A Comprehensive Review: Sphingolipid Metabolism and Implications of Disruption in Sphingolipid Homeostasis
Abstract
1. Introduction
2. Sphingolipid Structures
2.1. Sphingoid Bases and Simple Derivatives
2.2. Ceramide
2.3. Complex Sphingolipids
2.3.1. Phosphosphingolipids
2.3.2. Glycosphingolipids
2.3.3. Neutral Glycosphingolipids
2.3.4. Acidic Glycosphingolipids
Gangliosides
Glucuronoglycosphingolipids and Sulfatoglycosphingolipds
3. Sphingolipid Biosynthesis
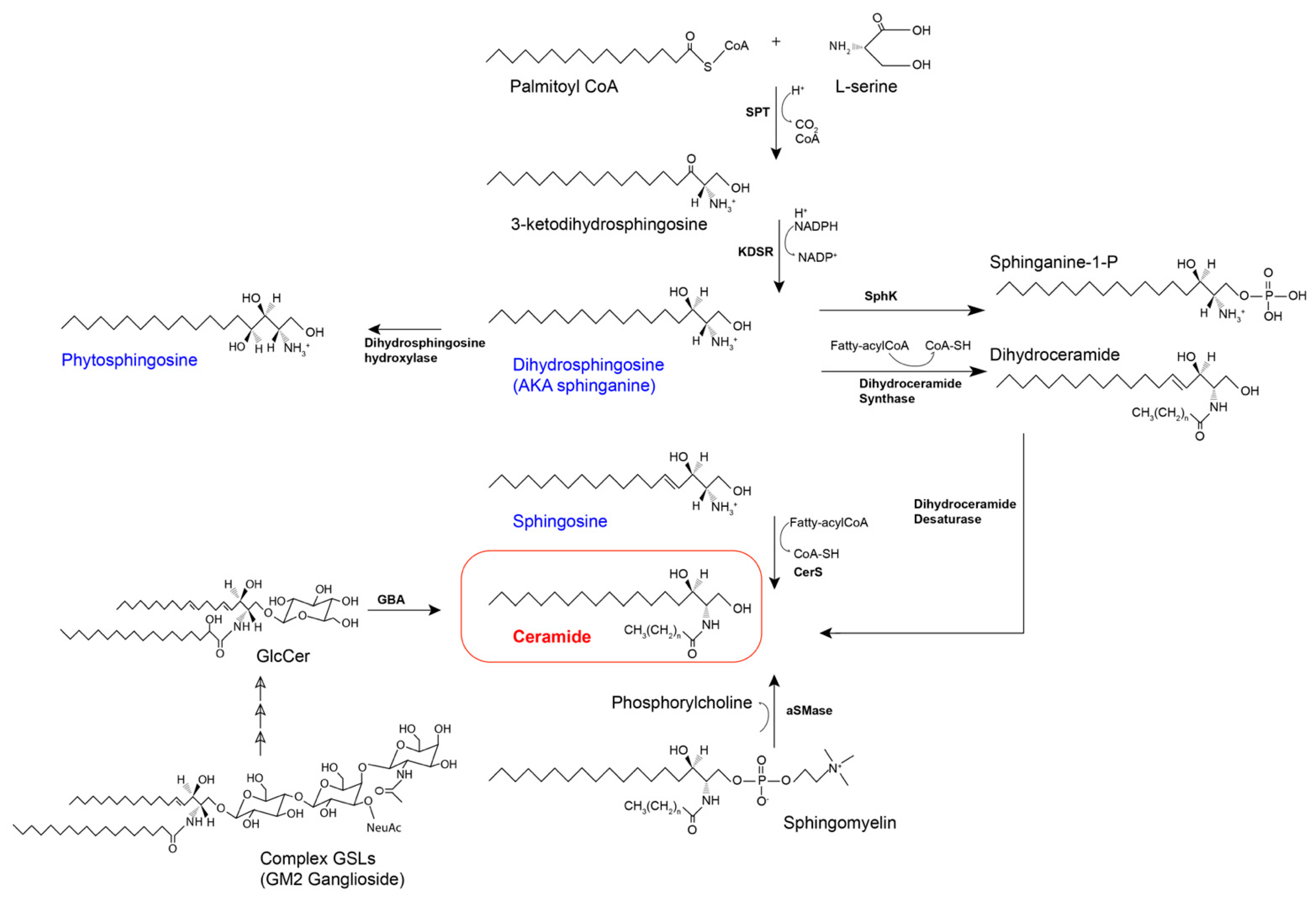
3.1. Biosynthesis of Sphingoid Bases and Ceramide via the de novo Synthetic Pathway
3.2. The Salvage Pathway
3.3. Formation of Complex Sphingolipids
3.3.1. Phosphosphingolipids
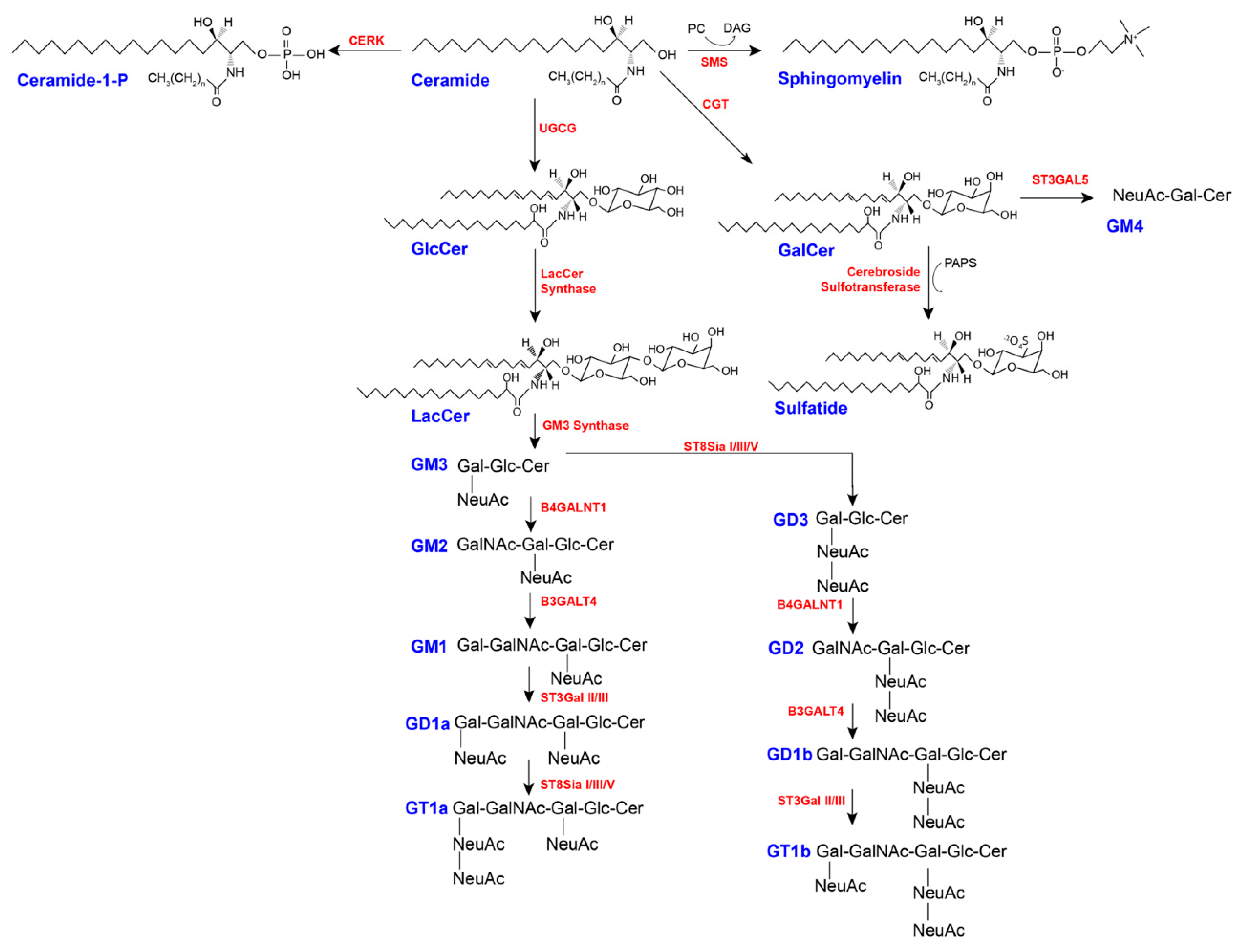
3.3.2. Glycosphingolipids
4. Sphingolipid Catabolism
4.1. Degradation of Complex Sphingolipids
4.1.1. Sphingomyelin Breakdown
4.1.2. Glycosphingolipid Breakdown
4.2. Ceramide Catabolism
4.3. Sphingosine-1-Phosphate—The Final Breakdown
5. Sphingolipid Functions
5.1. Membrane Domains and Signalling
5.2. Cell Death and Proliferation
5.3. Cell Migration and Invasiveness
5.4. Inflammation
5.5. CNS Development
6. Sphingolipids’ Role in Pathophysiology
6.1. Inflammatory Diseases
6.2. Cancer
6.2.1. Ceramide Synthesis
6.2.2. Ceramide Transport
6.2.3. Ceramide Metabolism
6.3. Metabolic Diseases
6.4. Neurodegenerative Diseases
6.5. Lysosomal Storage Disorders
6.5.1. Gaucher Disease
6.5.2. Niemann–Pick Disease
6.5.3. GM1 Gangliosidoses
6.5.4. GM2 Gangliosidoses
7. Conclusions
Funding
Conflicts of Interest
Abbreviations
| SMPDL3b | sphingomyelin phosphodiesterase acid-like 3b |
| LSDs | lysosomal storage disorders |
| LCB | long-chain bases |
| ER | endoplasmic reticulum |
| NADPH | nicotinamide adenine dinucleotide phosphate |
| FAD | flavin adenine dinucleotide |
| PHS | phytosphingosine |
| S1P | sphingosine-1-phosphate |
| PSLs | phosphosphingolipid |
| GSLs | glycosphingolipids |
| SM | sphingomyelin |
| C1P | ceramide-1-phosphate |
| GlcCer | glucosylceramide; glucocerebroside |
| GalCer | galactosylceramide; galactocerebroside |
| LacCer | lactosylceramide |
| Gal | galactose |
| GalNAc | N-acetylgalactosamine |
| Kdn | 2-keto-3-deoxy-nononic acid |
| Neu5Ac | N-acetylneuraminic acid |
| Neu5Gc | N-glycolylneuraminic |
| CoA | coenzyme-A |
| SPT | serine palmitoyltransferase |
| ORMDL | orosomucoid-like protein |
| KDSR | 3-ketodihydrosphingosine reductase |
| CerS | ceramide synthase |
| SMase | sphingomyelinase |
| CDase | ceramidase |
| SphK | sphingosine kinase |
| CERT | ceramide transport protein |
| SMS | sphingomyelin synthase |
| DAG | diacylglycerol |
| ERK | extracellular signal-regulated kinase |
| CERK | ceramide kinase |
| CPTP | ceramide phosphate transfer protein |
| UDP-Gal | uridine diphosphate galactose |
| UGCG | UDP-glucose ceramide glucosyltransferase |
| BMP | bis(monoacylglycerol)phosphate |
| aSMase | acid sphingomyelinase |
| L-aSMase | lysosomal acid sphingomyelinase |
| S-aSMase | secretory acid sphingomyelinase |
| nSMase | neutral sphingomyelinase |
| alkSMase | alkaline sphingomyelinase |
| NPP | nucleotide pyrophosphatase/phosphodiesterase |
| LLBP | lysosomal lipid binding proteins |
| GM2A | GM2 activator protein |
| GH | glycosyl hydrolase |
| HexA | β-hexosaminidase A |
| aCDase | acid ceramidase |
| nCDase | neutral ceramidase |
| alkCDase | alkaline ceramidase |
| LPP1-3 | lipid phosphate phosphatases |
| SPL | sphingosine-1-phosphate lyase |
| PLP | pyridoxal 5′-phosphate |
| GPCR | G-protein coupled receptor proteins |
| CT | cholera toxin |
| SV40 | simian virus 40 |
| JNK | c-Jun N-terminal kinases |
| BMDM | primary bone-marrow-derived macrophages |
| mTOR | mammalian target of rapamycin |
| GFR | growth factor receptors |
| EGFR | epidermal GFR |
| ERM | ezrin, radixin, and moesin |
| CF | cystic fibrosis |
| COPD | chronic obstructive pulmonary disease |
| SHRSP | spontaneously hypertensive stroke-prone |
| RTK | receptor tyrosine kinases |
| LT | leukotriene |
| 5-LO | 5-lipoxygenase |
| AA | arachidonic acid |
| PLA2 | phospholipase A2 activity |
| IBD | inflammatory bowel disease |
| HDAC1 | histone deacetylase 1 |
| PMA | horbol 12-myristate 13-acetate |
| EGF | epidermal growth factor |
| AD | Alzheimer’s disease |
| PD | Parkinson’s disease |
| Aβ | β-amyloid peptide |
| APP | amyloid precursor protein |
| α-syn | alpha-synuclein |
| GD | Gaucher disease |
| NPD | Niemann–Pick disease |
References
- Thudichum, J.L.W. A Treatise on the Chemical Constitution of the Brain: Based Throughout upon Original Researches. Glasg. Med. J. 1884, 22, 363–364. [Google Scholar]
- Schnaar, R.L.; Kinoshita, T. Glycosphingolipids. In Essentials of Glycobiology; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., Schnaar, R.L., Seeberger, P.H., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2015. [Google Scholar]
- Young, M.M.; Kester, M.; Wang, H.-G. Sphingolipids: Regulators of Crosstalk between Apoptosis and Autophagy. J. Lipid Res. 2013, 54, 5–19. [Google Scholar] [CrossRef] [PubMed]
- Dadsena, S.; Bockelmann, S.; Mina, J.G.M.; Hassan, D.G.; Korneev, S.; Razzera, G.; Jahn, H.; Niekamp, P.; Müller, D.; Schneider, M.; et al. Ceramides Bind VDAC2 to Trigger Mitochondrial Apoptosis. Nat. Commun. 2019, 10, 1832. [Google Scholar] [CrossRef]
- Takehara, M.; Bandou, H.; Kobayashi, K.; Nagahama, M. Clostridium Perfringens α-Toxin Specifically Induces Endothelial Cell Death by Promoting Ceramide-Mediated Apoptosis. Anaerobe 2020, 65, 102262. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, C.; Childs, S.; Ohotski, J.; McGlynn, L.; Riddick, M.; MacFarlane, S.; Tasker, D.; Pyne, S.; Pyne, N.J.; Edwards, J.; et al. Regulation of Cell Survival by Sphingosine-1-Phosphate Receptor S1P 1 via Reciprocal ERK-Dependent Suppression of Bim and PI-3-Kinase/Protein Kinase C-Mediated Upregulation of Mcl-1. Cell Death Dis. 2013, 4, e927. [Google Scholar] [CrossRef]
- Dai, Y.; Zhang, M.; Shi, X.; Wang, K.; Gao, G.; Shen, L.; Sun, T. Kinetic Study of Aβ(1-42) Amyloidosis in the Presence of Ganglioside-Containing Vesicles. Colloids Surf. B Biointerfaces 2020, 185, 110615. [Google Scholar] [CrossRef]
- Orsini, M.; Chateauvieux, S.; Rhim, J.; Gaigneaux, A.; Cheillan, D.; Christov, C.; Dicato, M.; Morceau, F.; Diederich, M. Sphingolipid-Mediated Inflammatory Signaling Leading to Autophagy Inhibition Converts Erythropoiesis to Myelopoiesis in Human Hematopoietic Stem/Progenitor Cells. Cell Death Differ. 2019, 26, 1796–1812. [Google Scholar] [CrossRef] [PubMed]
- Ponnusamy, S.; Selvam, S.P.; Mehrotra, S.; Kawamori, T.; Snider, A.J.; Obeid, L.M.; Shao, Y.; Sabbadini, R.; Ogretmen, B. Communication between Host Organism and Cancer Cells Is Transduced by Systemic Sphingosine Kinase 1/Sphingosine 1-Phosphate Signalling to Regulate Tumour Metastasis. EMBO Mol. Med. 2012, 4, 761–775. [Google Scholar] [CrossRef]
- Tidhar, R.; Futerman, A.H. The Complexity of Sphingolipid Biosynthesis in the Endoplasmic Reticulum. Biochim. Biophys. Acta BBA Mol. Cell Res. 2013, 1833, 2511–2518. [Google Scholar] [CrossRef]
- Gao, Y.; He, X.; Ding, F.; Zhang, Y. Recent Progress in Chemical Syntheses of Sphingosines and Phytosphingosines. Synthesis 2016, 48, 4017–4037. [Google Scholar] [CrossRef]
- Merrill, A.H. Sphingolipid and Glycosphingolipid Metabolic Pathways in the Era of Sphingolipidomics. Chem. Rev. 2011, 111, 6387–6422. [Google Scholar] [CrossRef]
- Hamanaka, S.; Suzuki, A.; Hara, M.; Nishio, H.; Otsuka, F.; Uchida, Y. Human Epidermal Glucosylceramides Are Major Precursors of Stratum Corneum Ceramides. J. Investig. Dermatol. 2002, 119, 416–423. [Google Scholar] [CrossRef]
- Nishimura, K. Phytosphingosine Is a Characteristic Component of the Glycolipids in the Vertebrate Intestine. Comp. Biochem. Physiol. B 1987, 86, 149–154. [Google Scholar] [CrossRef]
- Kondo, N.; Ohno, Y.; Yamagata, M.; Obara, T.; Seki, N.; Kitamura, T.; Naganuma, T.; Kihara, A. Identification of the Phytosphingosine Metabolic Pathway Leading to Odd-Numbered Fatty Acids. Nat. Commun. 2014, 5, 5338. [Google Scholar] [CrossRef]
- Lee, T.C.; Ou, M.C.; Shinozaki, K.; Malone, B.; Snyder, F. Biosynthesis of N-Acetylsphingosine by Platelet-Activating Factor: Sphingosine CoA-Independent Transacetylase in HL-60 Cels. J. Biol. Chem. 1996, 271, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Haribowo, A.G.; Hannich, J.T.; Michel, A.H.; Megyeri, M.; Schuldiner, M.; Kornmann, B.; Riezman, H. Cytotoxicity of 1-Deoxysphingolipid Unraveled by Genome-Wide Genetic Screens and Lipidomics in Saccharomyces Cerevisiae. Mol. Biol. Cell 2019, 30, 2814–2826. [Google Scholar] [CrossRef]
- Alecu, I.; Othman, A.; Penno, A.; Saied, E.M.; Arenz, C.; von Eckardstein, A.; Hornemann, T. Cytotoxic 1-Deoxysphingolipids Are Metabolized by a Cytochrome P450-Dependent Pathway. J. Lipid Res. 2017, 58, 60–71. [Google Scholar] [CrossRef]
- Steiner, R.; Saied, E.M.; Othman, A.; Arenz, C.; Maccarone, A.T.; Poad, B.L.J.; Blanksby, S.J.; von Eckardstein, A.; Hornemann, T. Elucidating the Chemical Structure of Native 1-Deoxysphingosine. J. Lipid Res. 2016, 57, 1194–1203. [Google Scholar] [CrossRef] [PubMed]
- Saied, E.M.; Le, T.L.-S.; Hornemann, T.; Arenz, C. Synthesis and Characterization of Some Atypical Sphingoid Bases. Bioorg. Med. Chem. 2018, 26, 4047–4057. [Google Scholar] [CrossRef] [PubMed]
- Poad, B.L.J.; Maccarone, A.T.; Yu, H.; Mitchell, T.W.; Saied, E.M.; Arenz, C.; Hornemann, T.; Bull, J.N.; Bieske, E.J.; Blanksby, S.J. Differential-Mobility Spectrometry of 1-Deoxysphingosine Isomers: New Insights into the Gas Phase Structures of Ionized Lipids. Anal. Chem. 2018. [Google Scholar] [CrossRef]
- Lone, M.A.; Hülsmeier, A.J.; Saied, E.M.; Karsai, G.; Arenz, C.; von Eckardstein, A.; Hornemann, T. Subunit Composition of the Mammalian Serine-Palmitoyltransferase Defines the Spectrum of Straight and Methyl-Branched Long-Chain Bases. Proc. Natl. Acad. Sci. USA 2020, 117, 15591–15598. [Google Scholar] [CrossRef]
- Hannich, J.T.; Mellal, D.; Feng, S.; Zumbuehl, A.; Riezman, H. Structure and Conserved Function of Iso-Branched Sphingoid Bases from the Nematode Caenorhabditis Elegans. Chem. Sci. 2017, 8, 3676–3686. [Google Scholar] [CrossRef]
- Al Sazzad, M.A.; Yasuda, T.; Murata, M.; Slotte, J.P. The Long-Chain Sphingoid Base of Ceramides Determines Their Propensity for Lateral Segregation. Biophys. J. 2017, 112, 976–983. [Google Scholar] [CrossRef]
- Hannun, Y.A.; Obeid, L.M. Many Ceramides. J. Biol. Chem. 2011, 286, 27855–27862. [Google Scholar] [CrossRef]
- Dutagaci, B.; Becker-Baldus, J.; Faraldo-Gómez, J.D.; Glaubitz, C. Ceramide–Lipid Interactions Studied by MD Simulations and Solid-State NMR. Biochim. Biophys. Acta BBA Biomembr. 2014, 1838, 2511–2519. [Google Scholar] [CrossRef]
- Pullmannová, P.; Staňková, K.; Pospíšilová, M.; Školová, B.; Zbytovská, J.; Vávrová, K. Effects of Sphingomyelin/Ceramide Ratio on the Permeability and Microstructure of Model Stratum Corneum Lipid Membranes. Biochim. Biophys. Acta BBA Biomembr. 2014, 1838, 2115–2126. [Google Scholar] [CrossRef]
- Chang, K.-T.; Anishkin, A.; Patwardhan, G.A.; Beverly, L.J.; Siskind, L.J.; Colombini, M. Ceramide Channels: Destabilization by Bcl-XL and Role in Apoptosis. Biochim. Biophys. Acta BBA Biomembr. 2015, 1848, 2374–2384. [Google Scholar] [CrossRef]
- Mencarelli, C.; Martinez–Martinez, P. Ceramide Function in the Brain: When a Slight Tilt Is Enough. Cell. Mol. Life Sci. 2013, 70, 181–203. [Google Scholar] [CrossRef] [PubMed]
- Gault, C.; Obeid, L.; Hannun, Y. An Overview of Sphingolipid Metabolism: From Synthesis to Breakdown. Adv. Exp. Med. Biol. 2010, 688, 1–23. [Google Scholar]
- Farfel-Becker, T.; Vitner, E.B.; Kelly, S.L.; Bame, J.R.; Duan, J.; Shinder, V.; Merrill, A.H.; Dobrenis, K.; Futerman, A.H. Neuronal Accumulation of Glucosylceramide in a Mouse Model of Neuronopathic Gaucher Disease Leads to Neurodegeneration. Hum. Mol. Genet. 2014, 23, 843–854. [Google Scholar] [CrossRef]
- Sipione, S.; Monyror, J.; Galleguillos, D.; Steinberg, N.; Kadam, V. Gangliosides in the Brain: Physiology, Pathophysiology and Therapeutic Applications. Front. Neurosci. 2020, 14. [Google Scholar] [CrossRef]
- Varki, A.; Schauer, R. Sialic Acids. In Essentials of Glycobiology; Varki, A., Cummings, R.D., Esko, J.D., Freeze, H.H., Stanley, P., Bertozzi, C.R., Hart, G.W., Etzler, M.E., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2009; ISBN 978-0-87969-770-9. [Google Scholar]
- Diaz, S.L.; Padler-Karavani, V.; Ghaderi, D.; Hurtado-Ziola, N.; Yu, H.; Chen, X.; Linden, E.C.M.B.-V.d.; Varki, A.; Varki, N.M. Sensitive and Specific Detection of the Non-Human Sialic Acid N-Glycolylneuraminic Acid In Human Tissues and Biotherapeutic Products. PLoS ONE 2009, 4, e4241. [Google Scholar] [CrossRef]
- Altman, M.O.; Gagneux, P. Absence of Neu5Gc and Presence of Anti-Neu5Gc Antibodies in Humans—An Evolutionary Perspective. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Chou, H.-H.; Hayakawa, T.; Diaz, S.; Krings, M.; Indriati, E.; Leakey, M.; Paabo, S.; Satta, Y.; Takahata, N.; Varki, A. Inactivation of CMP-N-Acetylneuraminic Acid Hydroxylase Occurred Prior to Brain Expansion during Human Evolution. Proc. Natl. Acad. Sci. USA 2002, 99, 11736–11741. [Google Scholar] [CrossRef]
- Ariga, T.; Kohriyama, T.; Freddo, L.; Latov, N.; Saito, M.; Kon, K.; Ando, S.; Suzuki, M.; Hemling, M.E.; Rinehart, K.L. Characterization of Sulfated Glucuronic Acid Containing Glycolipids Reacting with IgM M-Proteins in Patients with Neuropathy. J. Biol. Chem. 1987, 262, 848–853. [Google Scholar] [CrossRef]
- Hirahara, Y.; Wakabayashi, T.; Mori, T.; Koike, T.; Yao, I.; Tsuda, M.; Honke, K.; Gotoh, H.; Ono, K.; Yamada, H. Sulfatide Species with Various Fatty Acid Chains in Oligodendrocytes at Different Developmental Stages Determined by Imaging Mass Spectrometry. J. Neurochem. 2017, 140, 435–450. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Harada, M.; Hashimoto, K.; Guo, R.; Nakajima, T.; Kashihara, T.; Yamada, M.; Aoyama, T.; Kamijo, Y. Impact of Chronic Kidney Dysfunction on Serum Sulfatides and Its Metabolic Pathway in Mice. Glycoconj. J. 2019, 36, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Norton, W.T.; Cammer, W. Isolation and Characterization of Myelin. In Myelin; Morell, P., Ed.; Springer: Boston, MA, USA, 1984; pp. 147–195. ISBN 978-1-4757-1830-0. [Google Scholar]
- Pasquet, L.; Camara, K.; Bloom, A.C.; Richardson, S.K.; Howell, A.R.; Terabe, M.; Berzofsky, J.A. The Ceramide Structure of Sulfatide-Analogues Influences the Functional Activity of Type II NKT Cells. J. Immunol. 2018, 200, 20–57. [Google Scholar]
- Stettner, P.; Bourgeois, S.; Marsching, C.; Traykova-Brauch, M.; Porubsky, S.; Nordström, V.; Hopf, C.; Koesters, R.; Sandhoff, R.; Wiegandt, H.; et al. Sulfatides Are Required for Renal Adaptation to Chronic Metabolic Acidosis. Proc. Natl. Acad. Sci. USA 2013, 110, 9998–10003. [Google Scholar] [CrossRef]
- Li, S.; Xie, T.; Liu, P.; Wang, L.; Gong, X. Structural Insights into the Assembly and Substrate Selectivity of Human SPT–ORMDL3 Complex. Nat. Struct. Mol. Biol. 2021. [Google Scholar] [CrossRef]
- Wang, Y.; Niu, Y.; Zhang, Z.; Gable, K.; Gupta, S.D.; Somashekarappa, N.; Han, G.; Zhao, H.; Myasnikov, A.G.; Kalathur, R.C.; et al. Structural Insights into the Regulation of Human Serine Palmitoyltransferase Complexes. Nat. Struct. Mol. Biol. 2021. [Google Scholar] [CrossRef]
- Han, G.; Gupta, S.D.; Gable, K.; Niranjanakumari, S.; Moitra, P.; Eichler, F.; Brown, R.H.; Harmon, J.M.; Dunn, T.M. Identification of Small Subunits of Mammalian Serine Palmitoyltransferase That Confer Distinct Acyl-CoA Substrate Specificities. Proc. Natl. Acad. Sci. USA 2009, 106, 8186–8191. [Google Scholar] [CrossRef] [PubMed]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and Their Metabolism in Physiology and Disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef]
- Bejaoui, K.; Wu, C.; Scheffler, M.D.; Haan, G.; Ashby, P.; Wu, L.; de Jong, P.; Brown, R.H. SPTLC1 Is Mutated in Hereditary Sensory Neuropathy, Type 1. Nat. Genet. 2001, 27, 261–262. [Google Scholar] [CrossRef]
- Dawkins, J.L.; Hulme, D.J.; Brahmbhatt, S.B.; Auer-Grumbach, M.; Nicholson, G.A. Mutations in SPTLC1, Encoding Serine Palmitoyltransferase, Long Chain Base Subunit-1, Cause Hereditary Sensory Neuropathy Type I. Nat. Genet. 2001, 27, 309–312. [Google Scholar] [CrossRef]
- Auer-Grumbach, M.; Bode, H.; Pieber, T.R.; Schabhüttl, M.; Fischer, D.; Seidl, R.; Graf, E.; Wieland, T.; Schuh, R.; Vacariu, G.; et al. Mutations at Ser331 in the HSN Type I Gene SPTLC1 Are Associated with a Distinct Syndromic Phenotype. Eur. J. Med. Genet. 2013, 56, 266–269. [Google Scholar] [CrossRef]
- Rotthier, A.; Penno, A.; Rautenstrauss, B.; Auer-Grumbach, M.; Stettner, G.M.; Asselbergh, B.; Van Hoof, K.; Sticht, H.; Lévy, N.; Timmerman, V.; et al. Characterization of Two Mutations in the SPTLC1 Subunit of Serine Palmitoyltransferase Associated with Hereditary Sensory and Autonomic Neuropathy Type I. Hum. Mutat. 2011, 32, E2211–E2225. [Google Scholar] [CrossRef] [PubMed]
- Murphy, S.M.; Ernst, D.; Wei, Y.; Laurà, M.; Liu, Y.-T.; Polke, J.; Blake, J.; Winer, J.; Houlden, H.; Hornemann, T.; et al. Hereditary Sensory and Autonomic Neuropathy Type 1 (HSANI) Caused by a Novel Mutation in SPTLC2. Neurology 2013, 80, 2106–2111. [Google Scholar] [CrossRef]
- Ernst, D.; Murphy, S.M.; Sathiyanadan, K.; Wei, Y.; Othman, A.; Laurá, M.; Liu, Y.-T.; Penno, A.; Blake, J.; Donaghy, M.; et al. Novel HSAN1 Mutation in Serine Palmitoyltransferase Resides at a Putative Phosphorylation Site That Is Involved in Regulating Substrate Specificity. Neuromol. Med. 2015, 17, 47–57. [Google Scholar] [CrossRef]
- Wattenberg, B.W. Kicking off Sphingolipid Biosynthesis: Structures of the Serine Palmitoyltransferase Complex. Nat. Struct. Mol. Biol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Park, K.-H.; Ye, Z.-W.; Zhang, J.; Hammad, M.; Townsend, D.M.; Rockey, D.C.; Kim, S.-H. 3-Ketodihydrosphingosine Reductase Mutation Induces Steatosis and Hepatic Injury in Zebrafish. Sci. Rep. 2019. [Google Scholar] [CrossRef] [PubMed]
- Schick, A.; Kolter, T.; Giannis, A.; Sandhoff, K. Synthesis of Phosphonate Analogues of Sphinganine-1-Phosphate and Sphingosine-1-Phosphate. Tetrahedron 1995, 51, 11207–11218. [Google Scholar] [CrossRef]
- Levy, M.; Futerman, A.H. Critical Review Mammalian Ceramide Synthases. IUBMB Life 2010. [Google Scholar] [CrossRef] [PubMed]
- Pewzner-Jung, Y.; Ben-Dor, S.; Futerman, A.H. When Do Lasses (Longevity Assurance Genes) Become CerS (Ceramide Synthases)? Insights Into The Regulation Of Ceramide Synthesis. J. Biol. Chem. 2006. [Google Scholar] [CrossRef]
- Pewzner-Jung, Y.; Brenner, O.; Braun, S.; Laviad, E.L.; Ben-Dor, S.; Feldmesser, E.; Horn-Saban, S.; Amann-Zalcenstein, D.; Raanan, C.; Berkutzki, T.; et al. A Critical Role for Ceramide Synthase 2 in Liver Homeostasis II. Insights into molecular changes leading to hepatopathy. We Have Generated a Mouse That Cannot Synthesize Very Long Acyl Chain (C22–C24) Ceramides. J. Biol. Chem. 2010, 285, 10911–10923. [Google Scholar] [CrossRef] [PubMed]
- Merrill, A.H. De Novo Sphingolipid Biosynthesis: A Necessary, but Dangerous, Pathway. J. Biol. Chem. 2002. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, G.; Capasso, S.; Sticco, L.; Russo, D. Glycosphingolipids: Synthesis and Functions. FEBS J. 2013, 280, 6338–6353. [Google Scholar] [CrossRef]
- Schulze, H.; Sandhoff, K. Lysosomal Lipid Storage Diseases. Cold Spring Harbor Perspect. Biol. 2011. [Google Scholar] [CrossRef]
- Kitatani, K.; Idkowiak-Baldys, J.; Hannun, Y.A. The Sphingolipid Salvage Pathway in Ceramide Metabolism and Signaling. Cell. Signal. 2008, 20, 1010–1018. [Google Scholar] [CrossRef]
- Leipelt, M.; Merrill, A.H. Sphingolipid Biosynthesis. In Encyclopedia of Biological Chemistry; Elsevier: Amsterdam, The Netherlands, 2004; pp. 76–81. [Google Scholar]
- Hait, N.C.; Maiti, A. The Role of Sphingosine-1-Phosphate and Ceramide-1-Phosphate in Inflammation and Cancer. Mediat. Inflamm. 2017. [Google Scholar] [CrossRef]
- Hanada, K.; Kumagai, K.; Yasuda, S.; Miura, Y.; Kawano, M.; Fukasawa, M.; Nishijima, M. Molecular Machinery for Non-Vesicular Trafficking of Ceramide. Nature 2003, 426, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Huitema, K.; van den Dikkenberg, J.; Brouwers, J.F.H.M.; Holthuis, J.C.M. Identification of a Family of Animal Sphingomyelin Synthases. EMBO J. 2004, 23, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Cabukusta, B.; Kol, M.; Kneller, L.; Hilderink, A.; Bickert, A.; Mina, J.G.M.; Korneev, S.; Holthuis, J.C.M. ER Residency of the Ceramide Phosphoethanolamine Synthase SMSr Relies on Homotypic Oligomerization Mediated by Its SAM Domain. Sci. Rep. 2017, 7, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Vacaru, A.M.; Tafesse, F.G.; Ternes, P.; Kondylis, V.; Hermansson, M.; Brouwers, J.F.H.M.; Somerharju, P.; Rabouille, C.; Holthuis, J.C.M. Sphingomyelin Synthase-Related Protein SMSr Controls Ceramide Homeostasis in the ER. J. Cell Biol. 2009, 185, 1013–1027. [Google Scholar] [CrossRef] [PubMed]
- Strub, G.M.; Maceyka, M.; Hait, N.C.; Milstien, S.; Spiegel, S. Extracellular and Intracellular Actions of Sphingosine-1-Phosphate. Adv. Exp. Med. Biol. 2010, 688, 141–155. [Google Scholar] [CrossRef]
- Hait, N.C.; Allegood, J.; Maceyka, M.; Strub, G.M.; Harikumar, K.B.; Singh, S.K.; Luo, C.; Marmorstein, R.; Kordula, T.; Milstien, S.; et al. Regulation of Histone Acetylation in the Nucleus by Sphingosine-1-Phosphate. Science 2009, 325, 1254–1257. [Google Scholar] [CrossRef]
- Strub, G.M.; Paillard, M.; Liang, J.; Gomez, L.; Allegood, J.C.; Hait, N.C.; Maceyka, M.; Price, M.M.; Chen, Q.; Simpson, D.C.; et al. Sphingosine-1-phosphate Produced by Sphingosine Kinase 2 in Mitochondria Interacts with Prohibitin 2 to Regulate Complex IV Assembly and Respiration. FASEB J. 2011, 25, 600–612. [Google Scholar] [CrossRef]
- Sugiura, M.; Kono, K.; Liu, H.; Shimizugawa, T.; Minekura, H.; Spiegel, S.; Kohama, T. Ceramide Kinase, a Novel Lipid Kinase molecular cloning and functional characterization. J. Biol. Chem. 2002. [Google Scholar] [CrossRef]
- Kolter, T.; Proia, R.L.; Sandhoff, K. Combinatorial Ganglioside Biosynthesis. J. Biol. Chem. 2002, 277, 25859–25862. [Google Scholar] [CrossRef]
- Burdick, M.M.; Deaglio, S.; Farnoud, A.M.; Zhang, T.; De Waard, A.A.; Wuhrer, M.; Spaapen, R.M. The Role of Glycosphingolipids in Immune Cell Functions. Front. Immunol. 2019, 1, 90. [Google Scholar] [CrossRef]
- Allende, M.L.; Proia, R.L. Simplifying Complexity: Genetically Resculpting Glycosphingolipid Synthesis Pathways in Mice to Reveal Function. Glycoconj. J. 2014, 31, 613–622. [Google Scholar] [CrossRef]
- Takematsu, H.; Yamamoto, H.; Naito-Matsui, Y.; Fujinawa, R.; Tanaka, K.; Okuno, Y.; Tanaka, Y.; Kyogashima, M.; Kannagi, R.; Kozutsumi, Y. Quantitative Transcriptomic Profiling of Branching in a Glycosphingolipid Biosynthetic Pathway. J. Biol. Chem. 2011, 286, 27214–27224. [Google Scholar] [CrossRef]
- Mullen, T.D.; Hannun, Y.A.; Obeid, L.M. Ceramide Synthases at the Centre of Sphingolipid Metabolism and Biology. Biochem. J. 2012, 441, 789–802. [Google Scholar] [CrossRef]
- Olsen, A.S.B.; Færgeman, N.J. Sphingolipids: Membrane Microdomains in Brain Development, Function and Neurological Diseases. Open Biol. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane Lipids: Where They Are and How They Behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef]
- Hullin-Matsuda, F.; Luquain-Costaz, C.; Bouvier, J.; Delton-Vandenbroucke, I. Bis(Monoacylglycero)Phosphate, a Peculiar Phospholipid to Control the Fate of Cholesterol: Implications in Pathology. Prostaglandins Leukot. Essent. Fatty Acids 2009. [Google Scholar] [CrossRef] [PubMed]
- Schissel, S.L.; Keesler, G.A.; Schuchman, E.H.; Williams, K.J.; Tabas, I. The Cellular Trafficking and Zinc Dependence of Secretory and Lysosomal Sphingomyelinase, Two Products of the Acid Sphingomyelinase Gene. J. Biol. Chem. 1998, 273, 18250–18259. [Google Scholar] [CrossRef]
- Mallela, S.K.; Mitrofanova, A.; Merscher, S.; Fornoni, A. Regulation of the Amount of Ceramide-1-Phosphate Synthesized in Differentiated Human Podocytes. Biochim. Biophys. Acta BBA-Mol. Cell Biol. Lipids 2019, 1864, 158517. [Google Scholar] [CrossRef] [PubMed]
- Mitrofanova, A.; Mallela, S.K.; Ducasa, G.M.; Yoo, T.H.; Rosenfeld-Gur, E.; Zelnik, I.D.; Molina, J.; Varona Santos, J.; Ge, M.; Sloan, A.; et al. SMPDL3b Modulates Insulin Receptor Signaling in Diabetic Kidney Disease. Nat. Commun. 2019, 10, 2692. [Google Scholar] [CrossRef]
- Tan, L.H.R.; Tan, A.J.R.; Ng, Y.Y.; Chua, J.J.E.; Chew, W.S.; Muralidharan, S.; Torta, F.; Dutta, B.; Sze, S.K.; Herr, D.R.; et al. Enriched Expression of Neutral Sphingomyelinase 2 in the Striatum Is Essential for Regulation of Lipid Raft Content and Motor Coordination. Mol. Neurobiol. 2018, 55, 5741–5756. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.J.; Snook, C.F.; Tani, M.; Matmati, N.; Marchesini, N.; Hannun, Y.A. The Extended Family of Neutral Sphingomyelinases. Biochemistry 2006, 45, 11247–11256. [Google Scholar] [CrossRef]
- Clarke, C.J.; Guthrie, J.M.; Hannun, Y.A. Regulation of Neutral Sphingomyelinase-2 (NSMase2) by Tumor Necrosis Factor-α Involves Protein Kinase C-δ in Lung Epithelial Cells. Mol. Pharmacol. 2008, 74, 1022–1032. [Google Scholar] [CrossRef]
- Duan, R.-D. Alkaline Sphingomyelinase (NPP7) in Hepatobiliary Diseases: A Field That Needs to Be Closely Studied. World J. Hepatol. 2018. [Google Scholar] [CrossRef]
- Darmoise, A.; Maschmeyer, P.; Winau, F. The Immunological Functions of Saposins. In Advances in Immunology; Elsevier: Amsterdam, The Netherlands, 2010; Volume 105, pp. 25–62. ISBN 978-0-12-381302-2. [Google Scholar]
- Condori, J.; Acosta, W.; Ayala, J.; Katta, V.; Flory, A.; Martin, R.; Radin, J.; Cramer, C.L.; Radin, D.N. Enzyme Replacement for GM1-Gangliosidosis: Uptake, Lysosomal Activation, and Cellular Disease Correction Using a Novel β-Galactosidase: RTB Lectin Fusion. Mol. Genet. Metab. 2016, 117, 199–209. [Google Scholar] [CrossRef]
- O’Brien, J.S. Molecular Genetics of GM1β-Galactosidase. Clin. Genet. 2008, 8, 303–313. [Google Scholar] [CrossRef]
- Sandhoff, K.; Harzer, K. Gangliosides and Gangliosidoses: Principles of Molecular and Metabolic Pathogenesis. J. Neurosci. 2013, 33, 10195–10208. [Google Scholar] [CrossRef]
- Yu, R.K.; Tsai, Y.-T.; Ariga, T.; Yanagisawa, M. Structures, Biosynthesis, and Functions of Gangliosides—An Overview. J. Oleo Sci. 2011, 60, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Sergi, C. Sialidosis: A Review of Morphology and Molecular Biology of a Rare Pediatric Disorder. Diagnostics 2018, 8, 29. [Google Scholar] [CrossRef]
- Krieg, S.I.; Krägeloh-Mann, I.; Groeschel, S.; Beck-Wödl, S.; Husain, R.A.; Schöls, L.; Kehrer, C. Natural History of Krabbe Disease—A Nationwide Study in Germany Using Clinical and MRI Data. Orphanet J. Rare Dis. 2020, 15, 243. [Google Scholar] [CrossRef]
- Zschoche, A.; Furst, W.; Schwarzmann, G.; Sandhoff, K. Hydrolysis of Lactosylceramide by Human Galactosylceramidase and GM1-Beta-Galactosidase in a Detergent-Free System and Its Stimulation by Sphingolipid Activator Proteins, Sap-B and Sap-C Activator Proteins Stimulate Lactosylceramide Hydrolysis. Eur. J. Biochem. 1994, 222, 83–90. [Google Scholar] [CrossRef]
- Mao, C.; Obeid, L.M. Ceramidases: Regulators of Cellular Responses Mediated by Ceramide, Sphingosine, and Sphingosine-1-Phosphate. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2008, 1781, 424–434. [Google Scholar] [CrossRef]
- Duarte, C.; Akkaoui, J.; Yamada, C.; Ho, A.; Mao, C.; Movila, A. Elusive Roles of the Different Ceramidases in Human Health, Pathophysiology, and Tissue Regeneration. Cells 2020, 9, 1379. [Google Scholar] [CrossRef]
- Vasiliauskaité-Brooks, I.; Sounier, R.; Rochaix, P.; Bellot, G.; Fortier, M.; Hoh, F.; De Colibus, L.; Bechara, C.; Saied, E.M.; Arenz, C.; et al. Structural Insights into Adiponectin Receptors Suggest Ceramidase Activity. Nature 2017, 544, 120–123. [Google Scholar] [CrossRef]
- Allende, M.L.; Bektas, M.; Lee, B.G.; Bonifacino, E.; Kang, J.; Tuymetova, G.; Chen, W.P.; Saba, J.D.; Proia, R.L. Sphingosine-1-Phosphate Lyase Deficiency Produces a pro-Inflammatory Response While Impairing Neutrophil Trafficking. J. Biol. Chem. 2011, 286, 7348–7358. [Google Scholar] [CrossRef] [PubMed]
- Pike, L.J. Report Rafts Defined: A Report on the Keystone Symposium on Lipid Rafts and Cell Function. J. Lipid Res. 2006. [Google Scholar] [CrossRef] [PubMed]
- Varshney, P.; Yadav, V.; Saini, N. Lipid Rafts in Immune Signalling: Current Progress and Future Perspective. Immunology 2016. [Google Scholar] [CrossRef] [PubMed]
- Simons, K.; Toomre, D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000, 1, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Pike, L.J. Lipid Rafts: Heterogeneity on the High Seas. Biochem. J. 2004, 378, 281–292. [Google Scholar] [CrossRef]
- Razani, B.; Woodman, S.E.; Lisanti, M.P. Caveolae: From Cell Biology to Animal Physiology. Pharmacol. Rev. 2002, 54, 431–467. [Google Scholar] [CrossRef]
- Dart, C. Symposium Review: Lipid Microdomains and the Regulation of Ion Channel Function: Channels and Rafts. J. Physiol. 2010, 588, 3169–3178. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, C.; Bagatolli, L.A.; Volovyk, Z.N.; Thompson, N.L.; Levi, M.; Jacobson, K.; Gratton, E. Lipid Rafts Reconstituted in Model Membranes. Biophys. J. 2001, 80, 1417–1428. [Google Scholar] [CrossRef]
- Lingwood, D.; Simons, K. Lipid Rafts As a Membrane-Organizing Principle. Science 2010, 327, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Pinto, S.N.; Silva, L.C.; Futerman, A.H.; Prieto, M. Effect of Ceramide Structure on Membrane Biophysical Properties: The Role of Acyl Chain Length and Unsaturation. Biochim. Biophys. Acta BBA Biomembr. 2011, 1808, 2753–2760. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, T.; Al Sazzad, M.A.; Jäntti, N.Z.; Pentikäinen, O.T.; Slotte, J.P. The Influence of Hydrogen Bonding on Sphingomyelin/Colipid Interactions in Bilayer Membranes. Biophys. J. 2016, 110, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Goñi, F.M.; Sot, J.; Alonso, A. Biophysical Properties of Sphingosine, Ceramides and Other Simple Sphingolipids. Biochem. Soc. Trans. 2014, 42, 1401–1408. [Google Scholar] [CrossRef] [PubMed]
- Santos, T.C.B.; Vaz, A.; Ventura, A.E.; Saied, E.; Arenz, C.; Fedorov, A.; Prieto, M.; Silva, L.C. Canonical and 1-Deoxy(Methyl) Sphingoid Bases: Tackling the Effect of the Lipid Structure on Membrane Biophysical Properties. Langmuir 2020, 36, 6007–6016. [Google Scholar] [CrossRef]
- Venable, R.M.; Sodt, A.J.; Rogaski, B.; Rui, H.; Hatcher, E.; MacKerell, A.D.; Pastor, R.W.; Klauda, J.B. CHARMM All-Atom Additive Force Field for Sphingomyelin: Elucidation of Hydrogen Bonding and of Positive Curvature. Biophys. J. 2014, 107, 134–145. [Google Scholar] [CrossRef]
- Schengrund, C.L. Gangliosides: Glycosphingolipids Essential for Normal Neural Development and Function. Trends Biochem. Sci. 2015, 40, 397–406. [Google Scholar] [CrossRef]
- Goose, J.E.; Sansom, M.S.P. Reduced Lateral Mobility of Lipids and Proteins in Crowded Membranes. PLoS Comput. Biol. 2013, 9, e1003033. [Google Scholar] [CrossRef] [PubMed]
- Kwik, J.; Boyle, S.; Fooksman, D.; Margolis, L.; Sheetz, M.P.; Edidin, M. Membrane Cholesterol, Lateral Mobility, and the Phosphatidylinositol 4,5-Bisphosphate-Dependent Organization of Cell Actin. Proc. Natl. Acad. Sci. USA 2003, 100, 13964–13969. [Google Scholar] [CrossRef]
- Milescu, M.; Bosmans, F.; Lee, S.; Alabi, A.A.; Kim, J.I.; Swartz, K.J. Interactions between Lipids and Voltage Sensor Paddles Detected with Tarantula Toxins. Nat. Struct. Mol. Biol. 2009, 16, 1080–1085. [Google Scholar] [CrossRef]
- Xu, Y.; Ramu, Y.; Lu, Z. Removal of Phospho-Head Groups of Membrane Lipids Immobilizes Voltage Sensors of K+ Channels. Nature 2008, 451, 826–829. [Google Scholar] [CrossRef]
- Melkonian, K.A.; Ostermeyer, A.G.; Chen, J.Z.; Roth, M.G.; Brown, D.A. Role of Lipid Modifications in Targeting Proteins to Detergent-Resistant Membrane Rafts. J. Biol. Chem. 1999, 274, 3910–3917. [Google Scholar] [CrossRef]
- Yang, W.; Vizio, D.D.; Kirchner, M.; Freeman, M.R. Proteome Scale Characterization of Human S-Acylated Proteins in Lipid Raft-Enriched and Non-Raft Membranes. Mol. Cell. Proteom. 2010, 9, 54–70. [Google Scholar] [CrossRef]
- Nitabach, M.N.; Llamas, D.A.; Araneda, R.C.; Intile, J.L.; Thompson, I.J.; Zhou, Y.I.; Holmes, T.C. A Mechanism for Combinatorial Regulation of Electrical Activity: Potassium Channel Subunits Capable of Functioning as Src Homology 3-Dependent Adaptors. Proc. Natl. Acad. Sci. USA 2001, 98, 705–710. [Google Scholar] [CrossRef]
- Presa, N.; Gomez-Larrauri, A.; Dominguez-Herrera, A.; Trueba, M.; Gomez-Muñoz, A. BBA-Molecular and Cell Biology of Lipids Novel Signaling Aspects of Ceramide 1-Phosphate. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020. [Google Scholar] [CrossRef] [PubMed]
- Canals, D.; Roddy, P.; Hannun, Y.A. Protein Phosphatase 1α Mediates Ceramide-Induced ERM Protein Dephosphorylation: A Novel Mechanism Independent of Phosphatidylinositol 4,5-Biphosphate (PIP 2) and Myosin/ERM Phosphatase. J. Biol. Chem. 2012, 287, 10145–10155. [Google Scholar] [CrossRef] [PubMed]
- Hannun, Y.A. The Sphingomyelin Cycle and the Second Messenger Function of Ceramide. J. Biol. Chem. 1994, 269, 3125–3128. [Google Scholar] [CrossRef]
- Gomez-Muñoz, A.; Martin, A.; O’Brien, L.; Brindley, D.N. Cell-Permeable Ceramides Inhibit the Stimulation of DNA Synthesis and Phospholipase D Activity by Phosphatidate and Lysophosphatidate in Rat Fibroblasts. J. Biol. Chem. 1994, 269, 8937–8943. [Google Scholar] [CrossRef]
- Mathias, S.; Dressler, K.A.; Kolesnick, R.N. Characterization of a Ceramide-Activated Protein Kinase: Stimulation by Tumor Necrosis Factor a. Proc. Natl. Acad. Sci. USA 1991, 88, 10009–10013. [Google Scholar] [CrossRef]
- Iwabuchi, K.; Masuda, H.; Kaga, N.; Nakayama, H.; Matsumoto, R.; Iwahara, C.; Yoshizaki, F.; Tamaki, Y.; Kobayashi, T.; Hayakawa, T.; et al. Properties and Functions of Lactosylceramide from Mouse Neutrophils. Glycobiology 2015, 25, 655–668. [Google Scholar] [CrossRef]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Supporting Online Material Ceramide Triggers Budding of Exosome Vesicles into Multivesicular Endosomes. Science 2007. [Google Scholar] [CrossRef]
- Guo, B.B.; Bellingham, S.A.; Hill, A.F. The Neutral Sphingomyelinase Pathway Regulates Packaging of the Prion Protein into Exosomes. J. Biol. Chem. 2015, 290, 3455–3467. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Yoshioka, Y.; Takeshita, F.; Matsuki, Y.; Ochiya, T. Secretory Mechanisms and Intercellular Transfer of MicroRNAs in Living Cells. J. Biol. Chem. 2010, 285, 17442–17452. [Google Scholar] [CrossRef] [PubMed]
- Yuyama, K.; Sun, H.; Mitsutake, S.; Igarashi, Y. Sphingolipid-Modulated Exosome Secretion Promotes Clearance of Amyloid-β by Microglia. J. Biol. Chem. 2012, 287, 10977–10989. [Google Scholar] [CrossRef]
- Contreras, F.-X.; Ernst, A.M.; Haberkant, P.; Björkholm, P.; Lindahl, E.; Gönen, B.; Tischer, C.; Elofsson, A.; von Heijne, G.; Thiele, C.; et al. Molecular Recognition of a Single Sphingolipid Species by a Protein’s Transmembrane Domain. Nature 2012, 481, 525–529. [Google Scholar] [CrossRef] [PubMed]
- Botto, L.; Cunati, D.; Coco, S.; Sesana, S.; Bulbarelli, A.; Biasini, E.; Colombo, L.; Negro, A.; Chiesa, R.; Masserini, M.; et al. Role of Lipid Rafts and GM1 in the Segregation and Processing of Prion Protein. PLoS ONE 2014, 9, e98344. [Google Scholar] [CrossRef][Green Version]
- Jobling, M.G.; Yang, Z.; Kam, W.R.; Lencer, W.I.; Holmes, R.K. A Single Native Ganglioside GM1-Binding Site Is Sufficient for Cholera Toxin To Bind to Cells and Complete the Intoxication Pathway. mBio 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Ewers, H.; Römer, W.; Smith, A.E.; Bacia, K.; Dmitrieff, S.; Chai, W.; Mancini, R.; Kartenbeck, J.; Chambon, V.; Berland, L.; et al. GM1 Structure Determines SV40-Induced Membrane Invagination and Infection. Nat. Cell Biol. 2010, 12, 11–18. [Google Scholar] [CrossRef]
- Senkal, C.E.; Ponnusamy, S.; Bielawski, J.; Hannun, Y.A.; Ogretmen, B. Antiapoptotic Roles of Ceramide-Synthase-6-Generated C16-Ceramide via Selective Regulation of the ATF6/CHOP Arm of ER-Stress-Response Pathways. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2010, 24, 296–308. [Google Scholar] [CrossRef]
- Cuvillier, O.; Pirianov, G.; Kleuser, B.; Vanek, P.G.; Coso, O.A.; Gutkind, J.S.; Spiegel, S. Suppression of Ceramide-Mediated Programmed Cell Death by Sphingosine-1-Phosphate. Nature 1996, 381, 800–803. [Google Scholar] [CrossRef] [PubMed]
- Park, W.; Park, J. The Role of Sphingolipids in Endoplasmic Reticulum Stress. FEBS Lett. 2020, 594, 3632–3651. [Google Scholar] [CrossRef]
- Liu, H.; Toman, R.E.; Goparaju, S.K.; Maceyka, M.; Nava, V.E.; Sankala, H.; Payner, S.G.; Bektas, M.; Ishii, I.; Chun, J.; et al. Sphingosine Kinase Type 2 Is a Putative BH3-Only Protein That Induces Apoptosis. J. Biol. Chem. 2003, 278, 40330–40336. [Google Scholar] [CrossRef]
- Okada, T.; Ding, G.; Sonoda, H.; Kajimoto, T.; Haga, Y.; Khosrowbeygi, A.; Gao, S.; Miwa, N.; Jahangeer, S.; Nakamura, S.I. Involvement of N-Terminal-Extended Form of Sphingosine Kinase 2 in Serum-Dependent Regulation of Cell Proliferation and Apoptosis. J. Biol. Chem. 2005, 280, 36318–36325. [Google Scholar] [CrossRef] [PubMed]
- Chua, X.Y.; Chai, Y.L.; Chew, W.S.; Chong, J.R.; Ang, H.L.; Xiang, P.; Camara, K.; Howell, A.R.; Torta, F.; Wenk, M.R.; et al. Immunomodulatory Sphingosine-1-Phosphates as Plasma Biomarkers of Alzheimer’s Disease and Vascular Cognitive Impairment. Alzheimers Res. Ther. 2020, 12, 122. [Google Scholar] [CrossRef]
- Grassi, S.; Mauri, L.; Prioni, S.; Cabitta, L.; Sonnino, S.; Prinetti, A.; Giussani, P. Sphingosine 1-Phosphate Receptors and Metabolic Enzymes as Druggable Targets for Brain Diseases. Front. Pharmacol. 2019, 10. [Google Scholar] [CrossRef]
- Spiegel, S.; Maczis, M.A.; Maceyka, M.; Milstien, S. New Insights into Functions of the Sphingosine-1-Phosphate Transporter SPNS2. J. Lipid Res. 2019, 60, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Gangoiti, P.; Granado, M.H.; Wang, S.W.; Kong, J.Y.; Steinbrecher, U.P.; Gómez-Muñoz, A. Ceramide 1-Phosphate Stimulates Macrophage Proliferation through Activation of the PI3-Kinase/PKB, JNK and ERK1/2 Pathways. Cell. Signal. 2008, 20, 726–736. [Google Scholar] [CrossRef]
- Gangoiti, P.; Arana, L.; Ouro, A.; Granado, M.H.; Trueba, M.; Gómez-Muñoz, A. Activation of MTOR and RhoA Is a Major Mechanism by Which Ceramide 1-Phosphate Stimulates Macrophage Proliferation. Cell. Signal. 2011, 23, 27–34. [Google Scholar] [CrossRef]
- Miljan, E.A.; Meuillet, E.J.; Mania-Farnell, B.; George, D.; Yamamoto, H.; Simon, H.G.; Bremer, E.G. Interaction of the Extracellular Domain of the Epidermal Growth Factor Receptor with Gangliosides. J. Biol. Chem. 2002, 277, 10108–10113. [Google Scholar] [CrossRef]
- Dam, D.H.M.; Wang, X.Q.; Sheu, S.; Vijay, M.; Shipp, D.; Miller, L.; Paller, A.S. Ganglioside GM3 Mediates Glucose-Induced Suppression of IGF-1 Receptor–Rac1 Activation to Inhibit Keratinocyte Motility. J. Investig. Dermatol. 2017, 137, 440–448. [Google Scholar] [CrossRef]
- Adada, M.M.; Canals, D.; Jeong, N.; Kelkar, A.D.; Hernandez-Corbacho, M.; Pulkoski-Gross, M.J.; Donaldson, J.C.; Hannun, Y.A.; Obeid, L.M. Intracellular Sphingosine Kinase 2-Derived Sphingosine-1- Phosphate Mediates Epidermal Growth Factor-Induced Ezrin-Radixin-Moesin Phosphorylation and Cancer Cell Invasion. FASEB J. 2015, 29, 4654–4669. [Google Scholar] [CrossRef]
- Bretscher, A.; Edwards, K.; Fehon, R.G. ERM Proteins and Merlin: Integrators at the Cell Cortex. Nat. Rev. Mol. Cell Biol. 2002, 3, 586–599. [Google Scholar] [CrossRef]
- Arana, L.; Gangoiti, P.; Ouro, A.; Trueba, M.; Gómez-Muñoz, A. Ceramide and Ceramide 1-Phosphate in Health and Disease. Lipids Health Dis. 2010, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bodas, M.; Min, T.; Mazur, S.; Vij, N. Critical Modifier Role of Membrane-Cystic Fibrosis Transmembrane Conductance Regulator-Dependent Ceramide Signaling in Lung Injury and Emphysema. J. Immunol. 2011, 186, 602–613. [Google Scholar] [CrossRef]
- De Wit, N.M.; Den Hoedt, S.; Martinez-Martinez, P.; Rozemuller, A.J.; Mulder, M.T.; De Vries, H.E. Astrocytic Ceramide as Possible Indicator of Neuroinflammation. J. Neuroinflamm. 2019, 16, 48. [Google Scholar] [CrossRef] [PubMed]
- Yogi, A.; Callera, G.E.; Aranha, A.B.; Antunes, T.T.; Graham, D.; Mcbride, M.; Dominiczak, A.; Touyz, R.M. Sphingosine-1-Phosphate-Induced Inflammation Involves Receptor Tyrosine Kinase Transactivation in Vascular Cells Upregulation in Hypertension. Hypertension 2011. [Google Scholar] [CrossRef] [PubMed]
- Fettel, J.; Kühn, B.; Guillen, N.A.; Sürün, D.; Peters, M.; Bauer, R.; Angioni, C.; Geisslinger, G.; Schnütgen, F.; Heringdorf, D.M.; et al. Sphingosine-1-phosphate (S1P) Induces Potent Anti-inflammatory Effects in Vitro and in Vivo by S1P Receptor 4-mediated Suppression of 5-lipoxygenase Activity. FASEB J. 2019, 33, 1711–1726. [Google Scholar] [CrossRef]
- Dragusin, M.; Wehner, S.; Kelly, S.; Wang, E.; Merrill, A.H.; Kalff, J.C.; Echten-Deckert, G.; Dragusin, M.; Wehner, S.; Kelly, S.; et al. Effects of Sphingosine-1-phosphate and Ceramide-1-phosphate on Rat Intestinal Smooth Muscle Cells: Implications for Postoperative Ileus. FASEB J. 2006, 20, 1930–1932. [Google Scholar] [CrossRef]
- Pettus, B.J.; Bielawska, A.; Spiegel, S.; Roddy, P.; Hannun, Y.A.; Chalfant, C.E. Ceramide Kinase Mediates Cytokine- and Calcium Ionophore-Induced Arachidonic Acid Release. J. Biol. Chem. 2003, 278, 38206–38213. [Google Scholar] [CrossRef] [PubMed]
- Piccinini, M.; Scandroglio, F.; Prioni, S.; Buccinnà, B.; Loberto, N.; Aureli, M.; Chigorno, V.; Lupino, E.; DeMarco, G.; Lomartire, A.; et al. Deregulated Sphingolipid Metabolism and Membrane Organization in Neurodegenerative Disorders. Mol. Neurobiol. 2010, 41, 314–340. [Google Scholar] [CrossRef] [PubMed]
- van Echten-Deckert, G.; Herget, T. Sphingolipid Metabolism in Neural Cells. Biochim. Biophys. Acta BBA Biomembr. 2006, 1758, 1978–1994. [Google Scholar] [CrossRef] [PubMed]
- Baumann, N.; Pham-Dinh, D. Biology of Oligodendrocyte and Myelin in the Mammalian Central Nervous System. Physiol. Rev. 2001, 81, 871–927. [Google Scholar] [CrossRef]
- Posse de Chaves, E.; Sipione, S. Sphingolipids and Gangliosides of the Nervous System in Membrane Function and Dysfunction. FEBS Lett. 2010, 584, 1748–1759. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, S.; Ray, S.K. Diverse Biological Functions of Sphingolipids in the CNS: Ceramide and Sphingosine Regulate Myelination in Developing Brain but Stimulate Demyelination during Pathogenesis of Multiple Sclerosis. J. Neurol. Psychol. 2017, 5, 01–07. [Google Scholar] [CrossRef]
- Teichgräber, V.; Ulrich, M.; Endlich, N.; Riethmüller, J.; Wilker, B.; De Oliveira–Munding, C.C.; van Heeckeren, A.M.; Barr, M.L.; von Kürthy, G.; Schmid, K.W.; et al. Ceramide Accumulation Mediates Inflammation, Cell Death and Infection Susceptibility in Cystic Fibrosis. Nat. Med. 2008, 14, 382–391. [Google Scholar] [CrossRef]
- Becker, K.A.; Riethmüller, J.; Lüth, A.; Döring, G.; Kleuser, B.; Gulbins, E. Acid Sphingomyelinase Inhibitors Normalize Pulmonary Ceramide and Inflammation in Cystic Fibrosis. Am. J. Respir. Cell Mol. Biol. 2010, 42, 716–724. [Google Scholar] [CrossRef]
- Bodas, M.; Min, T.; Vij, N. Lactosylceramide-Accumulation in Lipid-Rafts Mediate Aberrant-Autophagy, Inflammation and Apoptosis in Cigarette Smoke Induced Emphysema. Apoptosis 2015, 20, 725–739. [Google Scholar] [CrossRef]
- Ammit, A.J.; Hastie, A.T.; Edsall, L.C.; Hoffman, R.K.; Amrani, Y.; Krymskaya, V.P.; Kane, S.A.; Peters, S.P.; Penn, R.B.; Spiegel, S.; et al. Sphingosine 1-Phosphate Modulates Human Airway Smooth Muscle Cell Functions That Promote Inflammation and Airway Remodeling in Asthma. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2001, 15, 1212–1214. [Google Scholar] [CrossRef]
- Jolly, P.S.; Rosenfeldt, H.M.; Milstien, S.; Spiegel, S. The Roles of Sphingosine-1-Phosphate in Asthma. Mol. Immunol. 2002, 38, 1239–1245. [Google Scholar] [CrossRef]
- Halayko, A.J.; Amrani, Y. Mechanisms of Inflammation-Mediated Airway Smooth Muscle Plasticity and Airways Remodeling in Asthma. Respir. Physiol. Neurobiol. 2003, 137, 209–222. [Google Scholar] [CrossRef]
- Hong Choi, O.; Kim, J.-H.; Kinet, J.-P. Calcium Mobilization via Sphingosine Kinase in Signalling by the FcɛRI Antigen Receptor. Nature 1996, 380, 634–636. [Google Scholar] [CrossRef]
- Prieschl, E.E.; Csonga, R.; Novotny, V.; Kikuchi, G.E.; Baumruker, T. The Balance between Sphingosine and Sphingosine-1-Phosphate Is Decisive for Mast Cell Activation after Fc Epsilon Receptor I Triggering. J. Exp. Med. 1999, 190, 1–8. [Google Scholar] [CrossRef]
- Takabe, K.; Paugh, S.W.; Milstien, S.; Spiegel, S. “Inside-out” Signaling of Sphingosine-1-Phosphate: Therapeutic Targets. Pharmacol. Rev. 2008, 60, 181–195. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.J.; Van Brocklyn, J.R.; Thangada, S.; Liu, C.H.; Hand, A.R.; Menzeleev, R.; Spiegel, S.; Hla, T. Sphingosine-1-Phosphate as a Ligand for the G Protein-Coupled Receptor EDG-1. Science 1998, 279, 1552–1555. [Google Scholar] [CrossRef] [PubMed]
- Jolly, P.S.; Bektas, M.; Olivera, A.; Gonzalez-Espinosa, C.; Proia, R.L.; Rivera, J.; Milstien, S.; Spiegel, S. Transactivation of Sphingosine-1–Phosphate Receptors by FcɛRI Triggering Is Required for Normal Mast Cell Degranulation and Chemotaxis. J. Exp. Med. 2004, 199, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Mitselou, A.; Grammeniatis, V.; Varouktsi, A.; Papadatos, S.S.; Katsanos, K.; Galani, V. Proinflammatory Cytokines in Irritable Bowel Syndrome: A Comparison with Inflammatory Bowel Disease. Intest. Res. 2020, 18, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Braun, A.; Treede, I.; Gotthardt, D.; Tietje, A.; Zahn, A.; Ruhwald, R.; Schoenfeld, U.; Welsch, T.; Kienle, P.; Erben, G.; et al. Alterations of Phospholipid Concentration and Species Composition of the Intestinal Mucus Barrier in Ulcerative Colitis: A Clue to Pathogenesis. Inflamm. Bowel Dis. 2009, 15, 1705–1720. [Google Scholar] [CrossRef] [PubMed]
- Fischbeck, A.; Leucht, K.; Frey-Wagner, I.; Bentz, S.; Pesch, T.; Kellermeier, S.; Krebs, M.; Fried, M.; Rogler, G.; Hausmann, M.; et al. Sphingomyelin Induces Cathepsin D-Mediated Apoptosis in Intestinal Epithelial Cells and Increases Inflammation in DSS Colitis. Gut 2011, 60, 55–65. [Google Scholar] [CrossRef]
- Qi, Y.; Jiang, C.; Tanaka, N.; Krausz, K.W.; Brocker, C.N.; Fang, Z.-Z.; Bredell, B.X.; Shah, Y.M.; Gonzalez, F.J. PPARα-Dependent Exacerbation of Experimental Colitis by the Hypolipidemic Drug Fenofibrate. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 307, G564–G573. [Google Scholar] [CrossRef]
- Scharl, M.; Leucht, K.; Frey-Wagner, I.; Zeitz, J.; Hausmann, M.; Fischbeck, A.; Liebisch, G.; Kellermeier, S.; Pesch, T.; Arikkat, J.; et al. Knock-out of β-Glucosidase 2 Has No Influence on Dextran Sulfate Sodium-Induced Colitis. Digestion 2011, 84, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Snider, A.J.; Wu, B.X.; Jenkins, R.W.; Sticca, J.A.; Kawamori, T.; Hannun, Y.A.; Obeid, L.M. Loss of Neutral Ceramidase Increases Inflammation in a Mouse Model of Inflammatory Bowel Disease. Prostaglandins Other Lipid Mediat. 2012, 99, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Snider, A.J.; Kawamori, T.; Bradshaw, S.G.; Orr, K.A.; Gilkeson, G.S.; Hannun, Y.A.; Obeid, L.M. A Role for Sphingosine Kinase 1 in Dextran Sulfate Sodium-Induced Colitis. FASEB J. 2009, 23, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Ogretmen, B. Sphingolipid Metabolism in Cancer Signalling and Therapy. Nat. Rev. Cancer 2018, 18, 33–50. [Google Scholar] [CrossRef]
- Meyers-Needham, M.; Ponnusamy, S.; Gencer, S.; Jiang, W.; Thomas, R.J.; Senkal, C.E.; Ogretmen, B. Concerted Functions of HDAC1 and MicroRNA-574-5p Repress Alternatively Spliced Ceramide Synthase 1 Expression in Human Cancer Cells. EMBO Mol. Med. 2012, 4, 78–92. [Google Scholar] [CrossRef]
- Koybasi, S.; Senkal, C.E.; Sundararaj, K.; Spassieva, S.; Bielawski, J.; Osta, W.; Day, T.A.; Jiang, J.C.; Jazwinski, S.M.; Hannun, Y.A.; et al. Defects in Cell Growth Regulation by C18:0-Ceramide and Longevity Assurance Gene 1 in Human Head and Neck Squamous Cell Carcinomas. J. Biol. Chem. 2004, 279, 44311–44319. [Google Scholar] [CrossRef]
- Senkal, C.E.; Ponnusamy, S.; Manevich, Y.; Meyers-Needham, M.; Saddoughi, S.A.; Mukhopadyay, A.; Dent, P.; Bielawski, J.; Ogretmen, B. Alteration of Ceramide Synthase 6/C16-Ceramide Induces Activating Transcription Factor 6-Mediated Endoplasmic Reticulum (ER) Stress and Apoptosis via Perturbation of Cellular Ca2+ and ER/Golgi Membrane Network. J. Biol. Chem. 2011, 286, 42446–42458. [Google Scholar] [CrossRef]
- Fekry, B.; Jeffries, K.A.; Esmaeilniakooshkghazi, A.; Ogretmen, B.; Krupenko, S.A.; Krupenko, N.I. CerS6 Is a Novel Transcriptional Target of P53 Protein Activated by Non-Genotoxic Stress. J. Biol. Chem. 2016, 291, 16586–16596. [Google Scholar] [CrossRef]
- Park, W.-J.; Brenner, O.; Kogot-Levin, A.; Saada, A.; Merrill, A.H.; Pewzner-Jung, Y.; Futerman, A.H. Development of Pheochromocytoma in Ceramide Synthase 2 Null Mice. Endocr. Relat. Cancer 2015, 22, 623–632. [Google Scholar] [CrossRef]
- Airola, M.V.; Shanbhogue, P.; Shamseddine, A.A.; Guja, K.E.; Senkal, C.E.; Maini, R.; Bartke, N.; Wu, B.X.; Obeid, L.M.; Garcia-Diaz, M.; et al. Structure of Human NSMase2 Reveals an Interdomain Allosteric Activation Mechanism for Ceramide Generation. Proc. Natl. Acad. Sci. USA 2017, 114, E5549–E5558. [Google Scholar] [CrossRef] [PubMed]
- Gorelik, A.; Illes, K.; Heinz, L.X.; Superti-Furga, G.; Nagar, B. Crystal Structure of Mammalian Acid Sphingomyelinase. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef]
- Santana, P.; Peña, L.A.; Haimovitz-Friedman, A.; Martin, S.; Green, D.; McLoughlin, M.; Cordon-Cardo, C.; Schuchman, E.H.; Fuks, Z.; Kolesnick, R. Acid Sphingomyelinase-Deficient Human Lymphoblasts and Mice Are Defective in Radiation-Induced Apoptosis. Cell 1996, 86, 189–199. [Google Scholar] [CrossRef]
- Carpinteiro, A.; Becker, K.A.; Japtok, L.; Hessler, G.; Keitsch, S.; Požgajovà, M.; Schmid, K.W.; Adams, C.; Müller, S.; Kleuser, B.; et al. Regulation of Hematogenous Tumor Metastasis by Acid Sphingomyelinase. EMBO Mol. Med. 2015, 7, 714–734. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, P.; Xu, S.-C.; Yang, L.; Voss, U.; Ekblad, E.; Wu, Y.; Min, Y.; Hertervig, E.; Nilsson, Å.; et al. Enhanced Colonic Tumorigenesis in Alkaline Sphingomyelinase (NPP7) Knockout Mice. Mol. Cancer Ther. 2015, 14, 259–267. [Google Scholar] [CrossRef]
- Heering, J.; Weis, N.; Holeiter, M.; Neugart, F.; Staebler, A.; Fehm, T.N.; Bischoff, A.; Schiller, J.; Duss, S.; Schmid, S.; et al. Loss of the Ceramide Transfer Protein Augments EGF Receptor Signaling in Breast Cancer. Cancer Res. 2012, 72, 2855–2866. [Google Scholar] [CrossRef]
- Hullin-Matsuda, F.; Tomishige, N.; Sakai, S.; Ishitsuka, R.; Ishii, K.; Makino, A.; Greimel, P.; Abe, M.; Laviad, E.L.; Lagarde, M.; et al. Limonoid Compounds Inhibit Sphingomyelin Biosynthesis by Preventing CERT Protein-Dependent Extraction of Ceramides from the Endoplasmic Reticulum. J. Biol. Chem. 2012, 287, 24397–24411. [Google Scholar] [CrossRef] [PubMed]
- Samaha, D.; Hamdo, H.H.; Cong, X.; Schumacher, F.; Banhart, S.; Aglar, Ö.; Möller, H.M.; Heuer, D.; Kleuser, B.; Saied, E.M.; et al. Liposomal FRET Assay Identifies Potent Drug-Like Inhibitors of the Ceramide Transport Protein (CERT). Chem. Weinh. Bergstr. Ger. 2020, 26, 16616–16621. [Google Scholar] [CrossRef] [PubMed]
- Payne, A.W.; Pant, D.K.; Pan, T.-C.; Chodosh, L.A. Ceramide Kinase Promotes Tumor Cell Survival and Mammary Tumor Recurrence. Cancer Res. 2014, 74, 6352–6363. [Google Scholar] [CrossRef] [PubMed]
- Pastukhov, O.; Schwalm, S.; Zangemeister-Wittke, U.; Fabbro, D.; Bornancin, F.; Japtok, L.; Kleuser, B.; Pfeilschifter, J.; Huwiler, A. The Ceramide Kinase Inhibitor NVP-231 Inhibits Breast and Lung Cancer Cell Proliferation by Inducing M Phase Arrest and Subsequent Cell Death. Br. J. Pharmacol. 2014, 171, 5829–5844. [Google Scholar] [CrossRef] [PubMed]
- Beckham, T.H.; Cheng, J.C.; Lu, P.; Shao, Y.; Troyer, D.; Lance, R.; Marrison, S.T.; Norris, J.S.; Liu, X. Acid Ceramidase Induces Sphingosine Kinase 1/S1P Receptor 2-Mediated Activation of Oncogenic Akt Signaling. Oncogenesis 2013, 2, e49. [Google Scholar] [CrossRef] [PubMed]
- Camacho, L.; Meca-Cortés, Ó.; Abad, J.L.; García, S.; Rubio, N.; Díaz, A.; Celià-Terrassa, T.; Cingolani, F.; Bermudo, R.; Fernández, P.L.; et al. Acid Ceramidase as a Therapeutic Target in Metastatic Prostate Cancer. J. Lipid Res. 2013, 54, 1207–1220. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.C.; Bai, A.; Beckham, T.H.; Marrison, S.T.; Yount, C.L.; Young, K.; Lu, P.; Bartlett, A.M.; Wu, B.X.; Keane, B.J.; et al. Radiation-Induced Acid Ceramidase Confers Prostate Cancer Resistance and Tumor Relapse. J. Clin. Investig. 2013, 123, 4344–4358. [Google Scholar] [CrossRef]
- Clifford, R.E.; Govindarajah, N.; Bowden, D.; Sutton, P.; Glenn, M.; Darvish-Damavandi, M.; Buczacki, S.; McDermott, U.; Szulc, Z.; Ogretmen, B.; et al. Targeting Acid Ceramidase to Improve the Radiosensitivity of Rectal Cancer. Cells 2020, 9, 2693. [Google Scholar] [CrossRef] [PubMed]
- Roh, J.-L.; Park, J.Y.; Kim, E.H.; Jang, H.J. Targeting Acid Ceramidase Sensitises Head and Neck Cancer to Cisplatin. Eur. J. Cancer 2016, 52, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.-F.; Liu, X.; Broeg, K.; Fox, T.E.; Feith, D.J.; Loughran, T.P., Jr. Acid Ceramidase Inhibition Impairs Tumor Progression in a Rat Model of LGL Leukemia. Blood 2015, 126, 1246. [Google Scholar] [CrossRef]
- Vijayan, Y.; Lankadasari, M.B.; Harikumar, K.B. Acid Ceramidase: A Novel Therapeutic Target in Cancer. Curr. Top. Med. Chem. 2019, 19, 1512–1520. [Google Scholar] [CrossRef]
- Realini, N.; Palese, F.; Pizzirani, D.; Pontis, S.; Basit, A.; Bach, A.; Ganesan, A.; Piomelli, D. Acid Ceramidase in Melanoma: Expression, Localization, And Effects Of Pharmacological Inhibition. J. Biol. Chem. 2016, 291, 2422–2434. [Google Scholar] [CrossRef]
- García-Barros, M.; Coant, N.; Kawamori, T.; Wada, M.; Snider, A.J.; Truman, J.-P.; Wu, B.X.; Furuya, H.; Clarke, C.J.; Bialkowska, A.B.; et al. Role of Neutral Ceramidase in Colon Cancer. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2016, 30, 4159–4171. [Google Scholar] [CrossRef] [PubMed]
- Coant, N.; García-Barros, M.; Zhang, Q.; Obeid, L.M.; Hannun, Y.A. AKT as a Key Target for Growth Promoting Functions of Neutral Ceramidase in Colon Cancer Cells. Oncogene 2018, 37, 3852–3863. [Google Scholar] [CrossRef]
- Stratford, S.; Hoehn, K.L.; Liu, F.; Summers, S.A. Regulation of Insulin Action by Ceramide: Dual Mechanisms Linking Ceramide Accumulation to the Inhibition of Akt/Protein Kinase B. J. Biol. Chem. 2004, 279, 36608–36615. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, C.; Jin, Y.; He, Q.; Liu, Z.; Ai, Q.; Lei, Y.; Li, Y.; Song, F.; Bu, Y.; et al. Alkaline Ceramidase 2 Is a Novel Direct Target of P53 and Induces Autophagy and Apoptosis through ROS Generation. Sci. Rep. 2017, 7, 44573. [Google Scholar] [CrossRef]
- Xu, R.; Garcia-Barros, M.; Wen, S.; Li, F.; Lin, C.-L.; Hannun, Y.A.; Obeid, L.M.; Mao, C. Tumor Suppressor P53 Links Ceramide Metabolism to DNA Damage Response through Alkaline Ceramidase 2. Cell Death Differ. 2017, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Wang, K.; Mileva, I.; Hannun, Y.A.; Obeid, L.M.; Mao, C. Alkaline Ceramidase 2 and Its Bioactive Product Sphingosine Are Novel Regulators of the DNA Damage Response. Oncotarget 2016, 7, 18440–18457. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Xiao, J.; Dong, M.; Qiu, Z.; Jin, J. Human Alkaline Ceramidase 2 Promotes the Growth, Invasion, and Migration of Hepatocellular Carcinoma Cells via Sphingomyelin Phosphodiesterase Acid-like 3B. Cancer Sci. 2020, 111, 2259–2274. [Google Scholar] [CrossRef]
- Hu, W.; Xu, R.; Sun, W.; Szulc, Z.M.; Bielawski, J.; Obeid, L.M.; Mao, C. Alkaline Ceramidase 3 (ACER3) Hydrolyzes Unsaturated Long-Chain Ceramides, and Its down-Regulation Inhibits Both Cell Proliferation and Apoptosis. J. Biol. Chem. 2010, 285, 7964–7976. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Xu, M.; Gao, J.; Li, M. Alkaline Ceramidase 3 Promotes Growth of Hepatocellular Carcinoma Cells via Regulating S1P/S1PR2/PI3K/AKT Signaling. Pathol. Res. Pract. 2018, 214, 1381–1387. [Google Scholar] [CrossRef]
- Vasiliauskaité-Brooks, I.; Healey, R.D.; Rochaix, P.; Saint-Paul, J.; Sounier, R.; Grison, C.; Waltrich-Augusto, T.; Fortier, M.; Hoh, F.; Saied, E.M.; et al. Structure of a Human Intramembrane Ceramidase Explains Enzymatic Dysfunction Found in Leukodystrophy. Nat. Commun. 2018, 9, 5437. [Google Scholar] [CrossRef]
- Nagahashi, M.; Ramachandran, S.; Kim, E.Y.; Allegood, J.C.; Rashid, O.M.; Yamada, A.; Zhao, R.; Milstien, S.; Zhou, H.; Spiegel, S.; et al. Sphingosine-1-Phosphate Produced by Sphingosine Kinase 1 Promotes Breast Cancer Progression by Stimulating Angiogenesis and Lymphangiogenesis. Cancer Res. 2012, 72, 726–735. [Google Scholar] [CrossRef]
- Kawamori, T.; Kaneshiro, T.; Okumura, M.; Maalouf, S.; Uflacker, A.; Bielawski, J.; Hannun, Y.A.; Obeid, L.M. Role for Sphingosine Kinase 1 in Colon Carcinogenesis. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2009, 23, 405–414. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Wan, Z.; Liu, S.; Cao, Y.; Zeng, Z. Sphingosine Kinase 1 and Cancer: A Systematic Review and Meta-Analysis. PLoS ONE 2014, 9, e90362. [Google Scholar] [CrossRef]
- Fang, Z.; Lin, M.; Li, C.; Liu, H.; Gong, C. A Comprehensive Review of the Roles of E2F1 in Colon Cancer. Am. J. Cancer Res. 2020, 10, 757–768. [Google Scholar] [PubMed]
- Postepska-Igielska, A.; Giwojna, A.; Gasri-Plotnitsky, L.; Schmitt, N.; Dold, A.; Ginsberg, D.; Grummt, I. LncRNA Khps1 Regulates Expression of the Proto-Oncogene SPHK1 via Triplex-Mediated Changes in Chromatin Structure. Mol. Cell 2015, 60, 626–636. [Google Scholar] [CrossRef]
- Wang, Q.; Li, J.; Li, G.; Li, Y.; Xu, C.; Li, M.; Xu, G.; Fu, S. Prognostic Significance of Sphingosine Kinase 2 Expression in Non-Small Cell Lung Cancer. Tumour Biol. 2014, 35, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Hait, N.C.; Bellamy, A.; Milstien, S.; Kordula, T.; Spiegel, S. Sphingosine Kinase Type 2 Activation by ERK-Mediated Phosphorylation. J. Biol. Chem. 2007, 282, 12058–12065. [Google Scholar] [CrossRef]
- Świderska, E.; Strycharz, J.; Wróblewski, A.; Szemraj, J.; Drzewoski, J.; Sliwinska, A. Role of PI3K/AKT Pathway in Insulin-Mediated Glucose Uptake. In Blood Glucose Levels; IntechOpen: London, UK, 2018; ISBN 978-1-78985-525-8. [Google Scholar]
- Holland, W.L.; Knotts, T.A.; Chavez, J.A.; Wang, L.-P.; Hoehn, K.L.; Summers, S.A. Lipid Mediators of Insulin Resistance. Nutr. Rev. 2007, 65, S39–S46. [Google Scholar] [CrossRef] [PubMed]
- Boon, J.; Hoy, A.J.; Stark, R.; Brown, R.D.; Meex, R.C.; Henstridge, D.C.; Schenk, S.; Meikle, P.J.; Horowitz, J.F.; Kingwell, B.A.; et al. Ceramides Contained in LDL Are Elevated in Type 2 Diabetes and Promote Inflammation and Skeletal Muscle Insulin Resistance. Diabetes 2013, 62, 401–410. [Google Scholar] [CrossRef]
- Turpin, S.M.; Nicholls, H.T.; Willmes, D.M.; Mourier, A.; Brodesser, S.; Wunderlich, C.M.; Mauer, J.; Xu, E.; Hammerschmidt, P.; Brönneke, H.S.; et al. Obesity-Induced CerS6-Dependent C16:0 Ceramide Production Promotes Weight Gain and Glucose Intolerance. Cell Metab. 2014, 20, 678–686. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.Y.; Holland, W.L.; Kusminski, C.M.; Sun, K.; Sharma, A.X.; Pearson, M.J.; Sifuentes, A.J.; McDonald, J.G.; Gordillo, R.; Scherer, P.E. Targeted Induction of Ceramide Degradation Leads to Improved Systemic Metabolism and Reduced Hepatic Steatosis. Cell Metab. 2015, 22, 266–278. [Google Scholar] [CrossRef]
- Chen, J.; Wang, W.; Qi, Y.; Kaczorowski, D.; McCaughan, G.W.; Gamble, J.R.; Don, A.S.; Gao, X.; Vadas, M.A.; Xia, P. Deletion of Sphingosine Kinase 1 Ameliorates Hepatic Steatosis in Diet-Induced Obese Mice: Role of PPARγ. Biochim. Biophys. Acta 2016, 1861, 138–147. [Google Scholar] [CrossRef] [PubMed]
- de Mello, V.D.F.; Lankinen, M.; Schwab, U.; Kolehmainen, M.; Lehto, S.; Seppänen-Laakso, T.; Oresic, M.; Pulkkinen, L.; Uusitupa, M.; Erkkilä, A.T. Link between Plasma Ceramides, Inflammation and Insulin Resistance: Association with Serum IL-6 Concentration in Patients with Coronary Heart Disease. Diabetologia 2009, 52, 2612–2615. [Google Scholar] [CrossRef]
- Maedler, K.; Oberholzer, J.; Bucher, P.; Spinas, G.A.; Donath, M.Y. Monounsaturated Fatty Acids Prevent the Deleterious Effects of Palmitate and High Glucose on Human Pancreatic β-Cell Turnover and Function. Diabetes 2003, 52, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Sjöholm, Å. Ceramide Inhibits Pancreatic β-Cell Insulin Production and Mitogenesis and Mimics the Actions of Interleukin-1β. FEBS Lett. 1995, 367, 283–286. [Google Scholar] [CrossRef]
- Taguchi, Y.; Allende, M.L.; Mizukami, H.; Cook, E.K.; Gavrilova, O.; Tuymetova, G.; Clarke, B.A.; Chen, W.; Olivera, A.; Proia, R.L. Sphingosine-1-Phosphate Phosphatase 2 Regulates Pancreatic Islet β-Cell Endoplasmic Reticulum Stress and Proliferation. J. Biol. Chem. 2016, 291, 12029–12038. [Google Scholar] [CrossRef]
- Kurek, K.; Wiesiołek-Kurek, P.; Piotrowska, D.M.; Łukaszuk, B.; Chabowski, A.; Żendzian-Piotrowska, M. Inhibition of Ceramide De Novo Synthesis with Myriocin Affects Lipid Metabolism in the Liver of Rats with Streptozotocin-Induced Type 1 Diabetes. BioMed Res. Int. 2014, 2014, e980815. [Google Scholar] [CrossRef]
- Chaurasia, B.; Summers, S.A. Ceramides—Lipotoxic Inducers of Metabolic Disorders. Trends Endocrinol. Metab. TEM 2015, 26, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, H.; Liu, J.; Liang, C.-P.; Li, Y.; Li, Y.; Teitelman, G.; Beyer, T.; Bui, H.H.; Peake, D.A.; et al. Reducing Plasma Membrane Sphingomyelin Increases Insulin Sensitivity. Mol. Cell. Biol. 2011, 31, 4205–4218. [Google Scholar] [CrossRef]
- Yano, M.; Yamamoto, T.; Nishimura, N.; Gotoh, T.; Watanabe, K.; Ikeda, K.; Garan, Y.; Taguchi, R.; Node, K.; Okazaki, T.; et al. Increased Oxidative Stress Impairs Adipose Tissue Function in Sphingomyelin Synthase 1 Null Mice. PLoS ONE 2013, 8, e61380. [Google Scholar] [CrossRef]
- Chavez, J.A.; Siddique, M.M.; Wang, S.T.; Ching, J.; Shayman, J.A.; Summers, S.A. Ceramides and Glucosylceramides Are Independent Antagonists of Insulin Signaling. J. Biol. Chem. 2014, 289, 723–734. [Google Scholar] [CrossRef]
- Othman, A.; Saely, C.H.; Muendlein, A.; Vonbank, A.; Drexel, H.; von Eckardstein, A.; Hornemann, T. Plasma 1-Deoxysphingolipids Are Predictive Biomarkers for Type 2 Diabetes Mellitus. BMJ Open Diabetes Res. Care 2015, 3. [Google Scholar] [CrossRef]
- Wei, N.; Pan, J.; Pop-Busui, R.; Othman, A.; Alecu, I.; Hornemann, T.; Eichler, F.S. Altered Sphingoid Base Profiles in Type 1 Compared to Type 2 Diabetes. Lipids Health Dis. 2014, 13, 161. [Google Scholar] [CrossRef]
- Mahfoud, R.; Garmy, N.; Maresca, M.; Yahi, N.; Puigserver, A.; Fantini, J. Identification of a Common Sphingolipid-Binding Domain in Alzheimer, Prion, and HIV-1 Proteins. J. Biol. Chem. 2002, 277, 11292–11296. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, K.; Odaka, A.; Suzuki, N.; Ihara, Y. GM1 Ganglioside-Bound Amyloid Beta-Protein (A Beta): A Possible Form of Preamyloid in Alzheimer’s Disease. Nat. Med. 1995, 1, 1062–1066. [Google Scholar] [CrossRef] [PubMed]
- Asiimwe, N.; Yeo, S.G.; Kim, M.-S.; Jung, J.; Jeong, N.Y. Nitric Oxide: Exploring the Contextual Link with Alzheimer’s Disease. Oxid. Med. Cell. Longev. 2016, 2016, e7205747. [Google Scholar] [CrossRef] [PubMed]
- Castillo, S.S.; Levy, M.; Wang, C.; Thaikoottathil, J.V.; Khan, E.; Goldkorn, T. Nitric Oxide-Enhanced Caspase-3 and Acidic Sphingomyelinase Interaction: A Novel Mechanism by Which Airway Epithelial Cells Escape Ceramide-Induced Apoptosis. Exp. Cell Res. 2007, 313, 816–823. [Google Scholar] [CrossRef]
- Huwiler, A.; Dorsch, S.; Briner, V.A.; van den Bosch, H.; Pfeilschifter, J. Nitric Oxide Stimulates Chronic Ceramide Formation in Glomerular Endothelial Cells. Biochem. Biophys. Res. Commun. 1999, 258, 60–65. [Google Scholar] [CrossRef]
- Takeda, Y.; Tashima, M.; Takahashi, A.; Uchiyama, T.; Okazaki, T. Ceramide Generation in Nitric Oxide-Induced Apoptosis. Activation of Magnesium-Dependent Neutral Sphingomyelinase via Caspase-3. J. Biol. Chem. 1999, 274, 10654–10660. [Google Scholar] [CrossRef]
- Malaplate-Armand, C.; Florent-Béchard, S.; Youssef, I.; Koziel, V.; Sponne, I.; Kriem, B.; Leininger-Muller, B.; Olivier, J.-L.; Oster, T.; Pillot, T. Soluble Oligomers of Amyloid-Beta Peptide Induce Neuronal Apoptosis by Activating a CPLA2-Dependent Sphingomyelinase-Ceramide Pathway. Neurobiol. Dis. 2006, 23, 178–189. [Google Scholar] [CrossRef]
- He, X.; Huang, Y.; Li, B.; Gong, C.-X.; Schuchman, E.H. Deregulation of Sphingolipid Metabolism in Alzheimer’s Disease. Neurobiol. Aging 2010, 31, 398–408. [Google Scholar] [CrossRef]
- Lin, G.; Lee, P.-T.; Chen, K.; Mao, D.; Tan, K.L.; Zuo, Z.; Lin, W.-W.; Wang, L.; Bellen, H.J. Phospholipase PLA2G6, a Parkinsonism-Associated Gene, Affects Vps26 and Vps35, Retromer Function, and Ceramide Levels, Similar to α-Synuclein Gain. Cell Metab. 2018, 28, 605–618. [Google Scholar] [CrossRef]
- Galvagnion, C. The Role of Lipids Interacting with α-Synuclein in the Pathogenesis of Parkinson’s Disease. J. Park. Dis. 2017, 7, 433–450. [Google Scholar] [CrossRef]
- Zunke, F.; Moise, A.C.; Belur, N.R.; Gelyana, E.; Stojkovska, I.; Dzaferbegovic, H.; Toker, N.J.; Jeon, S.; Fredriksen, K.; Mazzulli, J.R. Reversible Conformational Conversion of α-Synuclein into Toxic Assemblies by Glucosylceramide. Neuron 2018, 97, 92–107. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, V.E.; Nikolopoulou, G.; Antoniadou, I.; Karachaliou, A.; Arianoglou, G.; Emmanouilidou, E.; Sardi, S.P.; Stefanis, L.; Vekrellis, K. Modulation of β-Glucocerebrosidase Increases α-Synuclein Secretion and Exosome Release in Mouse Models of Parkinson’s Disease. Hum. Mol. Genet. 2018, 27, 1696–1710. [Google Scholar] [CrossRef] [PubMed]
- Ando, A.; Oka, M.; Satomi, Y. Deoxysphingolipids and Ether-Linked Diacylglycerols Accumulate in the Tissues of Aged Mice. Cell Biosci. 2019, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Onyenwoke, R.U.; Brenman, J.E. Lysosomal Storage Diseases—Regulating Neurodegeneration. J. Exp. Neurosci. 2016, 9, 81–91. [Google Scholar] [CrossRef]
- Sun, A. Lysosomal Storage Disease Overview. Ann. Transl. Med. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Vaccaro, A.M.; Salvioli, R.; Tatti, M.; Ciaffoni, F. Saposins and Their Interaction with Lipids. Neurochem. Res. 1999, 24, 307–314. [Google Scholar] [CrossRef]
- Ho, M.W.; O’Brien, J.S. Gaucher’s Disease: Deficiency of “acid” -Glucosidase and Reconstitution of Enzyme Activity in Vitro. Proc. Natl. Acad. Sci. USA 1971, 68, 2810–2813. [Google Scholar] [CrossRef]
- Vaccaro, A.M.; Motta, M.; Tatti, M.; Scarpa, S.; Masuelli, L.; Bhat, M.; Vanier, M.T.; Tylki-Szymanska, A.; Salvioli, R. Saposin C Mutations in Gaucher Disease Patients Resulting in Lysosomal Lipid Accumulation, Saposin C Deficiency, but Normal Prosaposin Processing and Sorting. Hum. Mol. Genet. 2010, 19, 2987–2997. [Google Scholar] [CrossRef]
- Sun, Y.; Qi, X.; Grabowski, G.A. Saposin C Is Required for Normal Resistance of Acid Beta-Glucosidase to Proteolytic Degradation. J. Biol. Chem. 2003, 278, 31918–31923. [Google Scholar] [CrossRef]
- Sun, Y.; Liou, B.; Ran, H.; Skelton, M.R.; Williams, M.T.; Vorhees, C.V.; Kitatani, K.; Hannun, Y.A.; Witte, D.P.; Xu, Y.-H.; et al. Neuronopathic Gaucher Disease in the Mouse: Viable Combined Selective Saposin C Deficiency and Mutant Glucocerebrosidase (V394L) Mice with Glucosylsphingosine and Glucosylceramide Accumulation and Progressive Neurological Deficits. Hum. Mol. Genet. 2010, 19, 1088–1097. [Google Scholar] [CrossRef]
- Aharon-Peretz, J.; Badarny, S.; Rosenbaum, H.; Gershoni-Baruch, R. Mutations in the Glucocerebrosidase Gene and Parkinson Disease: Phenotype-Genotype Correlation. Neurology 2005, 65, 1460–1461. [Google Scholar] [CrossRef] [PubMed]
- Clark, L.N.; Ross, B.M.; Wang, Y.; Mejia-Santana, H.; Harris, J.; Louis, E.D.; Cote, L.J.; Andrews, H.; Fahn, S.; Waters, C.; et al. Mutations in the Glucocerebrosidase Gene Are Associated with Early-Onset Parkinson Disease. Neurology 2007, 69, 1270–1277. [Google Scholar] [CrossRef] [PubMed]
- Sato, C.; Morgan, A.; Lang, A.E.; Salehi-Rad, S.; Kawarai, T.; Meng, Y.; Ray, P.N.; Farrer, L.A.; St George-Hyslop, P.; Rogaeva, E. Analysis of the Glucocerebrosidase Gene in Parkinson’s Disease. Mov. Disord. Off. J. Mov. Disord. Soc. 2005, 20, 367–370. [Google Scholar] [CrossRef]
- Sidransky, E.; Nalls, M.A.; Aasly, J.O.; Aharon-Peretz, J.; Annesi, G.; Barbosa, E.R.; Bar-Shira, A.; Berg, D.; Bras, J.; Brice, A.; et al. Multi-Center Analysis of Glucocerebrosidase Mutations in Parkinson Disease. N. Engl. J. Med. 2009, 361, 1651–1661. [Google Scholar] [CrossRef] [PubMed]
- Mazzulli, J.R.; Xu, Y.-H.; Sun, Y.; Knight, A.L.; McLean, P.J.; Caldwell, G.A.; Sidransky, E.; Grabowski, G.A.; Krainc, D. Gaucher Disease Glucocerebrosidase and α-Synuclein Form a Bidirectional Pathogenic Loop in Synucleinopathies. Cell 2011, 146, 37–52. [Google Scholar] [CrossRef]
- Buccinnà, B.; Piccinini, M.; Prinetti, A.; Scandroglio, F.; Prioni, S.; Valsecchi, M.; Votta, B.; Grifoni, S.; Lupino, E.; Ramondetti, C.; et al. Alterations of Myelin-Specific Proteins and Sphingolipids Characterize the Brains of Acid Sphingomyelinase-Deficient Mice, an Animal Model of Niemann-Pick Disease Type A. J. Neurochem. 2009, 109, 105–115. [Google Scholar] [CrossRef]
- Galvan, C.; Camoletto, P.G.; Cristofani, F.; Van Veldhoven, P.P.; Ledesma, M.D. Anomalous Surface Distribution of Glycosyl Phosphatidyl Inositol–Anchored Proteins in Neurons Lacking Acid Sphingomyelinase. Mol. Biol. Cell 2008, 19, 509–522. [Google Scholar] [CrossRef]
- Scandroglio, F.; Venkata, J.; Loberto, N.; Prioni, S.; Schuchman, E.; Chigorno, V.; Prinetti, A.; Sonnino, S. Lipid Content of Brain, Brain Membrane Lipid Domains, and Neurons from Acid Sphingomyelinase Deficient Mice. J. Neurochem. 2008, 107, 329–338. [Google Scholar] [CrossRef]
- Higgins, M.E.; Davies, J.P.; Chen, F.W.; Ioannou, Y.A. Niemann-Pick C1 Is a Late Endosome-Resident Protein That Transiently Associates with Lysosomes and the Trans-Golgi Network. Mol. Genet. Metab. 1999, 68, 1–13. [Google Scholar] [CrossRef]
- Naureckiene, S.; Sleat, D.E.; Lackland, H.; Fensom, A.; Vanier, M.T.; Wattiaux, R.; Jadot, M.; Lobel, P. Identification of HE1 as the Second Gene of Niemann-Pick C Disease. Science 2000, 290, 2298–2301. [Google Scholar] [CrossRef]
- Sleat, D.E.; Wiseman, J.A.; El-Banna, M.; Price, S.M.; Verot, L.; Shen, M.M.; Tint, G.S.; Vanier, M.T.; Walkley, S.U.; Lobel, P. Genetic Evidence for Nonredundant Functional Cooperativity between NPC1 and NPC2 in Lipid Transport. Proc. Natl. Acad. Sci. USA 2004, 101, 5886–5891. [Google Scholar] [CrossRef]
- Subramanian, K.; Balch, W.E. NPC1/NPC2 Function as a Tag Team Duo to Mobilize Cholesterol. Proc. Natl. Acad. Sci. USA 2008, 105, 15223–15224. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.J.; Lowenthal, A.; Ceuterick, C.; Vanier, M.T. Juvenile Dystonic Lipidosis (Variant of Niemann-Pick Disease Type C). J. Neurol. Sci. 1984, 66, 33–45. [Google Scholar] [CrossRef]
- Shlomovitz, R.; Schick, M. Model of a Raft in Both Leaves of an Asymmetric Lipid Bilayer. Biophys. J. 2013, 105, 1406–1413. [Google Scholar] [CrossRef]
- Skočaj, M.; Resnik, N.; Grundner, M.; Ota, K.; Rojko, N.; Hodnik, V.; Anderluh, G.; Sobota, A.; Maček, P.; Veranič, P.; et al. Tracking Cholesterol/Sphingomyelin-Rich Membrane Domains with the Ostreolysin A-MCherry Protein. PLoS ONE 2014, 9, e92783. [Google Scholar] [CrossRef]
- Sandhoff, K. Metabolic and Cellular Bases of Sphingolipidoses. Biochem. Soc. Trans. 2013, 41, 1562–1568. [Google Scholar] [CrossRef] [PubMed]
- Malbon, K. Atlas of Metabolic Diseases, 2nd Edition. Arch. Dis. Child. 2006, 91, 203. [Google Scholar] [CrossRef][Green Version]
- Norden, A.G.W.; O’Brien, J.S. Ganglioside GM1 β-Galactosidase: Studies in Human Liver and Brain. Arch. Biochem. Biophys. 1973, 159, 383–392. [Google Scholar] [CrossRef]
- Yoshida, K.; Oshima, A.; Sakuraba, H.; Nakano, T.; Yanagisawa, N.; Inui, K.; Okada, S.; Uyama, E.; Namba, R.; Kondo, K. GM1 Gangliosidosis in Adults: Clinical and Molecular Analysis of 16 Japanese Patients. Ann. Neurol. 1992, 31, 328–332. [Google Scholar] [CrossRef]
- Ryckman, A.E.; Brockhausen, I.; Walia, J.S. Metabolism of Glycosphingolipids and Their Role in the Pathophysiology of Lysosomal Storage Disorders. Int. J. Mol. Sci. 2020, 21, 6881. [Google Scholar] [CrossRef] [PubMed]
- Mahuran, D.J. Biochemical Consequences of Mutations Causing the GM2 Gangliosidoses. Biochim. Biophys. Acta 1999, 1455, 105–138. [Google Scholar] [CrossRef]
- Tada, E.; Toyomura, K.; Nakamura, H.; Sasaki, H.; Saito, T.; Kaneko, M.; Okuma, Y.; Murayama, T. Activation of Ceramidase and Ceramide Kinase by Vanadate via a Tyrosine Kinase-Mediated Pathway. J. Pharmacol. Sci. 2010, 114, 420–432. [Google Scholar] [CrossRef] [PubMed]
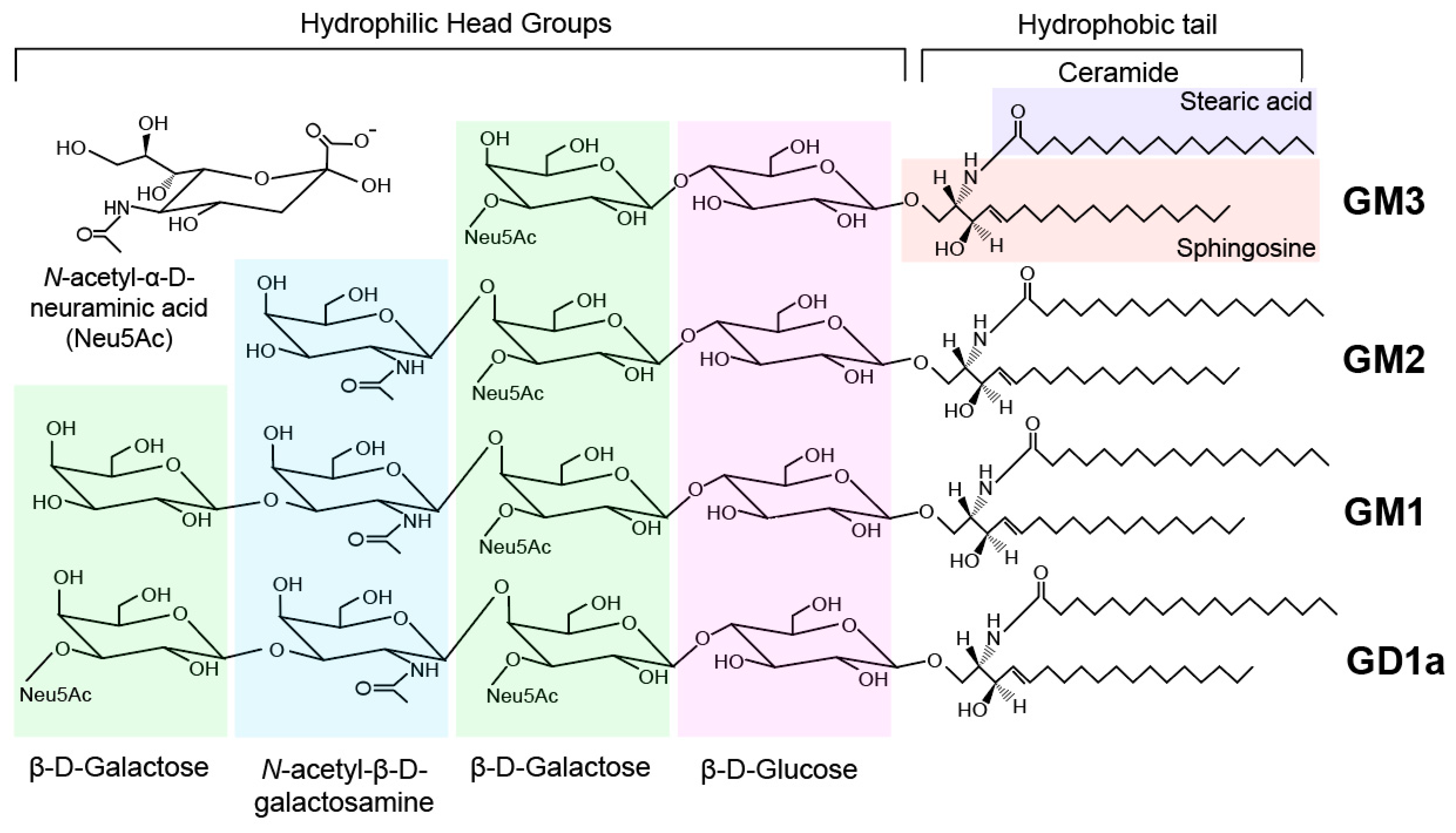
| Classification | Sphingolipid | IUPAC Name | Structure |
|---|---|---|---|
| Sphingoid base | Sphingosine | (2S,3R)-2-aminooctadec-4-trans-ene-1,3-diol | 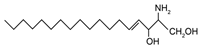 |
| Dihydrosphingosine (Sphinganine) | (2R,3S)-2-aminooctadecane-1,3-diol | 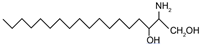 | |
| Phytosphingosine | (2S,3S,4R)-2-aminooctadecane-1,3,4-triol | 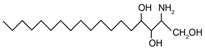 | |
| Sphingoid base derivatives | Sphingosine-1-phosphate | {[(4E)-2-amino-3-hydroxyoctadec-4-en-1-yl]oxy} phosphonic acid | 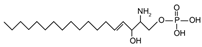 |
| Sphinganine-1-phosphate | {[(2S,3R)-2-amino-3-hydroxyoctadecyl]oxy}phosphonic acid | 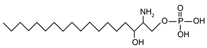 | |
| Phytosphingosine-1-phosphate | {[(2S,3S,4R)-2-amino-3,4-dihydroxyoctadecyl]oxy}phosphonic acid | 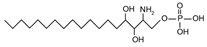 | |
| Deoxysphingolipids | 1-Deoxysphinganine | [(2S,3R)-3-hydroxyoctadecan-2-yl]azanium | 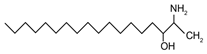 |
| 1-Deoxysphingosine (Hypothesized structure) | 2S-amino-4E-octadecen-3R-ol | 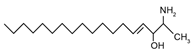 | |
| 1-Deoxysphingosine (Confirmed structure by Steiner et al., 2016) | [(2S,3R,14Z)-2-amino-14-octadecen-3-ol] | 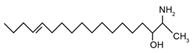 | |
| 1-Deoxymethylsphinganine | (2R)-1-aminoheptadecan-2-ol | 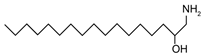 | |
| Methyl-branched sphingoid base | meC18SO (discovered by Lone et al., 2020) | 16S-methyl-sphingosine | 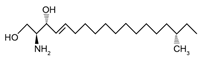 |
| Disease | Protein(s) Involved | Gene(s) | Affected Sphingolipid | MIM |
|---|---|---|---|---|
| Cystic Fibrosis | Cystic transmembrane conductance regulator | Cftr | Ceramide | 219700 |
| Asthma | ORMDL3 sphingolipid biosynthesis regulator 3 | ORMDL3 | Sphingosine-1-phosphate and ceramide | 600807 |
| Irritable bowel disease | Interleukin-6 | IL6 | Sphingomyelin, sphingosine, ceramide, sphingosine-1-phosphate, ceramide-1-phosphate | 612244 |
| Disease | Protein(s) Involved in Ceramide Synthesis and Tumor Suppression | Protein(s) Involved in Ceramide Metabolism and Pro-Survival | MIM |
|---|---|---|---|
| Breast Cancer | CERS6, aSMase | CERT, CERK, GCS, SphK2 | 114480 |
| Colorectal Cancer | SPL | - | 114500 |
| Head and neck cancer | CERS1, CERS6, | - | 275355 |
| Lung cancer | CERS6 | SphK1, SphK2 | 211980 |
| Leukemia | CERS1, aSMase | - | 613065 |
| Pheochromocytoma | CERS2 | - | 171300 |
| Disease | Protein(s) Involved | Gene(s) | Affected Sphingolipid | MIM |
|---|---|---|---|---|
| Diabetes type 1 | Tumor necrosis factor α, interleukins | INS | Ceramide, sphingosine-1-phosphate | 222100 |
| Diabetes type 2 | Protein kinase B (Akt) | AKT1 | Ceramide, sphingosine-1-phosphate | 125853 |
| Disease | Protein(s) Involved | Gene(s) | Affected Sphingolipid | MIM |
|---|---|---|---|---|
| Alzheimer’s Disease | β-amyloid | APP; PSEN1; PSEN2 | Sphingomyelin, galactosylceramide, GM1 ganglioside, ceramide, sphingosine-1-phosphate | 104300 |
| Parkinson’s Disease | α-synuclein | SNCA | ceramide, glycosylceramide | 168600 |
| Disease | Protein(s)/Enzymes | Gene(s) | Affected Sphingolipid | MIM |
|---|---|---|---|---|
| Gaucher Disease | Glucocerebrosidase | GBA1 | Glucosylceramide | 230800 (Type 1); 230900 (Type 2); 231000 (Type 3) |
| Niemann–Pick Disease (Type A and B) | Acid Sphingomyelinase | SMPD1 | Sphingomyelin | 257200 (Type A); 607616 (Type B) |
| Niemann–Pick Disease (Type C) | NPC1 or NPC2 | NPC1 or NPC2 | Sphingomyelin, GM2 ganglioside, GM3 ganglioside | 607623 (NPC1); 607625 (NPC2) |
| GM1 Gangliosidoses | β-galactosidase | GLB1 | GM1 ganglioside | 230500 (Type 1); 230600 (Type 2); 230650 (Type 3) |
| GM2 Gangliosidoses | Hex A (Tay-Sachs and Sandhoff) or GM2AP (AB-Variant) | HEXA, HEXB or GM2A | GM2 ganglioside | 272800 (Tay-Sachs); 268800 (Sandhoff); 272750 (AB-Variant) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quinville, B.M.; Deschenes, N.M.; Ryckman, A.E.; Walia, J.S. A Comprehensive Review: Sphingolipid Metabolism and Implications of Disruption in Sphingolipid Homeostasis. Int. J. Mol. Sci. 2021, 22, 5793. https://doi.org/10.3390/ijms22115793
Quinville BM, Deschenes NM, Ryckman AE, Walia JS. A Comprehensive Review: Sphingolipid Metabolism and Implications of Disruption in Sphingolipid Homeostasis. International Journal of Molecular Sciences. 2021; 22(11):5793. https://doi.org/10.3390/ijms22115793
Chicago/Turabian StyleQuinville, Brianna M., Natalie M. Deschenes, Alex E. Ryckman, and Jagdeep S. Walia. 2021. "A Comprehensive Review: Sphingolipid Metabolism and Implications of Disruption in Sphingolipid Homeostasis" International Journal of Molecular Sciences 22, no. 11: 5793. https://doi.org/10.3390/ijms22115793
APA StyleQuinville, B. M., Deschenes, N. M., Ryckman, A. E., & Walia, J. S. (2021). A Comprehensive Review: Sphingolipid Metabolism and Implications of Disruption in Sphingolipid Homeostasis. International Journal of Molecular Sciences, 22(11), 5793. https://doi.org/10.3390/ijms22115793





