Molecular Insights into the Role of Cysteine-Rich Peptides in Induced Resistance to Fusarium oxysporum Infection in Tomato Based on Transcriptome Profiling
Abstract
1. Introduction
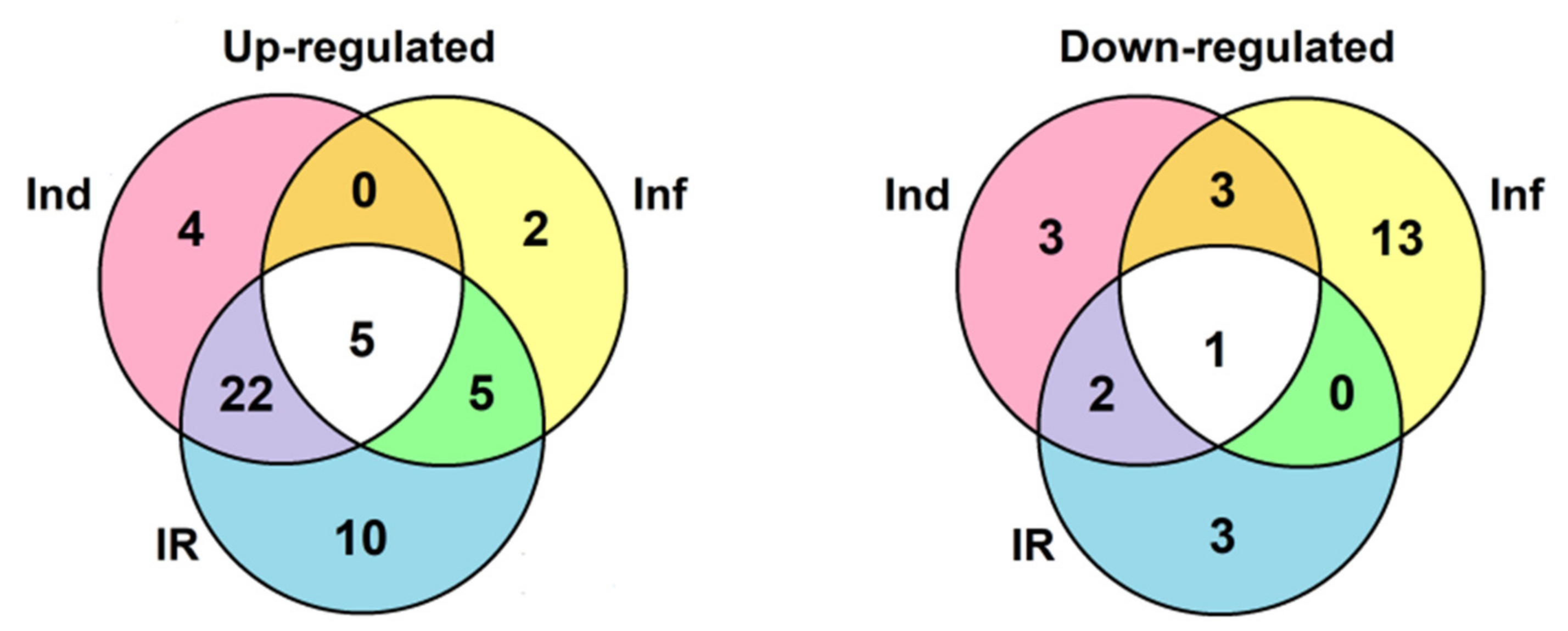
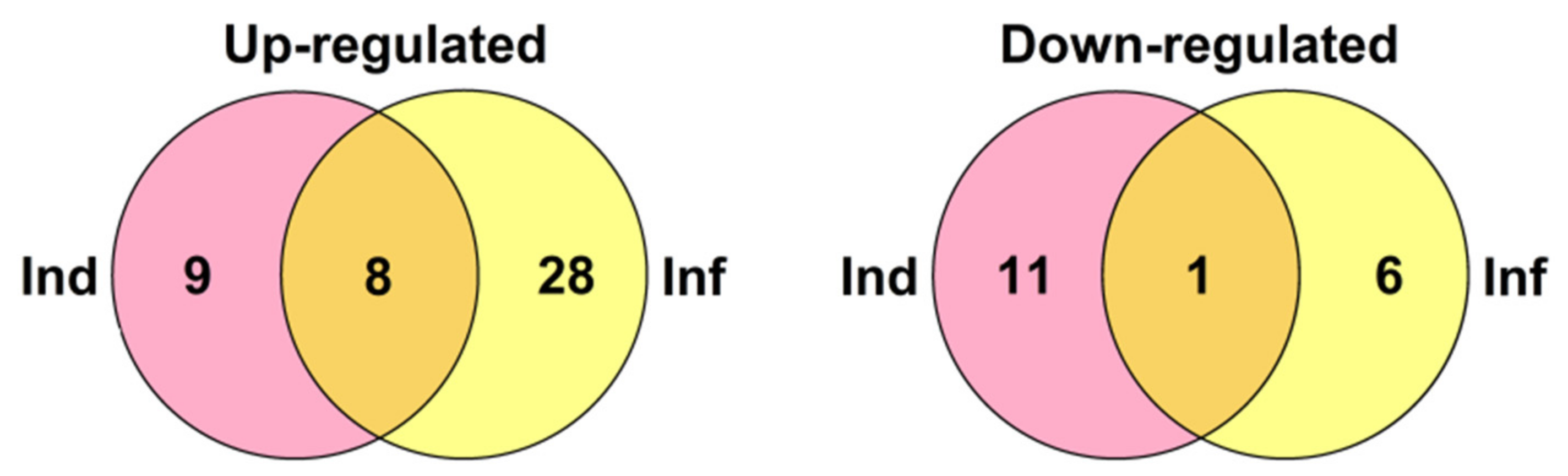
2. Results
2.1. F. sambucinum FS-94 Elicitors Protect Tomato Seeds and Seedlings from F. oxysporum F37 Infection
2.2. Transcriptome Sequencing and Assembly
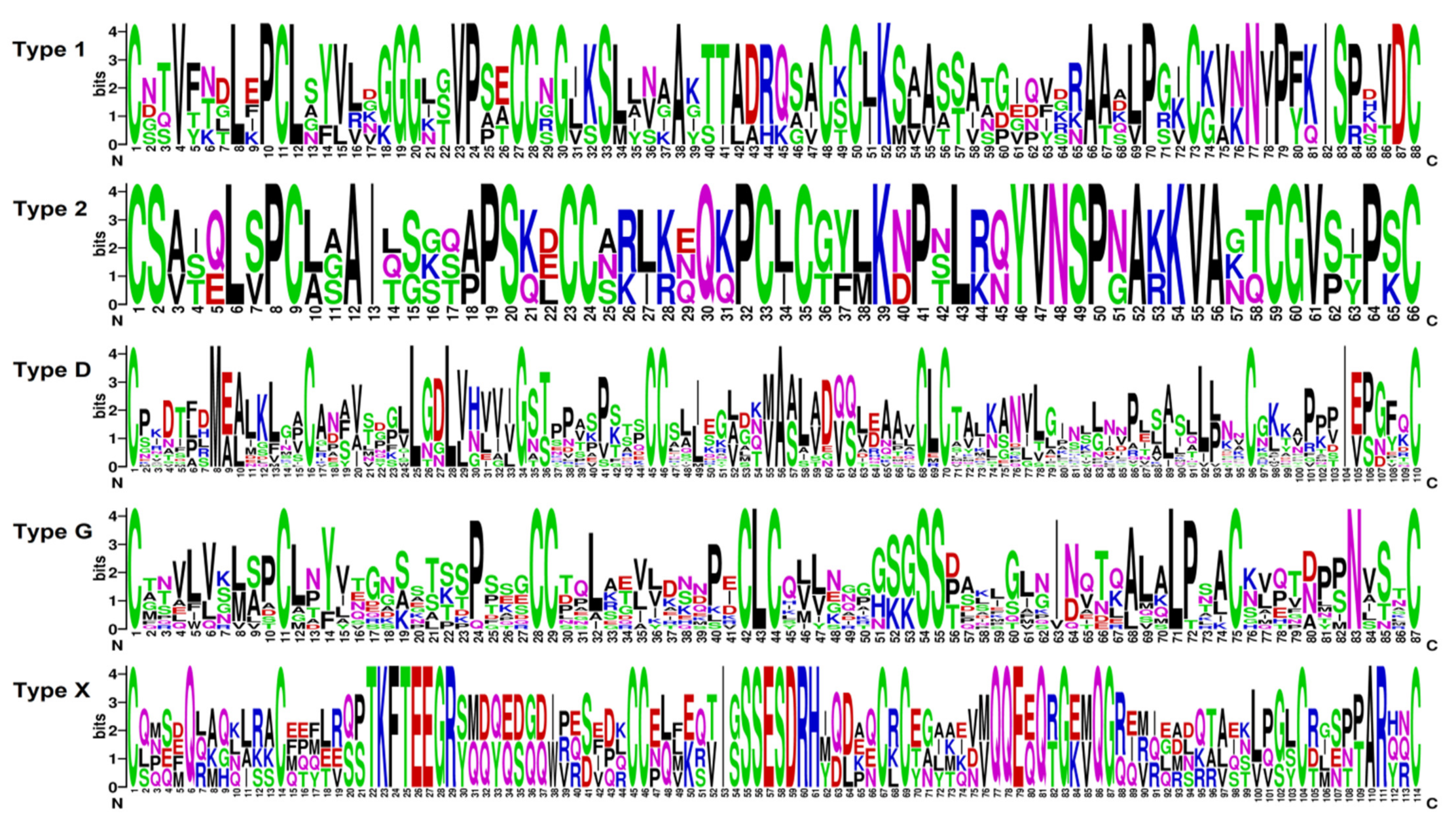
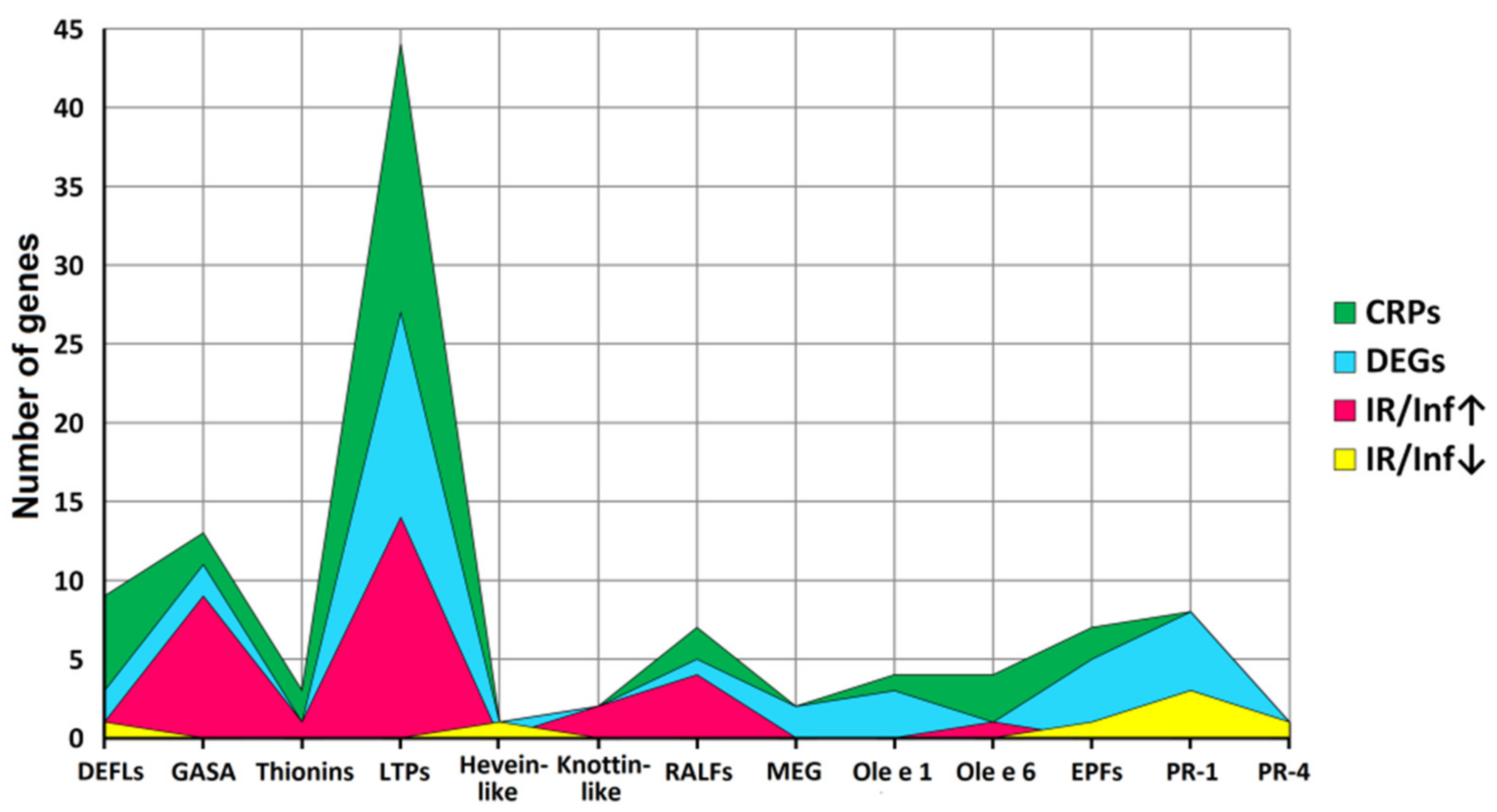
2.3. Identification of CRP Precursors in Tomato Transcriptomes
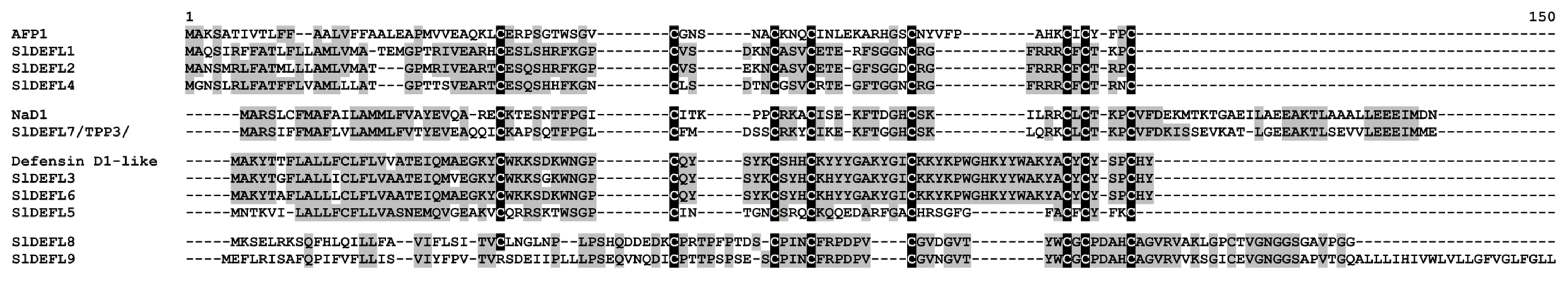
2.3.1. Defensins
2.3.2. Non-Specific Lipid-Transfer Proteins
2.3.3. Snakin/Gibberellic Acid Stimulated-like (GASA)
2.3.4. Thionins
2.3.5. Hevein-Like Peptides
2.3.6. Knottin-Like Peptides
2.3.7. RALFs
2.3.8. MEG (Maternally Expressed Gene)
2.3.9. Ole e 1 and Ole e 6
2.3.10. EPF-Like Peptides
2.3.11. PR-1 Proteins
2.3.12. PR-4 Proteins
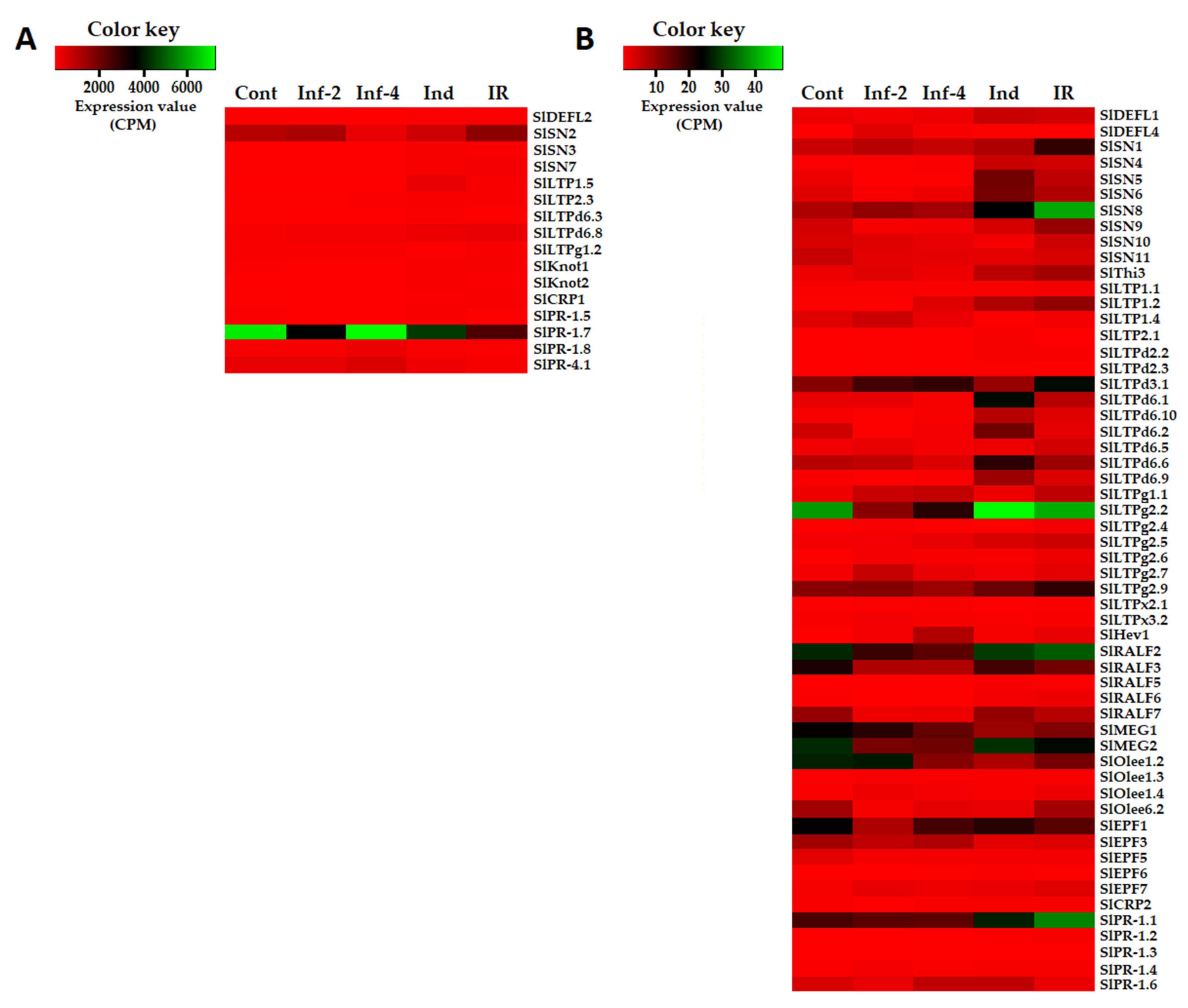
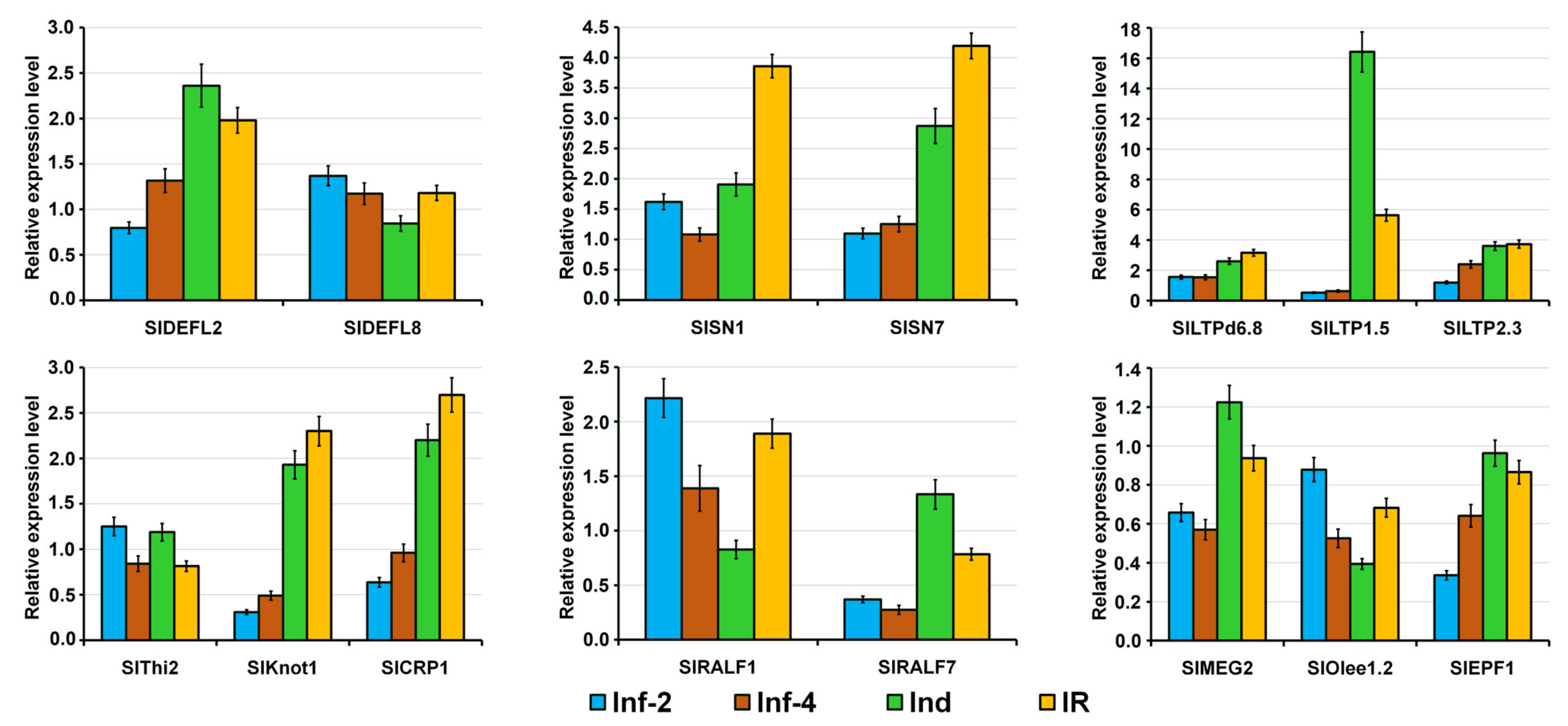
2.4. Validation of RNA-Seq Data by RT-PCR and qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Biological Material
4.2. Isolation of FS-94 Elicitors
4.3. Plant Protection Assay
4.4. Experimental Design
4.5. RNA Isolation
4.6. Library Construction and NGS
4.7. Sequencing Data Analysis
4.8. Identification of CRP Precursors in Tomato Transcriptomes
4.9. Differential Gene Expression Analysis
4.10. RT-PCR Validation of SlHev1 Expression
4.11. Real-Time PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Avery, S.V.; Singleton, I.; Magan, N.; Goldman, G.H. The fungal threat to global food security. Fungal Biol. 2019, 123, 555–557. [Google Scholar] [CrossRef] [PubMed]
- Kettles, G.J.; Luna, E. Food security in 2044: How do we control the fungal threat? Fungal Biol. 2019, 123, 558–564. [Google Scholar] [CrossRef] [PubMed]
- Velivelli, S.L.; De Vos, P.; Kromann, P.; Declerck, S.; Prestwich, B.D. Biological control agents: From field to market, problems, and challenges. Trends Biotechnol. 2014, 32, 493–496. [Google Scholar] [CrossRef]
- Burketova, L.; Trda, L.; Ott, P.G.; Valentova, O. Bio-based resistance inducers for sustainable plant protection against pathogens. Biotechnol. Adv. 2015, 33, 994–1004. [Google Scholar] [CrossRef]
- Goyal, S.; Lambert, C.; Cluzet, S.; Mérillon, J.M.; Ramawat, K.G. Secondary Metabolites and Plant Defence. In Plant Defence: Biological Control; Mérillon, J.M., Ramawat, K.G., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 109–138. [Google Scholar] [CrossRef]
- Tavormina, P.; De Coninck, B.; Nikonorova, N.; De Smet, I.; Cammue, B.P. The Plant peptidome: An expanding repertoire of structural features and biological functions. Plant Cell 2015, 27, 2095–2118. [Google Scholar] [CrossRef]
- Silverstein, K.A.; Moskal, W.A., Jr.; Wu, H.C.; Underwood, B.A.; Graham, M.A.; Town, C.D.; VandenBosch, K.A. Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 2007, 51, 262–280. [Google Scholar] [CrossRef]
- Heber, D.; Lu, Q.Y. Overview of mechanisms of action of lycopene. Exp. Biol. Med. 2002, 227, 920–923. [Google Scholar] [CrossRef]
- Srinivas, C.; Nirmala Devi, D.; Narasimha Murthy, K.; Mohan, C.D.; Lakshmeesha, T.R.; Singh, B.; Kalagatur, N.K.; Niranjana, S.R.; Hashem, A.; Alqarawi, A.A.; et al. Fusarium oxysporum f. sp. lycopersici causal agent of vascular wilt disease of tomato: Biology to diversity—A review. Saudi J. Biol. Sci. 2019, 26, 1315–1324. [Google Scholar] [CrossRef]
- Perincherry, L.; Lalak-Kańczugowska, J.; Stępień, Ł. Fusarium-Produced Mycotoxins in Plant-Pathogen Interactions. Toxins 2019, 11, 664. [Google Scholar] [CrossRef]
- Shcherbakova, L.A.; Nazarova, T.A.; Mikityuk, O.D.; Fravel, D.R. Fusarium sambucinum isolate FS-94 induces resistance against fusarium wilt of tomato via activation and priming of a salicylic acid-dependent signaling system. Russ. J. Plant Physiol. 2011, 58, 808–818. [Google Scholar] [CrossRef]
- Shcherbakova, L.; Odintsova, T.; Stakheev, A.A.; Fravel, D.R.; Zavriev, S.K. Identification of a novel small cysteine-rich protein in the fraction from the biocontrol Fusarium oxysporum strain CS-20 that mitigates fusarium wilt symptoms and triggers defense responses in tomato. Front. Plant Sci. 2016, 6, 1207. [Google Scholar] [CrossRef]
- Bushmanova, E.; Antipov, D.; Lapidus, A.; Prjibelski, A.D. rnaSPAdes: A de novo transcriptome assembler and its application to RNA-Seq data. GigaScience 2019, 8, giz100. [Google Scholar] [CrossRef]
- Parisi, K.; Shafee, T.M.A.; Quimbar, P.; Van der Weerden, N.L.; Bleackley, M.R.; Anderson, M.A. The evolution, function and mechanisms of action for plant defensins. Semin. Cell Dev. Biol. 2019, 88, 107–118. [Google Scholar] [CrossRef]
- Istomina, E.A.; Slezina, M.P.; Kovtun, A.S.; Odintsova, T.I. In silico identification of gene families encoding cysteine-rich peptides in Solanum lycopersicum L. Russ. J. Genet. 2020, 56, 572–579. [Google Scholar] [CrossRef]
- Yount, N.Y.; Yeaman, M.R. Multidimensional signatures in antimicrobial peptides. Proc. Natl. Acad. Sci. USA 2004, 101, 7363–7368. [Google Scholar] [CrossRef]
- Sagaram, U.S.; Pandurangi, R.; Kaur, J.; Smith, T.J.; Shah, D.M. Structure-activity determinants in antifungal plant defensins MsDef1 and MtDef4 with different modes of action against Fusarium graminearum. PLoS ONE 2011, 6, e18550. [Google Scholar] [CrossRef]
- Salminen, T.A.; Blomqvist, K.; Edqvist, J. Lipid transfer proteins: Classification, nomenclature, structure, and function. Planta 2016, 244, 971–997. [Google Scholar] [CrossRef]
- Kader, J.C. Lipid-transfer proteins: A puzzling family of plant proteins. Trends Plant Sci. 1997, 2, 66–70. [Google Scholar] [CrossRef]
- Quevillon, E.; Silventoinen, V.; Pilai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domain identifier. Nucleic Acids Res. 2005, 33, 116–120. [Google Scholar] [CrossRef]
- Odintsova, T.I.; Slezina, M.P.; Istomina, E.A.; Korostyleva, T.V.; Kovtun, A.S.; Kasianov, A.S.; Shcherbakova, L.A.; Kudryavtsev, A.M. Non-specific lipid transfer proteins in Triticum kiharae Dorof. et Migush.: Identification, characterization and expression profiling in response to pathogens and resistance inducers. Pathogens 2019, 8, 221. [Google Scholar] [CrossRef] [PubMed]
- Berrocal-Lobo, M.; Segura, A.; Moreno, M.; López, G.; García-Olmedo, F.; Molina, A. Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant Physiol. 2002, 128, 951–961. [Google Scholar] [CrossRef]
- Segura, A.; Moreno, M.; Madueño, F.; Molina, A.; García-Olmedo, F. Snakin-1, a peptide from potato that is active against plant pathogens. Mol. Plant Microbe Interact. 1999, 12, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Almasia, N.I.; Bazzini, A.A.; Hopp, H.E.; Vazquez-Rovere, C. Overexpression of snakin-1 gene enhances resistance to Rhizoctonia solani and Erwinia carotovora in transgenic potato plants. Mol. Plant Pathol. 2008, 9, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Nahirñak, V.; Almasia, N.I.; Fernandez, P.V.; Hopp, H.E.; Estevez, J.M.; Carrari, F.; Vazquez-Rovere, C. Potato snakin-1 gene silencing affects cell division, primary metabolism, and cell wall composition. Plant Physiol. 2012, 158, 252–263. [Google Scholar] [CrossRef]
- Stec, B. Plant thionins—The structural perspective. Cell. Mol. Life Sci. 2006, 63, 1370–1385. [Google Scholar] [CrossRef]
- Slavokhotova, A.A.; Shelenkov, A.A.; Andreev, Y.A.; Odintsova, T.I. Hevein-like antimicrobial peptides of plants. Biochemistry 2017, 82, 1659–1674. [Google Scholar] [CrossRef]
- Huang, R.H.; Xiang, Y.; Liu, X.Z.; Zhang, Y.; Hu, Z.; Wang, D.C. Two novel antifungal peptides distinct with a five-disulfide motif from the bark of Eucommia ulmoides Oliv. FEBS Lett. 2002, 521, 87–90. [Google Scholar] [CrossRef]
- Van den Bergh, K.P.; Proost, P.; Van Damme, J.; Coosemans, J.; Van Damme, E.J.; Peumans, W.J. Five disulfide bridges stabilize a hevein-type antimicrobial peptide from the bark of spindle tree (Euonymus europaeus L.). FEBS Lett. 2002, 530, 181–185. [Google Scholar] [CrossRef]
- Odintsova, T.I.; Vassilevski, A.A.; Slavokhotova, A.A.; Musolyamov, A.K.; Finkina, E.I.; Khadeeva, N.V.; Rogozhin, E.A.; Korostyleva, T.V.; Pukhalsky, V.A.; Grishin, E.V.; et al. A novel antifungal hevein-type peptide from Triticum kiharae seeds with a unique 10-cysteine motif. FEBS J. 2009, 276, 4266–4275. [Google Scholar] [CrossRef]
- Van den Bergh, K.P.; Rougé, P.; Proost, P.; Coosemans, J.; Krouglova, T.; Engelborghs, Y.; Peumans, W.J.; Van Damme, E.J. Synergistic antifungal activity of two chitin-binding proteins from spindle tree (Euonymus europaeus L.). Planta 2004, 219, 221–232. [Google Scholar] [CrossRef]
- Slavokhotova, A.A.; Naumann, T.A.; Price, N.P.; Rogozhin, E.A.; Andreev, Y.A.; Vassilevski, A.A.; Odintsova, T.I. Novel mode of action of plant defense peptides—Hevein-like antimicrobial peptides from wheat inhibit fungal metalloproteases. FEBS J. 2014, 281, 4754–4764. [Google Scholar] [CrossRef]
- Nielsen, K.K.; Nielsen, J.E.; Madrid, S.M.; Mikkelsen, J.D. Characterization of a new antifungal chitin-binding peptide from sugar beet leaves. Plant Physiol. 1997, 113, 83–91. [Google Scholar] [CrossRef][Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Tam, J.P.; Wang, S.; Wong, K.H.; Tan, W.L. Antimicrobial peptides from plants. Pharmaceuticals 2015, 8, 711–757. [Google Scholar] [CrossRef]
- Gruber, C.W.; Cemazar, M.; Anderson, M.A.; Craik, D.J. Insecticidal plant cyclotides and related cystine knot toxins. Toxicon 2007, 49, 561–575. [Google Scholar] [CrossRef]
- Cotabarren, J.; Tellechea, M.E.; Tanco, S.M.; Lorenzo, J.; Garcia-Pardo, J.; Avilés, F.X.; Obregón, W.D. Biochemical and MALDI-TOF mass spectrometric characterization of a novel native and recombinant cystine knot miniprotein from Solanum tuberosum subsp. andigenum cv. Churqueña. Int. J. Mol. Sci. 2018, 19, 678. [Google Scholar] [CrossRef]
- Pearce, G.; Moura, D.S.; Stratmann, J.; Ryan, C.A., Jr. RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc. Natl. Acad. Sci. USA 2001, 98, 12843–12847. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, M.R.; Haruta, M.; Moura, D.S. Twenty Years of Progress in Physiological and Biochemical Investigation of RALF Peptides. Plant Physiol. 2020, 182, 1657–1666. [Google Scholar] [CrossRef]
- Matos, J.L.; Fiori, C.S.; Silva-Filho, M.C.; Moura, D.S. A conserved dibasic site is essential for correct processing of the peptide hormone AtRALF1 in Arabidopsis thaliana. FEBS Lett. 2008, 582, 3343–3347. [Google Scholar] [CrossRef]
- Srivastava, R.; Liu, J.X.; Guo, H.; Yin, Y.; Howell, S.H. Regulation and processing of a plant peptide hormone, AtRALF23, in Arabidopsis. Plant J. 2009, 59, 930–939. [Google Scholar] [CrossRef] [PubMed]
- Stegmann, M.; Monaghan, J.; Smakowska-Luzan, E.; Rovenich, H.; Lehner, A.; Holton, N.; Belkhadir, Y.; Zipfel, C. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 2017, 355, 287–289. [Google Scholar] [CrossRef] [PubMed]
- Pearce, G.; Yamaguchi, Y.; Munske, G.; Ryan, C.A. Structure-activity studies of RALF, Rapid Alkalinization Factor, reveal an essential—YISY—motif. Peptides 2010, 31, 1973–1977. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Marcos, J.F.; Costa, L.M.; Biderre-Petit, C.; Khbaya, B.; O’Sullivan, D.M.; Wormald, M.; Perez, P.; Dickinson, H.G. maternally expressed gene1 Is a novel maize endosperm transfer cell-specific gene with a maternal parent-of-origin pattern of expression. Plant Cell 2004, 16, 1288–1301. [Google Scholar] [CrossRef]
- Quiralte, J.; Palacios, L.; Rodríguez, R.; Cárdaba, B.; Arias de Saavedra, J.M.; Villalba, M.; Florido, J.F.; Lahoz, C. Modelling diseases: The allergens of Olea europaea pollen. J. Invest. Allergol. Clin. Immunol. 2007, 17, 76–82. [Google Scholar]
- Villalba, M.; Batanero, E.; Monsalve, R.I.; González de la Peña, M.A.; Lahoz, C.; Rodríguez, R. Cloning and expression of Ole e I, the major allergen from olive tree pollen: Polymorphism analysis and tissue specificity. J. Biol. Chem. 1994, 269, 15217–15222. [Google Scholar] [CrossRef]
- Treviño, M.A.; García-Mayoral, M.F.; Barral, P.; Villalba, M.; Santoro, J.; Rico, M.; Rodríguez, R.; Bruix, M. NMR solution structure of Ole e 6, a major allergen from olive tree pollen. J. Biol. Chem. 2004, 279, 39035–39041. [Google Scholar] [CrossRef]
- Takata, N.; Yokota, K.; Ohki, S.; Mori, M.; Taniguchi, T.; Kurita, M. Evolutionary relationship and structural characterization of the EPF/EPFL gene family. PLoS ONE 2013, 8, e65183. [Google Scholar] [CrossRef]
- Hunt, L.; Bailey, K.J.; Gray, J.E. The signalling peptide EPFL9 is a positive regulator of stomatal development. New Phytol. 2010, 186, 609–614. [Google Scholar] [CrossRef]
- Hara, K.; Kajita, R.; Torii, K.U.; Bergmann, D.C.; Kakimoto, T. The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 2007, 21, 1720–1725. [Google Scholar] [CrossRef]
- van Loon, L.C.; Rep, M.; Pieterse, C.M.J. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef]
- Breen, S.; Williams, S.J.; Outram, M.; Kobe, B.; Solomon, P.S. Emerging insights into the functions of pathogenesis-related protein 1. Trends Plant Sci. 2017, 22, 871–879. [Google Scholar] [CrossRef]
- Niki, T.; Mitsuhara, I.; Seo, S.; Ohtsubo, N.; Ohashi, Y. Antagonistic effect of salicylic acid and jasmonic acid on the expression of pathogenesis-related (PR) protein genes in wounded mature tobacco leaves. Plant Cell Physiol. 1998, 39, 500–507. [Google Scholar] [CrossRef]
- Hosokawa, D.; Ohashi, Y. Immunochemical localization of pathogenesis-related proteins secreted into the intercellular spaces of salicylate-treated tobacco leaves. Plant Cell Physiol. 1988, 29, 1035–1040. [Google Scholar] [CrossRef]
- Ali, S.; Ganai, B.A.; Kamili, A.N.; Bhat, A.A.; Mir, Z.A.; Bhat, J.A.; Tyagi, A.; Islam, S.T.; Mushtaq, M.; Yadav, P.; et al. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 2018, 212–213, 29–37. [Google Scholar] [CrossRef]
- Svensson, B.; Svendsen, I.; Højrup, P.; Roepstorff, P.; Ludvigsen, S.; Poulsen, F.M. Primary structure of barwin: A barley seed protein closely related to the C-terminal domain of proteins encoded by wound-induced plant genes. Biochemistry 1992, 31, 8767–8770. [Google Scholar] [CrossRef]
- Caruso, C.; Nobile, M.; Leonardi, L.; Bertini, L.; Buonocore, V.; Caporale, C. Isolation and amino acid sequence of two new PR-4 proteins from wheat. J. Protein Chem. 2001, 20, 327–335. [Google Scholar] [CrossRef]
- Caporale, C.; Facchiano, A.; Bertini, L.; Leonardi, L.; Chilosi, G.; Buonocore, V.; Caruso, C. Comparing the modeled structures of PR-4 proteins from wheat. J. Mol. Model. 2003, 9, 9–15. [Google Scholar] [CrossRef]
- Bertini, L.; Cascone, A.; Tucci, M.; D’Amore, R.; Di Berardino, I.; Buonocore, V.; Caporale, C.; Caruso, C. Molecular and functional analysis of new members of the wheat PR4 gene family. Biol. Chem. 2006, 387, 1101–1111. [Google Scholar] [CrossRef]
- Ludvigsen, S.; Poulsen, F.M. Three-dimensional structure in solution of barwin, a protein from barley seed. Biochemistry 1992, 31, 8783–8789. [Google Scholar] [CrossRef]
- Hejgaard, J.; Jacobsen, S.; Bjørn, S.E.; Kragh, K.M. Antifungal activity of chitin-binding PR-4 type proteins from barley grain and stressed leaf. FEBS Lett. 1992, 307, 389–392. [Google Scholar] [CrossRef]
- Stintzi, A.; Heitz, T.; Prasad, V.; Wiedemann-Merdinoglu, S.; Kauffmann, S.; Geoffroy, P.; Legrand, M.; Fritig, B. Plant ‘pathogenesis-related’ proteins and their role in defense against pathogens. Biochimie 1993, 75, 687–706. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.; Bakker, P.A. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [PubMed]
- Zamioudis, C.; Pieterse, C.M. Modulation of host immunity by beneficial microbes. Mol. Plant Microbe Interact. 2012, 25, 139–150. [Google Scholar] [CrossRef]
- Alexandersson, E.; Mulugeta, T.; Lankinen, Å.; Liljeroth, E.; Andreasson, E. Plant resistance inducers against pathogens in Solanaceae species—From molecular mechanisms to field application. Int. J. Mol. Sci. 2016, 17, 1673. [Google Scholar] [CrossRef]
- Odintsova, T.I.; Slezina, M.P.; Istomina, E.A.; Korostyleva, T.V.; Kasianov, A.S.; Kovtun, A.S.; Makeev, V.J.; Shcherbakova, L.A.; Kudryavtsev, A.M. Defensin-like peptides in wheat analyzed by whole-transcriptome sequencing: A focus on structural diversity and role in induced resistance. PeerJ 2019, 7, e6125. [Google Scholar] [CrossRef]
- Farrokhi, N.; Whitelegge, J.P.; Brusslan, J.A. Plant peptides and peptidomics. Plant Biotechnol. J. 2008, 6, 105–134. [Google Scholar] [CrossRef]
- Goyal, R.K.; Mattoo, A.K. Multitasking antimicrobial peptides in plant development and host defense against biotic/abiotic stress. Plant Sci. 2014, 228, 135–149. [Google Scholar] [CrossRef]
- Hirakawa, Y.; Torii, K.U.; Uchida, N. Mechanisms and strategies shaping plant peptide hormones. Plant Cell Physiol. 2017, 58, 1313–1318. [Google Scholar] [CrossRef]
- Slavokhotova, A.A.; Shelenkov, A.A.; Korostyleva, T.V.; Rogozhin, E.A.; Melnikova, N.V.; Kudryavtseva, A.V.; Odintsova, T.I. Defense peptide repertoire of Stellaria media predicted by high throughput next generation sequencing. Biochimie 2017, 135, 15–27. [Google Scholar] [CrossRef]
- Champigny, M.J.; Isaacs, M.; Carella, P.; Faubert, J.; Fobert, P.R.; Cameron, R.K. Long distance movement of DIR1 and investigation of the role of DIR1-like during systemic acquired resistance in Arabidopsis. Front. Plant Sci. 2013, 4, 230. [Google Scholar] [CrossRef]
- Chen, Y.L.; Lee, C.Y.; Cheng, K.T.; Chang, W.H.; Huang, R.N.; Nam, H.G.; Chen, Y.R. Quantitative peptidomics study reveals that a wound-induced peptide from PR-1 regulates immune signaling in tomato. Plant Cell 2014, 26, 4135–4148. [Google Scholar] [CrossRef]
- Slezina, M.P.; Istomina, E.A.; Korostyleva, T.V.; Odintsova, T.I. WAMP homologues in grasses. unpublished.
- Shcherbakova, L.A.; Nazarova, T.A.; Mikityuk, O.D.; Istomina, E.A.; Odintsova, T.I. An extract purified from the mycelium of a tomato wilt-controlling strain of Fusarium sambucinum can protect wheat against fusarium and common root rots. Pathogens 2018, 7, 61. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bendtsen, J.D.; Nielsen, H.; von Heijne, G.; Brunak, S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004, 340, 783–795. [Google Scholar] [CrossRef]
- Waghu, F.H.; Barai, R.S.; Gurung, P.; Idicula-Thomas, S. CAMPR3: A database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res. 2015, 44, D1094–D1097. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Eisenhaber, B.; Wildpaner, M.; Schultz, C.J.; Borner, G.H.H.; Dupree, P.; Eisenhaber, F. Glycosylphosphatidylinositol lipid anchoring of plant proteins. Sensitive prediction from sequence- and genome-wide studies for Arabidopsis and rice. Plant Physiol. 2003, 133, 1691–1701. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using realtime quantitative PCR and the 2−ΔΔCT method. Methods 2011, 25, 402–408. [Google Scholar] [CrossRef]














| Elicitor Fraction 1, µg/mL | Efficacy of Seedling Protection, % Relative to Control (M ± SE) 2 | |
|---|---|---|
| 5 dpi | 10 dpi | |
| Seed soaking | ||
| 200 | 76.4 ± 4.05 | 57.7 ± 2.43 |
| 100 | 58.5 ± 1.16 | 52.6 ± 2.13 |
| 50 | 46.5 ± 0.33 | 37.5 ± 0.96 |
| Root immersion | ||
| 200 | 74.0 ± 8.14 | 53.2 ± 3.86 |
| № | Variant | Incubation with FS-94 Elicitors | Inoculation with F37 Conidia |
|---|---|---|---|
| 1 | Non-inoculated control (Cont) | 0 h | Without inoculation |
| 2 | Inoculation with F. oxysporum (Inf-4) | 0 h | Immediate for 48 h |
| 3 | Treatment with elicitors (Ind) | Immediate for 48 h | Without inoculation |
| 4 | Inoculation with F.oxysporum (Inf-2) | 0 h | After 48 h for 48 h |
| 5 | Treatment with elicitors followed by inoculation with F. oxysporum (IR) | 48 h | After 48 h for 48 h |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Slezina, M.P.; Istomina, E.A.; Korostyleva, T.V.; Kovtun, A.S.; Kasianov, A.S.; Konopkin, A.A.; Shcherbakova, L.A.; Odintsova, T.I. Molecular Insights into the Role of Cysteine-Rich Peptides in Induced Resistance to Fusarium oxysporum Infection in Tomato Based on Transcriptome Profiling. Int. J. Mol. Sci. 2021, 22, 5741. https://doi.org/10.3390/ijms22115741
Slezina MP, Istomina EA, Korostyleva TV, Kovtun AS, Kasianov AS, Konopkin AA, Shcherbakova LA, Odintsova TI. Molecular Insights into the Role of Cysteine-Rich Peptides in Induced Resistance to Fusarium oxysporum Infection in Tomato Based on Transcriptome Profiling. International Journal of Molecular Sciences. 2021; 22(11):5741. https://doi.org/10.3390/ijms22115741
Chicago/Turabian StyleSlezina, Marina P., Ekaterina A. Istomina, Tatyana V. Korostyleva, Alexey S. Kovtun, Artem S. Kasianov, Alexey A. Konopkin, Larisa A. Shcherbakova, and Tatyana I. Odintsova. 2021. "Molecular Insights into the Role of Cysteine-Rich Peptides in Induced Resistance to Fusarium oxysporum Infection in Tomato Based on Transcriptome Profiling" International Journal of Molecular Sciences 22, no. 11: 5741. https://doi.org/10.3390/ijms22115741
APA StyleSlezina, M. P., Istomina, E. A., Korostyleva, T. V., Kovtun, A. S., Kasianov, A. S., Konopkin, A. A., Shcherbakova, L. A., & Odintsova, T. I. (2021). Molecular Insights into the Role of Cysteine-Rich Peptides in Induced Resistance to Fusarium oxysporum Infection in Tomato Based on Transcriptome Profiling. International Journal of Molecular Sciences, 22(11), 5741. https://doi.org/10.3390/ijms22115741






