The Odd Faces of Oligomers: The Case of TRAF2-C, A Trimeric C-Terminal Domain of TNF Receptor-Associated Factor
Abstract
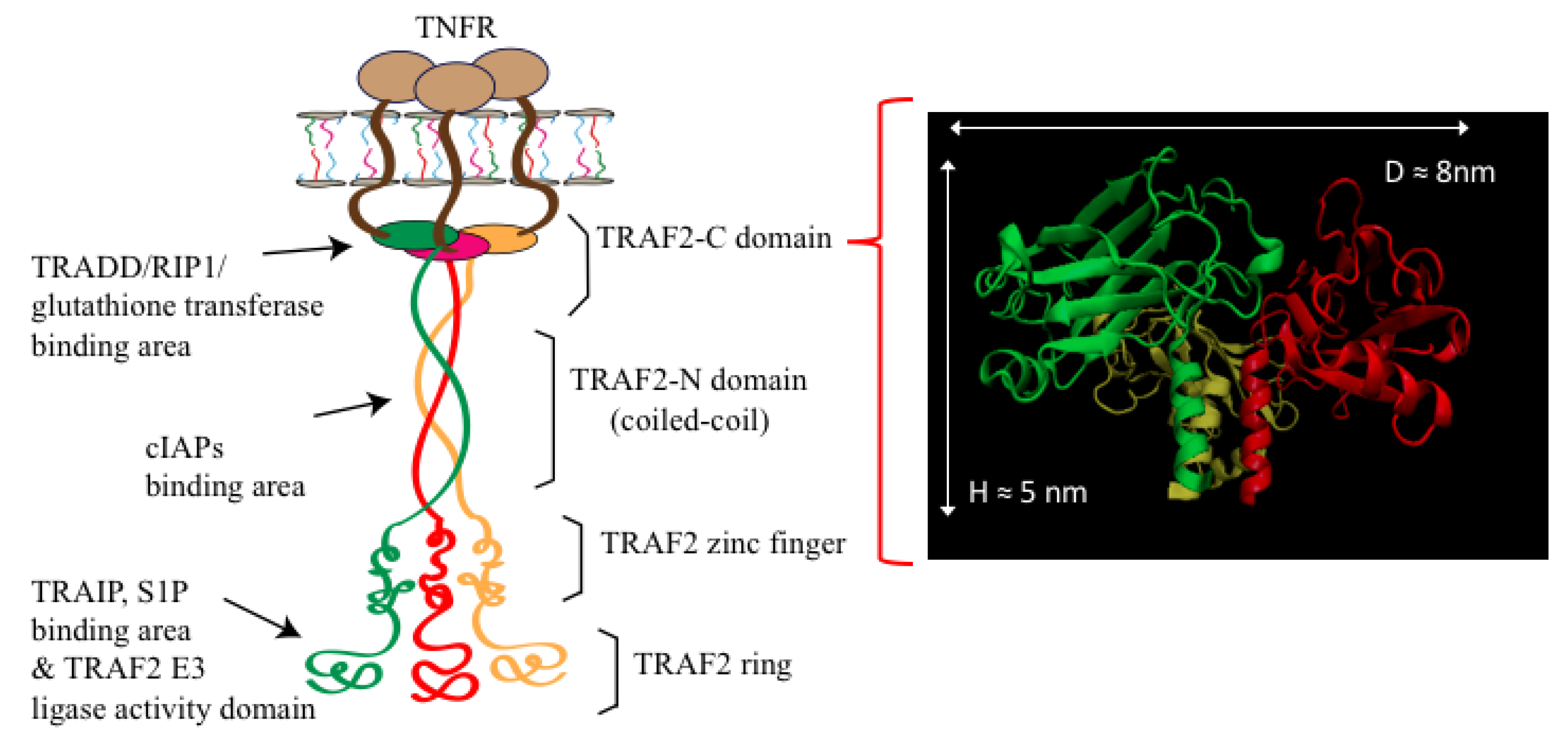
1. Introduction
2. Results
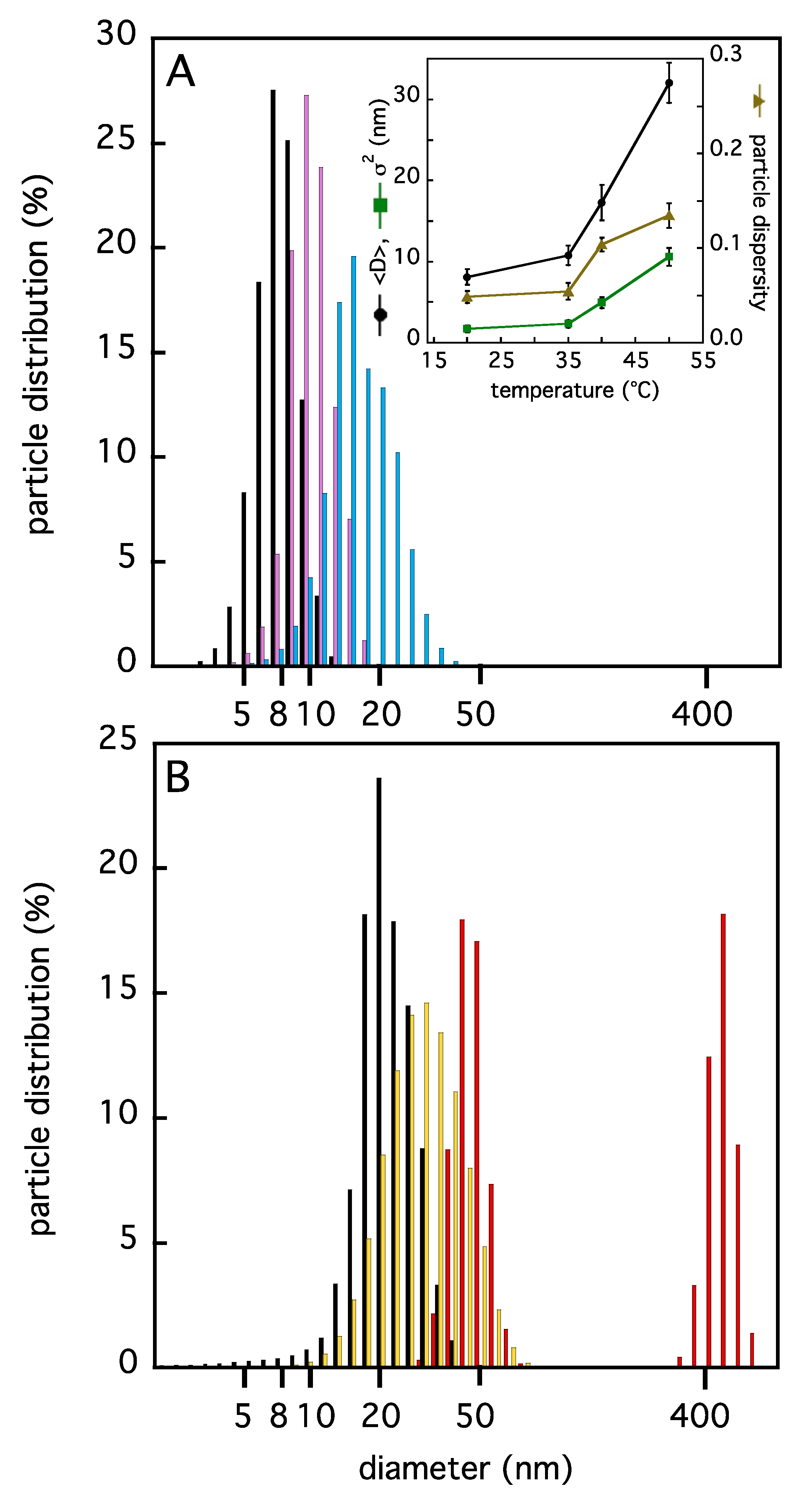
2.1. Insights on TRAF2-C Aggregation State and Secondary Structure Content at Different Temperatures
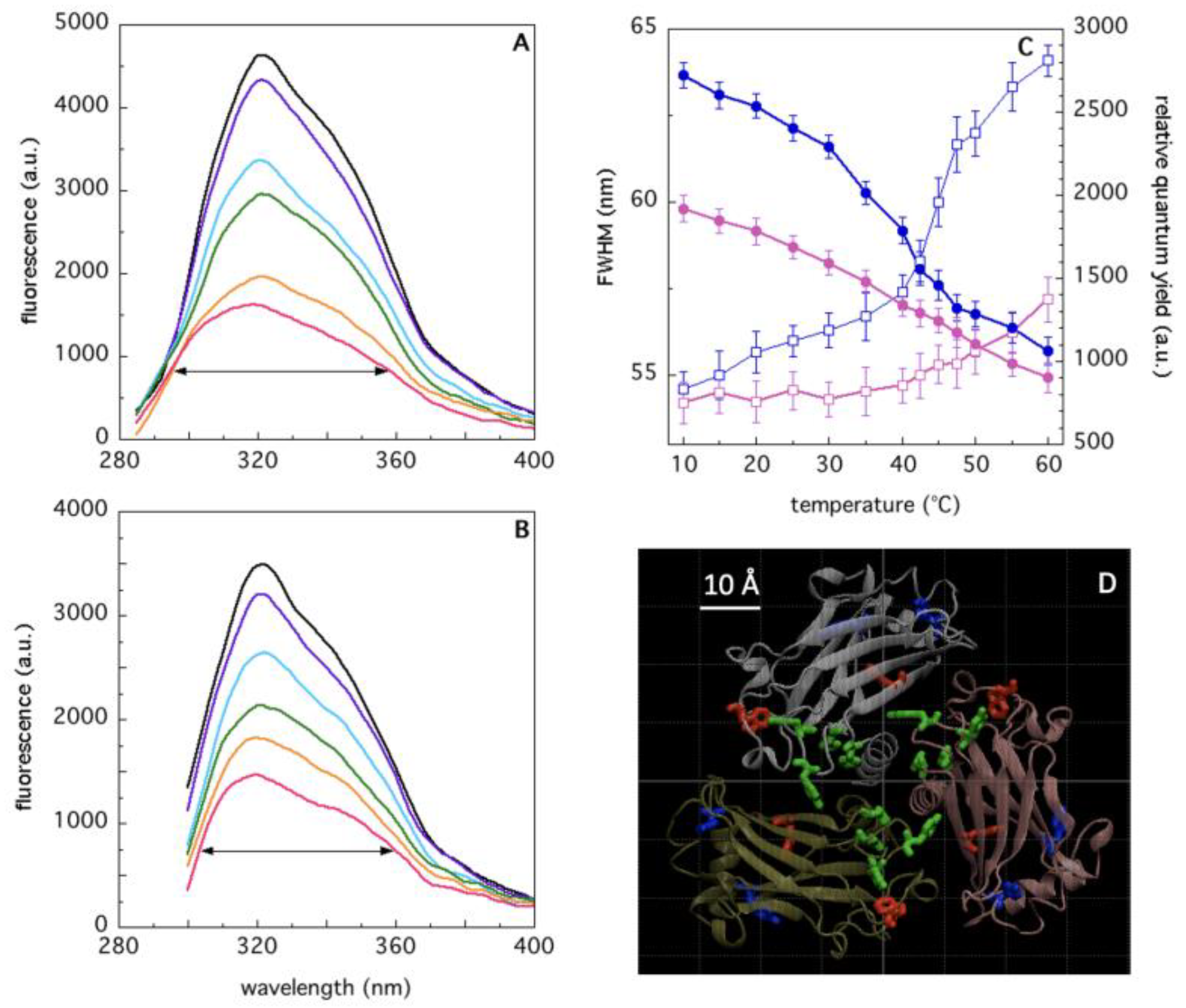
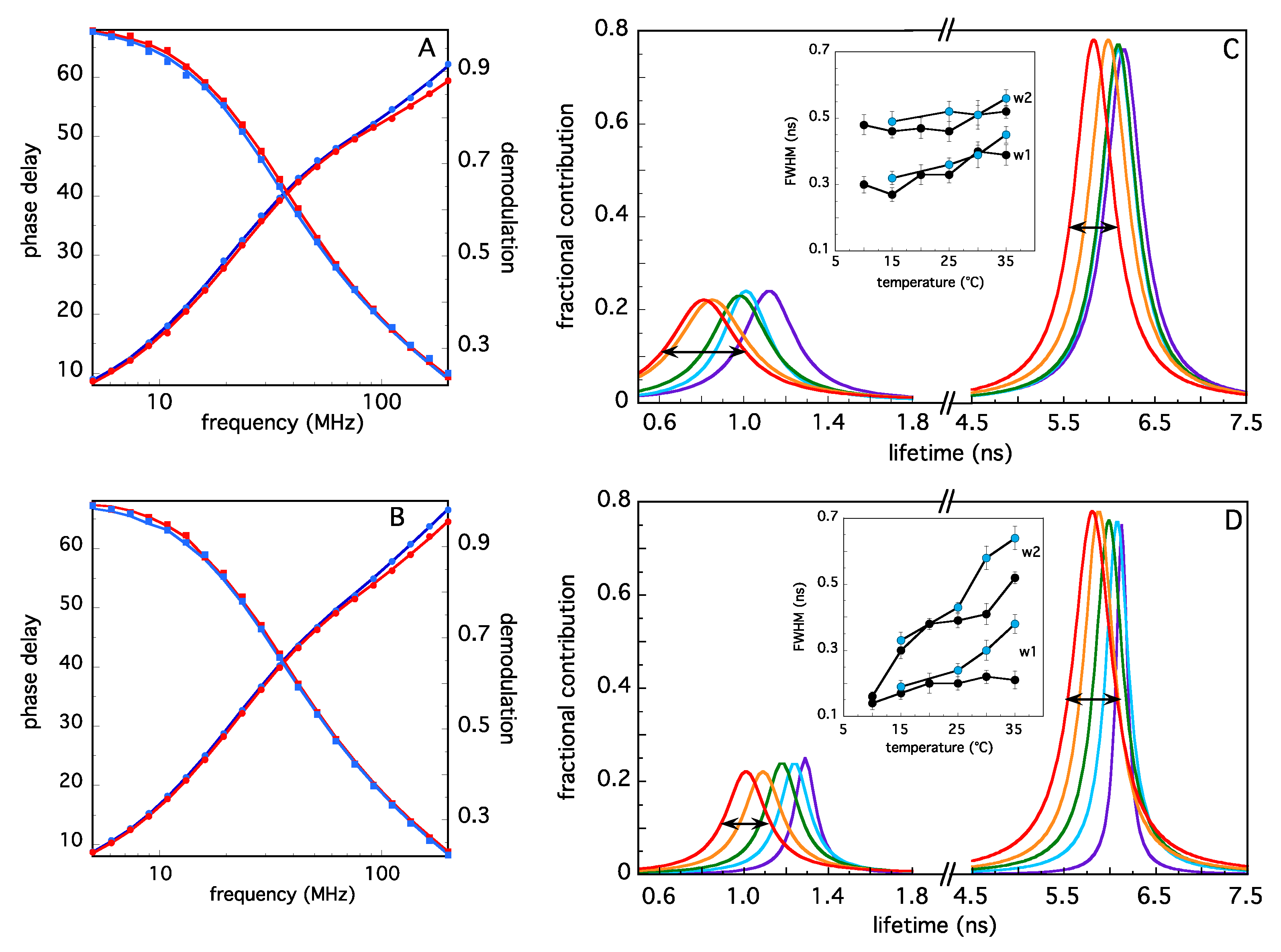
2.2. Analysis of TRAF2-C Tertiary Structure and Temperature-Induced Conformational Changes by Fluorecence Spectroscopy
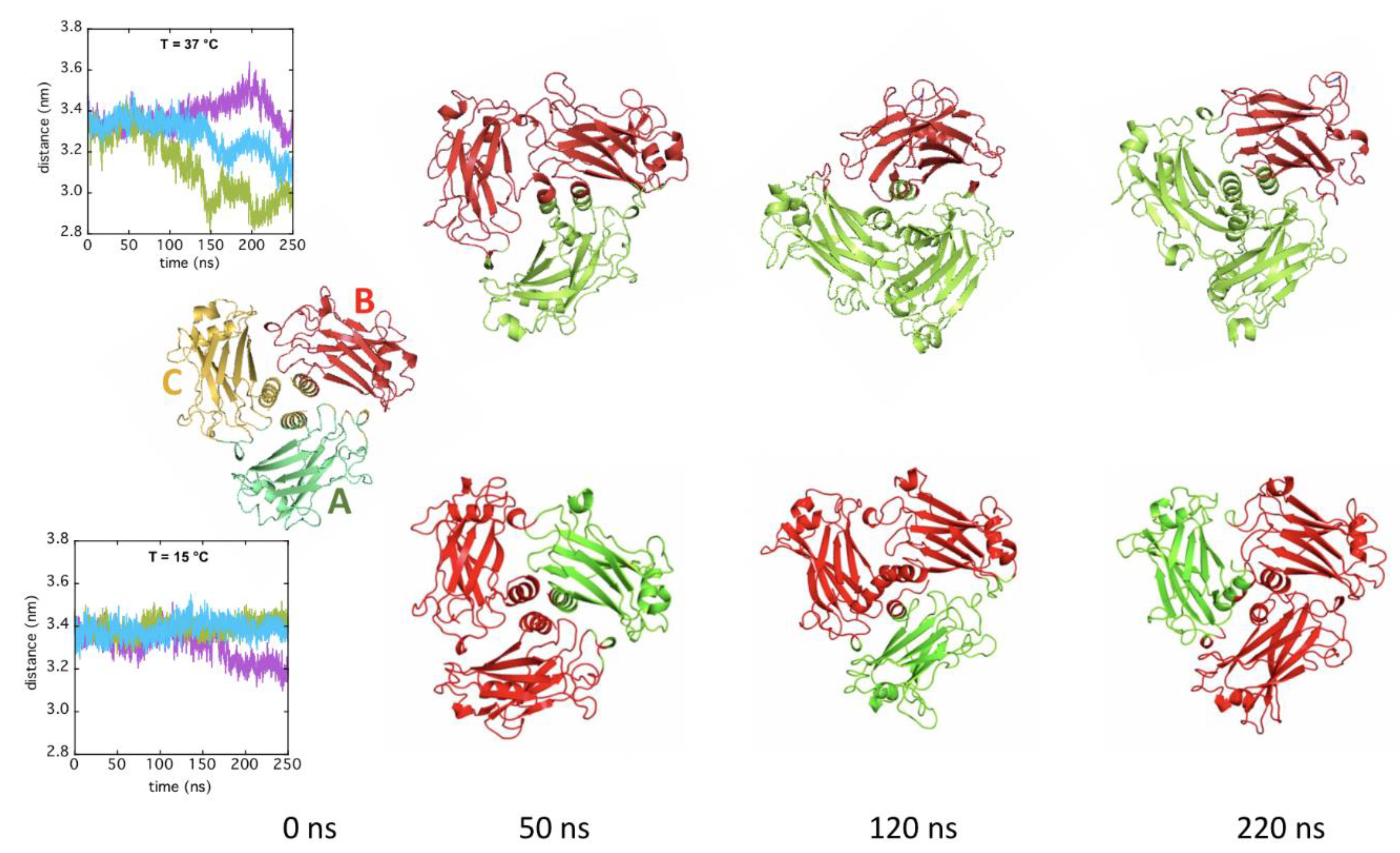
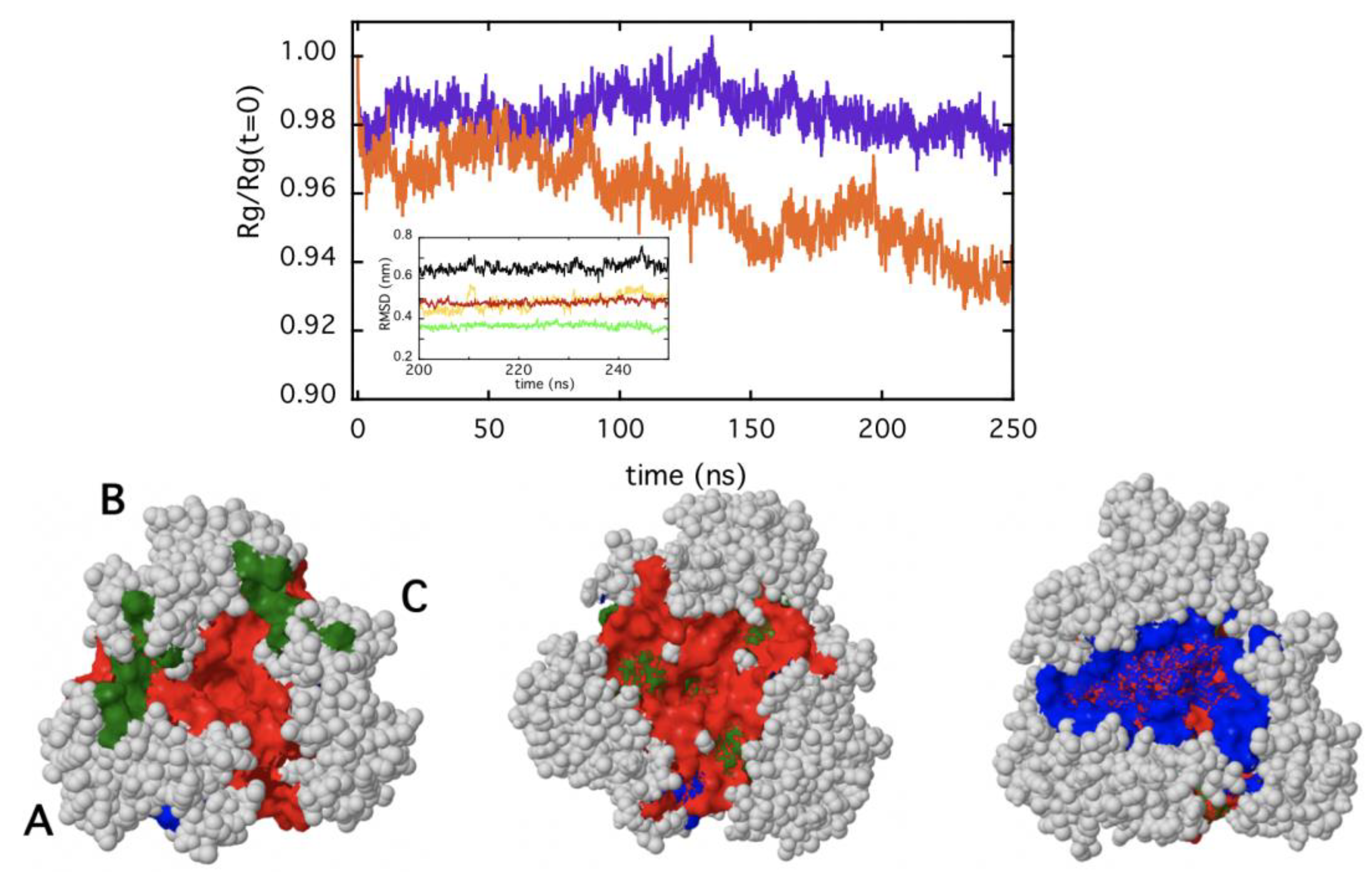
2.3. In Silico Modeling of TRAF2-C Structural Dynamics and Cluster Analysis
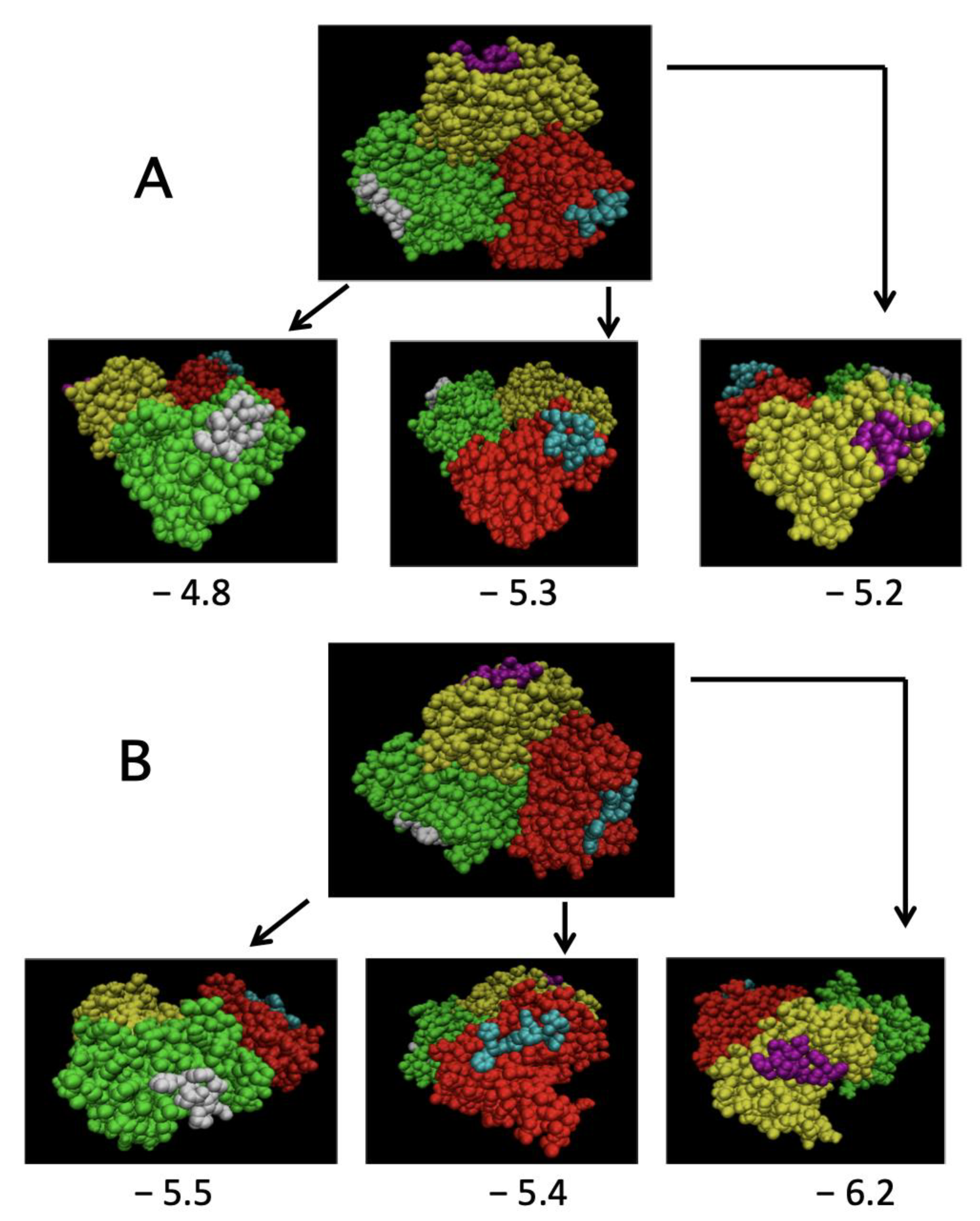
2.4. Simulation of TRAF2-C Binding to Peptide Receptor through Molecular Docking
3. Discussion
4. Materials and Methods
4.1. Sample Preparation
4.2. Spectroscopy Measurements
4.3. Molecular Dynamics
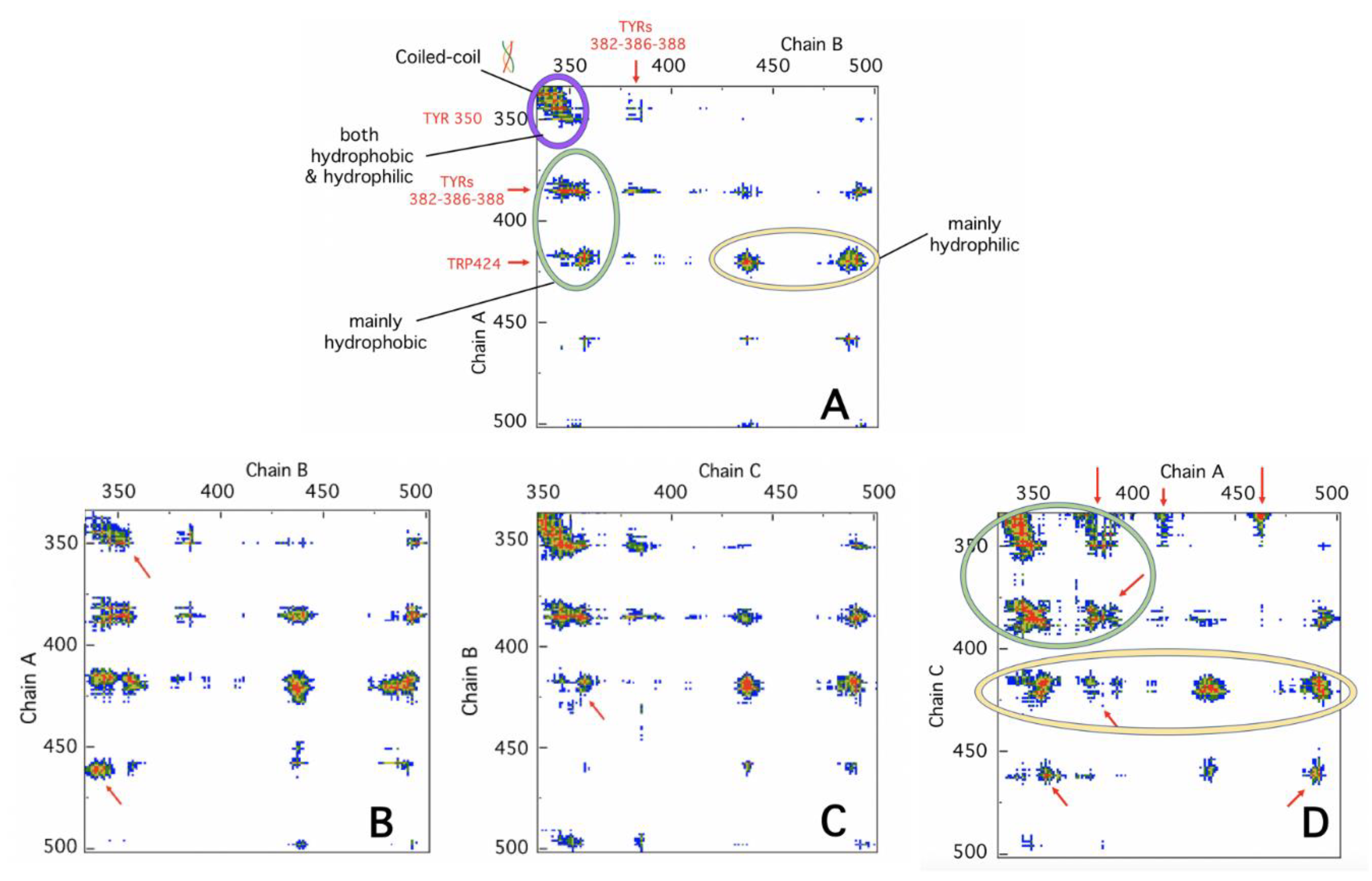
4.4. Protein Contact Networks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MDPI | Multidisciplinary Digital Publishing Institute |
| DOAJ | Directory of open access journals |
| MD | Molecular Dynamics |
| CD | Circular dichroism |
| DLS | Dynamic light scattering |
| TNF | Tumor necrosis factor |
| TNFR | Tumor necrosis factor receptor |
| TRAF(s) | TNF receptor-associated factors |
| RMSD | Root mean square deviation |
| TRAF2-C | C-terminal domain truncated form of TRAF2 |
References
- Mehrabi, P.; Di Pietrantonio, C.; Kim, T.H.; Sljoka, A.; Taverner, K.; Ing, C.; Kruglyak, N.; Pomès, R.; Pai, E.F.; Prosser, R.S. Substrate-Based Allosteric Regulation of a Homodimeric Enzyme. J. Am. Chem. Soc. 2019, 141, 11540–11556. [Google Scholar] [CrossRef]
- Finazzi Agrò, A.; Mei, G. Allostery: The Rebound of Proteins. Methods Mol. Biol. 2021, 2253, 1–6. [Google Scholar] [PubMed]
- Doyle, C.M.; Rumfeldt, J.A.; Broom, H.R.; Broom, A.; Stathopulos, P.B.; Vassall, K.A.; Almey, J.J.; Meiering, E.M. Energetics of oligomeric protein folding and association. Arch. Biochem. Biophys. 2013, 531, 44–64. [Google Scholar] [CrossRef] [PubMed]
- Miles, E.W.; Rhee, S.; Davies, D.R. The molecular basis of substrate channeling. J. Biol. Chem. 1999, 274, 12193–12196. [Google Scholar] [CrossRef]
- Huang, X.; Holden, H.M.; Raushel, F.M. Channeling of substrates and intermediates in enzyme-catalyzed reactions. Annu. Rev. Biochem. 2001, 70, 149–180. [Google Scholar] [CrossRef] [PubMed]
- Mei, G.; Di Venere, A.; Rosato, N.; Finazzi-Agrò, A. The importance of being dimeric. FEBS J. 2005, 272, 16–27. [Google Scholar] [CrossRef]
- Rumfeldt, J.A.; Galvagnion, C.; Vassall, K.A.; Meiering, E.M. Conformational stability and folding mechanisms of dimeric pro- teins. Prog. Biophys. Mol. Biol. 2008, 98, 61–84. [Google Scholar] [CrossRef]
- Perica, T.; Marsh, J.A.; Sousa, F.L.; Natan, E.; Colwell, L.J.; Ahnert, S.E.; Teichmann, S.A. The emergence of protein complexes: Quaternary structure, dynamics and allostery. Colworth Medal Lecture. Biochem. Soc. Trans. 2012, 40, 475–491. [Google Scholar] [CrossRef]
- Lukatsky, D.B.; Shakhnovich, B.E.; Mintseris, J.; Shakhnovich, E.I. Structural similarity enhances interaction propensity of proteins. J. Mol. Biol. 2007, 365, 1596–1606. [Google Scholar] [CrossRef] [PubMed]
- Levy, E.D.; Teichmann, S. Structural, evolutionary, and assembly principles of protein oligomerization. Prog. Mol. Biol. Transl. Sci. 2013, 117, 25–51. [Google Scholar]
- Levy, E.D.; Pereira-Leal, J.B.; Chothia, C.; Teichmann, S.A. 3D complex: A structural classification of protein complexes. PLoS Comput. Biol. 2006, 2, e155. [Google Scholar] [CrossRef]
- Ahnert, S.E.; Marsh, J.A.; Hernández, H.; Robinson, C.V.; Teichmann, S.A. Principles of assembly reveal a periodic table of protein complexes. Science 2015, 350, aaa2245. [Google Scholar] [CrossRef] [PubMed]
- Li, F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef]
- Kiessling, A.R.; Malik, A.; Goldman, A. Recent advances in the understanding of trimeric autotransporter adhesins. Med. Microbiol. Immunol. 2020, 209, 233–242. [Google Scholar] [CrossRef]
- Ben, J.; Zhu, X.; Zhang, H.; Chen, Q. Class A1 scavenger receptors in cardiovascular diseases. Br. J. Pharmacol. 2015, 172, 5523–5530. [Google Scholar] [CrossRef] [PubMed]
- Kucka, K.; Wajant, H. Receptor Oligomerization and its relevance for signaling by receptors of the tumor necrosis factor receptor superfamily. Front. Cell Dev. Biol. 2021, 8, 615141. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yang, C.; Xu, X.F.; Xu, W.; Liu, S.W. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef]
- Bremer, E. Targeting of the tumor necrosis factor receptor superfamily for cancer immunotherapy. ISRN Oncol. 2013, 2013, 371854. [Google Scholar] [CrossRef]
- Chen, G.; Goeddel, D.V. TNF-R1 signaling: A beautiful pathway. Science 2002, 296, 1634–1635. [Google Scholar] [CrossRef]
- Ye, H.; Park, Y.C.; Kreishman, M.; Kieff, E.; Wu, H. The structural basis for the recognition of diverse receptor sequences by TRAF2. Mol. Cell. 1999, 4, 321–430. [Google Scholar] [CrossRef]
- Park, Y.C.; Ye, H.; Hsia, C.; Segal, D.; Rich, R.L.; Liou, H.C.; Myszka, D.G.; Wu, H. A novel mechanism of TRAF signaling revealed by structural and functional analyses of the TRADD-TRAF2 interaction. Cell 2000, 101, 777–787. [Google Scholar] [CrossRef]
- Zheng, C.; Kabaleeswaran, V.; Wang, Y.; Cheng, G.; Wu, H. Crystal structures of the TRAF2: cIAP2 and the TRAF1: TRAF2: cIAP2 complexes—Affinity, specificity and regulation. Mol. Cell. 2010, 38, 101–113. [Google Scholar] [CrossRef] [PubMed]
- De Luca, A.; Mei, G.; Rosato, N.; Nicolai, E.; Federici, L.; Palumbo, C.; Pastore, A.; Serra, M.; Caccuri, A.M. The fine-tuning of TRAF2-GSTP1-1 interaction: Effect of ligand binding and in situ detection of the complex. Cell. Death. Dis. 2014, 5, e1015. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Soetandyo, N.; Wang, Q.; Ye, Y. The zinc finger protein A20 targets TRAF2 to the lysosomes for degradation. Biochim. Biophys. Acta 2009, 1793, 346–353. [Google Scholar] [CrossRef]
- Webster, J.D.; Vucic, D. The Balance of TNF Mediated Pathways Regulates Inflammatory Cell Death Signaling in Healthy and Diseased Tissues. Front. Cell Dev. Biol. 2020, 8, 365. [Google Scholar] [CrossRef]
- Shi, J.H.; Sun, S.C. Tumor Necrosis Factor Receptor-Associated Factor Regulation of Nuclear Factor κB and Mitogen-Activated Protein Kinase Pathways. Front. Immunol. 2018, 9, 1849. [Google Scholar] [CrossRef]
- Ceccarelli, A.; Di Venere, A.; Nicolai, E.; De Luca, A.; Rosato, N.; Gratton, E.; Mei, G.; Caccuri, A.M. New insight into the interaction of TRAF2 C-terminal domain with lipid raft microdomains. Biochim. Biophys. Acta 2017, 1862, 813–822. [Google Scholar] [CrossRef]
- Di Venere, A.; Nicolai, E.; Sinibaldi, F.; Di Pierro, D.; Caccuri, A.M.; Mei, G. Studying the TRAF2 binding to model membranes: The role of subunits dissociation. Biotechnol. Appl. Biochem. 2018, 65, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Minicozzi, V.; Di Venere, A.; Nicolai, E.; Giuliani, A.; Caccuri, A.M.; Di Paola, L.; Mei, G. Non-symmetrical structural behavior of a symmetric protein: The case of homo-trimeric TRAF2 (tumor necrosis factor-receptor associated factor 2). J. Biomol. Struct. Dyn. 2021, 39, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Di Paola, L.; De Ruvo, M.; Paci, P.; Santoni, D.; Giuliani, A. Protein contact networks: An emerging paradigm in chemistry. Chem. Rev. 2013, 113, 1598–1613. [Google Scholar] [CrossRef]
- Cantor, C.R.; Schimmel, P.R. Fluorescence spectroscopy. In Biophysical Chemistry; Kluwer, W.H., Ed.; Freeman and Company Publishers: San Francisco, CA, USA, 1980; pp. 433–465. [Google Scholar]
- Ceccarelli, A.; Di Venere, A.; Nicolai, E.; De Luca, A.; Minicozzi, V.; Rosato, N.; Caccuri, A.M.; Mei, G. TNFR-Associated Factor-2 (TRAF2): Not only a Trimer. Biochemistry 2015, 54, 6153–6161. [Google Scholar] [CrossRef]
- Park, Y.C.; Burkitt, V.; Villa, A.R.; Tong, L.; Wu, H. Structural basis for self-association and receptor recognition of human TRAF2. Nature 1999, 398, 533–538. [Google Scholar] [CrossRef]
- Vangone, A.; Spinelli, R.; Scarano, V.; Cavallo, L.; Oliva, R. COCOMAPS: A web application to analyse and visualize contacts at the interface of biomolecular complexes. Bioinformatic 2011, 27, 2915–2916. [Google Scholar] [CrossRef] [PubMed]
- La, D.; Esquivel-Rodriguez, J.; Venkatraman, V.; Li, B.; Sael, L.; Ueng, S.; Ahrendt, S.; Kihara, D. 3D-SURFER: Software for high throughput protein surface comparison and analysis. Bioinformatics 2009, 25, 2843–2844. [Google Scholar] [CrossRef]
- McWhirter, S.M.; Pullen, S.S.; Holton, J.M.; Crute, J.J.; Kehry, M.R.; Alber, T. Crystallographic analysis of CD40 recognition and signaling by human TRAF2. Proc. Natl. Acad. Sci. USA 1999, 96, 8408–8413. [Google Scholar] [CrossRef] [PubMed]
- Nilofer, C.; Sukhwal, A.; Mohanapriya, A.; Kangueane, P. Protein-protein interfaces are vdW dominant with selective H-bonds and (or) electrostatics towards broad functional specificity. Bioinformation 2017, 13, 164–173. [Google Scholar] [CrossRef]
- Jayashree, S.; Murugavel, P.; Sowdhamini, R.; Srinivasan, N. Interface residues of transient protein-protein complexes have extensive intra-protein interactions apart from inter-protein interactions. Biol. Direct 2019, 14, 1. [Google Scholar] [CrossRef] [PubMed]
- Mayes, M.L.; Perreault, L. Probing the Nature of Noncovalent Interactions in Dimers of Linear Tyrosine-Based Dipeptides. ACS Omega 2019, 4, 911–919. [Google Scholar] [CrossRef]
- Yan, C.; Wu, F.; Jernigan, R.L.; Dobbs, D.; Honavar, V. Characterization of Protein–Protein Interfaces. Protein J. 2008, 27, 59–70. [Google Scholar] [CrossRef]
- Silke, J.; Brink, R. Regulation of TNFRSF and innate immune signalling complexes by TRAFs and cIAPs. Cell. Death Differ. 2010, 17, 35–45. [Google Scholar] [CrossRef]
- Roy, S.; Jaiswar, A.; Sarkar, R. Dynamic Asymmetry Exposes 2019-nCoV Prefusion Spike. J. Phys. Chem. Lett. 2020, 17, 7021–7027. [Google Scholar] [CrossRef]
- Di Paola, L.; Hadi-Alijanvand, H.; Song, X.; Hu, G.; Giuliani, A. The Discovery of a Putative Allosteric Site in the SARS-CoV-2 Spike Protein Using an Integrated Structural/Dynamic Approach. J. Proteome Res. 2020, 19, 4576–4586. [Google Scholar] [CrossRef]
- Noce, A.; Santoro, M.L.; Marrone, G.; D’Agostini, C.; Amelio, I.; Duggento, A.; Tesauro, M.; Di Daniele, N. Serological determinants of COVID-19. Biol. Direct 2020, 15, 21. [Google Scholar] [CrossRef]
- Ehsani, S. COVID-19 and iron dysregulation: Distant sequence similarity between hepcidin and the novel coronavirus spike glycoprotein. Biol. Direct 2020, 15, 19. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. AMDock: A versatile graphical tool for assisting molecular docking with Autodock Vina and Autodock4. Biol. Direct 2020, 15, 12. [Google Scholar] [CrossRef] [PubMed]
- Di Paola, L.; Mei, G.; Di Venere, A.; Giuliani, A. Disclosing Allostery Through Protein Contact Networks. Methods Mol. Biol. 2021, 2253, 7–20. [Google Scholar] [PubMed]
- Viloira, J.S.; Allega, M.F.; Lambrughi, M.; Papaleo, E. An optimal distance cutoff for contact-based Protein Structure Networks using side-chain centers of mass. Sci. Rep. 2017, 7, 2838. [Google Scholar]
- Brinda, K.V.; Vishveshwara, S. A network representation of protein structures: Implications for protein stability. Biophys. J. 2005, 89, 4159–4170. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Venere, A.; Nicolai, E.; Minicozzi, V.; Caccuri, A.M.; Di Paola, L.; Mei, G. The Odd Faces of Oligomers: The Case of TRAF2-C, A Trimeric C-Terminal Domain of TNF Receptor-Associated Factor. Int. J. Mol. Sci. 2021, 22, 5871. https://doi.org/10.3390/ijms22115871
Di Venere A, Nicolai E, Minicozzi V, Caccuri AM, Di Paola L, Mei G. The Odd Faces of Oligomers: The Case of TRAF2-C, A Trimeric C-Terminal Domain of TNF Receptor-Associated Factor. International Journal of Molecular Sciences. 2021; 22(11):5871. https://doi.org/10.3390/ijms22115871
Chicago/Turabian StyleDi Venere, Almerinda, Eleonora Nicolai, Velia Minicozzi, Anna Maria Caccuri, Luisa Di Paola, and Giampiero Mei. 2021. "The Odd Faces of Oligomers: The Case of TRAF2-C, A Trimeric C-Terminal Domain of TNF Receptor-Associated Factor" International Journal of Molecular Sciences 22, no. 11: 5871. https://doi.org/10.3390/ijms22115871
APA StyleDi Venere, A., Nicolai, E., Minicozzi, V., Caccuri, A. M., Di Paola, L., & Mei, G. (2021). The Odd Faces of Oligomers: The Case of TRAF2-C, A Trimeric C-Terminal Domain of TNF Receptor-Associated Factor. International Journal of Molecular Sciences, 22(11), 5871. https://doi.org/10.3390/ijms22115871







