Regulation of Bone Cell Differentiation and Activation by Microbe-Associated Molecular Patterns
Abstract
1. Introduction
2. Microbe-Associated Molecular Patterns
2.1. Bacterial Polysaccharides
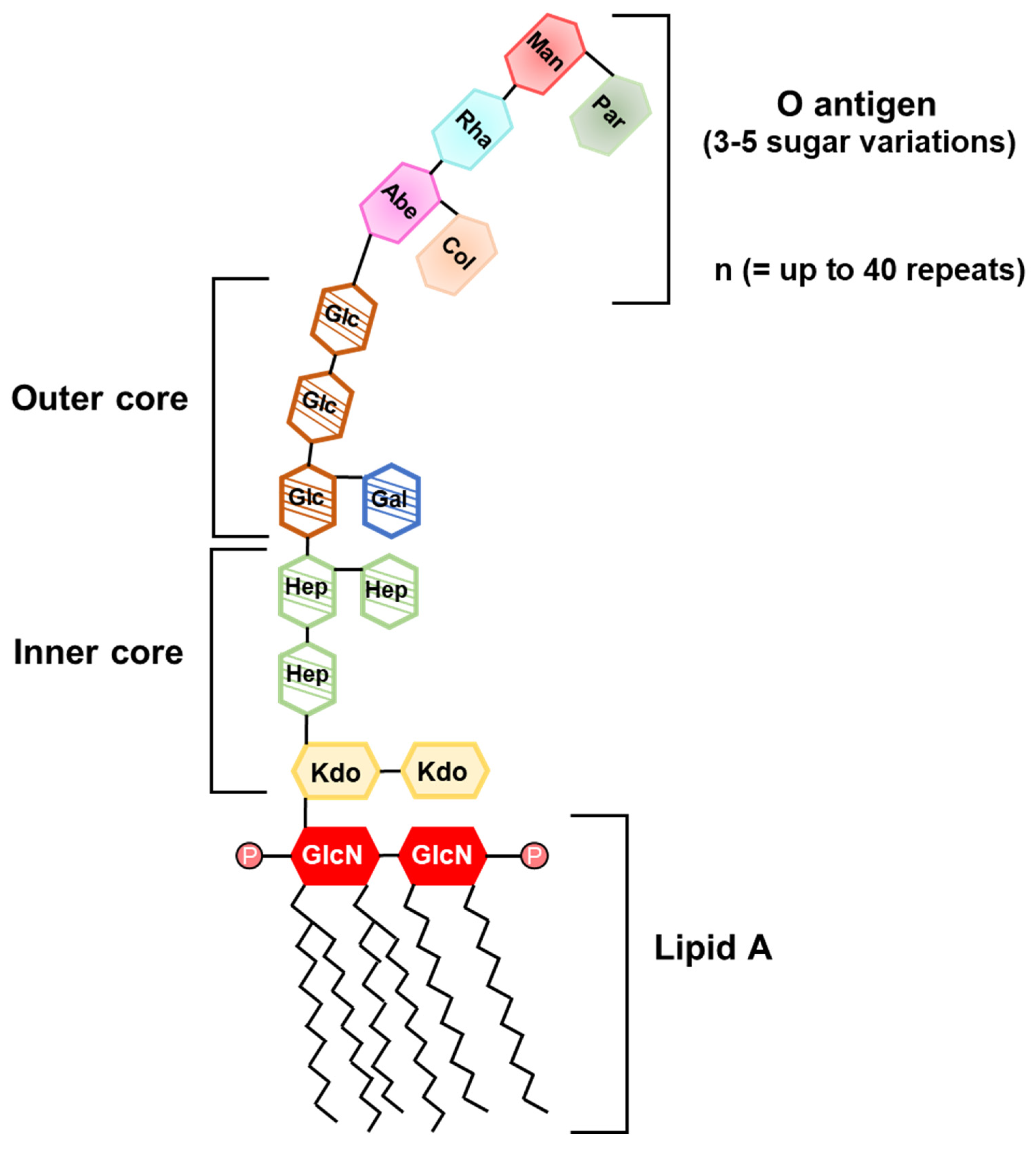
2.1.1. Lipopolysaccharide
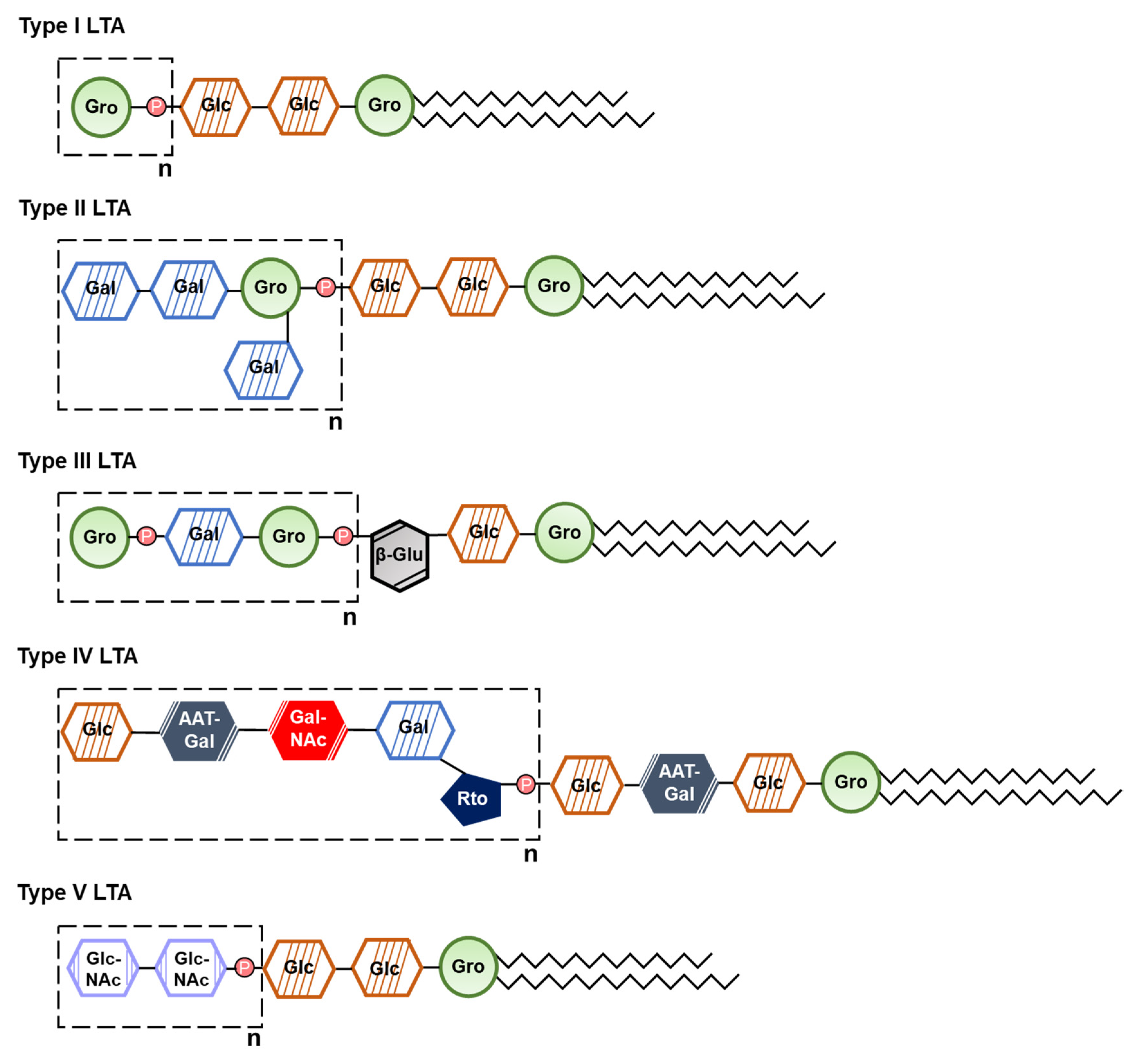
2.1.2. Lipoteichoic Acid
2.2. Surface Proteins
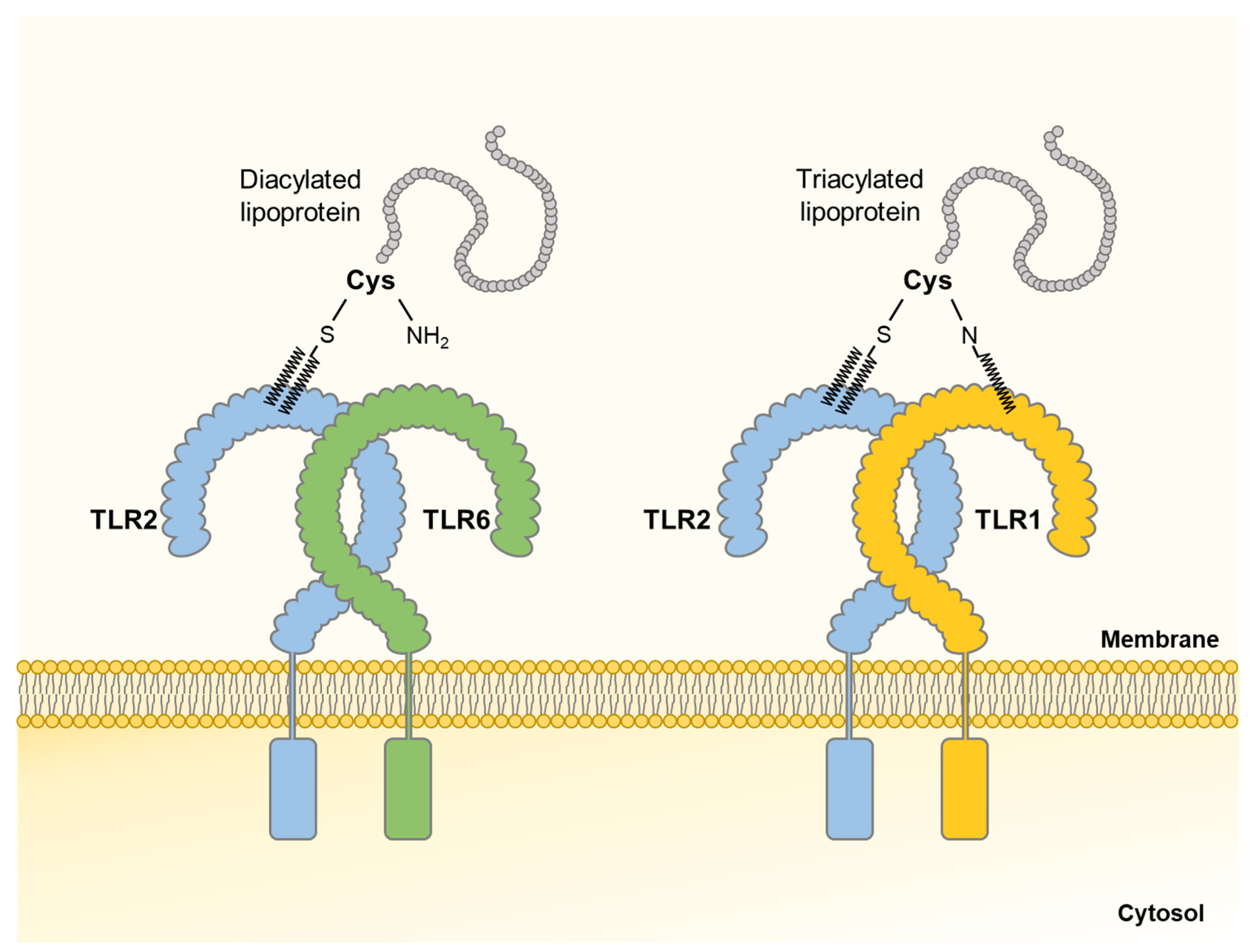
2.2.1. Lipoprotein
2.2.2. Adhesin
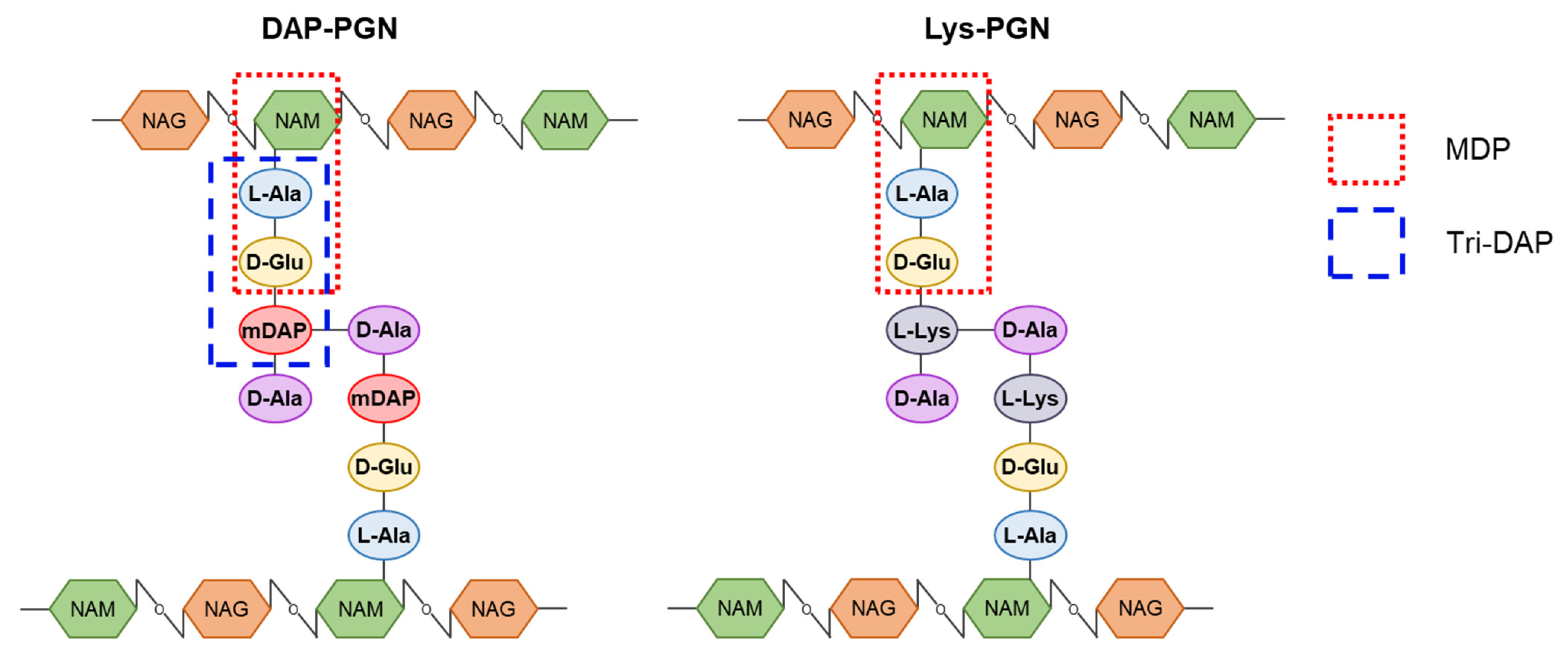
2.3. Peptidoglycan
2.4. Secretory Microbial Molecules
2.4.1. Short Chain Fatty Acid
2.4.2. Extracellular Vesicle
2.4.3. Extracellular Polysaccharide
2.4.4. Cyclic Dinucleotide
3. Therapeutics
3.1. Treatment of Microbe-Associated Molecular Patterns-Induced Bone Diseases
3.2. Probiotics as Therapeutic Agent for Bone Health
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kular, J.; Tickner, J.; Chim, S.M.; Xu, J. An overview of the regulation of bone remodelling at the cellular level. Clin. Biochem. 2012, 45, 863–873. [Google Scholar] [CrossRef]
- Robling, A.G.; Castillo, A.B.; Turner, C.H. Biomechanical and molecular regulation of bone remodeling. Annu. Rev. Biomed. Eng. 2006, 8, 455–498. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; McDonald, J.M. Disorders of bone remodeling. Annu. Rev. Pathol. 2011, 6, 121–145. [Google Scholar] [CrossRef] [PubMed]
- Krauss, J.L.; Roper, P.M.; Ballard, A.; Shih, C.C.; Fitzpatrick, J.A.J.; Cassat, J.E.; Ng, P.Y.; Pavlos, N.J.; Veis, D.J. Staphylococcus aureus Infects Osteoclasts and Replicates Intracellularly. mBio 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Wright, J.A.; Nair, S.P. Interaction of staphylococci with bone. Int. J. Med. Microbiol. 2010, 300, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Martin, T.R.; Mathison, J.C.; Tobias, P.S.; Leturcq, D.J.; Moriarty, A.M.; Maunder, R.J.; Ulevitch, R.J. Lipopolysaccharide binding protein enhances the responsiveness of alveolar macrophages to bacterial lipopolysaccharide. Implications for cytokine production in normal and injured lungs. J. Clin. Investig. 1992, 90, 2209–2219. [Google Scholar] [CrossRef]
- Charles, J.F.; Nakamura, M.C. Bone and the innate immune system. Curr. Osteoporos. Rep. 2014, 12, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, A.; Okahashi, N.; Nakagawa, I.; Kawabata, S.; Amano, A.; Ooshima, T.; Hamada, S. Streptococcus pyogenes infection induces septic arthritis with increased production of the receptor activator of the NF-kappaB ligand. Infect. Immun. 2003, 71, 6019–6026. [Google Scholar] [CrossRef]
- Binder Gallimidi, A.; Fischman, S.; Revach, B.; Bulvik, R.; Maliutina, A.; Rubinstein, A.M.; Nussbaum, G.; Elkin, M. Periodontal pathogens Porphyromonas gingivalis and Fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget 2015, 6, 22613–22623. [Google Scholar] [CrossRef]
- Parvaneh, K.; Jamaluddin, R.; Karimi, G.; Erfani, R. Effect of probiotics supplementation on bone mineral content and bone mass density. Sci. World J. 2014, 2014, 595962. [Google Scholar] [CrossRef]
- Li, J.Y.; Chassaing, B.; Tyagi, A.M.; Vaccaro, C.; Luo, T.; Adams, J.; Darby, T.M.; Weitzmann, M.N.; Mulle, J.G.; Gewirtz, A.T.; et al. Sex steroid deficiency-associated bone loss is microbiota dependent and prevented by probiotics. J. Clin. Investig. 2016, 126, 2049–2063. [Google Scholar] [CrossRef]
- McCabe, L.R.; Irwin, R.; Schaefer, L.; Britton, R.A. Probiotic use decreases intestinal inflammation and increases bone density in healthy male but not female mice. J. Cell. Physiol. 2013, 228, 1793–1798. [Google Scholar] [CrossRef]
- Kobayashi, R.; Kobayashi, T.; Sakai, F.; Hosoya, T.; Yamamoto, M.; Kurita-Ochiai, T. Oral administration of Lactobacillus gasseri SBT2055 is effective in preventing Porphyromonas gingivalis-accelerated periodontal disease. Sci. Rep. 2017, 7, 545. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, T.; Hajishengallis, G. Topical treatment with probiotic Lactobacillus brevis CD2 inhibits experimental periodontal inflammation and bone loss. J. Periodontal Res. 2014, 49, 785–791. [Google Scholar] [CrossRef]
- Chu, H.; Mazmanian, S.K. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat. Immunol. 2013, 14, 668–675. [Google Scholar] [CrossRef] [PubMed]
- Iwami, K.; Moriyama, T. Effects of short chain fatty acid, sodium butyrate, on osteoblastic cells and osteoclastic cells. Int. J. Biochem. 1993, 25, 1631–1635. [Google Scholar] [CrossRef]
- Song, M.K.; Kim, H.Y.; Choi, B.K.; Kim, H.H. Filifactor alocis-derived extracellular vesicles inhibit osteogenesis through TLR2 signaling. Mol. Oral Microbiol. 2020, 35, 202–210. [Google Scholar] [CrossRef]
- Kwon, Y.; Park, O.J.; Kim, J.; Cho, J.H.; Yun, C.H.; Han, S.H. Cyclic Dinucleotides Inhibit Osteoclast Differentiation Through STING-Mediated Interferon-beta Signaling. J. Bone Miner. Res. 2019, 34, 1366–1375. [Google Scholar] [CrossRef]
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef]
- Choi, H.W.; Klessig, D.F. DAMPs, MAMPs, and NAMPs in plant innate immunity. BMC Plant Biol. 2016, 16, 232. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef]
- Sun, M.; Wu, W.; Liu, Z.; Cong, Y. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J. Gastroenterol. 2017, 52, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Brubaker, S.W.; Bonham, K.S.; Zanoni, I.; Kagan, J.C. Innate immune pattern recognition: A cell biological perspective. Annu. Rev. Immunol. 2015, 33, 257–290. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. TLR signaling. Semin. Immunol. 2007, 19, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Platnich, J.M.; Muruve, D.A. NOD-like receptors and inflammasomes: A review of their canonical and non-canonical signaling pathways. Arch. Biochem. Biophys. 2019, 670, 4–14. [Google Scholar] [CrossRef]
- Chatterjee, S.; Jungraithmayr, W.; Bagchi, D. Immunity and Inflammation in Health and Disease: Emerging Roles of Nutraceuticals and Functional Foods in Immune Support; Academic Press: London, UK, 2018; pp. 175–187. [Google Scholar]
- D’Amelio, P.; Sassi, F. Gut Microbiota, Immune System, and Bone. Calcif. Tissue Int. 2018, 102, 415–425. [Google Scholar] [CrossRef]
- Chen, M.F.; Chang, C.H.; Hu, C.C.; Wu, Y.Y.; Chang, Y.; Ueng, S.W.N. Periprosthetic Joint Infection Caused by Gram-Positive Versus Gram-Negative Bacteria: Lipopolysaccharide, but not Lipoteichoic Acid, Exerts Adverse Osteoclast-Mediated Effects on the Bone. J. Clin. Med. 2019, 8, 1289. [Google Scholar] [CrossRef]
- Ridwan, R.D.; Sidarningsih, T.K.; Salim, S. Effect of lipopolysaccharide derived from surabaya isolates of Actinobacillus actinomycetemcomitans on alveolar bone destruction. Vet. World 2018, 11, 161–166. [Google Scholar] [CrossRef]
- Nishihara, T.; Ishihara, Y.; Koseki, T.; Boutsi, E.A.; Senpuku, H.; Hanada, N. Membrane-associated interleukin-1 on macrophages stimulated with Actinobacillus actinomycetemcomitans lipopolysaccharide induces osteoclastic bone resorption in vivo. Cytobios 1995, 81, 229–237. [Google Scholar] [PubMed]
- Zou, W.; Bar-Shavit, Z. Dual modulation of osteoclast differentiation by lipopolysaccharide. J. Bone Miner. Res. 2002, 17, 1211–1218. [Google Scholar] [CrossRef]
- Liu, J.; Wang, S.; Zhang, P.; Said-Al-Naief, N.; Michalek, S.M.; Feng, X. Molecular mechanism of the bifunctional role of lipopolysaccharide in osteoclastogenesis. J. Biol. Chem. 2009, 284, 12512–12523. [Google Scholar] [CrossRef]
- Kadono, H.; Kido, J.; Kataoka, M.; Yamauchi, N.; Nagata, T. Inhibition of osteoblastic cell differentiation by lipopolysaccharide extract from Porphyromonas gingivalis. Infect. Immun. 1999, 67, 2841–2846. [Google Scholar] [CrossRef]
- Tomomatsu, N.; Aoki, K.; Alles, N.; Soysa, N.S.; Hussain, A.; Nakachi, H.; Kita, S.; Shimokawa, H.; Ohya, K.; Amagasa, T. LPS-induced inhibition of osteogenesis is TNF-alpha dependent in a murine tooth extraction model. J. Bone Miner. Res. 2009, 24, 1770–1781. [Google Scholar] [CrossRef]
- Bandow, K.; Maeda, A.; Kakimoto, K.; Kusuyama, J.; Shamoto, M.; Ohnishi, T.; Matsuguchi, T. Molecular mechanisms of the inhibitory effect of lipopolysaccharide (LPS) on osteoblast differentiation. Biochem. Biophys. Res. Commun. 2010, 402, 755–761. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Park, O.J.; Kim, J.; Baik, J.E.; Yun, C.H.; Han, S.H. Lipoteichoic Acid of Enterococcus faecalis Inhibits the Differentiation of Macrophages into Osteoclasts. J. Endod. 2016, 42, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Heng, B.C.; Qiu, S.; Deng, J.; Shun Pan Cheung, G.; Jin, L.; Zhao, B.; Zhang, C. Lipoteichoic acid of Enterococcus faecalis inhibits osteoclastogenesis via transcription factor RBP-J. Innate Immun. 2019, 25, 13–21. [Google Scholar] [CrossRef]
- Yang, J.; Ryu, Y.H.; Yun, C.H.; Han, S.H. Impaired osteoclastogenesis by staphylococcal lipoteichoic acid through Toll-like receptor 2 with partial involvement of MyD88. J. Leukoc. Biol. 2009, 86, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, Y.; Cao, Z.; Dou, C.; Bai, Y.; Liu, C.; Dong, S.; Fei, J. Staphylococcal lipoteichoic acid promotes osteogenic differentiation of mouse mesenchymal stem cells by increasing autophagic activity. Biochem. Biophys. Res. Commun. 2017, 485, 421–426. [Google Scholar] [CrossRef]
- Hu, C.C.; Chang, C.H.; Hsiao, Y.M.; Chang, Y.; Wu, Y.Y.; Ueng, S.W.N.; Chen, M.F. Lipoteichoic Acid Accelerates Bone Healing by Enhancing Osteoblast Differentiation and Inhibiting Osteoclast Activation in a Mouse Model of Femoral Defects. Int. J. Mol. Sci. 2020, 21, 5550. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Yang, J.; Park, O.J.; Kang, S.S.; Kim, W.S.; Kurokawa, K.; Yun, C.H.; Kim, H.H.; Lee, B.L.; Han, S.H. Lipoproteins are an important bacterial component responsible for bone destruction through the induction of osteoclast differentiation and activation. J. Bone Miner. Res. 2013, 28, 2381–2391. [Google Scholar] [CrossRef]
- Sato, N.; Takahashi, N.; Suda, K.; Nakamura, M.; Yamaki, M.; Ninomiya, T.; Kobayashi, Y.; Takada, H.; Shibata, K.; Yamamoto, M.; et al. MyD88 but not TRIF is essential for osteoclastogenesis induced by lipopolysaccharide, diacyl lipopeptide, and IL-1alpha. J. Exp. Med. 2004, 200, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Souza, J.A.C.; Magalhaes, F.A.C.; Oliveira, G.; RS, D.E.M.; Zuanon, J.A.; Souza, P.P.C. Pam2CSK4 (TLR2 agonist) induces periodontal destruction in mice. Braz. Oral Res. 2020, 34, e012. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, H.; Watanabe, K.; Toyama, T.; Koyata, Y.; Hamada, N. Porphyromonas gulae 41-kDa fimbriae induced osteoclast differentiation and cytokine production. J. Vet. Med. Sci. 2015, 77, 265–271. [Google Scholar] [CrossRef]
- Hiramine, H.; Watanabe, K.; Hamada, N.; Umemoto, T. Porphyromonas gingivalis 67-kDa fimbriae induced cytokine production and osteoclast differentiation utilizing TLR2. FEMS Microbiol. Lett. 2003, 229, 49–55. [Google Scholar] [CrossRef]
- Kawata, Y.; Hanazawa, S.; Amano, S.; Murakami, Y.; Matsumoto, T.; Nishida, K.; Kitano, S. Porphyromonas gingivalis fimbriae stimulate bone resorption in vitro. Infect. Immun. 1994, 62, 3012–3016. [Google Scholar] [CrossRef] [PubMed]
- Hanazawa, S.; Kawata, Y.; Murakami, Y.; Naganuma, K.; Amano, S.; Miyata, Y.; Kitano, S. Porphyromonas gingivalis fimbria-stimulated bone resorption in vitro is inhibited by a tyrosine kinase inhibitor. Infect. Immun. 1995, 63, 2374–2377. [Google Scholar] [CrossRef]
- Zhang, W.; Ju, J.; Rigney, T.; Tribble, G.D. Fimbriae of Porphyromonas gingivalis are important for initial invasion of osteoblasts, but not for inhibition of their differentiation and mineralization. J. Periodontol. 2011, 82, 909–916. [Google Scholar] [CrossRef]
- Zhang, W.; Ju, J.; Rigney, T.; Tribble, G. Integrin alpha5beta1-fimbriae binding and actin rearrangement are essential for Porphyromonas gingivalis invasion of osteoblasts and subsequent activation of the JNK pathway. BMC Microbiol. 2013, 13, 5. [Google Scholar] [CrossRef]
- Kishimoto, T.; Kaneko, T.; Ukai, T.; Yokoyama, M.; Ayon Haro, R.; Yoshinaga, Y.; Yoshimura, A.; Hara, Y. Peptidoglycan and lipopolysaccharide synergistically enhance bone resorption and osteoclastogenesis. J. Periodontal Res. 2012, 47, 446–454. [Google Scholar] [CrossRef]
- Ozaki, Y.; Kishimoto, T.; Yamashita, Y.; Kaneko, T.; Higuchi, K.; Mae, M.; Oohira, M.; Mohammad, A.I.; Yanagiguchi, K.; Yoshimura, A. Expression of osteoclastogenic and anti-osteoclastogenic cytokines differs in mouse gingiva injected with lipopolysaccharide, peptidoglycan, or both. Arch. Oral Biol. 2021, 122, 104990. [Google Scholar] [CrossRef]
- Ishida, M.; Kitaura, H.; Kimura, K.; Sugisawa, H.; Aonuma, T.; Takada, H.; Takano-Yamamoto, T. Muramyl dipeptide enhances lipopolysaccharide-induced osteoclast formation and bone resorption through increased RANKL expression in stromal cells. J. Immunol. Res. 2015, 2015, 132765. [Google Scholar] [CrossRef]
- Sato, T.; Watanabe, K.; Kumada, H.; Toyama, T.; Tani-Ishii, N.; Hamada, N. Peptidoglycan of Actinomyces naeslundii induces inflammatory cytokine production and stimulates osteoclastogenesis in alveolar bone resorption. Arch. Oral Biol. 2012, 57, 1522–1528. [Google Scholar] [CrossRef] [PubMed]
- Chaves de Souza, J.A.; Frasnelli, S.C.; Curylofo-Zotti, F.A.; Avila-Campos, M.J.; Spolidorio, L.C.; Zamboni, D.S.; Graves, D.T.; Rossa, C., Jr. NOD1 in the modulation of host-microbe interactions and inflammatory bone resorption in the periodontal disease model. Immunology 2016, 149, 374–385. [Google Scholar] [CrossRef]
- Kitaura, H.; Ishida, M.; Kimura, K.; Sugisawa, H.; Kishikawa, A.; Shima, K.; Ogawa, S.; Qi, J.; Shen, W.R. Role of Muramyl Dipeptide in Lipopolysaccharide-Mediated Biological Activity and Osteoclast Activity. Anal. Cell. Pathol. 2018, 2018, 8047610. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Darzi, Y.; Tawaratsumida, K.; Marchesan, J.T.; Hasegawa, M.; Moon, H.; Chen, G.Y.; Nunez, G.; Giannobile, W.V.; Raes, J.; et al. Induction of bone loss by pathobiont-mediated Nod1 signaling in the oral cavity. Cell Host Microbe 2013, 13, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Park, O.J.; Kim, J.; Yang, J.; Yun, C.H.; Han, S.H. Muramyl Dipeptide, a Shared Structural Motif of Peptidoglycans, Is a Novel Inducer of Bone Formation through Induction of Runx2. J. Bone Miner. Res. 2017, 32, 1455–1468. [Google Scholar] [CrossRef]
- Rietschel, E.T.; Kirikae, T.; Schade, F.U.; Mamat, U.; Schmidt, G.; Loppnow, H.; Ulmer, A.J.; Zahringer, U.; Seydel, U.; Di Padova, F.; et al. Bacterial endotoxin: Molecular relationships of structure to activity and function. FASEB J. 1994, 8, 217–225. [Google Scholar] [CrossRef]
- Kabanov, D.S.; Prokhorenko, I.R. Structural analysis of lipopolysaccharides from Gram-negative bacteria. Biochemistry 2010, 75, 383–404. [Google Scholar] [CrossRef]
- Chow, J.C.; Young, D.W.; Golenbock, D.T.; Christ, W.J.; Gusovsky, F. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J. Biol. Chem. 1999, 274, 10689–10692. [Google Scholar] [CrossRef]
- Shimazu, R.; Akashi, S.; Ogata, H.; Nagai, Y.; Fukudome, K.; Miyake, K.; Kimoto, M. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J. Exp. Med. 1999, 189, 1777–1782. [Google Scholar] [CrossRef]
- Su, G.L.; Klein, R.D.; Aminlari, A.; Zhang, H.Y.; Steinstraesser, L.; Alarcon, W.H.; Remick, D.G.; Wang, S.C. Kupffer cell activation by lipopolysaccharide in rats: Role for lipopolysaccharide binding protein and toll-like receptor 4. Hepatology 2000, 31, 932–936. [Google Scholar] [CrossRef] [PubMed]
- Plociennikowska, A.; Hromada-Judycka, A.; Borzecka, K.; Kwiatkowska, K. Co-operation of TLR4 and raft proteins in LPS-induced pro-inflammatory signaling. Cell. Mol. Life Sci. 2015, 72, 557–581. [Google Scholar] [CrossRef]
- Fitzgerald, K.A.; Palsson-McDermott, E.M.; Bowie, A.G.; Jefferies, C.A.; Mansell, A.S.; Brady, G.; Brint, E.; Dunne, A.; Gray, P.; Harte, M.T.; et al. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature 2001, 413, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.X.; Kirschning, C.J.; Mancinelli, R.; Xu, X.P.; Jin, Y.; Faure, E.; Mantovani, A.; Rothe, M.; Muzio, M.; Arditi, M. Bacterial lipopolysaccharide activates nuclear factor-kappaB through interleukin-1 signaling mediators in cultured human dermal endothelial cells and mononuclear phagocytes. J. Biol. Chem. 1999, 274, 7611–7614. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.H.; Jung, S.H.; Jung, S. Beet root (Beta vulgaris) protects lipopolysaccharide and alcohol-induced liver damage in rat. Toxicol. Res. 2020, 36, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Nishihara, T.; Ohsaki, Y.; Ueda, N.; Saito, N.; Mundy, G.R. Mouse interleukin-1 receptor antagonist induced by Actinobacillus actinomycetemcomitans lipopolysaccharide blocks the effects of interleukin-1 on bone resorption and osteoclast-like cell formation. Infect. Immun. 1994, 62, 390–397. [Google Scholar] [CrossRef]
- Takami, M.; Kim, N.; Rho, J.; Choi, Y. Stimulation by toll-like receptors inhibits osteoclast differentiation. J. Immunol. 2002, 169, 1516–1523. [Google Scholar] [CrossRef] [PubMed]
- Park, O.J.; Yang, J.; Kim, J.; Yun, C.H.; Han, S.H. Enterococcus faecalis attenuates the differentiation of macrophages into osteoclasts. J. Endod. 2015, 41, 658–662. [Google Scholar] [CrossRef]
- Ishimi, Y.; Miyaura, C.; Jin, C.H.; Akatsu, T.; Abe, E.; Nakamura, Y.; Yamaguchi, A.; Yoshiki, S.; Matsuda, T.; Hirano, T.; et al. IL-6 is produced by osteoblasts and induces bone resorption. J. Immunol. 1990, 145, 3297–3303. [Google Scholar]
- Keeting, P.E.; Rifas, L.; Harris, S.A.; Colvard, D.S.; Spelsberg, T.C.; Peck, W.A.; Riggs, B.L. Evidence for interleukin-1 beta production by cultured normal human osteoblast-like cells. J. Bone Miner. Res. 1991, 6, 827–833. [Google Scholar] [CrossRef]
- Sakuma, Y.; Tanaka, K.; Suda, M.; Komatsu, Y.; Yasoda, A.; Miura, M.; Ozasa, A.; Narumiya, S.; Sugimoto, Y.; Ichikawa, A.; et al. Impaired bone resorption by lipopolysaccharide in vivo in mice deficient in the prostaglandin E receptor EP4 subtype. Infect. Immun. 2000, 68, 6819–6825. [Google Scholar] [CrossRef]
- Kikuchi, T.; Matsuguchi, T.; Tsuboi, N.; Mitani, A.; Tanaka, S.; Matsuoka, M.; Yamamoto, G.; Hishikawa, T.; Noguchi, T.; Yoshikai, Y. Gene expression of osteoclast differentiation factor is induced by lipopolysaccharide in mouse osteoblasts via Toll-like receptors. J. Immunol. 2001, 166, 3574–3579. [Google Scholar] [CrossRef] [PubMed]
- Percy, M.G.; Grundling, A. Lipoteichoic acid synthesis and function in gram-positive bacteria. Annu. Rev. Microbiol. 2014, 68, 81–100. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.S.; Sim, J.R.; Yun, C.H.; Han, S.H. Lipoteichoic acids as a major virulence factor causing inflammatory responses via Toll-like receptor 2. Arch. Pharm. Res. 2016, 39, 1519–1529. [Google Scholar] [CrossRef] [PubMed]
- Chien, H.F.; Yeh, K.Y.; Jiang-Shieh, Y.F.; Wei, I.H.; Chang, C.Y.; Chang, M.L.; Wu, C.H. Signal transduction pathways of nitric oxide release in primary microglial culture challenged with gram-positive bacterial constituent, lipoteichoic acid. Neuroscience 2005, 133, 423–436. [Google Scholar] [CrossRef]
- Kang, S.S.; Ryu, Y.H.; Baik, J.E.; Yun, C.H.; Lee, K.; Chung, D.K.; Han, S.H. Lipoteichoic acid from Lactobacillus plantarum induces nitric oxide production in the presence of interferon-gamma in murine macrophages. Mol. Immunol. 2011, 48, 2170–2177. [Google Scholar] [CrossRef]
- Hong, S.W.; Baik, J.E.; Kang, S.S.; Yun, C.H.; Seo, D.G.; Han, S.H. Lipoteichoic acid of Streptococcus mutans interacts with Toll-like receptor 2 through the lipid moiety for induction of inflammatory mediators in murine macrophages. Mol. Immunol. 2014, 57, 284–291. [Google Scholar] [CrossRef]
- Hashimoto, M.; Tawaratsumida, K.; Kariya, H.; Kiyohara, A.; Suda, Y.; Krikae, F.; Kirikae, T.; Gotz, F. Not lipoteichoic acid but lipoproteins appear to be the dominant immunobiologically active compounds in Staphylococcus aureus. J. Immunol. 2006, 177, 3162–3169. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.T.; Uebele, J.; Kumari, N.; Nakayama, H.; Peter, L.; Ticha, O.; Woischnig, A.K.; Schmaler, M.; Khanna, N.; Dohmae, N.; et al. Lipid moieties on lipoproteins of commensal and non-commensal staphylococci induce differential immune responses. Nat. Commun. 2017, 8, 2246. [Google Scholar] [CrossRef]
- Akira, S. Mammalian Toll-like receptors. Curr. Opin. Immunol. 2003, 15, 5–11. [Google Scholar] [CrossRef]
- Kurokawa, K.; Lee, H.; Roh, K.B.; Asanuma, M.; Kim, Y.S.; Nakayama, H.; Shiratsuchi, A.; Choi, Y.; Takeuchi, O.; Kang, H.J.; et al. The Triacylated ATP Binding Cluster Transporter Substrate-binding Lipoprotein of Staphylococcus aureus Functions as a Native Ligand for Toll-like Receptor 2. J. Biol. Chem. 2009, 284, 8406–8411. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.S.; Kim, S.E.; Heo, J.Y.; Lee, M.E.; Kim, H.M.; Paik, S.G.; Lee, H.; Lee, J.O. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell 2007, 130, 1071–1082. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.Y.; Nan, X.; Jin, M.S.; Youn, S.J.; Ryu, Y.H.; Mah, S.; Han, S.H.; Lee, H.; Paik, S.G.; Lee, J.O. Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity 2009, 31, 873–884. [Google Scholar] [CrossRef] [PubMed]
- Verstak, B.; Nagpal, K.; Bottomley, S.P.; Golenbock, D.T.; Hertzog, P.J.; Mansell, A. MyD88 adapter-like (Mal)/TIRAP interaction with TRAF6 is critical for TLR2- and TLR4-mediated NF-kappaB proinflammatory responses. J. Biol. Chem. 2009, 284, 24192–24203. [Google Scholar] [CrossRef] [PubMed]
- Vinod, V.; Vijayrajratnam, S.; Vasudevan, A.K.; Biswas, R. The cell surface adhesins of Mycobacterium tuberculosis. Microbiol. Res. 2020, 232, 126392. [Google Scholar] [CrossRef] [PubMed]
- Solanki, V.; Tiwari, M.; Tiwari, V. Host-bacteria interaction and adhesin study for development of therapeutics. Int. J. Biol. Macromol. 2018, 112, 54–64. [Google Scholar] [CrossRef]
- Pizarro-Cerda, J.; Cossart, P. Bacterial adhesion and entry into host cells. Cell 2006, 124, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, W.; Blanot, D.; de Pedro, M.A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008, 32, 149–167. [Google Scholar] [CrossRef]
- Swaminathan, C.P.; Brown, P.H.; Roychowdhury, A.; Wang, Q.; Guan, R.; Silverman, N.; Goldman, W.E.; Boons, G.J.; Mariuzza, R.A. Dual strategies for peptidoglycan discrimination by peptidoglycan recognition proteins (PGRPs). Proc. Natl. Acad. Sci. USA 2006, 103, 684–689. [Google Scholar] [CrossRef]
- Jeon, D.I.; Park, S.R.; Ahn, M.Y.; Ahn, S.G.; Park, J.H.; Yoon, J.H. NOD1 and NOD2 stimulation triggers innate immune responses of human periodontal ligament cells. Int. J. Mol. Med. 2012, 29, 699–703. [Google Scholar] [CrossRef]
- Inohara, N.; Koseki, T.; Lin, J.; del Peso, L.; Lucas, P.C.; Chen, F.F.; Ogura, Y.; Nunez, G. An induced proximity model for NF-kappa B activation in the Nod1/RICK and RIP signaling pathways. J. Biol. Chem. 2000, 275, 27823–27831. [Google Scholar] [CrossRef]
- Girardin, S.E.; Tournebize, R.; Mavris, M.; Page, A.L.; Li, X.; Stark, G.R.; Bertin, J.; DiStefano, P.S.; Yaniv, M.; Sansonetti, P.J.; et al. CARD4/Nod1 mediates NF-kappaB and JNK activation by invasive Shigella flexneri. EMBO Rep. 2001, 2, 736–742. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N. Oral Microbiome Metabolism: From “Who Are They?” to “What Are They Doing?”. J. Dent. Res. 2015, 94, 1628–1637. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.H. Short chain fatty acids in the human colon. Gut 1981, 22, 763–779. [Google Scholar] [CrossRef]
- Bergman, E.N. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 1990, 70, 567–590. [Google Scholar] [CrossRef]
- Wootten, D.; Christopoulos, A.; Marti-Solano, M.; Babu, M.M.; Sexton, P.M. Mechanisms of signalling and biased agonism in G protein-coupled receptors. Nat. Rev. Mol. Cell Biol. 2018, 19, 638–653. [Google Scholar] [CrossRef]
- Priyadarshini, M.; Kotlo, K.U.; Dudeja, P.K.; Layden, B.T. Role of Short Chain Fatty Acid Receptors in Intestinal Physiology and Pathophysiology. Compr. Physiol. 2018, 8, 1091–1115. [Google Scholar] [CrossRef]
- Stoddart, L.A.; Smith, N.J.; Jenkins, L.; Brown, A.J.; Milligan, G. Conserved polar residues in transmembrane domains V, VI, and VII of free fatty acid receptor 2 and free fatty acid receptor 3 are required for the binding and function of short chain fatty acids. J. Biol. Chem. 2008, 283, 32913–32924. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.H.; Nishimura, Y.; Hishikawa, D.; Tsuzuki, H.; Miyahara, H.; Gotoh, C.; Choi, K.C.; Feng, D.D.; Chen, C.; Lee, H.G.; et al. Acetate and propionate short chain fatty acids stimulate adipogenesis via GPCR43. Endocrinology 2005, 146, 5092–5099. [Google Scholar] [CrossRef]
- Futosi, K.; Fodor, S.; Mocsai, A. Neutrophil cell surface receptors and their intracellular signal transduction pathways. Int. Immunopharmacol. 2013, 17, 638–650. [Google Scholar] [CrossRef]
- Wang, X.; Iyer, A.; Lyons, A.B.; Korner, H.; Wei, W. Emerging Roles for G-protein Coupled Receptors in Development and Activation of Macrophages. Front. Immunol. 2019, 10, 2031. [Google Scholar] [CrossRef]
- Luo, J.; Sun, P.; Siwko, S.; Liu, M.; Xiao, J. The role of GPCRs in bone diseases and dysfunctions. Bone Res. 2019, 7, 19. [Google Scholar] [CrossRef]
- Yi, S.J.; Lee, H.; Lee, J.; Lee, K.; Kim, J.; Kim, Y.; Park, J.I.; Kim, K. Bone Remodeling: Histone Modifications as Fate Determinants of Bone Cell Differentiation. Int. J. Mol. Sci. 2019, 20, 3147. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Kim, H.Y.; Kim, M.G.; Jeong, S.; Yun, C.H.; Han, S.H. Short-chain Fatty Acids Inhibit Staphylococcal Lipoprotein-induced Nitric Oxide Production in Murine Macrophages. Immune Netw. 2019, 19, e9. [Google Scholar] [CrossRef]
- Sim, J.R.; Kang, S.S.; Lee, D.; Yun, C.H.; Han, S.H. Killed Whole-Cell Oral Cholera Vaccine Induces CCL20 Secretion by Human Intestinal Epithelial Cells in the Presence of the Short-Chain Fatty Acid, Butyrate. Front. Immunol. 2018, 9, 55. [Google Scholar] [CrossRef]
- Yan, J.; Takakura, A.; Zandi-Nejad, K.; Charles, J.F. Mechanisms of gut microbiota-mediated bone remodeling. Gut Microbes 2018, 9, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Kukita, A.; Kukita, T.; Shobuike, T.; Nakamura, T.; Kohashi, O. Two histone deacetylase inhibitors, trichostatin A and sodium butyrate, suppress differentiation into osteoclasts but not into macrophages. Blood 2003, 101, 3451–3459. [Google Scholar] [CrossRef]
- Chang, M.C.; Chen, Y.J.; Lian, Y.C.; Chang, B.E.; Huang, C.C.; Huang, W.L.; Pan, Y.H.; Jeng, J.H. Butyrate Stimulates Histone H3 Acetylation, 8-Isoprostane Production, RANKL Expression, and Regulated Osteoprotegerin Expression/Secretion in MG-63 Osteoblastic Cells. Int. J. Mol. Sci. 2018, 19, 4071. [Google Scholar] [CrossRef]
- Wauquier, F.; Philippe, C.; Leotoing, L.; Mercier, S.; Davicco, M.J.; Lebecque, P.; Guicheux, J.; Pilet, P.; Miot-Noirault, E.; Poitout, V.; et al. The free fatty acid receptor G protein-coupled receptor 40 (GPR40) protects from bone loss through inhibition of osteoclast differentiation. J. Biol. Chem. 2013, 288, 6542–6551. [Google Scholar] [CrossRef] [PubMed]
- Montalvany-Antonucci, C.C.; Duffles, L.F.; de Arruda, J.A.A.; Zicker, M.C.; de Oliveira, S.; Macari, S.; Garlet, G.P.; Madeira, M.F.M.; Fukada, S.Y.; Andrade, I., Jr.; et al. Short-chain fatty acids and FFAR2 as suppressors of bone resorption. Bone 2019, 125, 112–121. [Google Scholar] [CrossRef]
- Morozumi, A. High concentration of sodium butyrate suppresses osteoblastic differentiation and mineralized nodule formation in ROS17/2.8 cells. J. Oral Sci. 2011, 53, 509–516. [Google Scholar] [CrossRef]
- Chang, M.C.; Tsai, Y.L.; Liou, E.J.; Tang, C.M.; Wang, T.M.; Liu, H.C.; Liao, M.W.; Yeung, S.Y.; Chan, C.P.; Jeng, J.H. Effect of Butyrate on Collagen Expression, Cell Viability, Cell Cycle Progression and Related Proteins Expression of MG-63 Osteoblastic Cells. PLoS ONE 2016, 11, e0165438. [Google Scholar] [CrossRef]
- Lucas, S.; Omata, Y.; Hofmann, J.; Bottcher, M.; Iljazovic, A.; Sarter, K.; Albrecht, O.; Schulz, O.; Krishnacoumar, B.; Kronke, G.; et al. Short-chain fatty acids regulate systemic bone mass and protect from pathological bone loss. Nat. Commun. 2018, 9, 55. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367. [Google Scholar] [CrossRef] [PubMed]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- Toyofuku, M.; Nomura, N.; Eberl, L. Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 2019, 17, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.Y.; Bang, J.Y.; Park, G.W.; Choi, D.S.; Kang, J.S.; Kim, H.J.; Park, K.S.; Lee, J.O.; Kim, Y.K.; Kwon, K.H.; et al. Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics 2007, 7, 3143–3153. [Google Scholar] [CrossRef] [PubMed]
- Klieve, A.V.; Yokoyama, M.T.; Forster, R.J.; Ouwerkerk, D.; Bain, P.A.; Mawhinney, E.L. Naturally occurring DNA transfer system associated with membrane vesicles in cellulolytic Ruminococcus spp. of ruminal origin. Appl. Environ. Microbiol. 2005, 71, 4248–4253. [Google Scholar] [CrossRef]
- Macia, L.; Nanan, R.; Hosseini-Beheshti, E.; Grau, G.E. Host- and Microbiota-Derived Extracellular Vesicles, Immune Function, and Disease Development. Int. J. Mol. Sci. 2019, 21, 107. [Google Scholar] [CrossRef]
- Codemo, M.; Muschiol, S.; Iovino, F.; Nannapaneni, P.; Plant, L.; Wai, S.N.; Henriques-Normark, B. Immunomodulatory Effects of Pneumococcal Extracellular Vesicles on Cellular and Humoral Host Defenses. mBio 2018, 9. [Google Scholar] [CrossRef]
- Zupan, J.; Jeras, M.; Marc, J. Osteoimmunology and the influence of pro-inflammatory cytokines on osteoclasts. Biochem. Med. 2013, 23, 43–63. [Google Scholar] [CrossRef] [PubMed]
- Schmid, J.; Sieber, V.; Rehm, B. Bacterial exopolysaccharides: Biosynthesis pathways and engineering strategies. Front. Microbiol. 2015, 6, 496. [Google Scholar] [CrossRef] [PubMed]
- Nwodo, U.U.; Green, E.; Okoh, A.I. Bacterial exopolysaccharides: Functionality and prospects. Int. J. Mol. Sci. 2012, 13, 14002–14015. [Google Scholar] [CrossRef]
- Taylor, C.M.; Roberts, I.S. Capsular polysaccharides and their role in virulence. Contrib. Microbiol. 2005, 12, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.S.; Mody, K.; Jha, B. Bacterial exopolysaccharides—A perception. J. Basic Microbiol. 2007, 47, 103–117. [Google Scholar] [CrossRef]
- Wallimann, A.; Hildebrand, M.; Groeger, D.; Stanic, B.; Akdis, C.A.; Zeiter, S.; Richards, R.G.; Moriarty, T.F.; O’Mahony, L.; Thompson, K. An Exopolysaccharide Produced by Bifidobacterium longum 35624(R) Inhibits Osteoclast Formation via a TLR2-Dependent Mechanism. Calcif. Tissue Int. 2021, 108, 654–666. [Google Scholar] [CrossRef] [PubMed]
- Zanchetta, P.; Lagarde, N.; Guezennec, J. A new bone-healing material: A hyaluronic acid-like bacterial exopolysaccharide. Calcif. Tissue Int. 2003, 72, 74–79. [Google Scholar] [CrossRef]
- Velasco, C.R.; Baud’huin, M.; Sinquin, C.; Maillasson, M.; Heymann, D.; Colliec-Jouault, S.; Padrines, M. Effects of a sulfated exopolysaccharide produced by Altermonas infernus on bone biology. Glycobiology 2011, 21, 781–795. [Google Scholar] [CrossRef]
- Nishihara, T.; Ueda, N.; Amano, K.; Ishihara, Y.; Hayakawa, H.; Kuroyanagi, T.; Ohsaki, Y.; Nagata, K.; Noguchi, T. Actinobacillus actinomycetemcomitans Y4 capsular-polysaccharide-like polysaccharide promotes osteoclast-like cell formation by interleukin-1 alpha production in mouse marrow cultures. Infect. Immun. 1995, 63, 1893–1898. [Google Scholar] [CrossRef]
- Ueda, N.; Nishihara, T.; Ishihara, Y.; Amano, K.; Kuroyanagi, T.; Noguchi, T. Role of prostaglandin in the formation of osteoclasts induced by capsular-like polysaccharide antigen of Actinobacillus actinomycetemcomitans strain Y4. Oral Microbiol. Immunol. 1995, 10, 69–75. [Google Scholar] [CrossRef]
- Yamamoto, S.; Mogi, M.; Kinpara, K.; Ishihara, Y.; Ueda, N.; Amano, K.; Nishihara, T.; Noguchi, T.; Togari, A. Anti-proliferative capsular-like polysaccharide antigen from Actinobacillus actinomycetemcomitans induces apoptotic cell death in mouse osteoblastic MC3T3-E1 cells. J. Dent. Res. 1999, 78, 1230–1237. [Google Scholar] [CrossRef]
- Gonzalez, D.; Tzianabos, A.O.; Genco, C.A.; Gibson, F.C., 3rd. Immunization with Porphyromonas gingivalis capsular polysaccharide prevents P. gingivalis-elicited oral bone loss in a murine model. Infect. Immun. 2003, 71, 2283–2287. [Google Scholar] [CrossRef]
- Ross, P.; Weinhouse, H.; Aloni, Y.; Michaeli, D.; Weinberger-Ohana, P.; Mayer, R.; Braun, S.; de Vroom, E.; van der Marel, G.A.; van Boom, J.H.; et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 1987, 325, 279–281. [Google Scholar] [CrossRef]
- Danilchanka, O.; Mekalanos, J.J. Cyclic dinucleotides and the innate immune response. Cell 2013, 154, 962–970. [Google Scholar] [CrossRef]
- Corrigan, R.M.; Grundling, A. Cyclic di-AMP: Another second messenger enters the fray. Nat. Rev. Microbiol. 2013, 11, 513–524. [Google Scholar] [CrossRef]
- Gjermansen, M.; Ragas, P.; Tolker-Nielsen, T. Proteins with GGDEF and EAL domains regulate Pseudomonas putida biofilm formation and dispersal. FEMS Microbiol. Lett. 2006, 265, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Gries, C.M.; Bruger, E.L.; Moormeier, D.E.; Scherr, T.D.; Waters, C.M.; Kielian, T. Cyclic di-AMP Released from Staphylococcus aureus Biofilm Induces a Macrophage Type I Interferon Response. Infect. Immun. 2016, 84, 3564–3574. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Barber, G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 2008, 455, 674–678. [Google Scholar] [CrossRef]
- Barker, J.R.; Koestler, B.J.; Carpenter, V.K.; Burdette, D.L.; Waters, C.M.; Vance, R.E.; Valdivia, R.H. STING-dependent recognition of cyclic di-AMP mediates type I interferon responses during Chlamydia trachomatis infection. mBio 2013, 4, e00018-13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Shang, G.; Gui, X.; Zhang, X.; Bai, X.C.; Chen, Z.J. Structural basis of STING binding with and phosphorylation by TBK1. Nature 2019, 567, 394–398. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Chen, Z.J. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 2012, 5, ra20. [Google Scholar] [CrossRef]
- Choe, C.H.; Park, I.S.; Park, J.; Yu, K.Y.; Jang, H.; Kim, J.; Jang, Y.S. Transmembrane protein 173 inhibits RANKL-induced osteoclast differentiation. FEBS Lett. 2015, 589, 836–841. [Google Scholar] [CrossRef] [PubMed]
- Baum, R.; Sharma, S.; Organ, J.M.; Jakobs, C.; Hornung, V.; Burr, D.B.; Marshak-Rothstein, A.; Fitzgerald, K.A.; Gravallese, E.M. STING Contributes to Abnormal Bone Formation Induced by Deficiency of DNase II in Mice. Arthritis Rheumatol. 2017, 69, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Takayanagi, H.; Kim, S.; Matsuo, K.; Suzuki, H.; Suzuki, T.; Sato, K.; Yokochi, T.; Oda, H.; Nakamura, K.; Ida, N.; et al. RANKL maintains bone homeostasis through c-Fos-dependent induction of interferon-beta. Nature 2002, 416, 744–749. [Google Scholar] [CrossRef] [PubMed]
- Dunphy, G.; Flannery, S.M.; Almine, J.F.; Connolly, D.J.; Paulus, C.; Jonsson, K.L.; Jakobsen, M.R.; Nevels, M.M.; Bowie, A.G.; Unterholzner, L. Non-canonical Activation of the DNA Sensing Adaptor STING by ATM and IFI16 Mediates NF-kappaB Signaling after Nuclear DNA Damage. Mol. Cell 2018, 71, 745–760. [Google Scholar] [CrossRef]
- Cortes-Penfield, N.W.; Kulkarni, P.A. The History of Antibiotic Treatment of Osteomyelitis. Open Forum Infect. Dis. 2019, 6, ofz181. [Google Scholar] [CrossRef]
- Handel, A.; Margolis, E.; Levin, B.R. Exploring the role of the immune response in preventing antibiotic resistance. J. Theor. Biol. 2009, 256, 655–662. [Google Scholar] [CrossRef]
- Drake, M.T.; Clarke, B.L.; Khosla, S. Bisphosphonates: Mechanism of action and role in clinical practice. Mayo Clin. Proc. 2008, 83, 1032–1045. [Google Scholar] [CrossRef]
- Hanley, D.A.; Adachi, J.D.; Bell, A.; Brown, V. Denosumab: Mechanism of action and clinical outcomes. Int. J. Clin. Pract. 2012, 66, 1139–1146. [Google Scholar] [CrossRef]
- Nikitovic, D.; Kavasi, R.M.; Berdiaki, A.; Papachristou, D.J.; Tsiaoussis, J.; Spandidos, D.A.; Tsatsakis, A.M.; Tzanakakis, G.N. Parathyroid hormone/parathyroid hormone-related peptide regulate osteosarcoma cell functions: Focus on the extracellular matrix (Review). Oncol. Rep. 2016, 36, 1787–1792. [Google Scholar] [CrossRef][Green Version]
- Kyrgidis, A.; Toulis, K.A. Denosumab-related osteonecrosis of the jaws. Osteoporos. Int. 2011, 22, 369–370. [Google Scholar] [CrossRef]
- Woo, T.; Adachi, J.D. Role of bisphosphonates and calcitonin in the prevention and treatment of osteoporosis. Best Pract. Res. Clin. Rheumatol. 2001, 15, 469–481. [Google Scholar] [CrossRef]
- Park, O.J.; Kwon, Y.; Park, C.; So, Y.J.; Park, T.H.; Jeong, S.; Im, J.; Yun, C.H.; Han, S.H. Streptococcus gordonii: Pathogenesis and Host Response to Its Cell Wall Components. Microorganisms 2020, 8, 1852. [Google Scholar] [CrossRef] [PubMed]
- Alves, C.H.; Farrell, E.; Vis, M.; Colin, E.M.; Lubberts, E. Animal Models of Bone Loss in Inflammatory Arthritis: From Cytokines in the Bench to Novel Treatments for Bone Loss in the Bedside-a Comprehensive Review. Clin. Rev. Allergy Immunol. 2016, 51, 27–47. [Google Scholar] [CrossRef] [PubMed]
- Axmann, R.; Bohm, C.; Kronke, G.; Zwerina, J.; Smolen, J.; Schett, G. Inhibition of interleukin-6 receptor directly blocks osteoclast formation in vitro and in vivo. Arthritis Rheum. 2009, 60, 2747–2756. [Google Scholar] [CrossRef]
- Gao, W.; Xiong, Y.; Li, Q.; Yang, H. Inhibition of Toll-Like Receptor Signaling as a Promising Therapy for Inflammatory Diseases: A Journey from Molecular to Nano Therapeutics. Front. Physiol. 2017, 8, 508. [Google Scholar] [CrossRef] [PubMed]
- Souza, P.P.C.; Lerner, U.H. Finding a Toll on the Route: The Fate of Osteoclast Progenitors After Toll-Like Receptor Activation. Front. Immunol. 2019, 10, 1663. [Google Scholar] [CrossRef]
- Jung, K.; Lee, J.E.; Kim, H.Z.; Kim, H.M.; Park, B.S.; Hwang, S.I.; Lee, J.O.; Kim, S.C.; Koh, G.Y. Toll-like receptor 4 decoy, TOY, attenuates gram-negative bacterial sepsis. PLoS ONE 2009, 4, e7403. [Google Scholar] [CrossRef] [PubMed]
- Andresen, L.; Theodorou, K.; Grunewald, S.; Czech-Zechmeister, B.; Konnecke, B.; Luhder, F.; Trendelenburg, G. Evaluation of the Therapeutic Potential of Anti-TLR4-Antibody MTS510 in Experimental Stroke and Significance of Different Routes of Application. PLoS ONE 2016, 11, e0148428. [Google Scholar] [CrossRef]
- Heinbockel, L.; Weindl, G.; Martinez-de-Tejada, G.; Correa, W.; Sanchez-Gomez, S.; Barcena-Varela, S.; Goldmann, T.; Garidel, P.; Gutsmann, T.; Brandenburg, K. Inhibition of Lipopolysaccharide- and Lipoprotein-Induced Inflammation by Antitoxin Peptide Pep19-2.5. Front. Immunol. 2018, 9, 1704. [Google Scholar] [CrossRef]
- Tsuzuki, H.; Tani, T.; Ueyama, H.; Kodama, M. Lipopolysaccharide: Neutralization by polymyxin B shuts down the signaling pathway of nuclear factor kappaB in peripheral blood mononuclear cells, even during activation. J. Surg. Res. 2001, 100, 127–134. [Google Scholar] [CrossRef]
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef]
- Schepper, J.D.; Collins, F.L.; Rios-Arce, N.D.; Raehtz, S.; Schaefer, L.; Gardinier, J.D.; Britton, R.A.; Parameswaran, N.; McCabe, L.R. Probiotic Lactobacillus reuteri Prevents Postantibiotic Bone Loss by Reducing Intestinal Dysbiosis and Preventing Barrier Disruption. J. Bone Miner. Res. 2019, 34, 681–698. [Google Scholar] [CrossRef]
- Britton, R.A.; Irwin, R.; Quach, D.; Schaefer, L.; Zhang, J.; Lee, T.; Parameswaran, N.; McCabe, L.R. Probiotic L. reuteri treatment prevents bone loss in a menopausal ovariectomized mouse model. J. Cell. Physiol. 2014, 229, 1822–1830. [Google Scholar] [CrossRef]
- Zhang, J.; Motyl, K.J.; Irwin, R.; MacDougald, O.A.; Britton, R.A.; McCabe, L.R. Loss of Bone and Wnt10b Expression in Male Type 1 Diabetic Mice Is Blocked by the Probiotic Lactobacillus reuteri. Endocrinology 2015, 156, 3169–3182. [Google Scholar] [CrossRef]
- Gatej, S.M.; Marino, V.; Bright, R.; Fitzsimmons, T.R.; Gully, N.; Zilm, P.; Gibson, R.J.; Edwards, S.; Bartold, P.M. Probiotic Lactobacillus rhamnosus GG prevents alveolar bone loss in a mouse model of experimental periodontitis. J. Clin. Periodontol. 2018, 45, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Chiang, S.S.; Pan, T.M. Antiosteoporotic effects of Lactobacillus-fermented soy skim milk on bone mineral density and the microstructure of femoral bone in ovariectomized mice. J. Agric. Food Chem. 2011, 59, 7734–7742. [Google Scholar] [CrossRef]
- Wang, Z.; Xue, K.; Bai, M.; Deng, Z.; Gan, J.; Zhou, G.; Qian, H.; Bao, N.; Zhao, J. Probiotics protect mice from CoCrMo particles-induced osteolysis. Int. J. Nanomed. 2017, 12, 5387–5397. [Google Scholar] [CrossRef] [PubMed]
- Azizieh, F.Y.; Shehab, D.; Jarallah, K.A.; Gupta, R.; Raghupathy, R. Circulatory Levels of RANKL, OPG, and Oxidative Stress Markers in Postmenopausal Women With Normal or Low Bone Mineral Density. Biomark. Insights 2019, 14, 1177271919843825. [Google Scholar] [CrossRef]
- Zaiss, M.M.; Jones, R.M.; Schett, G.; Pacifici, R. The gut-bone axis: How bacterial metabolites bridge the distance. J. Clin. Investig. 2019, 129, 3018–3028. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Patel, M.S.; Levasseur, R.; Lobov, I.; Chang, B.H.; Glass, D.A., 2nd; Hartmann, C.; Li, L.; Hwang, T.H.; Brayton, C.F.; et al. Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J. Cell Biol. 2002, 157, 303–314. [Google Scholar] [CrossRef] [PubMed]
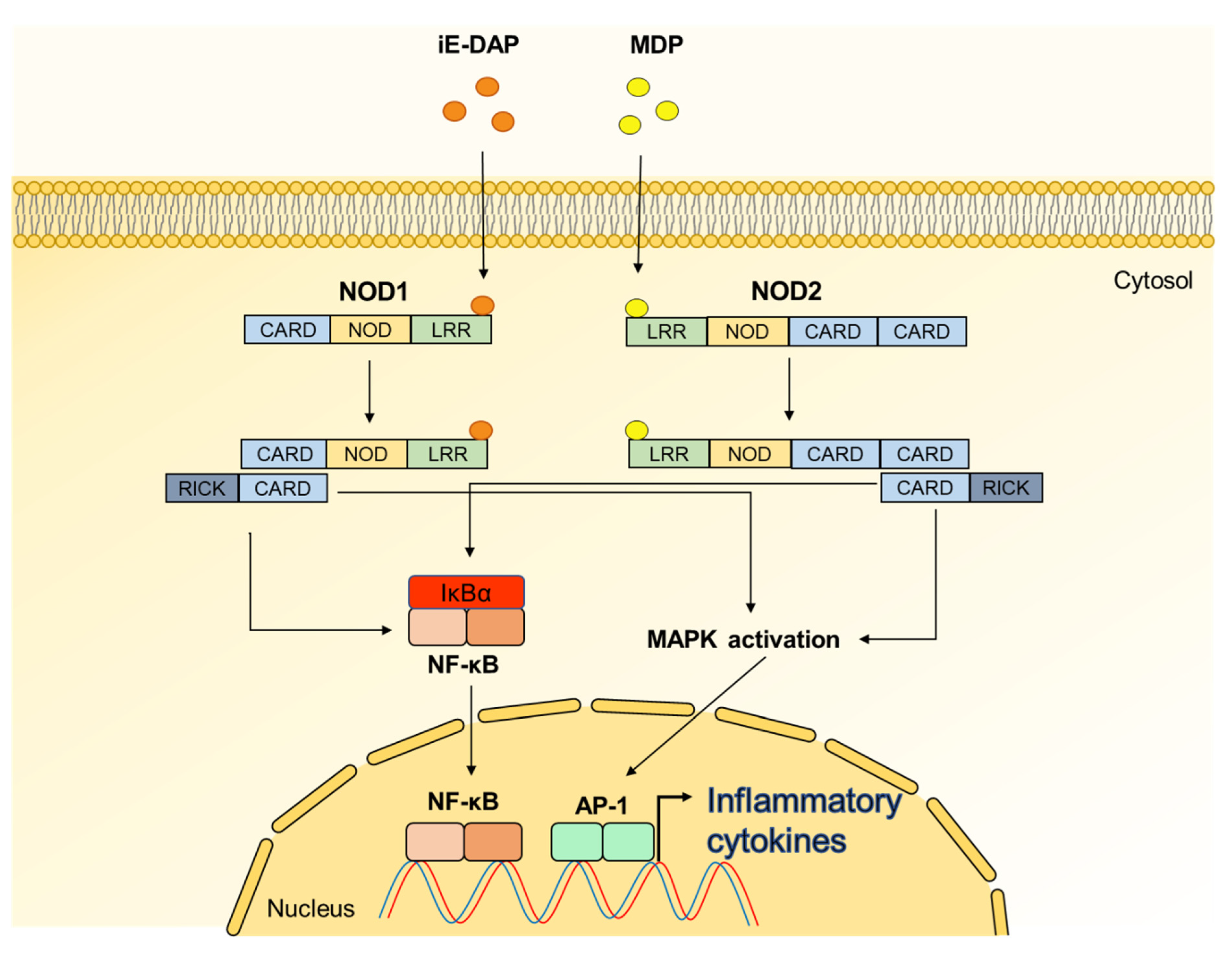
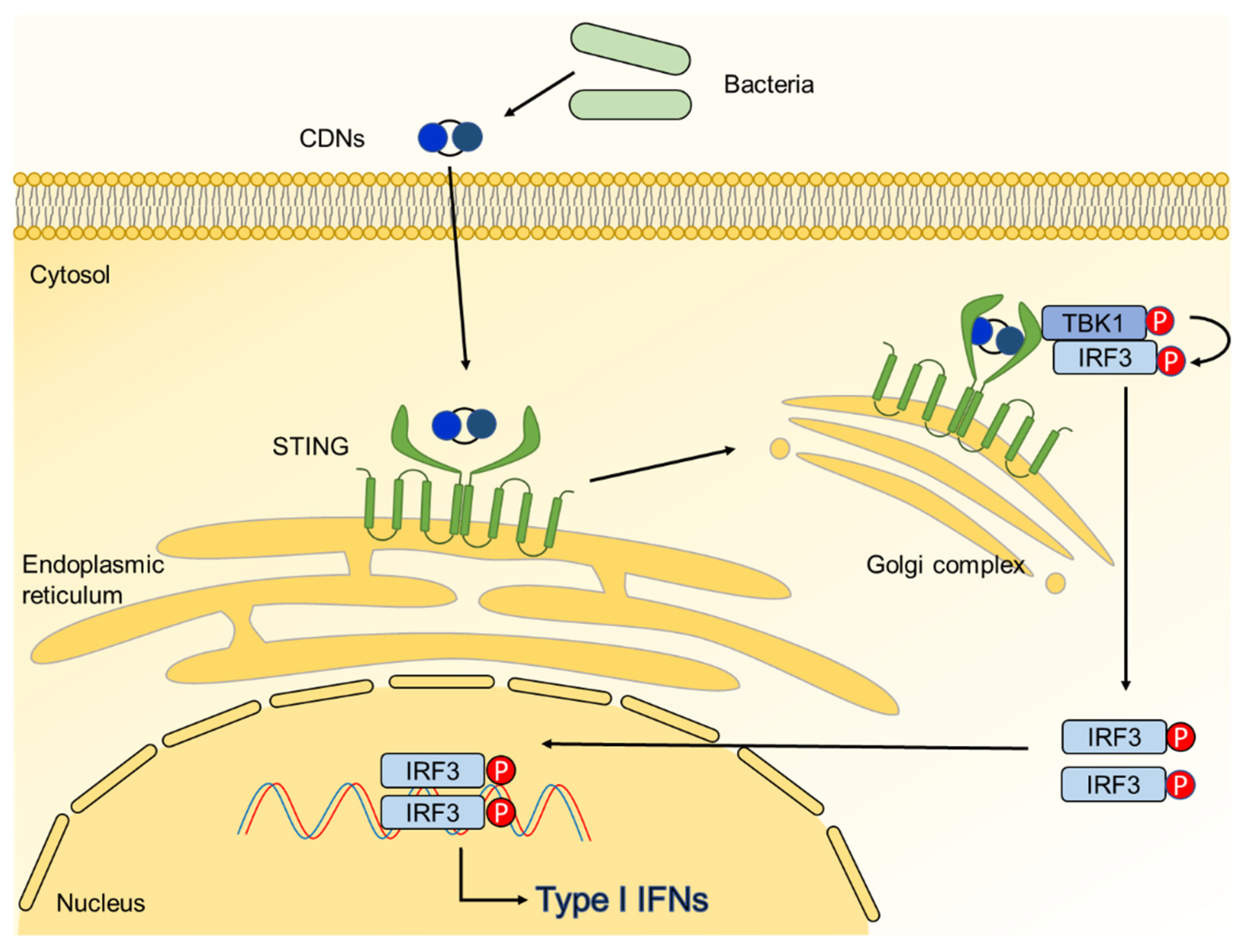
| MAMPs | Receptor | Effects | References |
|---|---|---|---|
| Lipopolysaccharide | TLR4 | Inducing bone loss Inhibiting osteoclastogenesis on macrophages Facilitating osteoclast differentiation on committed osteoclasts Downregulating osteoblast differentiation | [28,29,30,31,32,33,34,35] |
| Lipoteichoic acid | TLR2 | Healing femoral fractures in mice Attenuating osteoclast differentiation and activating phagocytosis Upregulating osteogenic markers and osteoblastogenesis | [36,37,38,39,40] |
| Lipoprotein | TLR2 | Promoting bone resorption Upregulating osteoclast differentiation Stimulating osteoblasts to elevate RANKL/OPG ratio | [41,42,43] |
| Fimbria | TLR4 | Inducing osteoclastogenesis and bone resorption | [44,45,46,47,48,49] |
| Peptidoglycan | NOD1 | Enhancing osteoclastogenesis and bone resorption Triggering osteoclast differentiation synergistically with LPS | [50,51,52,53,54,55,56] |
| NOD2 | Upregulation of bone density Facilitating osteoblast differentiation Diminishing osteoclastogenesis by reducing RANKL/OPG ratio | [54,57] |
| MAMPs | Mechanism | Effects on Bone Metabolism | References |
|---|---|---|---|
| Short chain fatty acids | Activation of GPCRs Inhibition of histone deacetylases | Inhibited osteoclast differentiation and function Upregulated osteogenic factors in low dose Attenuated osteoblast differentiation and mineralization Prevented bone loss in various mouse models | [109,111,112,114] |
| Extracellular vesicles | Activation of TLR2 Induction of pro-inflammatory cytokines | Downregulated osteoblast differentiation and activity Regulated RANKL and OPG expression in mesenchymal cells | [17] |
| Extracellular polysaccharides | Activation of TLR2 | Inhibited osteoclast differentiation from macrophages, but some EPS increased collagenolytic activity of osteoclasts Enhanced osteoblast differentiation, but oral pathogen-derived CPS decreased proliferation of osteoblasts | [127,128,129,132] |
| Cyclic dinucleotides | Induction of STING-mediated IFN-β | Inhibited differentiation of macropahges into mature osteoclasts Alleviated RANKL-induced bone destruction | [18] |
| Probiotics | Bone Effects | Animal Model | References | |
|---|---|---|---|---|
| Increase | Decrease | |||
| L. reuteri ATCC 6475 | BV/TV, Tb.N, Tb.Th, OCN | Normal | [12] | |
| BV/TV, Tb.N, Tb.Th | RANKL, TRAP5 | Ovariectomy | [165] | |
| BV/TV, OCN, Wnt10b | Diabetic osteoporosis | [166] | ||
| L. rhamnosus GG | BV/TV, OCN | RANKL, TNF-α, IL-17 | Ovariectomy | [11] |
| Bone loss, Inflammation | Periodontitis | [167] | ||
| L. paracasei and L. plantarum | BV/TV, Tb.N, Cortical bone | Ovariectomy | [168] | |
| L. casei | Osteolysis | Calvarial resorption | [169] | |
| L. gasseri SBT2055 | Bone loss, Inflammation | Periodontitis | [13] | |
| L. brevis CD2 | Bone loss, Inflammation | Periodontitis | [14] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwon, Y.; Park, C.; Lee, J.; Park, D.H.; Jeong, S.; Yun, C.-H.; Park, O.-J.; Han, S.H. Regulation of Bone Cell Differentiation and Activation by Microbe-Associated Molecular Patterns. Int. J. Mol. Sci. 2021, 22, 5805. https://doi.org/10.3390/ijms22115805
Kwon Y, Park C, Lee J, Park DH, Jeong S, Yun C-H, Park O-J, Han SH. Regulation of Bone Cell Differentiation and Activation by Microbe-Associated Molecular Patterns. International Journal of Molecular Sciences. 2021; 22(11):5805. https://doi.org/10.3390/ijms22115805
Chicago/Turabian StyleKwon, Yeongkag, Chaeyeon Park, Jueun Lee, Dong Hyun Park, Sungho Jeong, Cheol-Heui Yun, Ok-Jin Park, and Seung Hyun Han. 2021. "Regulation of Bone Cell Differentiation and Activation by Microbe-Associated Molecular Patterns" International Journal of Molecular Sciences 22, no. 11: 5805. https://doi.org/10.3390/ijms22115805
APA StyleKwon, Y., Park, C., Lee, J., Park, D. H., Jeong, S., Yun, C.-H., Park, O.-J., & Han, S. H. (2021). Regulation of Bone Cell Differentiation and Activation by Microbe-Associated Molecular Patterns. International Journal of Molecular Sciences, 22(11), 5805. https://doi.org/10.3390/ijms22115805






