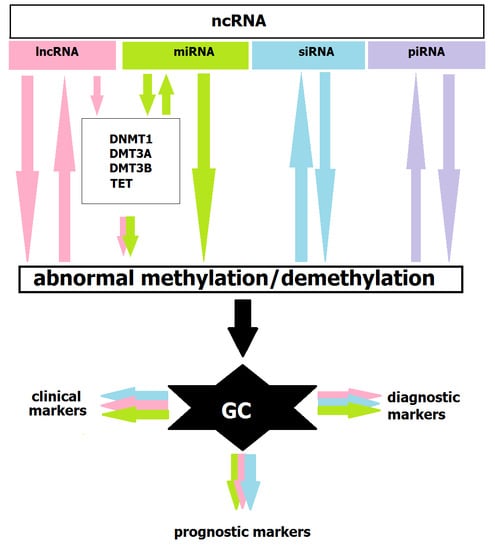
Methylation and Noncoding RNAs in Gastric Cancer: Everything Is Connected
Abstract
1. Introduction
2. Methylation and miRNAs: Feedback Loops
3. DNA Methylation and lncRNAs: Extending the lncRNA–miRNA–mRNA Pathway
4. DNA Methylation and siRNAs: Retrotransposon Silencing
5. DNA Methylation and piRNAs: Novel Candidate?
6. Methylation and ncRNAs in GC Diagnostics and Treatment
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Sitarz, R.; Skierucha, M.; Mielko, J.; Offerhaus, J.; Maciejewski, R.; Polkowski, W. Gastric cancer: Epidemiology, prevention, classification, and treatment. Cancer Manag. Res. 2018, 10, 239–248. [Google Scholar] [CrossRef]
- Necula, L.; Matei, L.; Dragu, D.; Neagu, A.I.; Mambet, C.; Nedeianu, S.; Bleotu, C.; Diaconu, C.C.; Chivu-Economescu, M. Recent advances in gastric cancer early diagnosis. World J. Gastroenterol. 2019, 25, 2029–2044. [Google Scholar] [CrossRef]
- Zhou, Z.; Lin, Z.; Pang, X.; Tariq, M.A.; Ao, X.; Li, P.; Wang, J. Epigenetic regulation of long non-coding RNAs in gastric cancer. Oncotarget 2017, 9, 19443–19458. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, R.; Suzuki, H.; Yamamoto, E.; Imai, K.; Shinomura, Y. Emerging links between epigenetic alterations and dysregulation of noncoding RNAs in cancer. Tumor Biol. 2012, 33, 277–285. [Google Scholar] [CrossRef]
- Fattahi, S.; Kosari-Monfared, M.; Ghadami, E.; Golpour, M.; Khodadadi, P.; Ghasemiyan, M.; Akhavan-Niaki, H. Infection-associated epigenetic alterations in gastric cancer: New insight in cancer therapy. J. Cell Physiol. 2018, 233, 9261–9270. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Zhu, S.; Xiong, S.; Xue, X.; Zhou, X. MicroRNAs and the PTEN/PI3K/Akt pathway in gastric cancer (Review). Oncol. Rep. 2019, 41, 1439–1454. [Google Scholar] [CrossRef] [PubMed]
- Skierucha, M.; Na Milne, A.; Offerhaus, G.J.A.; Polkowski, W.P.; Maciejewski, R.; Sitarz, R. Molecular alterations in gastric cancer with special reference to the early-onset subtype. World J. Gastroenterol. 2016, 22, 2460–2474. [Google Scholar] [CrossRef]
- Fang, X.-Y.; Pan, H.-F.; Leng, R.-X.; Ye, D.-Q. Long noncoding RNAs: Novel insights into gastric cancer. Cancer Lett. 2015, 356, 357–366. [Google Scholar] [CrossRef]
- Puneet; Kazmi, H.R.; Kumari, S.; Tiwari, S.; Khanna, A.; Narayan, G. Epigenetic Mechanisms and Events in Gastric Cancer-Emerging Novel Biomarkers. Pathol. Oncol. Res. 2018, 24, 757–770. [Google Scholar] [CrossRef]
- Sonohara, F.; Inokawa, Y.; Hayashi, M.; Kodera, Y.; Nomoto, S. Epigenetic modulation associated with carcinogenesis and prognosis of human gastric cancer. Oncol. Lett. 2017, 13, 3363–3368. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jaenisch, R.; Bird, A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003, 33, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Liu, Y. DNA methylation in human diseases. Genes Dis. 2018, 5, 1–8. [Google Scholar] [CrossRef]
- Field, A.E.; Robertson, N.; Wang, T.; Havas, A.; Ideker, T.; Adams, P.D. DNA Methylation Clocks in Aging: Categories, Causes, and Consequences. Mol. Cell 2018, 71, 882–895. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, M.V.C.; Bourc’His, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef]
- Pfeifer, G.P. Defining Driver DNA Methylation Changes in Human Cancer. Int. J. Mol. Sci. 2018, 19, 1166. [Google Scholar] [CrossRef]
- Lee, H.; Zhang, Z.; Krause, H.M. Long Noncoding RNAs and Repetitive Elements: Junk or Intimate Evolutionary Partners? Trends Genet. 2019, 35, 892–902. [Google Scholar] [CrossRef]
- 6 Non-coding RNA characterization. Nature 2019. [CrossRef]
- Haller, F.; Zaletaev, D.V. Coding and Non-coding: Molecular Portrait of GIST and its Clinical Implication. Curr. Mol. Med. 2018, 18, 252–259. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 20190027. [Google Scholar] [CrossRef]
- Wei, J.-W.; Huang, K.; Yang, C.; Kang, C.-S. Non-coding RNAs as regulators in epigenetics. Oncol. Rep. 2016, 37, 3–9. [Google Scholar] [CrossRef]
- Arif, K.M.T.; Elliott, E.K.; Haupt, L.M.; Griffiths, L.R. Regulatory Mechanisms of Epigenetic miRNA Relationships in Human Cancer and Potential as Therapeutic Targets. Cancers 2020, 12, 2922. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Chen, Y.; Zhou, X. The roles of microRNAs in epigenetic regulation. Curr. Opin. Chem. Biol. 2019, 51, 11–17. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Ivey, K.N.; Srivastava, D. microRNAs as Developmental Regulators. Cold Spring Harb. Perspect. Biol. 2015, 7, a008144. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Gebert, L.F.R.; Macrae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.Q.; Ahmed, E.I.; Elareer, N.R.; Junejo, K.; Steinhoff, M.; Uddin, S. Role of miRNA-Regulated Cancer Stem Cells in the Pathogenesis of Human Malignancies. Cells 2019, 8, 840. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.L.; Magalhães, L.; Moreira, F.C.; Reis-Das-Mercês, L.; Vidal, A.F.; Ribeiro-Dos-Santos, A.M.; Demachki, S.; Anaissi, A.K.M.; Burbano, R.M.R.; Albuquerque, P.; et al. Epigenetic Field Cancerization in Gastric Cancer: microRNAs as Promising Biomarkers. J. Cancer 2019, 10, 1560–1569. [Google Scholar] [CrossRef]
- Kurata, A.; Yamada, M.; Ohno, S.-I.; Inoue, S.; Hashimoto, H.; Fujita, K.; Takanashi, M.; Kuroda, M. Expression level of microRNA-200c is associated with cell morphology in vitro and histological differentiation through regulation of ZEB1/2 and E-cadherin in gastric carcinoma. Oncol. Rep. 2018, 39, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Bure, I.V.; Nemtsova, M.V.; Zaletaev, D.V. Roles of E-cadherin and Noncoding RNAs in the Epithelial–mesenchymal Transition and Progression in Gastric Cancer. Int. J. Mol. Sci. 2019, 20, 2870. [Google Scholar] [CrossRef] [PubMed]
- Fuso, A.; Raia, T.; Orticello, M.; Lucarelli, M. The complex interplay between DNA methylation and miRNAs in gene expression regulation. Biochimie 2020, 173, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Romero-Cordoba, S.L.; Salido-Guadarrama, I.; Rodriguez-Dorantes, M.; Hidalgo-Miranda, A. miRNA biogenesis: Biological impact in the development of cancer. Cancer Biol. Ther. 2014, 15, 1444–1455. [Google Scholar] [CrossRef]
- Tsai, K.-W.; Wu, C.-W.; Hu, L.-Y.; Li, S.-C.; Liao, Y.-L.; Lai, C.-H.; Kao, H.-W.; Fang, W.-L.; Huang, K.-H.; Chan, W.-C.; et al. Epigenetic regulation of miR-34b and miR-129 expression in gastric cancer. Int. J. Cancer 2011, 129, 2600–2610. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.-W.; Liao, Y.-L.; Wu, C.-W.; Hu, L.-Y.; Li, S.-C.; Chan, W.-C.; Ho, M.-R.; Lai, C.-H.; Kao, H.-W.; Fang, W.-L.; et al. Aberrant hypermethylation of miR-9 genes in gastric cancer. Epigenetics 2011, 6, 1189–1197. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Young, J.; Prabhala, H.; Pan, E.; Mestdagh, P.; Muth, D.; Teruya-Feldstein, J.; Reinhardt, F.; Onder, T.; Valastyan, S.; et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 2010, 12, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Selcuklu, S.D.; Donoghue, M.T.A.; Rehmet, K.; Gomes, M.D.S.; Fort, A.; Kovvuru, P.; Muniyappa, M.K.; Kerin, M.J.; Enright, A.; Spillane, C. MicroRNA-9 Inhibition of Cell Proliferation and Identification of Novel miR-9 Targets by Transcriptome Profiling in Breast Cancer Cells. J. Biol. Chem. 2012, 287, 29516–29528. [Google Scholar] [CrossRef]
- Cai, M.; Chen, Q.; Shen, J.; Lv, C.; Cai, L. Retracted: Epigenetic silenced miR-125a-5p could be self-activated through targeting Suv39H1 in gastric cancer. J. Cell. Mol. Med. 2018, 22, 4721–4731, retracted in J. Cell. Mol. Med. 2021, 25, 2285–2285. [Google Scholar] [CrossRef]
- Nishida, N.; Mimori, K.; Fabbri, M.; Yokobori, T.; Sudo, T.; Tanaka, F.; Shibata, K.; Ishii, H.; Doki, Y.; Mori, M. MicroRNA-125a-5p Is an Independent Prognostic Factor in Gastric Cancer and Inhibits the Proliferation of Human Gastric Cancer Cells in Combination with Trastuzumab. Clin. Cancer Res. 2011, 17, 2725–2733. [Google Scholar] [CrossRef]
- Dai, J.; Wang, J.; Yang, L.; Xiao, Y.; Ruan, Q. miR-125a regulates angiogenesis of gastric cancer by targeting vascular endothelial growth factor A. Int. J. Oncol. 2015, 47, 1801–1810. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Huang, Z.; Liu, Y. Reduced miR-125a-5p expression is associated with gastric carcinogenesis through the targeting of E2F3. Mol. Med. Rep. 2014, 10, 2601–2608. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, B.; Liu, Y.; Shi, L.; Li, H.; Lu, S. MiR125a-5p acting as a novel Gab2 suppressor inhibits invasion of glioma. Mol. Carcinog. 2016, 55, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Wang, Z.; Wang, Z.; Yang, Y.; Fu, C.; Zhu, J.; Jiang, D. MicroRNA-31 Function as a Suppressor Was Regulated by Epigenetic Mechanisms in Gastric Cancer. BioMed Res. Int. 2017, 2017, 5348490. [Google Scholar] [CrossRef]
- Ando, T.; Yoshida, T.; Enomoto, S.; Asada, K.; Tatematsu, M.; Ichinose, M.; Sugiyama, T.; Ushijima, T. DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: Its possible involvement in the formation of epigenetic field defect. Int. J. Cancer 2009, 124, 2367–2374. [Google Scholar] [CrossRef]
- Roscigno, G.; Quintavalle, C.; Donnarumma, E.; Puoti, I.; Diaz-Lagares, A.; Iaboni, M.; Fiore, D.; Russo, V.; Todaro, M.; Romano, G.; et al. MiR-221 promotes stemness of breast cancer cells by targeting DNMT3b. Oncotarget 2015, 7, 580–592. [Google Scholar] [CrossRef]
- Ng, E.K.-O.; Tsang, W.P.; Ng, S.S.M.; Jin, H.; Yu, J.; Li, J.J.; Röcken, C.; Ebert, M.P.A.; Kwok, T.T.; Sung, J.J.Y. MicroRNA-143 targets DNA methyltransferases 3A in colorectal cancer. Br. J. Cancer 2009, 101, 699–706. [Google Scholar] [CrossRef]
- Fabbri, M.; Garzon, R.; Cimmino, A.; Liu, Z.; Zanesi, N.; Callegari, E.; Liu, S.; Alder, H.; Costinean, S.; Fernandez-Cymering, C.; et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. USA 2007, 104, 15805–15810. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Liu, S.; Fabbri, M.; Liu, Z.; Heaphy, C.E.; Callegari, E.; Schwind, S.; Pang, J.; Yu, J.; Muthusamy, N.; et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood 2009, 113, 6411–6418. [Google Scholar] [CrossRef]
- Xu, Q.; Jiang, Y.; Yin, Y.; Li, Q.; He, J.; Jing, Y.; Qi, Y.-T.; Xu, Q.; Li, W.; Lu, B.; et al. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J. Mol. Cell Biol. 2012, 5, 3–13. [Google Scholar] [CrossRef]
- Zhu, A.; Xia, J.; Zuo, J.; Jin, S.; Zhou, H.; Yao, L.; Huang, H.; Han, Z. MicroRNA-148a is silenced by hypermethylation and interacts with DNA methyltransferase 1 in gastric cancer. Med. Oncol. 2011, 29, 2701–2709. [Google Scholar] [CrossRef]
- Wada, R.; Akiyama, Y.; Hashimoto, Y.; Fukamachi, H.; Yuasa, Y. miR-212 is downregulated and suppresses methyl-CpG-binding protein MeCP2 in human gastric cancer. Int. J. Cancer 2010, 127, 1106–1114. [Google Scholar] [CrossRef]
- Sas-Chen, A.; Srivastava, S.; Yarden, Y. The short and the long: Non-coding RNAs and growth factors in cancer progression. Biochem. Soc. Trans. 2017, 45, 51–64. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, J.; Li, Y.; Song, T.; Wu, Y.; Fang, S.; Bu, D.; Li, H.; Sun, L.; Pei, D.; et al. NONCODEV6: An updated database dedicated to long non-coding RNA annotation in both animals and plants. Nucleic Acids Res. 2021, 49, D165–D171. [Google Scholar] [CrossRef]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.M.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nat. Cell Biol. 2009, 458, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Atianand, M.K.; Fitzgerald, K.A. Long non-coding RNAs and control of gene expression in the immune system. Trends Mol. Med. 2014, 20, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Melissari, M.-T.; Grote, P. Roles for long non-coding RNAs in physiology and disease. Pflügers Arch.-Eur. J. Physiol. 2016, 468, 945–958. [Google Scholar] [CrossRef]
- Lennox, K.A.; Behlke, M.A. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016, 44, 863–877. [Google Scholar] [CrossRef]
- Fatima, R.; Akhade, V.S.; Pal, D.; Rao, S.M. Long noncoding RNAs in development and cancer: Potential biomarkers and therapeutic targets. Mol. Cell. Ther. 2015, 3, 5. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, Y.; Huang, G.; Cui, P.; Zhang, W.; Zhang, Y. Long non-coding RNAs in gastric cancer: Versatile mechanisms and potential for clinical translation. Am. J. Cancer Res. 2015, 5, 907–927. [Google Scholar] [PubMed]
- Tam, C.; Wong, J.H.; Tsui, S.K.-W.; Zuo, T.; Chan, T.F.; Ng, T.B. LncRNAs with miRNAs in regulation of gastric, liver, and colorectal cancers: Updates in recent years. Appl. Microbiol. Biotechnol. 2019, 103, 4649–4677. [Google Scholar] [CrossRef] [PubMed]
- Endo, H.; Shiroki, T.; Nakagawa, T.; Yokoyama, M.; Tamai, K.; Yamanami, H.; Fujiya, T.; Sato, I.; Yamaguchi, K.; Tanaka, N.; et al. Enhanced Expression of Long Non-Coding RNA HOTAIR Is Associated with the Development of Gastric Cancer. PLoS ONE 2013, 8, e77070. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-H.; Sun, M.; Nie, F.-Q.; Ge, Y.-B.; Zhang, E.-B.; Yin, D.-D.; Kong, R.; Xia, R.; Lu, K.-H.; Li, J.-H.; et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol. Cancer 2014, 13, 92. [Google Scholar] [CrossRef]
- Liu, Y.-w.; Sun, M.; Xia, R.; Zhang, E.-b.; Liu, X.-h.; Zhang, Z.-h.; Xu, T.-p.; De, W.; Liu, B.-r.; Wang, Z.-x. LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis. 2015, 6, e1802. [Google Scholar] [CrossRef]
- Zhang, Z.-Z.; Shen, Z.-Y.; Shen, Y.-Y.; Zhao, E.-H.; Wang, M.; Wang, C.-J.; Cao, H.; Xu, J. HOTAIR Long Noncoding RNA Promotes Gastric Cancer Metastasis through Suppression of Poly r(C)-Binding Protein (PCBP) 1. Mol. Cancer Ther. 2015, 14, 1162–1170. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Jiang, Q.; Jiang, L.; Wu, J.; Zhang, Q.; Wang, J.; Feng, H.; Zang, P. Lnc-SGK1 induced by Helicobacter pylori infection and highsalt diet promote Th2 and Th17 differentiation in human gastric cancer by SGK1/Jun B signaling. Oncotarget 2016, 7, 20549–20560. [Google Scholar] [CrossRef]
- Zhou, X.; Chen, H.; Zhu, L.; Hao, B.; Zhang, W.; Hua, J.; Gu, H.; Jin, W.; Zhang, G. Helicobacter pylori infection related long noncoding RNA (lncRNA) AF147447 inhibits gastric cancer proliferation and invasion by targeting MUC2 and up-regulating miR-34c. Oncotarget 2016, 7, 82770–82782. [Google Scholar] [CrossRef]
- Li, T.; Mo, X.; Fu, L.; Xiao, B.; Guo, J. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget 2016, 7, 8601–8612. [Google Scholar] [CrossRef]
- Guo, W.; Dong, Z.; Shi, Y.; Liu, S.; Liang, J.; Guo, Y.; Guo, X.; Shen, S.; Wang, G. Methylation-mediated downregulation of long noncoding RNA LOC100130476 in gastric cardia adenocarcinoma. Clin. Exp. Metastasis 2016, 33, 497–508. [Google Scholar] [CrossRef]
- Xie, M.; Nie, F.-Q.; Sun, M.; Xia, R.; Liu, Y.-W.; Zhou, P.; De, W.; Liu, X.-H. Decreased long noncoding RNA SPRY4-IT1 contributing to gastric cancer cell metastasis partly via affecting epithelial-mesenchymal transition. J. Transl. Med. 2015, 13, 250. [Google Scholar] [CrossRef]
- Cao, S.; Lin, L.; Xia, X.; Wu, H. lncRNA SPRY4-IT1 Regulates Cell Proliferation and Migration by Sponging miR-101-3p and Regulating AMPK Expression in Gastric Cancer. Mol. Ther. Nucleic Acids 2019, 17, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Xia, R.; Jin, F.; Xu, T.; Liu, Z.; De, W.; Liu, X. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumor Biol. 2013, 35, 1065–1073. [Google Scholar] [CrossRef]
- Sun, M.; Jin, F.-Y.; Xia, R.; Kong, R.; Li, J.-H.; Xu, T.-P.; Liu, Y.-W.; Zhang, E.-B.; Liu, X.-H.; De, W. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer 2014, 14, 319. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, A.-Y.; Wang, X.-K.; Sun, X.-M.; Xue, H.-Z. GAS5 is downregulated in gastric cancer cells by promoter hypermethylation and regulates adriamycin sensitivity. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3199–3205. [Google Scholar]
- Tsai, K.-W.; Tsai, C.-Y.; Chou, N.-H.; Wang, K.-C.; Kang, C.-H.; Li, S.-C.; Lao, Y.-H.; Chang, H.-T. Aberrant DNA Hypermethylation Silenced LncRNA Expression in Gastric Cancer. Anticancer. Res. 2019, 39, 5381–5391. [Google Scholar] [CrossRef]
- Song, Y.; Wang, R.; Li, L.-W.; Liu, X.; Wang, Y.; Wang, Q.-X.; Zhang, Q. Long non-coding RNA HOTAIR mediates the switching of histone H3 lysine 27 acetylation to methylation to promote epithelial-to-mesenchymal transition in gastric cancer. Int. J. Oncol. 2018, 54, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, X.; Zhang, H.; Sun, L.; Zhou, Y.; Jin, H.; Zhang, H.; Zhang, H.; Liu, J.; Guo, H.; et al. Hypoxia-inducible lncRNA-AK058003 promotes gastric cancer metastasis by targeting γ-synuclein. Neoplasia 2014, 16, 1094–1106. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, R.; Zhang, T.; Dong, X. Hypoxia/lncRNA-AK123072/EGFR pathway induced metastasis and invasion in gastric cancer. Int. J. Clin. Exp. Med. 2015, 8, 19954–19968. [Google Scholar] [PubMed]
- Wang, S.S.; Wuputra, K.; Liu, C.-J.; Lin, Y.-C.; Chen, Y.-T.; Chai, C.-Y.; Lin, C.-L.S.; Kuo, K.-K.; Tsai, M.-H.; Wang, S.-W.; et al. Oncogenic function of the homeobox A13-long noncoding RNA HOTTIP-insulin growth factor-binding protein 3 axis in human gastric cancer. Oncotarget 2016, 7, 36049–36064. [Google Scholar] [CrossRef]
- Sun, M.; Nie, F.; Wang, Y.; Zhang, Z.; Hou, J.; He, D.; Xie, M.; Xu, L.; De, W.; Wang, Z.; et al. LncRNA HOXA11-AS Promotes Proliferation and Invasion of Gastric Cancer by Scaffolding the Chromatin Modification Factors PRC2, LSD1, and DNMT1. Cancer Res. 2016, 76, 6299–6310. [Google Scholar] [CrossRef]
- Miao, H.; Wang, L.; Zhan, H.; Dai, J.; Chang, Y.; Wu, F.; Liu, T.; Liu, Z.; Gao, C.; Li, L.; et al. A long noncoding RNA distributed in both nucleus and cytoplasm operates in the PYCARD-regulated apoptosis by coordinating the epigenetic and translational regulation. PLoS Genet. 2019, 15, e1008144. [Google Scholar] [CrossRef]
- Quan, Y.; Zhang, Y.; Lin, W.; Shen, Z.; Wu, S.; Zhu, C.; Wang, X. Knockdown of long non-coding RNA MAP3K20 antisense RNA 1 inhibits gastric cancer growth through epigenetically regulating miR-375. Biochem. Biophys. Res. Commun. 2018, 497, 527–534. [Google Scholar] [CrossRef]
- Taft, R.J.; Kaplan, C.D.; Simons, C.; Mattick, J.S. Evolution, biogenesis and function of promoter-associated RNAs. Cell Cycle 2009, 8, 2332–2338. [Google Scholar] [CrossRef] [PubMed]
- Moazed, D. Small RNAs in transcriptional gene silencing and genome defence. Nat. Cell Biol. 2009, 457, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Li, L.-C. Chromatin remodeling by the small RNA machinery in mammalian cells. Epigenetics 2013, 9, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Wang, J.; Man, W.-Y.; Zhang, Q.-W.; Xu, W.-G. siRNA Silencing EZH2 Reverses Cisplatin-resistance of Human Non-small Cell Lung and Gastric Cancer Cells. Asian Pac. J. Cancer Prev. 2015, 16, 2425–2430. [Google Scholar] [CrossRef]
- Chalertpet, K.; Pin-On, P.; Aporntewan, C.; Patchsung, M.; Ingrungruanglert, P.; Israsena, N.; Mutirangura, A. Argonaute 4 as an Effector Protein in RNA-Directed DNA Methylation in Human Cells. Front. Genet. 2019, 10, 645. [Google Scholar] [CrossRef]
- Chen, L.; Dahlstrom, J.E.; Lee, S.-H.; Rangasamy, D. Naturally occurring endo-siRNA silences LINE-1 retrotransposons in human cells through DNA methylation. Epigenetics 2012, 7, 758–771. [Google Scholar] [CrossRef]
- Siomi, M.C.; Sato, K.; Pezic, D.; Aravin, A.A. PIWI-interacting small RNAs: The vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011, 12, 246–258. [Google Scholar] [CrossRef]
- Cabral, G.F.; Pinheiro, J.A.D.S.; Vidal, A.F.; Santos, S.; Ribeiro-Dos-Santos, Â. piRNAs in Gastric Cancer: A New Approach Towards Translational Research. Int. J. Mol. Sci. 2020, 21, 2126. [Google Scholar] [CrossRef] [PubMed]
- Chalbatani, G.M.; Dana, H.; Memari, F.; Gharagozlou, E.; Ashjaei, S.; Kheirandish, P.; Marmari, V.; Mahmoudzadeh, H.; Mozayani, F.; Maleki, A.R.; et al. Biological function and molecular mechanism of piRNA in cancer. Pr. Lab. Med. 2019, 13, e00113. [Google Scholar] [CrossRef]
- Iwasaki, Y.; Siomi, M.C.; Siomi, H. PIWI-Interacting RNA: Its Biogenesis and Functions. Annu. Rev. Biochem. 2015, 84, 405–433. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Dou, M.; Song, X.; Dong, Y.; Liu, S.; Liu, H.; Tao, J.; Li, W.; Yin, X.; Xu, W. The emerging role of the piRNA/piwi complex in cancer. Mol. Cancer 2019, 18, 123. [Google Scholar] [CrossRef] [PubMed]
- Esposito, T.; Magliocca, S.; Formicola, D.; Gianfrancesco, F. piR_015520 Belongs to Piwi-Associated RNAs Regulates Expression of the Human Melatonin Receptor 1A Gene. PLoS ONE 2011, 6, e22727. [Google Scholar] [CrossRef]
- Cheng, J.; Guo, J.-M.; Xiao, B.-X.; Miao, Y.; Jiang, Z.; Zhou, H.; Li, Q.-N. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin. Chim. Acta 2011, 412, 1621–1625. [Google Scholar] [CrossRef]
- Brennecke, J.; Malone, C.D.; Aravin, A.A.; Sachidanandam, R.; Stark, A.; Hannon, G.J. An Epigenetic Role for Maternally Inherited piRNAs in Transposon Silencing. Science 2008, 322, 1387–1392. [Google Scholar] [CrossRef]
- Kuramochi-Miyagawa, S.; Watanabe, T.; Gotoh, K.; Totoki, Y.; Toyoda, A.; Ikawa, M.; Asada, N.; Kojima, K.; Yamaguchi, Y.; Ijiri, T.W.; et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008, 22, 908–917. [Google Scholar] [CrossRef]
- Saito, Y.; Suzuki, H.; Tsugawa, H.; Nakagawa, I.; Matsuzaki, J.; Kanai, Y.; Hibi, T. Chromatin remodeling at Alu repeats by epigenetic treatment activates silenced microRNA-512-5p with downregulation of Mcl-1 in human gastric cancer cells. Oncogene 2009, 28, 2738–2744. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Akiyama, Y.; Otsubo, T.; Shimada, S.; Yuasa, Y. Involvement of epigenetically silenced microRNA-181c in gastric carcinogenesis. Carcinogenesis 2010, 31, 777–784. [Google Scholar] [CrossRef]
- Shen, R.; Pan, S.; Qi, S.; Lin, X.; Cheng, S. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 in gastric cancer. Biochem. Biophys. Res. Commun. 2010, 394, 1047–1052. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Chen, X.; Zhang, M.; Fan, Q.; Luo, S.; Cao, X. miR-137 Is Frequently Down-Regulated in Gastric Cancer and Is a Negative Regulator of Cdc42. Dig. Dis. Sci. 2011, 56, 2009–2016. [Google Scholar] [CrossRef]
- Bao, W.; Fu, H.; Xie, Q.; Wang, L.; Zhang, R.; Guo, Z.; Zhao, J.; Meng, Y.; Ren, X.; Wang, T.; et al. HER2 Interacts With CD44 to Up-regulate CXCR4 via Epigenetic Silencing of microRNA-139 in Gastric Cancer Cells. Gastroenterology 2011, 141, 2076–2087.e6. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.-Y.; Li, C.-L.; Nie, H.; Wang, M.; Su, L.-P.; Li, J.-F.; Yu, Y.-Y.; Yan, M.; Qu, Q.-L.; Zhu, Z.-G. microRNA-155 is downregulated in gastric cancer cells and involved in cell metastasis. Oncol. Rep. 2012, 27, 1960–1966. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Guo, Y.; Song, H.; Xiao, B.; Sun, W.; Liu, Z.; Yu, X.; Xia, T.; Cui, L.; Guo, J. MicroRNA-195 and microRNA-378 mediate tumor growth suppression by epigenetical regulation in gastric cancer. Gene 2013, 518, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Shen, J.; Xiao, Z.; Li, J.; Zhao, Y.; Zhao, Q.; Cho, C.H.; Li, M. An overview of the multifaceted roles of miRNAs in gastric cancer: Spotlight on novel biomarkers and therapeutic targets. Biochem. Pharmacol. 2019, 163, 425–439. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zhang, X.; Gu, L.; Zhou, J.; Deng, D. P16 methylation increases the sensitivity of cancer cells to the CDK4/6 inhibitor palbociclib. PLoS ONE 2019, 14, e0223084. [Google Scholar] [CrossRef]
- Link, A.; Kupcinskas, J. MicroRNAs as non-invasive diagnostic biomarkers for gastric cancer: Current insights and future perspectives. World J. Gastroenterol. 2018, 24, 3313–3329. [Google Scholar] [CrossRef]
- Ling, H.; Girnita, L.; Buda, O.; Calin, G.A. Non-coding RNAs: The cancer genome dark matter that matters! Clin. Chem. Lab. Med. 2017, 55, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.-H.; Chiu, C.-T.; Lee, C.; Chu, Y.-Y.; Cheng, H.-T.; Hsu, J.-T.; Wu, R.-C.; Yeh, T.-S.; Lin, K.-H. Circulating microRNA-22-3p Predicts the Malignant Progression of Precancerous Gastric Lesions from Intestinal Metaplasia to Early Adenocarcinoma. Dig. Dis. Sci. 2018, 63, 2301–2308. [Google Scholar] [CrossRef]
- Cai, H.; Yuan, Y.; Hao, Y.-F.; Guo, T.-K.; Wei, X.; Zhang, Y.-M. Plasma microRNAs serve as novel potential biomarkers for early detection of gastric cancer. Med. Oncol. 2013, 30, 452. [Google Scholar] [CrossRef]
- Zhang, L.; Guo, X.; Zhang, D.; Fan, Y.; Qin, L.; Dong, S. Upregulated miR-132 in Lgr5+gastric cancer stem cell-like cells contributes to cisplatin-resistance via SIRT1/CREB/ABCG2 signaling pathway. Mol. Carcinog. 2017, 56, 2022–2034. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, Q.; Cai, X. MicroRNA-106a induces multidrug resistance in gastric cancer by targeting RUNX3. FEBS Lett. 2013, 587, 3069–3075. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2015. CA Cancer J. Clin. 2015, 65, 5–29. [Google Scholar] [CrossRef]
- Zong, L.; Hattori, N.; Yasukawa, Y.; Kimura, K.; Mori, A.; Seto, Y.; Ushijima, T. LINC00162 confers sensitivity to 5-Aza-2′-deoxycytidine via modulation of an RNA splicing protein, HNRNPH1. Oncogene 2019, 38, 5281–5293. [Google Scholar] [CrossRef] [PubMed]
- Piekarz, R.L.; Bates, S.E. Epigenetic Modifiers: Basic Understanding and Clinical Development. Clin. Cancer Res. 2009, 15, 3918–3926. [Google Scholar] [CrossRef]
- Schneider, B.J.; Shah, M.A.; Klute, K.; Ocean, A.; Popa, E.; Altorki, N.; Lieberman, M.; Schreiner, A.; Yantiss, R.; Christos, P.J.; et al. Phase I Study of Epigenetic Priming with Azacitidine Prior to Standard Neoadjuvant Chemotherapy for Patients with Resectable Gastric and Esophageal Adenocarcinoma: Evidence of Tumor Hypomethylation as an Indicator of Major Histopathologic Response. Clin. Cancer Res. 2017, 23, 2673–2680. [Google Scholar] [CrossRef]
- Cheetham, S.; Gruhl, F.; Mattick, J.S.; Dinger, M.E. Long noncoding RNAs and the genetics of cancer. Br. J. Cancer 2013, 108, 2419–2425. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Jiang, C.; Ji, Y.; Xiao, C.; Song, H. Long Noncoding RNAs: New Regulators of Resistance to Systemic Therapies for Gastric Cancer. BioMed Res. Int. 2021, 2021, 8853269. [Google Scholar] [CrossRef]
- Yan, J.; Dang, Y.; Liu, S.; Zhang, Y.; Zhang, G. LncRNA HOTAIR promotes cisplatin resistance in gastric cancer by targeting miR-126 to activate the PI3K/AKT/MRP1 genes. Tumor Biol. 2016, 37, 16345–16355. [Google Scholar] [CrossRef]
- Lan, W.-G.; Xu, D.-H.; Xu, C.; Ding, C.-L.; Ning, F.-L.; Zhou, Y.-L.; Ma, L.-B.; Liu, C.-M.; Chang-Ling, D. Silencing of long non-coding RNA ANRIL inhibits the development of multidrug resistance in gastric cancer cells. Oncol. Rep. 2016, 36, 263–270. [Google Scholar] [CrossRef]
- Li, C.; Feng, Y.; Coukos, G.; Zhang, L. Therapeutic MicroRNA Strategies in Human Cancer. AAPS J. 2009, 11, 747–757. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Dorrance, A.M.; Neviani, P.; Ferenchak, G.J.; Huang, X.; Nicolet, D.; Maharry, K.S.; Ozer, H.G.; Hoellarbauer, P.; Khalife, J.; Hill, E.B.; et al. Targeting leukemia stem cells in vivo with antagomiR-126 nanoparticles in acute myeloid leukemia. Leukemia 2015, 29, 2143–2153. [Google Scholar] [CrossRef]
- Ebert, M.S.; Sharp, P.A. MicroRNA sponges: Progress and possibilities. RNA 2010, 16, 2043–2050. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.; Yeom, C.; Choi, Y.-S.; Kim, S.; Lee, E.; Park, M.J.; Kang, S.W.; Kim, S.B.; Chang, S. Simultaneous inhibition of multiple oncogenic miRNAs by a multi-potent microRNA sponge. Oncotarget 2015, 6, 20370–20387. [Google Scholar] [CrossRef] [PubMed]
- Dizaji, B.F. Strategies to target long non-coding RNAs in cancer treatment: Progress and challenges. Egypt. J. Med. Hum. Genet. 2020, 21, 41. [Google Scholar] [CrossRef]
- Ling, H. Non-coding RNAs: Therapeutic Strategies and Delivery Systems. Single Mol. Single Cell Seq. 2016, 937, 229–237. [Google Scholar] [CrossRef]
- Kurreck, J.; Wyszko, E.; Gillen, C.; Erdmann, V.A. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002, 30, 1911–1918. [Google Scholar] [CrossRef]
- Modarresi, F.; Faghihi, M.A.; Lopez-Toledano, M.A.; Fatemi, R.P.; Magistri, M.; Brothers, S.; Van Der Brug, M.P.; Wahlestedt, C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 2012, 30, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Kole, R.; Krainer, A.R.; Altman, S. RNA therapeutics: Beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012, 11, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Ye, F.; Yin, C.; Zhuang, Y.; Yue, G.; Zhang, G. The Interaction Between MiR-141 and lncRNA-H19 in Regulating Cell Proliferation and Migration in Gastric Cancer. Cell. Physiol. Biochem. 2015, 36, 1440–1452. [Google Scholar] [CrossRef] [PubMed]
- Han, T.-S.; Hur, K.; Xu, G.; Choi, B.; Okugawa, Y.; Toiyama, Y.; Oshima, H.; Oshima, M.; Lee, H.-J.; Kim, V.N.; et al. MicroRNA-29c mediates initiation of gastric carcinogenesis by directly targeting ITGB1. Gut 2015, 64, 203–214. [Google Scholar] [CrossRef]
- Garzon, R.; Marcucci, G.; Croce, C.M. Targeting microRNAs in cancer: Rationale, strategies and challenges. Nat. Rev. Drug Discov. 2010, 9, 775–789. [Google Scholar] [CrossRef]
- Davalos, V.; Esteller, M. MicroRNAs and cancer epigenetics: A macrorevolution. Curr. Opin. Oncol. 2010, 22, 35–45. [Google Scholar] [CrossRef]
- Abba, M.L.; Patil, N.; Leupold, J.H.; Moniuszko, M.; Utikal, J.; Niklinski, J.; Allgayer, H. MicroRNAs as novel targets and tools in cancer therapy. Cancer Lett. 2017, 387, 84–94. [Google Scholar] [CrossRef] [PubMed]
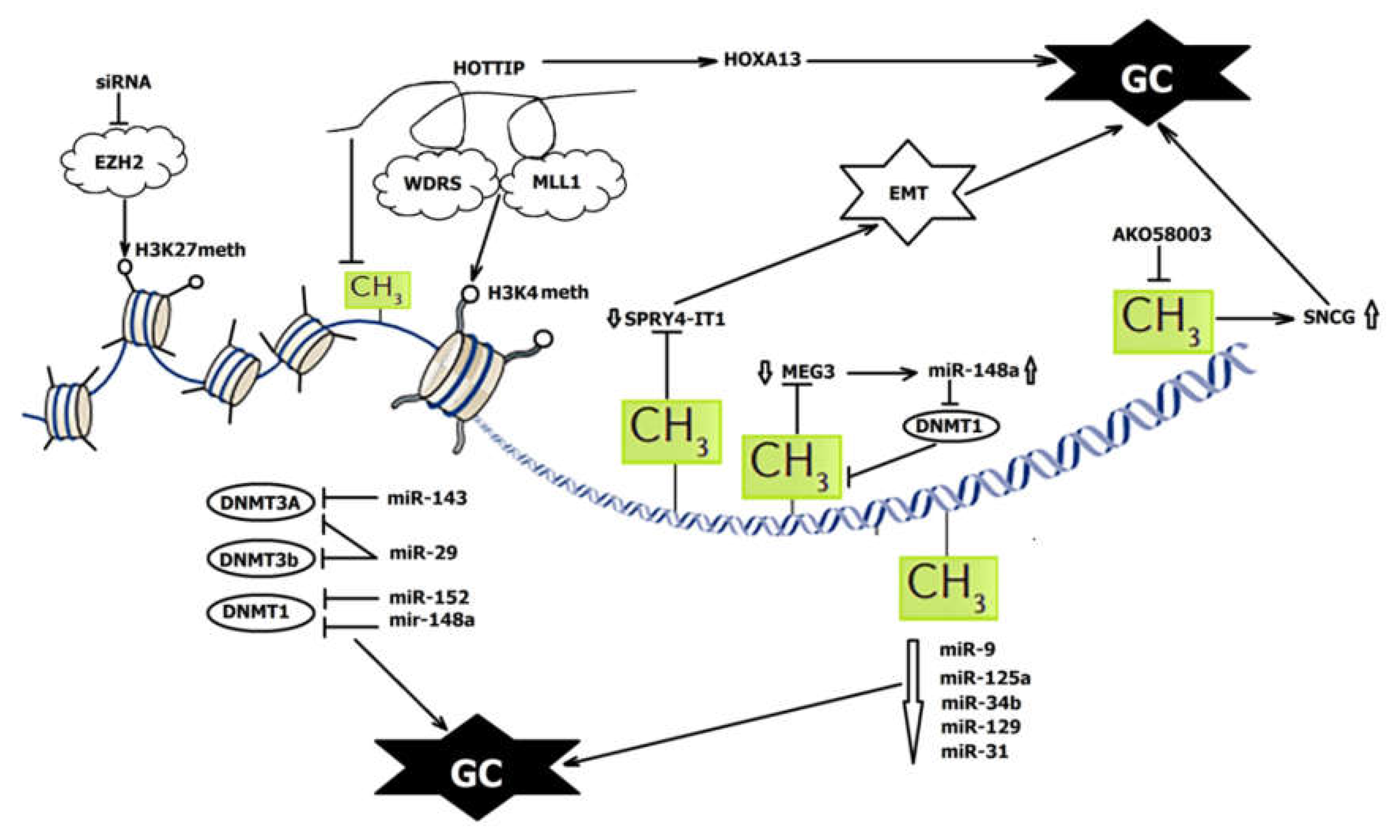
| NcRNA | Status in GC | Interaction with DNA Methylation, Function in GC | References |
|---|---|---|---|
| DNA methylation regulates ncRNAs | |||
| miRNA-34b miRNA-129-3p | Down | Methylated; associated with poor prognosis | [35] |
| miR-9 | Down | Methylated; regulates cell proliferation, migration, and invasion | [36,37,38] |
| miR-125a-5p | Down | Methylated; directly targets HDACs; regulates cell proliferation and migration | [39,40] |
| miR-31 | Down | Methylated; involved in epigenetic feedback loop through directly targeting oncogenic HDAC2; regulates proliferation and apoptosis | [44] |
| miR-124-1 miR-124-2 miR-124-3 | Down | Hypermethylated in GC mucosae with H. pylori infection | [45] |
| miR-512-5p | Down | Methylated; activated upon DNA demethylation at Alu repeats; suppresses Mcl-1, resulting in apoptosis | [99] |
| miR-181c | Down | Methylated; targets oncogenes NOTCH4 and KRAS | [100] |
| miR-129-2 | Down | Methylated; targets SOX4 and, thus, regulates apoptosis | [101] |
| miR-137 | Down | Methylated; targets Cdc42 and, upon reactivation, induces apoptosis and cell-cycle G1 arrest in gastric cancer cells | [102] |
| miR-139 | Down | Methylated; regulates metastases through HER2, CD44, and CXCR4 | [103] |
| miR-148a | Down | Methylated; targets DNMT1 | [51] |
| miR-155 | Down | Methylated; involved in cell metastasis | [104] |
| miR-195 | Down | Methylated; suppresses CDK6 and VEGF signaling | [105] |
| miR-378a | |||
| miRNA-212 | Down | Methylated; targets MYC and, thus, participates in tumorigenesis | [52] |
| SPRY4-IT1 | Down | Regulated by DNMT1-mediated DNA methylation; regulates proliferation, invasion, and EMT | [71] |
| MEG3 | Down | Promoter methylation; regulates proliferation and apoptosis; modulates p53 expression; correlates with invasion and tumor size | [73] |
| GAS5 | Down | Promoter methylation; regulates proliferation and adriamycin sensitivity; correlates with poor prognosis | [74,75] |
| HOXA11-AS | Up | DNA methylation; regulates proliferation and invasion by scaffolding PRC2, LSD1, and DNMT1 | [81] |
| LOC100130476 | Down | Methylation in the CpG islands; associated with pathological differentiation, TNM stage, and survival | [70] |
| NcRNAs regulate DNA methylation | |||
| miR-148a | Down | Regulator of a DNMT3b splice variant; directly targets DNMT1 | [50,51] |
| miR-143 | Directly targets DNMT3a | [47] | |
| miR-29 | Directly targets DNMT3a and DNMT3b; indirectly targets DNMT1 | [48,49] | |
| miR-152 | Directly targets DNMT1 | [50] | |
| HOTAIR | Up | H3K27 trimethylation; inhibits miR-34a and induces EMT; promotes metastasis; correlated with shorter survival | [64,65,66] |
| HOTTIP | Up | Methylation in the CpG islands H3K4 methylation HoxA13 suppression restores the recruitment of DNMT3b | [4,80] |
| AK058003 | Up | Regulates methylation of CpG islands in SNCG; promotes migration, invasion, and metastasis | [79] |
| AK123072 | Up | Regulates methylation of CpG islands in EGFR; promotes migration and invasion | [78] |
| PYCARD-AS1 | Down | Recruit DNMT1 and histone methyltransferase G9a to the PYCARD promoter to regulate apoptosis | [82] |
| MLK7-AS1 | Up | Regulates proliferation and apoptosis; interacts with DNMT1 and recruits it to miR-375, resulting in its hypermethylation and repression of miR-375; correlates with poorer prognosis | [83] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bure, I.V.; Nemtsova, M.V. Methylation and Noncoding RNAs in Gastric Cancer: Everything Is Connected. Int. J. Mol. Sci. 2021, 22, 5683. https://doi.org/10.3390/ijms22115683
Bure IV, Nemtsova MV. Methylation and Noncoding RNAs in Gastric Cancer: Everything Is Connected. International Journal of Molecular Sciences. 2021; 22(11):5683. https://doi.org/10.3390/ijms22115683
Chicago/Turabian StyleBure, Irina V., and Marina V. Nemtsova. 2021. "Methylation and Noncoding RNAs in Gastric Cancer: Everything Is Connected" International Journal of Molecular Sciences 22, no. 11: 5683. https://doi.org/10.3390/ijms22115683
APA StyleBure, I. V., & Nemtsova, M. V. (2021). Methylation and Noncoding RNAs in Gastric Cancer: Everything Is Connected. International Journal of Molecular Sciences, 22(11), 5683. https://doi.org/10.3390/ijms22115683