaging in the living brain. Proc. Natl. Acad. Sci. USA 2019, 116, 9586–9591. [Google Scholar] [CrossRef] [PubMed]
- Nowasad, C.R. Tunable dynamics of B cell selection in gut germinal centres. Nature 2020, 588, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Entenberg, D.; Voiculescu, S.; Guo, P.; Borriello, L.; Wang, Y.; Karagiannis, G.S.; Jones, J.; Baccay, F.; Oktay, M.; Condeelis, J. A permanent window for the murine lung enables high-resolution imaging of cancer metastasis. Nat. Methods 2018, 15, 73–80. [Google Scholar] [CrossRef]
- Rakhymzhan, A.; Reuter, L.; Raspe, R.; Bremer, D.; Günther, R.; Leben, R.; Heidelin, J.; Andresen, V.; Cheremukhin, S.; Schulz-Hildebrandt, H.; et al. Coregistered Spectral Optical Coherence Tomography and Two-Photon Microscopy for Multimodal Near-Instantaneous Deep-Tissue Imaging. Cytom. Part A 2020, 97, 515–527. [Google Scholar] [CrossRef]
- Snippert, H.J.; Van Der Flier, L.G.; Sato, T.; Van Es, J.H.; Born, M.V.D.; Kroon-Veenboer, C.; Barker, N.; Klein, A.M.; Van Rheenen, J.; Simons, B.; et al. Intestinal Crypt Homeostasis Results from Neutral Competition between Symmetrically Dividing Lgr5 Stem Cells. Cell 2010, 143, 134–144. [Google Scholar] [CrossRef]
- Seibler, J. Rapid generation of inducible mouse mutants. Nucleic Acids Res. 2003, 31, e12. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rakhymzhan, A.; Acs, A.; Hauser, A.E.; Winkler, T.H.; Niesner, R.A. Improvement of the Similarity Spectral Unmixing Approach for Multiplexed Two-Photon Imaging by Linear Dimension Reduction of the Mixing Matrix. Int. J. Mol. Sci. 2021, 22, 6046. https://doi.org/10.3390/ijms22116046
Rakhymzhan A, Acs A, Hauser AE, Winkler TH, Niesner RA. Improvement of the Similarity Spectral Unmixing Approach for Multiplexed Two-Photon Imaging by Linear Dimension Reduction of the Mixing Matrix. International Journal of Molecular Sciences. 2021; 22(11):6046. https://doi.org/10.3390/ijms22116046
Chicago/Turabian StyleRakhymzhan, Asylkhan, Andreas Acs, Anja E. Hauser, Thomas H. Winkler, and Raluca A. Niesner. 2021. "Improvement of the Similarity Spectral Unmixing Approach for Multiplexed Two-Photon Imaging by Linear Dimension Reduction of the Mixing Matrix" International Journal of Molecular Sciences 22, no. 11: 6046. https://doi.org/10.3390/ijms22116046
APA StyleRakhymzhan, A., Acs, A., Hauser, A. E., Winkler, T. H., & Niesner, R. A. (2021). Improvement of the Similarity Spectral Unmixing Approach for Multiplexed Two-Photon Imaging by Linear Dimension Reduction of the Mixing Matrix. International Journal of Molecular Sciences, 22(11), 6046. https://doi.org/10.3390/ijms22116046




