Abstract
The vegetative phase transition is a prerequisite for flowering in angiosperm plants. Mulberry miR156 has been confirmed to be a crucial factor in the vegetative phase transition in Arabidopsis thaliana. The over-expression of miR156 in transgenic Populus × canadensis dramatically prolongs the juvenile phase. Here, we find that the expression of mno-miR156 decreases with age in all tissues in mulberry, which led us to study the hierarchical action of miR156 in mulberry. Utilizing degradome sequencing and dual-luciferase reporter assays, nine MnSPLs were shown to be directly regulated by miR156. The results of yeast one-hybrid and dual-luciferase reporter assays also revealed that six MnSPLs could recognize the promoter sequences of mno-miR172 and activate its expression. Our results demonstrate that mno-miR156 performs its role by repressing MnSPL/mno-miR172 pathway expression in mulberry. This work uncovered a miR156/SPLs/miR172 regulation pathway in the development of mulberry and fills a gap in our knowledge about the molecular mechanism of vegetative phase transition in perennial woody plants.
1. Introduction
During their post-embryonic development, plants need to go through three vital developmental processes from seed to mature plant, including seed germination, vegetative phase transition (juvenile-to-adult transition), and flower transition [1]. Vegetative phase transition determines the time of plant vegetative growth, and plants must go through sufficient vegetative growth from juvenile to maturity and eventual reproductive growth [2].
SPL genes have been shown to be repressed by miR156, and the miR156/SPLs/miR172 regulation pathway in vegetative phase transition was systematically revealed in Arabidopsis thaliana [3]. From seedlings to mature plants, the expression level of miR156 gradually decreased, while the expression levels of SPLs and miR172 gradually increased in model annual forbs, such as A. thaliana [3], Nicotiana benthamiana [4,5], and rice [6,7]. In A. thaliana, the roots were not involved in vegetative phase transition regulation; however, sugar in the leaves could trigger the vegetative phase transition by the suppression of miR156 expression [4,8,9]. Perennial woody plants had significantly longer vegetative growth times than annual plants, thus it is of more pragmatic value to study the vegetative phase transition mechanism of perennial woody plants [10]. Compared with annual plants, miR156 and miR172 showed similar expression trends in vegetative phase growth in some perennial woody plants such as Acacia confusa, Acacia colei, Eucalyptus globulus, Hedera helix, Quercus acutissima, P. × canadensis and some tropical/subtropical trees, and the over-expression of miR156 in P. × canadensis delayed its vegetative phase change [11,12]. Researchers also found that the expression of SPLs decreased under heavy fruit load in Citrus clementina, and Citrus SPL was able to promote flowering independent of photoperiod in Arabidopsis [13]. EjSPL4, EjSPL5, and EjSPL9 could significantly activate the promoters of EjSOC1-1, EjLFY-1, and EjAP1-1 in loquat and the over-expression of EjSPL3, EjSPL4, EjSPL5, and EjSPL9 in A. thaliana could promote obvious flowering [14]. However, the hierarchical action of miR156 in vegetative phase growth in perennial woody plants is still unknown.
Mulberry is a typical perennial woody plant with very important economic and medicinal values [15,16]. Mulberry trees have been utilized by humans for thousands of years, the leaves are used for raising silkworms, and the fruit is consumed fresh [17]. The medicinal value of mulberry has been explored, and abundant flavonoids and polysaccharides have been detected, especially in the leaves and fruits [18,19]. Mulberry plants also show strong resistance to abiotic stresses, and this crop can significantly improve the eco-environment [20]. Like other perennial woody plant breeding [21], a long juvenile period limits the mulberry resource development and utilization. In recent years, the mulberry genome sequencing has been gradually completed [16,22] and high-throughput sequencing of small RNA also has been finished in some varieties of mulberry [23,24]. All of these have laid a foundation for basic research on mulberry.
The molecular mechanism of the juvenile-to-adult transition in mulberry, however, is completely unknown. In this study, seven miR156s (mno-miR156a to mno-miR156g) and six miR172s (mno-miR172a to mno-miR172f) were identified using small RNAs high-throughput sequencing. Fourteen MnSPL genes were cloned. Our data revealed that miR156c directly repressed the transcription levels of nine MnSPL genes, including MnSPL2, MnSPL15, and MnSPL16A. Six MnSPL genes (MnSPL2, MnSPL8, MnSPL10A, MnSPL10B, MnSPL15, and MnSPL16A) could recognize the GTAC cis-element in the promoter region of mno-miR172a and activate the expression of mno-miR172a based on yeast one-hybrid experiments. An existing miR156/SPLs/miR172 pathway involved in the mulberry vegetative phase transition has been proposed.
2. Results
2.1. Mulberry miR156 Is Differentially Expressed in Juvenile and Mature Phase
In mulberry, the miR156 family has seven members named miR156a–g with mno-miR156c having a prominently higher expression than other mno-miR156s (Table 1). Sequence alignment and phylogenetic tree analysis showed that mulberry miR156s shared high sequence similarity, except miR156a. For example, there are only four base differences in miR156b–g (Figure 1a,b).

Table 1.
miR156 and miR172 in mulberry.
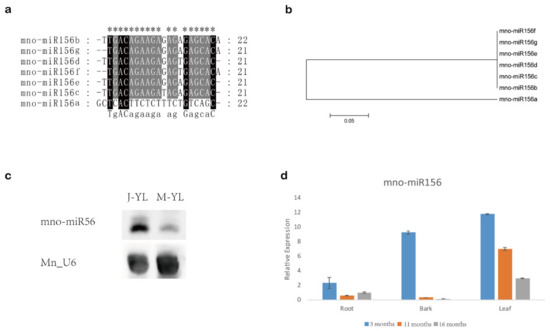
Figure 1.
miR156 in mulberry. (a) Sequence alignment of miR156 in mulberry. “*” shows conserved sites. (b) Neighbor-joining (N.J.) phylogenetic tree of miR156 in mulberry. One thousand bootstrap replicates were used. (c) Northern blot showing the expression of miR156 in young leaves of juvenile (J.-Y.L.) and mature (M.-Y.L.) mulberry. Mn_U6 was used as a reference gene. (d) The expression profile of miR156 in three tissues (roots, bark, and leaves) at different time periods (3, 11, and 16 months) in mulberry. Values represent the mean ± SD from three biological replicates.
The Northern blot showed that miR156 had higher expression levels in young leaves from juvenile phase mulberry than in young leaves from mature phase mulberry (Figure 1c). Small RNA RT-qPCR revealed that the expression of miR156 gradually decreased with age in the roots, barks, and leaves of mulberry (Figure 1d). These results suggested that miR156 might be associated with vegetative phase transition in mulberry.
2.2. Identification of the miR156 Target Genes in Mulberry
Our results from in silico prediction showed that 115 genes from the mulberry genes database (https://morus.swu.edu.cn/, accessed on 3 March 2019) were selected as the presumptive miR156 target genes (Table S1). GO analysis showed that over half of the presumptive miR156 target genes were clustered into the secondary level “binding” group in the first level molecular function group (Figure 2).
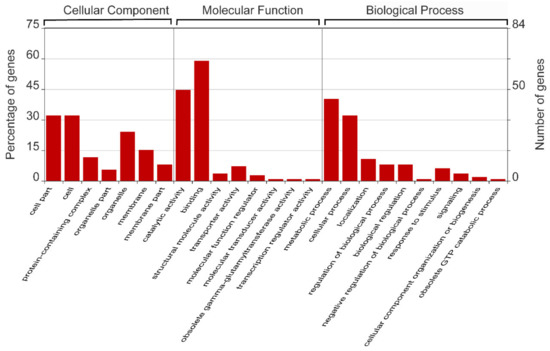
Figure 2.
Gene ontology analysis of the predicted target genes of miR156 in mulberry.
For further analysis of these presumptive target genes, we counted those genes whose expectation values were less than 2.5, and found seven SPL genes (Morus010792, Morus010123, Morus015493, Morus018032, Morus014488, Morus017456, and Morus026457), three WD40 repeat-like genes (Morus026689, Morus002371, and Morus011698), one fructose 1,6-diphosphatase gene (Morus017876) and two genes (Morus021290 and Morus006566) with no annotation (Table 2). The results from the degradome sequencing verified seven SPL genes, including MnSPL2, MnSPL4, MnSPL6, MnSPL13, MnSPL15, MnSPL16A, and MnSPL16B, with the p values less than 0.05. The other genes with p values more than 0.05 were not considered as miR156 target genes (Figure 3 and Table 3).

Table 2.
Predicted target genes of miR156 in mulberry (Expectation ≤ 2.5).
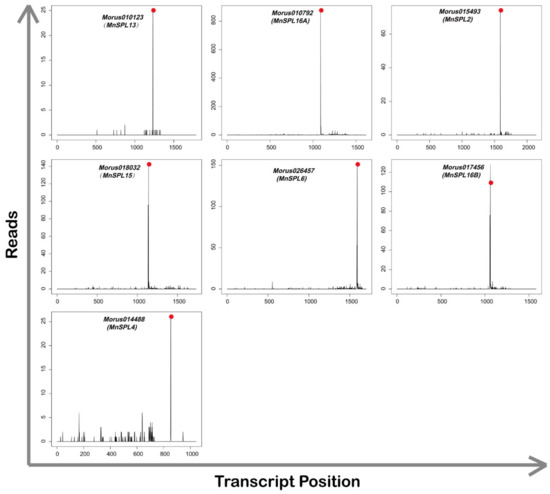
Figure 3.
T-plot diagram of miR156 target genes verified by degradome sequencing. Red dots show the splice site of miR156 target genes.

Table 3.
Target genes of miR156 verified by degradome sequencing.
Except for MnSPLs, no other genes were identified by degradome sequencing, which demonstrated that the SPLs genes were the main or exclusive target genes of miR156 in mulberry. Moreover, our results from the RT-qPCR showed that all of MnSPLs except for MnSPL2 had a higher expression level in elder leaves, suggesting the opposite expression profile to mno-miR156 (Figure 4a,b).
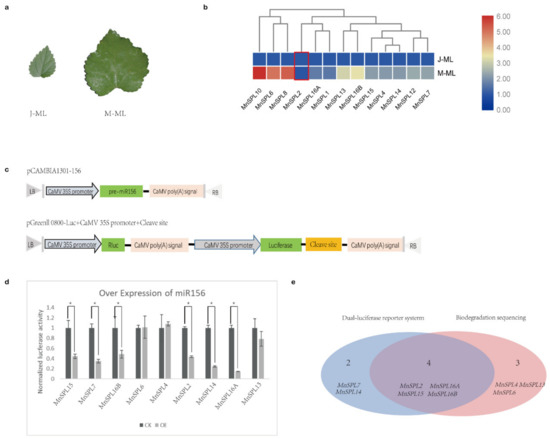
Figure 4.
Identification of miR156 target genes in mulberry. (a) Morphological photos of mature leaves of juvenile (J-ML) and mature (M-ML) mulberry. (b) The expression trend of MnSPLs in mature leaves of juvenile (J-ML) and mature (M-ML) mulberry measured by RT-qPCR. Red boxes indicate there were no significant differences between this sample. Heatmap was drawn using TBtools, and the min–max normalization method was used to normalize the data. (c) Schematic of plasmids used in dual-luciferase assays. (d) The target genes of miR156 identified by dual-luciferase assays. CK: control check, empty pGreenII 0800-Luc vector and pCAMBIA-156; OE: reconstructed pGreenII 0800-Luc vector and pCAMBIA-156. Values represent the mean ± SD from three biological replicates and were statistically analyzed (independent-samples t-test): *, p < 0.05. (e) Venn diagrams showing miR156 target genes in mulberry.
Next, all of MnSPL genes predicted as miR156 target genes by psRNATarget were tested using dual-luciferase assays. Compared to the CK group, the pGreenII 0800 LUC vector with the supposed miR156 splice site of six MnSPLs (MnSPL2, MnSPL7, MnSPL14, MnSPL15, MnSPL16A, and MnSPL16B) showed significantly lower luciferase activity when miR156 was over-expressed (Figure 4c,d). We integrated the results of degradome sequencing and dual-luciferase assays, and identified nine MnSPLs (MnSPL2, MnSPL4, MnSPL6, MnSPL7, MnSPL13, MnSPL14, MnSPL15, MnSPL16A, and MnSPL16B) as miR156 target genes in mulberry (Figure 4e).
2.3. MnSPLs Bound to the Promoter of mno-miR172a and Promoted Its Expression in Mulberry
The result of the BLASTN and HMMER search showed there were 15 MnSPL genes in mulberry (Figure S1a and Table S3). All of the 15 MnSPL genes had the SBP conserved domains (Figure S1a). The neighbor-joining (NJ) phylogenetic tree for the SPL genes from M. notabilis, P. trichocarpa, and A. thaliana showed that the MnSPL genes were separately distributed to five groups (Figure S1b). Tissue expression profiles showed that MnSPL genes were differentially expressed in five tissues (winter bud, male flower, root, branch bark, and leaf). MnSPL6, MnSPL8, MnSPL10, MnSPL15, and MnSPL16A had significantly high expression in winter buds, while MnSPL2, MnSPL7, and MnSPL16B had much higher expression in male flowers. MnSPL1, MnSPL13, and MnSPL14 were mainly expressed in the root, and MnSPL4 and MnSPL12 showed much higher expression in the leaf and branch bark, respectively (Figure S2). Six highly conserved mno-miR172s were identified in mulberry (Figure 5a,b), and the expression of mno-miR172a was significantly higher than the mno-miR172s in mulberry roots, barks, and leaves (Table 1).
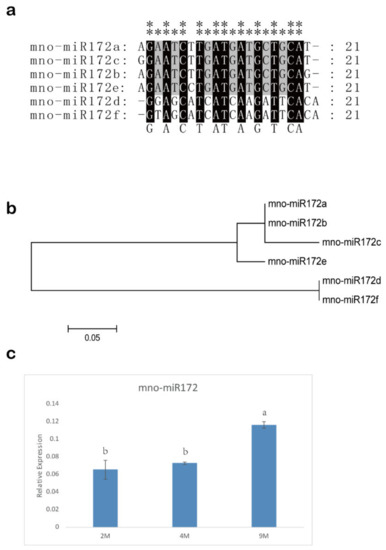
Figure 5.
miR172 in mulberry. (a) Sequence alignment of miR1172 in mulberry. “**” shows highly conserved sites with no base difference, “*” indicates low conservation sites with two base differences. (b) Neighbor-joining (NJ) phylogenetic tree of miR172 in mulberry. One thousand bootstrap replicates were used. (c) The expression profile of miR172 in leaves at 2 months (2 M), 4 months (4 M) and 9 months (9 M) in mulberry. Values represent the mean ± SD of three biological replicates and were statistically analyzed (one-way ANOVA test): p < 0.05.
The miR172 had a higher expression level in 9-month-old leaves than in younger leaves, which indicated that miR172 might be involved in the regulation of vegetative development in mulberry (Figure 5c). GUS activity was detected in CK, OE-156, and STTM156cd samples, indicating that genes were over-expressed by transient transgenic technology (Figure 6a,b). The over-expression of miR156 in mulberry leaves significantly decreased the expression of miR172 and MnSPL genes, except for MnSPL1 and MnSPL12, while the down regulation of miR156 by STTM in mulberry seedlings significantly increased the expression of miR172 and most MnSPL genes except for MnSPL1, MnSPL8, MnSPL12, and MnSPL14 (Figure 6).
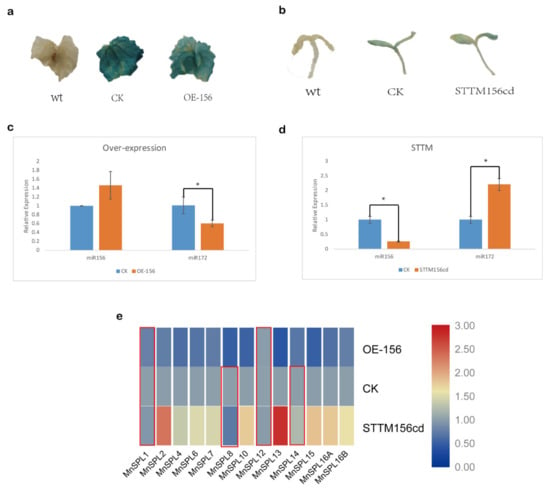
Figure 6.
Altered miR156 expression in mulberry. (a,b) GUS staining of mulberry leaves and seedlings, wt: wild type; CK: control check, the empty pCAMBIA1301 vector; OE-156: over-expression of miR156; STTM 156cd: repression of mno-miR156c and mno-miR156d by short tandem target mimic (STTM). (c,d) The expression level of miR156 and miR172 under over-expression and reduced expression of miR156 in mulberry. Values represent the mean ± SD of three biological replicates and were statistically analyzed (independent-samples t-test): *, p < 0.05. (e) The expression profile of SPL genes under over-expression and reduced expression of miR156 in mulberry. Red box indicates there were no significant differences between these samples. Heatmap was finished using TBtools, and the min–max normalization method was utilized to normalize the data.
We wondered if MnSPLs acted as upstream regulators of mno-miR172 and how MnSPLs functioned in this process. Thus, yeast one-hybrid assays were carried out to identify which MnSPL genes could recognize the predetermined GTAC elements in the mno-miR172a promoter. Except for three genes (MnSPL6, MnSPL8, and MnSPL12), which were toxic in yeast, the remaining 11 MnSPL genes were tested by the yeast one-hybrid assay, and six of those genes (MnSPL2, MnSPL7, MnSPL10A, MnSPL10B, MnSPL15, and MnSPL16A) could grow normally on the –Leu/A150 screening medium (Figure 7a,b).
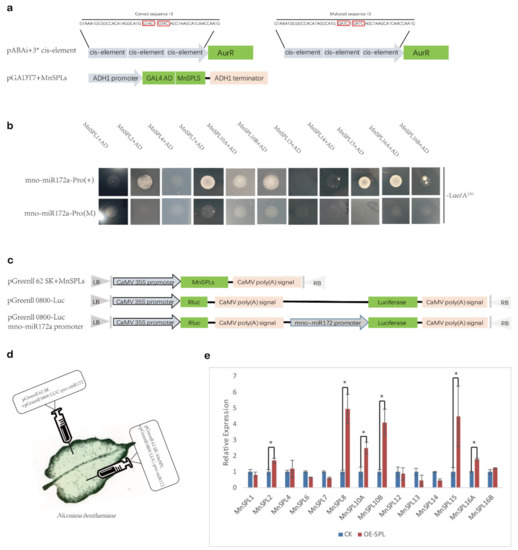
Figure 7.
MnSPLs promote miR172 expression in mulberry. (a) Schematic of plasmids used in yeast one-hybrid assays. Red boxes show the sites of cis-elements in the promoter of miR172, which can be recognized by MnSPLs. (b) Yeast one-hybrid assay identified the interaction between MnSPLs and the promoter of miR172. (+) means a correct sequence of the cis-element. (M) means a mutated sequence of the cis-element. (c) Schematic of plasmids used in dual-luciferase assays. (d) Schematic diagram of dual-luciferase assays. (e) Dual-luciferase assays identified the interaction between MnSPLs and the promoter of miR172. CK: control check. OE-SPL: over-expression of SPL genes. Values represent the mean ± SD from three biological replicates and were statistically analyzed (independent-samples t-test): *, p < 0.05.
Then, we used dual-luciferase assays to further verify these predicted miR156 target MnSPL genes (Table S2) and found that six MnSPL genes (MnSPL2, MnSPL8, MnSPL10A, MnSPL10B, MnSPL15, and MnSPL16A) could increase luciferase activity. Taking the results of the yeast one-hybrid and dual-luciferase assays into consideration, we confirmed that six MnSPL genes (MnSPL2, MnSPL8, MnSPL10A, MnSPL10B, MnSPL15, and MnSPL16A) directly bound to the promoter of mno-miR172a and promoted its expression in mulberry.
3. Discussion
Woody plants were favorable systems for studying vegetative phase change because the stability and prolonged duration of the various stages of shoot development make these phases easy to observe and characterize [25]. However, the stability and prolonged duration of the vegetative stages limit breeding and genetic research in perennial woody plants. The miR156 over-expression can suppress vegetative phase transition and promote vegetative growth in poplar [11]. To date, however, researchers are still unclear about the molecular mechanism of vegetative phase development in perennial woody plants due to the limitations of the genetic research. Therefore, understanding the molecular mechanism of this process in perennial woody plants has very important research and practical value.
The sequence of miR156 was shown to be highly conserved among angiosperms [26]. Small RNA sequencing in mulberry showed that the expression level of mno-miR156c in three tissues (leaves, bark and male flowers) was significantly higher than other miR156 family members, and mno-miR156c also was shown to have the same sequences as miR156 found in A. thaliana, G. max, and M. domestica [23]. Similar to the discovery in A. thaliana [3] and P. trichocarpa [11], miR156 was expressed at higher levels in juvenile mulberry tissues than in mature mulberry tissues, which suggested that this conserved miRNA was probably involved in the vegetative phase transition in mulberry. In A. thaliana, AtSPLs were downstream factors of miR156-regulated vegetative stage development [3,27,28]. The SPL family gene function as promoters of vegetative phase transition and are distributed across angiosperms [29,30,31,32]. In total, there are 16 AtSPL genes in A. thaliana and 10 members (AtSPL2, AtSPL3, AtSPL4, AtSPL5, AtSPL6, AtSPL9, AtSPL10, AtSPL11, AtSPL13, and AtSPL15) were the miR156 target genes [3]. A total of 15 MnSPL genes were predicted by BLASTN and HUMMER modeling in MorusDB, and 14 gene sequences with an SBP-box domain were cloned successfully (MnSPL10 has two alternative spliceosomes named MnSPL10A and MnSPL10B, but MnSPL3 and MnSPL5 were not cloned). A total of nine MnSPLs (MnSPL2, MnSPL4, MnSPL6, MnSPL7, MnSPL13, MnSPL14, MnSPL15, MnSPL16A, and MnSPL16B) were identified as miR156-targeted genes in mulberry in the present work. This finding implied that the SPL genes were the exclusive miR156 target genes across herbs and woody plants. The miR156/SPLs modules can also control the development of lateral roots and miR156-targeted SPL10-controlled root meristem activity in A. thaliana [29,33]. We also found three MnSPL genes (MnSPL1, MnSPL13, and MnSPL14) prominently expressed in the root, which implied that these genes might be involved in root development regulation.
As a conserved miRNA, miR172 participated in the regulation of flowering and was regulated by SBP-box transcription factors in A. thaliana [34,35]. Additionally, the miR172 expression level increased with age in A. thaliana and P. × canadensis [11,36]. In A. thaliana, SPL9/10 activates the vegetative phase transition by promoting miR172 expression [3], while the over-expression of miR172 in A. thaliana can increase the expression of SPL3/4/5 [37]. In mulberry, there were six mature miR172 transcripts, named mno-miR172a–f, and six MnSPL genes (MnSPL2, MnSPL8, MnSPL10A, MnSPL10B, MnSPL15, and MnSPL16A) were verified as the direct upstream regulators of mno-miR172a. Additional miR172-upstream MnSPL genes were identified in mulberry, which suggested there were more complex regulatory relationships between SPLs and miR172 in mulberry.
In summary, we found that the expression of miR156 subsequently reduced as mulberry aged, while SPLs and miR172 showed the opposite profile. A total of nine MnSPL genes as exclusive miR156 target genes were verified and six MnSPL genes could promote the expression of mno-miR172a in mulberry. The over-expression of miR156 reduced the expression levels of miR172 and SPL genes, except for two miR156 non-target genes (MnSPL1 and MnSPL12) and decreasing the expression level of miR156 by STTM increased the miR172 and SPL genes expression levels, except for three non-target genes of miR156 (MnSPL1, MnSPL12, and MnSPL14). All of these experimental results indicated that the miR156/SPLs/miR172 regulation network also existed in mulberry, which implied this regulation pathway was conserved across angiosperms (Figure 8).
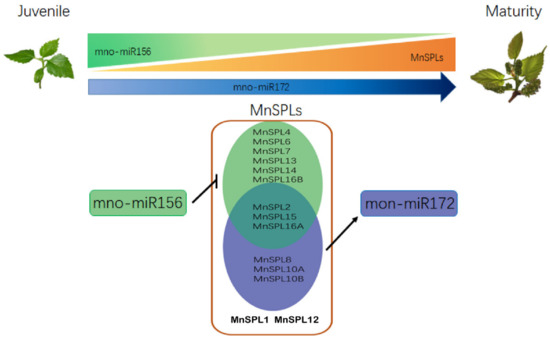
Figure 8.
The molecular mechanism of miR156 in mulberry development. The larger area and darker color indicate higher gene expression level.
Our work should facilitate the integration of information about vegetative phase transition across perennial woody plants and give significant insight into how to reduce the breeding cycle of perennial woody plants.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Tobacco seeds (N. tabacum L) were planted in sterilized soil and left at 4 °C for 2 d before transfer to a climate chamber at 25 °C under a long-day condition (16 h light/8 h dark), as were mulberry seedlings (Morus atropurpurea cv. Guiyou 62, abbreviated as GY62, which can bloom after 13 months). The older mulberry trees were grown on the open balcony of our laboratory. Plant age was measured from the date that seeds were transferred to the growth chamber.
Various mulberry tissues were used to analyze the expression of miRNAs and genes in this work. Young leaves, barks, and roots from 3-month-old (3 M), 11-month-old (11 M) and 16-month-old (16 M) mulberry were used to detect the expression of miR156 in mulberry by RT-qPCR. Young leaves from juvenile phase mulberry (J-YL, 3 months old) and young leaves of mature phase mulberry (M-YL, 16 months old) were used to analyze miR156 expression level by small RNA Northern blot assay. Young leaves of 2-month-old (2 M), 4-month-old (4 M), and 9-month-old (9 M) mulberry were used to detect the expression profile of miR172 in mulberry by RT-qPCR. Mature leaves from juvenile phase mulberry (J-ML) and mature leaves from mature phase mulberry (M-ML) were used to investigate the expression level of MnSPLs.
4.2. Bioinformatics Analysis of miR156, SPL Genes, and miR172 in Mulberry
We obtained sequence information from miR156s and miR172s in Morus notabilis from the NCBI/SRA database under accession number SRP032829. BLAST and HUMMER searches of SPL genes in MorusDB (https://morus.swu.edu.cn/hmmer, accessed on 3 March 2019) were carried out using AtSPL as the query sequence. An SPL gene structure diagram of mulberry was analyzed using Pfam (http://pfam.sanger.ac.uk, accessed on 19 March 2019) and GSDS 2.0 (http://gsds.cbi.pku.edu.cn/index.php, accessed on 19 March 2019). Additionally, 28 full-length PtSPLs [38], known AtSPL genes (TAIR, https://www.arabidopsis.org/, accessed on 30 March 2019), and 15 predicted MnSPLs were used to build a neighbor-joining (NJ) phylogenetic tree using MEGA5.1, and the branching reliability was assessed by the bootstrap re-sampling method using 1000 bootstrap replicates. The target genes of miR156 in mulberry were predicted by psRNATarget (http://plantgrn.noble.org/psRNATarget/, accessed on 3 March 2019) and the expectation value was limited to 4.0. GO analysis of predicted miR156 target genes was performed using the online software WEGO (https://wego.genomics.cn/, accessed on 23 December 2020).
4.3. Epression Analysis of miRNAs and Genes
Small RNAs (<200 bp) and total RNA were both extracted using the miRcute Plant miRNA Isolation Kit purchased from TIANGEN (DP504, Beijing, China, OD260/280 of all RNA samples were determined using a NanoDrop2000 (Thermo Scientific, Waltham, MA, USA), and agarose gel electrophoresis (AGE) was used to verify the integrity of RNA samples (data not shown). RNA probes were labeled with DIG by Riboprobe® Systems (P1440, Promoga, Madison, WI, USA) and purified using an EasyPure® RNA Purification Kit (ER701-1, TransGen Biotech, Beijing, China). Northern blots were performed as follows: twenty micrograms of small RNA were isolated on a 15% Ura-PAGE gel, transferred to a membrane using a Semi-Dry Transfer apparatus, cross-linked using ultraviolet crosslinking, and visualized using the protocol of a DIG DNA Labeling and Detection Kit (11093657910, Roche, Switzerland). Small RNA reverse transcriptions were performed using the miRNA First Strand cDNA Synthesis Kit (tailing reaction) (B532451, Sangon Biotech, Shanghai, China). The levels of mature miR156 and miR172 were quantified using the MicroRNAs qPCR Kit (SYBR Green method) (B532461, Sangon Biotech, Shanghai, China). mno-miR166b and MnU6 were selected as reference genes together in our small RNA RT-qPCRs. Heatmaps were made using TBbtools [39], and the mimax normalization method was utilized to normalize this data. The PrimeScript™ RT reagent Kit with gDNA Eraser (perfect real time) (RR047Q, Takara, Japan) was used to reverse transcribe total RNA to cDNA. Quantitative reverse transcription polymerase chain reaction (RT-qPCR) was performed to analyze the transcript abundance of various SPL genes using the TB Green® Premix Ex Taq™ II (Tli RNaseH Plus) (RR820Q, Takara, Japan). All primers and probes used in this work are listed in Table S4.
4.4. Transient Trnsgene Expression in Mulberry
An 800-bp-predicted pre-miR156 sequence was cloned from the genome of GY62 into the pCAMBIA1301 vector to over-express mature miR156. Short tandem target mimicking (STTM) was performed to repress the expression of mature miR156. The STTM vectors were constructed as described previously [40,41]. For STTMd vector, three extra bases (cta) were inserted into the reverse complement sequences of mno-miR156c and mno-miR156d between the 11th and 12th bases. These two modified sequences were inserted into the end of 88-bp spacer sequence which could form the hairpin structure. pCAMBIA1301 was used to over-express this construct. The protocol of transient transgene expression of pCAMBIA-156 and pCAMBIA-STTM156cd in mulberry was as described in [42]. For GUS staining aay, samples were incubated with staining buffer (G3061, Solarbio, Beijing, China) at 37 °C for 12 h. The stained samples were washed by 75% (v/v) ethanol and photographed using a Leica microscope. All the primers and probes used in this work are listed in Table S4.
4.5. Dual-Luciferase Repoter Assay
A 200-bp sequence containing the predicted cleavage of target SPL genes was cloned into the pGreenII 0800-LUC vector, which had the multiple cloning site after the 3′ end of the luciferase gene. The resultant vector was verified by sequencing. pCAMBIA-156 was used in dual-luciferase assays to over-express mature mno-miR156c. Vector structures are showed in (Figure 4c). Agrobacterium-mediated co-transformation of the pGreenII 0800-LUC and pCAMBIA-156 vectors into tobacco leaves was performed as described previously [43]. After infiltration fothree days, the ratio of LUC to REN activity was measured using the Dual-Luciferase Reporter Gene Assay Kit (11402ES80, Yeasen, Shanghai, China) on a configurable multi-mode microplate reader (Synergy™ H1, BioTek, Beijing, China). MnSPL geneloned into theGreenII 62-SK vector. A 1500-bp promoter sequence from mno-miR172a predicted by Promoter 2.0 Prediction Server (http://www.cbs.dtu.dk/services/Promoter/, accessed on 31 May 2020) was amplified and cloned into the pGreenII 0800-LUC vector. Vector structures are shown in (Figure 7c). Agrobacterium-mediated co-transformation and LUC/REN activity detection were processed as described above. All the primers and probes used in this work are listed in Table S4.
4.6. Yeast One-Hybrid Assay
The fulllength sequences of SPL genes were amplified and then cloned into the pGADT7 vector. Because the full length of three genes were over 3000 bp, only partial sequences (1000 bp) from Morus024784, Morus013868 and Morus025152 were amplified. A 50-bp sequence containing a predicted cis-acting element was repeated three times and cloned into the pAbAi vector. A mutant cis-acting element (modified from GTACTGTAC to GATCTGATC) was used as a negative control. A verified concentration (150 ng/mL) of Aureobasidin A (AbA) was used to screen positive yeast strains in a yeast one-hybrid system. The assays were performed using the Matchmaker® Gold Yeast One-Hybrid System (630491, Takara, Japan) according to the manufacturer’s instructions. All the primers and probes used in this work are listed in Table S4.
4.7. Degradome Sequencing
We selectedroots, leaves, and bark from juvenile mulberry and winter buds from mature mulberry to constructed a degradome library. The purified cDNA library was sequenced on an Illumina HiSeq2000 Sequencing System (LC-BIO Sciences, Beijing, China).
Supplementary Materials
The Supplementary Materials are available online at https://www.mdpi.com/article/10.3390/ijms22115550/s1.
Author Contributions
H.L. and N.H. conceived the study; N.H. conducted experiments; H.L., Y.L., J.H. and W.W. performed the main experiments. H.L., B.M., Z.L., H.H. and J.Y. analyzed the data; H.L. and N.H. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Key Research and Development Program of China (no. 2019YFD1001200), National Natural Science Foundation of China (no. 31572323), Fundamental Research Funds for the Central Universities (no. XDJK2019C088), and National Natural Science Foundation of China (no. 32001328).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: [SRP032829 in NCBI/SRA database, https://morus.swu.edu.cn/, https://www.arabidopsis.org/, http://www.phytozome.net/poplar.php.
Acknowledgments
We appreciate Han Li and Changying Liu for supporting the transient gene expression in mulberry. We are grateful for the technology support of degradome sequencing from the LC-Bio company.
Conflicts of Interest
We declare that we have no conflict of interest.
References
- Baurle, I.; Dean, C. The timing of developmental transitions in plants. Cell 2006, 125, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Poethig, R.S. Vegetative phase change and shoot maturation in plants. Curr. Top. Dev. Biol. 2013, 105, 125–152. [Google Scholar] [CrossRef]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef]
- Yang, L.; Conway, S.R.; Poethig, R.S. Vegetative phase change is mediated by a leaf-derived signal that represses the transcription of miR156. Development 2011, 138, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wang, J.; Zhou, C. The role of miR156 in developmental transitions in Nicotiana tabacum. Sci. China Life Sci. 2015, 58, 253–260. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xie, K.; Wu, C.; Xiong, L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 2006, 142, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, S.; Jin, J.; Fu, D.; Yang, X.; Weng, X.; Xu, C.; Li, X.; Xiao, J.; Zhang, Q. Coordinated regulation of vegetative and reproductive branching in rice. Proc. Natl. Acad. Sci. USA 2015, 112, 15504–15509. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Cao, L.; Zhou, C.M.; Zhang, T.Q.; Lian, H.; Sun, Y.; Wu, J.; Huang, J.; Wang, G.; Wang, J.W. Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. eLife 2013, 2, e00269. [Google Scholar] [CrossRef]
- Yang, L.; Xu, M.; Koo, Y.; He, J.; Poethig, R.S. Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. eLife 2013, 2, e00260. [Google Scholar] [CrossRef]
- Lawson, E.J.; Poethig, R.S. Shoot development in plants: Time for a change. Trends Genet. 1995, 11, 263–268. [Google Scholar] [CrossRef]
- Wang, J.W.; Park, M.Y.; Wang, L.J.; Koo, Y.; Chen, X.Y.; Weigel, D.; Poethig, R.S. miRNA control of vegetative phase change in trees. PLoS Genet. 2011, 7, e1002012. [Google Scholar] [CrossRef]
- Ahsan, M.U.; Hayward, A.; Irihimovitch, V.; Fletcher, S.; Tanurdzic, M.; Pocock, A.; Beveridge, C.A.; Mitter, N. Juvenility and Vegetative Phase Transition in Tropical/Subtropical Tree Crops. Front. Plant Sci. 2019, 10, 729. [Google Scholar] [CrossRef]
- Shalom, L.; Shlizerman, L.; Zur, N.; Doron-Faigenboim, A.; Blumwald, E.; Sadka, A. Molecular characterization of Squamosa Promoter Binding Protein-Like (SPL) gene family from Citrus and the effect of fruit load on their expression. Front. Plant Sci. 2015, 6, 389. [Google Scholar] [CrossRef]
- Jiang, Y.; Peng, J.; Wang, M.; Su, W.; Gan, X.; Jing, Y.; Yang, X. The Role of EjSPL3, EjSPL4, EjSPL5, and EjSPL9 in Regulating Flowering in Loquat (Eriobotrya japonica Lindl.). Int. J. Mol. Sci. 2019, 21, 248. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.P.; Ou, T.T.; Wang, C.J. Mulberry (sang shèn zǐ) and its bioactive compounds, the chemoprevention effects and molecular mechanisms in vitro and in vivo. J. Tradit. Complement. Med. 2013, 3, 7–15. [Google Scholar] [CrossRef] [PubMed]
- He, N.; Zhang, C.; Qi, X.; Zhao, S.; Tao, Y.; Yang, G.; Lee, T.H.; Wang, X.; Cai, Q.; Li, D.; et al. Draft genome sequence of the mulberry tree Morus notabilis. Nat. Commun. 2013, 4, 2445. [Google Scholar] [CrossRef]
- Yuan, Q.; Zhao, L. The Mulberry (Morus alba L.) Fruit-A Review of Characteristic Components and Health Benefits. J. Agric. Food Chem. 2017, 65, 10383–10394. [Google Scholar] [CrossRef]
- Chen, H.; Pu, J.; Liu, D.; Yu, W.; Shao, Y.; Yang, G.; Xiang, Z.; He, N. Anti-Inflammatory and Antinociceptive Properties of Flavonoids from the Fruits of Black Mulberry (Morus nigra L.). PLoS ONE 2016, 11, e0153080. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Yu, W.; Chen, G.; Meng, S.; Xiang, Z.; He, N. Antinociceptive and Antibacterial Properties of Anthocyanins and Flavonols from Fruits of Black and Non-Black Mulberries. Molecules 2017, 23, 4. [Google Scholar] [CrossRef]
- Liu, C.; Xu, Y.; Feng, Y.; Long, D.; Cao, B.; Xiang, Z.; Zhao, A. Ectopic Expression of Mulberry G-Proteins Alters Drought and Salt Stress Tolerance in Tobacco. Int. J. Mol. Sci. 2018, 20, 89. [Google Scholar] [CrossRef]
- Yamagishi, N.; Kishigami, R.; Yoshikawa, N. Reduced generation time of apple seedlings to within a year by means of a plant virus vector: A new plant-breeding technique with no transmission of genetic modification to the next generation. Plant Biotechnol. J. 2014, 12, 60–68. [Google Scholar] [CrossRef]
- Jiao, F.; Luo, R.; Dai, X.; Liu, H.; Yu, G.; Han, S.; Lu, X.; Su, C.; Chen, Q.; Song, Q.; et al. Chromosome-Level Reference Genome and Population Genomic Analysis Provide Insights into the Evolution and Improvement of Domesticated Mulberry (Morus alba). Mol. Plant 2020, 13, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Zhang, D.; Qi, X.; Ma, B.; Xiang, Z.; He, N. Identification of the conserved and novel miRNAs in Mulberry by high-throughput sequencing. PloS ONE 2014, 9, e104409. [Google Scholar] [CrossRef]
- Wu, P.; Han, S.; Zhao, W.; Chen, T.; Zhou, J.; Li, L. Genome-wide identification of abiotic stress-regulated and novel microRNAs in mulberry leaf. Plant Physiol Biochem 2015, 95, 75–82. [Google Scholar] [CrossRef]
- Poethig, R.S. The past, present, and future of vegetative phase change. Plant Physiol. 2010, 154, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J.; Bowman, J.L. Evolution of plant microRNAs and their targets. Trends Plant Sci. 2008, 13, 343–349. [Google Scholar] [CrossRef]
- Zheng, C.; Ye, M.; Sang, M.; Wu, R. A Regulatory Network for miR156-SPL Module in Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 166. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Hu, T.; Zhao, J. Developmental Functions of miR156-Regulated Squamosa Promoter Binding Protein-Like (SPL) Genes in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006263. [Google Scholar] [CrossRef] [PubMed]
- Barrera-Rojas, C.H.; Rocha, G.H.B.; Polverari, L.; Pinheiro Brito, D.A.; Batista, D.S.; Notini, M.M.; da Cruz, A.C.F.; Morea, E.G.O.; Sabatini, S.; Otoni, W.C.; et al. miR156-targeted SPL10 controls Arabidopsis root meristem activity and root-derived de novo shoot regeneration via cytokinin responses. J. Exp. Bot. 2020, 71, 934–950. [Google Scholar] [CrossRef] [PubMed]
- Nanda, S.; Hussain, S. Genome-wide identification of the SPL gene family in Dichanthelium oligosanthes. Bioinformation 2019, 15, 165–171. [Google Scholar] [CrossRef]
- Zhu, T.; Liu, Y.; Ma, L.; Wang, X.; Zhang, D.; Han, Y.; Ding, Q.; Ma, L. Genome-wide identification, phylogeny and expression analysis of the SPL gene family in wheat. BMC Plant Biol. 2020, 20, 420. [Google Scholar] [CrossRef]
- Shao, F.; Lu, Q.; Wilson, I.W.; Qiu, D. Genome-wide identification and characterization of the SPL gene family in Ziziphus jujuba. Gene 2017, 627, 315–321. [Google Scholar] [CrossRef]
- Yu, N.; Niu, Q.W.; Ng, K.H.; Chua, N.H. The role of miR156/SPLs modules in Arabidopsis lateral root development. Plant J. 2015, 83, 673–685. [Google Scholar] [CrossRef]
- Wang, J.W. Regulation of flowering time by the miR156-mediated age pathway. J. Exp. Bot. 2014, 65, 4723–4730. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 2004, 303, 2022–2025. [Google Scholar] [CrossRef] [PubMed]
- Aukerman, M.J.; Sakai, H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Seo, P.J.; Kang, S.K.; Park, C.M. miR172 signals are incorporated into the miR156 signaling pathway at the SPL3/4/5 genes in Arabidopsis developmental transitions. Plant Mol. Biol. 2011, 76, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Lu, S. Molecular characterization of the SPL gene family in Populus trichocarpa. BMC Plant Biol. 2014, 14, 131. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Yan, J.; Gu, Y.; Qiao, M.; Fan, R.; Mao, Y.; Tang, X. Construction of short tandem target mimic (STTM) to block the functions of plant and animal microRNAs. Methods 2012, 58, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, J.; Yan, J.; Gou, F.; Mao, Y.; Tang, G.; Botella, J.R. Short tandem target mimic rice lines uncover functions of miRNAs in regulating important agronomic traits. Proc. Natl. Acad. Sci. USA 2017, 114, 5277–5282. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, Z.; Zeng, Q.; Wang, S.; Luo, Y.; Huang, Y.; Xin, Y.; He, N. Abnormal expression of bHLH3 disrupts a flavonoid homeostasis network, causing differences in pigment composition among mulberry fruits. Hortic. Res. 2020, 7, 83. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhu, P.; Fan, W.; Feng, Y.; Kou, M.; Hu, J.; Zhao, A. Functional analysis of drought and salt tolerance mechanisms of mulberry RACK1 gene. Tree Physiol. 2019, 39, 2055–2069. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).