Expression of Cholinergic Markers and Characterization of Splice Variants during Ontogenesis of Rat Dorsal Root Ganglia Neurons
Abstract
1. Introduction
2. Results
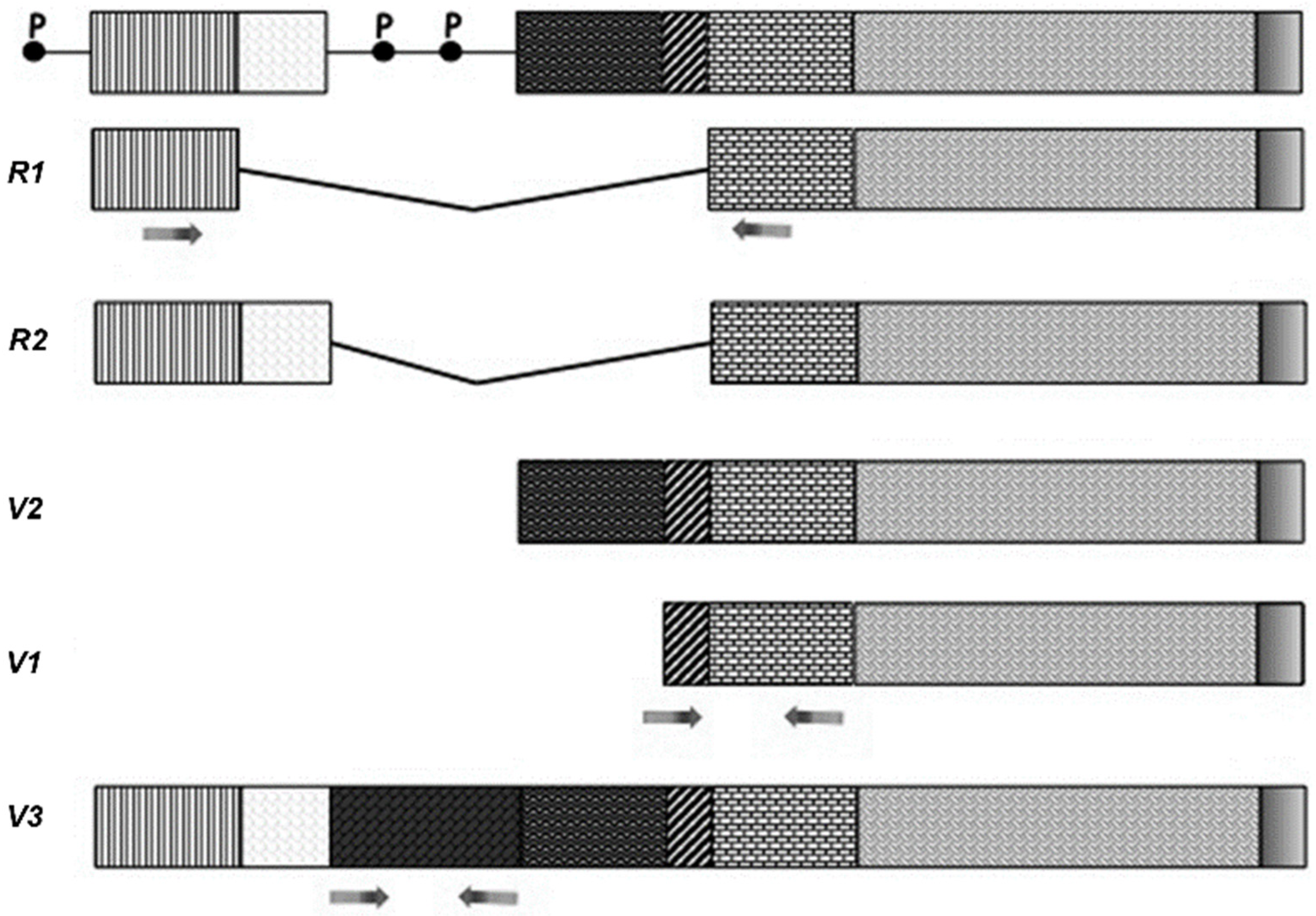
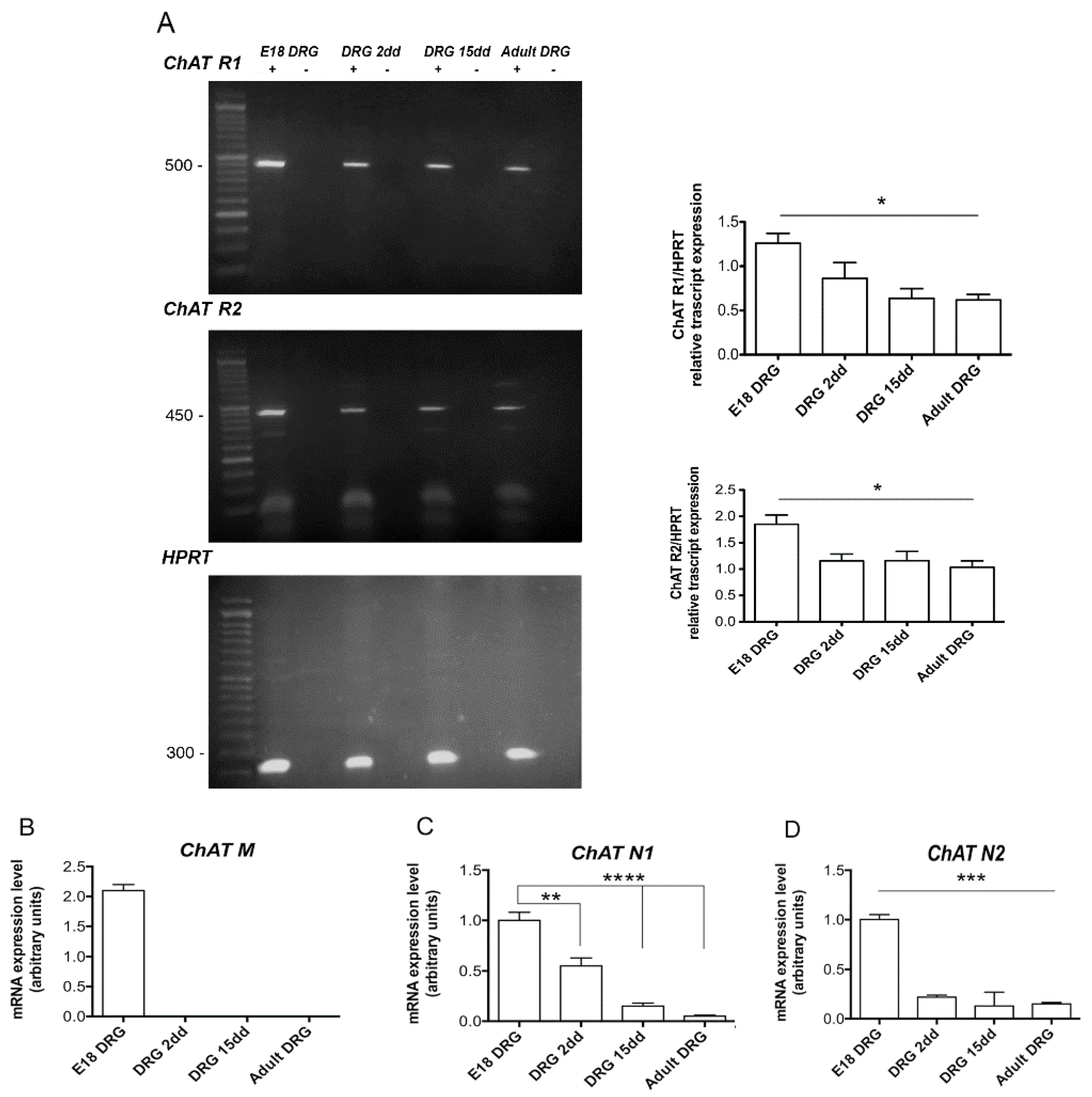
2.1. Analysis of VAChT and ChAT Splice Variants
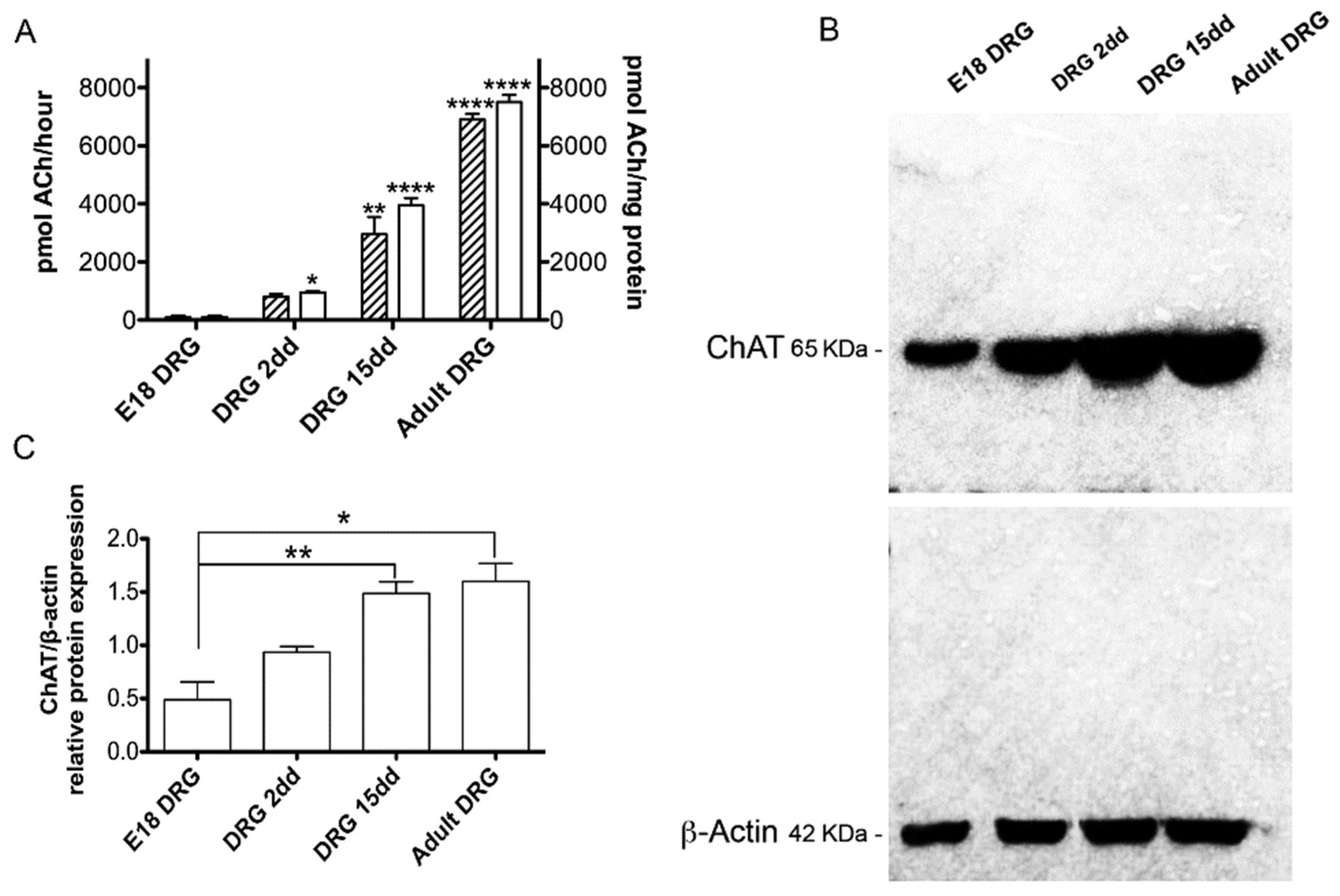
2.2. ChAT Protein Expression
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. RNA Extraction
4.3. RT-PCR and qRT- PCR Analysis
4.3.1. RT–PCR Analysis
4.3.2. qRT- PCR Analysis
4.3.3. Primers
4.4. Protein Extraction and Western Blot Analysis
4.5. ChAT Assay
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Ach | Acetylcholine |
| cAMP | Cyclic adenosine monophosphate |
| ChAT | Choline acetyltransferase |
| CGRP | Calcitonin gene related peptide |
| CNTF | Ciliary neurotrophic factor |
| DRG | Dorsal root ganglion |
| FBS | Fetal bovine serum |
| GAPDH | Glyceraldehyde phosphodehydrogenase |
| HPRT | Hypoxanthine phosphoribosyl transferase |
| NG | Neuroblastoma-glioma |
| NGF | Nerve growth factor |
| RA | Retinoic acid |
| SDS | Sodium dodecyl sulphate |
| VAChT | Vesicular acetylcholine transporter |
References
- Eglen, R.M. Muscarinic receptor subtypes in neuronal and non-neuronal cholinergic function. Auton. Autocoid Pharmacol. 2005, 26, 219–233. [Google Scholar] [CrossRef]
- Wessler, I.; Kirkpatrick, C.J. Acetylcholine beyond neurons: The non-neuronal cholinergic system in humans. Br. J. Pharmacol. 2008, 154, 1558–1571. [Google Scholar] [CrossRef]
- Horiuchi, Y.; Kimura, R.; Kato, N.; Fujii, T.; Seki, M.; Endo, T.; Kato, T.; Kawashima, K. Evolutional study on acetylcholine expression. Life Sci. 2003, 72, 1745–1756. [Google Scholar] [CrossRef]
- Biagioni, S.; Tata, A.M.; De Jaco, A.; Augusti-Tocco, G. Acetylcholine synthesis and neuron differentiation. Int. J. Dev. Biol. 2000, 44, 689–697. [Google Scholar] [PubMed]
- Abreu-Villaça, Y.; Filgueiras, C.C.; Manhães, A.C. Developmental aspects of the cholinergic system. Behav. Brain Res. 2011, 221, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Asrican, B.; Paez-Gonzalez, P.; Erb, J.; Kuo, C.T. Cholinergic circuits control of postnatal neurogenesis. Neurogenesis 2016, 3, e1127310. [Google Scholar] [CrossRef]
- Paez-Gonzalez, P.; Asrican, B.; Rodriguez, E.; Kuo, C.T. Identification of distinct ChAT+ neurons and activity dependent control of postnatal SVZ neurogenesis. Nat. Neurosci. 2014, 17, 934–942. [Google Scholar] [CrossRef]
- Tata, A.M.; De Stefano, M.E.; Srubek Tomassy, G.; Vilarò, M.T.; Levey, A.I.; Biagioni, S. Subpopulations of rat dorsal root ganglion neurons express active vesicular acetylcholine transporter. J. Neurosci. Res. 2004, 75, 194–202. [Google Scholar] [CrossRef]
- Tata, A.M.; Cursi, S.; Biagioni, S.; Augusti-Tocco, G. Cholinergic modulation of neurite outgrowth and neurofilament expression in developing chick sensory neurons. J. Neurosci. Res. 2003, 73, 227–234. [Google Scholar] [CrossRef]
- Salani, M.; Anelli, T.; Augusti-Tocco, G.; Lucarini, E.; Mozzetta, C.; Poiana, G.; Tata, A.M.; Biagioni, S. Acetylcholine-induced neuronal differentiation: Muscarinic receptor activation regulates EGR-1 and REST expression in neuroblastoma cells. J. Neurochem. 2009, 108, 821–834. [Google Scholar] [CrossRef] [PubMed]
- Corsetti, V.; Mozzetta, C.; Biagioni, S.; Augusti-Tocco, G.; Tata, A.M. Acetylcholine release: The mechanisms and the site of release during chick dorsal root ganglia ontogenesis. Life Sci. 2012, 91, 783–788. [Google Scholar] [CrossRef]
- Eiden, L.E. The cholinergic gene locus. J. Neurochem. 1998, 70, 2227–2240. [Google Scholar] [CrossRef]
- Mathews, E.A.; Mullen, G.P.; Manjarrez, J.R.; Rand, J.B. Unusual Regulation of Splicing of the Cholinergic Locus in Caenorhabditis elegans. Genetics 2015, 199, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Candiani, S.; Lacalli, T.C.; Parodi, M.; Oliveri, D.; Pestarino, M. The cholinergic gene locus in Amphioxus: Molecular characterization and developmental expression patterns. Dev. Dyn. 2008, 237, 1399–1411. [Google Scholar] [CrossRef] [PubMed]
- Berse, B.; Blusztajn, J.K. Coordinated up-regulation of choline acetyltransferase and vesicular acetylcholine transporter gene expression by the retinoic acid receptor alpha, cAMP, and leukemia inhibitory factor/ciliary neurotrophic factor signaling pathways in a murine septal cell line. J. Biol. Chem. 1995, 270, 22101–22104. [Google Scholar]
- Castell, X.; Cheviron, N.; Barnier, J.V.; Diebler, M.F. Exploring the regulation of the expression of VAChT and ChAT genes in NG108-15 cells: Implication of PKA and PI3K signalling pathways. Neurochem. Res. 2003, 28, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Yamamuro, Y.; Aizawa, S. Asymmetric regulation by estrogen at the cholinergic locus in differentiated NG108-15 neuronal cells. Life Sci. 2010, 86, 839–843. [Google Scholar] [CrossRef]
- Biagioni, S.; Tata, A.M.; Agrati, C.; Cianfarani, F.; Augusti-Tocco, G. Nerve growth factor modulation of cholinergic marker expression in dorsal root ganglia. J. Neurosci. Res. 2000, 62, 591–599. [Google Scholar] [CrossRef]
- Tata, A.M.; Plateroti, M.; Cibati, M.; Biagioni, S.; Augusti-Tocco, G. Cholinergic markers are expressed during development of chick dorsal root ganglia. J. Neurosci. Res. 1994, 37, 247–255. [Google Scholar] [CrossRef]
- Tata, A.M.; Tripiciano, A.; Filippini, A.; Biagioni, S.; Augusti-Tocco, G. Muscarinic receptors modulate intracellular calcium level in chick sensory neurons. Brain Res. 2000, 866, 65–72. [Google Scholar] [CrossRef]
- Tata, A.M.; Vilaró, M.T.; Mengod, G. Muscarinic receptor subtypes expression in rat and chick dorsal root ganglia. Brain Res. Mol. Brain Res. 2000, 82, 1–10. [Google Scholar] [CrossRef]
- Biagioni, S.; Tata, A.M.; Augusti-Tocco, G. Expression of cholinergic system components in dorsal root ganglion (DRG) neurons: Its possible dual role in development and nociception. Recent Res. Devel. Neurochem. 1999, 2, 443–461. [Google Scholar]
- Bernardini, N.; Srubek Tomassy, G.; Tata, A.M.; Augusti-Tocco, G.; Biagioni, S. Detection of basal and potassium-evoked acetylcholine release from embryonic DRG explants. J. Neurochem. 2004, 88, 1533–1539. [Google Scholar] [CrossRef] [PubMed]
- Bernardini, N.; De Stefano, M.E.; Tata, A.M.; Biagioni, S.; Augusti-Tocco, G. Neuronal and non-neuronal cell populations of the avian dorsal root ganglia express muscarinic acetylcholine receptors. Int. J. Dev. Neurosci. 1998, 16, 365–377. [Google Scholar] [CrossRef]
- Tata, A.M.; Vilaró, M.T.; Agrati, C.; Biagioni, S.; Mengod, G.; Augusti-Tocco, G. Expression of muscarinic M2 receptor mRNA in dorsal root ganglia of neonatal rat. Brain Res. 1999, 824, 63–70. [Google Scholar] [CrossRef][Green Version]
- Loreti, S.; Vilaró, M.T.; Visentin, S.; Rees, H.; Levey, A.I.; Tata, A.M. Rat Schwann cells express M1–M4 muscarinic receptor subtypes. J. Neurosci. Res. 2006, 84, 97–105. [Google Scholar] [CrossRef]
- Bellier, J.P.; Kimura, H. Acetylcholine synthesis by choline acetyltransferase of peripheral type as demonstrated in adult rat dorsal root ganglion. J. Neurochem. 2007, 101, 1607–1618. [Google Scholar] [CrossRef]
- Elnasharty, M.A.; Sayed-Ahmed, A. Expression and localization of pChAT as a novel method to study cholinergic innervation of rat adrenal gland. Acta Histochem. 2014, 116, 1382–1389. [Google Scholar] [CrossRef]
- Nadorp, B.; Soreq, H. Predicted overlapping microRNA regulators of acetylcholine packaging and degradation in neuroinflammation-related disorders. Front. Mol. Neurosci. 2014, 7, 9. [Google Scholar] [CrossRef]
- Greenberg, D.S.; Soreq, H. MicroRNA therapeutics in neurological disease. Curr. Pharm. Des. 2014, 20, 6022–6027. [Google Scholar] [CrossRef]
- Loreti, S.; Ricordy, R.; De Stefano, M.E.; Augusti-Tocco, G.; Tata, A.M. Acetylcholine inhibits cell cycle progression in cultured rat Schwann cells by activation of M2 receptor subtype. Neuron Glia Biol. 2007, 3, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Uggenti, C.; De Stefano, M.E.; Costantino, M.; Loreti, S.; Pisano, A.; Avallone, B.; Talora, C.; Magnaghi, V.; Tata, A.M. M2 muscarinic receptor activation addresses Schwann cell differentiation and myelin organization. Dev. Neurobiol. 2014, 74, 676–691. [Google Scholar] [CrossRef] [PubMed]
- Magnaghi, V.; Procacci, P.; Tata, A.M. Novel pharmacological approaches to Schwann cells as neuroprotective agents for peripheral nerve regeneration. Int. Rev. Neurobiol. 2009, 87, 295–315. [Google Scholar] [PubMed]
- De Angelis, F.; Marinelli, S.; Fioretti, B.; Catacuzzeno, L.; Franciolini, F.; Pavone, F.; Tata, A.M. M2 receptors exert analgesic action on DRG sensory neurons by negatively modulating VR1 activity. J. Cell Physiol. 2014, 229, 783–790. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, F.; Tata, A.M. Analgesic effects mediated by muscarinic receptors: Mechanisms and pharmacological approaches. Cent. Nerv. Syst. Agents Med. Chem. 2016, 16, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Bernardini, N.; Roza, C.; Sauer, S.K.; Gomeza, J.; Wess, J.; Reeh, P.W. Muscarinic M2 receptors on peripheral nerve endings: A molecular target of antinociception. J. Neurosci. 2002, 22, RC229. [Google Scholar] [CrossRef] [PubMed]
- Volpicelli, F.; Perrone-Capano, C.; Da Pozzo, P.; Colucci-D’Amato, L.; di Porzio, U. Modulation of Nurr-1 gene expression in mesenchephalic dopaminergic neurones. J. Neurochem. 2004, 88, 1283–1294. [Google Scholar] [CrossRef] [PubMed]
- Cervini, R.; Houhou, L.; Pradat, P.F.; Béjanin, S.; Mallet, J.; Berrard, S. Specific vesicular acetylcholine transporter promoters lie within the first intron of the rat choline acetyltransferase gene. J. Biol. Chem. 1995, 270, 24654–24657. [Google Scholar] [CrossRef] [PubMed]
- Kengaku, M.; Misawa, H.; Deguchi, T. Multiple mRNA species of choline acetyltransferase from rat spinal cord. Brain Res. Mol. Brain Res. 1993, 18, 71–76. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corsetti, V.; Perrone-Capano, C.; Salazar Intriago, M.S.; Botticelli, E.; Poiana, G.; Augusti-Tocco, G.; Biagioni, S.; Tata, A.M. Expression of Cholinergic Markers and Characterization of Splice Variants during Ontogenesis of Rat Dorsal Root Ganglia Neurons. Int. J. Mol. Sci. 2021, 22, 5499. https://doi.org/10.3390/ijms22115499
Corsetti V, Perrone-Capano C, Salazar Intriago MS, Botticelli E, Poiana G, Augusti-Tocco G, Biagioni S, Tata AM. Expression of Cholinergic Markers and Characterization of Splice Variants during Ontogenesis of Rat Dorsal Root Ganglia Neurons. International Journal of Molecular Sciences. 2021; 22(11):5499. https://doi.org/10.3390/ijms22115499
Chicago/Turabian StyleCorsetti, Veronica, Carla Perrone-Capano, Michael Sebastian Salazar Intriago, Elisabetta Botticelli, Giancarlo Poiana, Gabriella Augusti-Tocco, Stefano Biagioni, and Ada Maria Tata. 2021. "Expression of Cholinergic Markers and Characterization of Splice Variants during Ontogenesis of Rat Dorsal Root Ganglia Neurons" International Journal of Molecular Sciences 22, no. 11: 5499. https://doi.org/10.3390/ijms22115499
APA StyleCorsetti, V., Perrone-Capano, C., Salazar Intriago, M. S., Botticelli, E., Poiana, G., Augusti-Tocco, G., Biagioni, S., & Tata, A. M. (2021). Expression of Cholinergic Markers and Characterization of Splice Variants during Ontogenesis of Rat Dorsal Root Ganglia Neurons. International Journal of Molecular Sciences, 22(11), 5499. https://doi.org/10.3390/ijms22115499






