METTL8 mRNA Methyltransferase Enhances Cancer Cell Migration via Direct Binding to ARID1A
Abstract
1. Introduction
2. Results
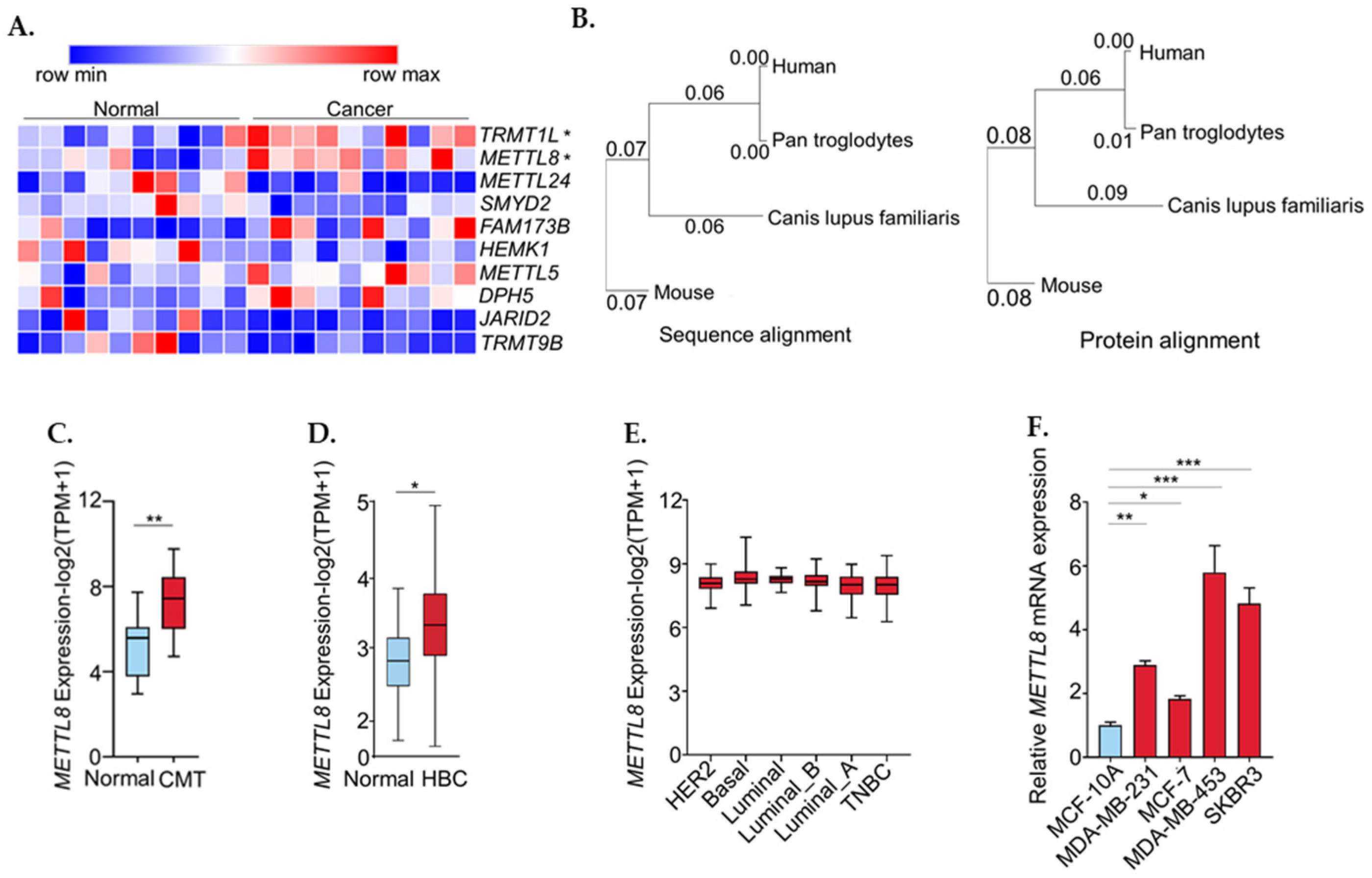
2.1. METTL8 Expression Was Up-Regulated in Breast Cancer and Canine Mammary Tumor
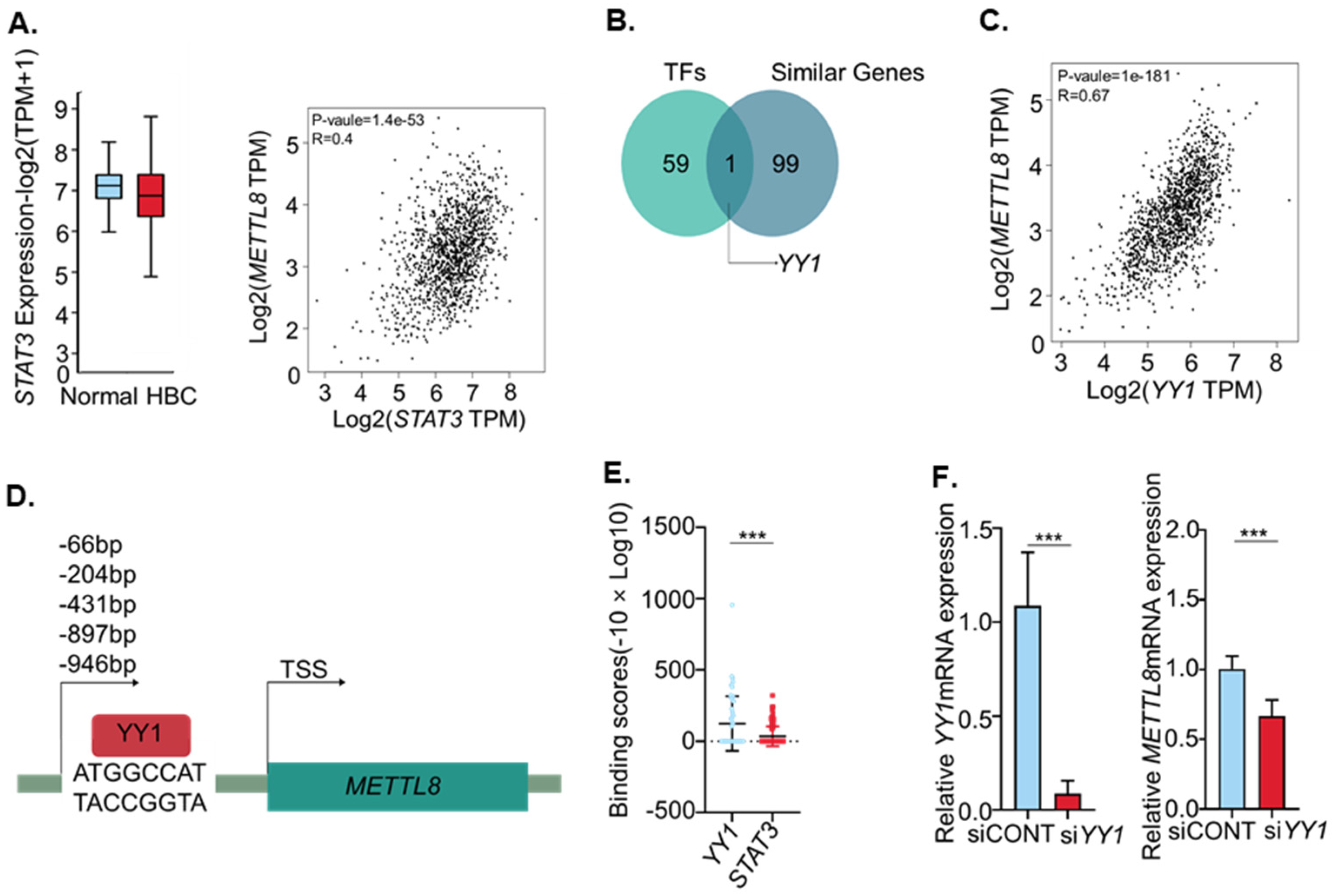
2.2. YY1 Is an Important Transcription Factor of METTL8 in Breast Cancer
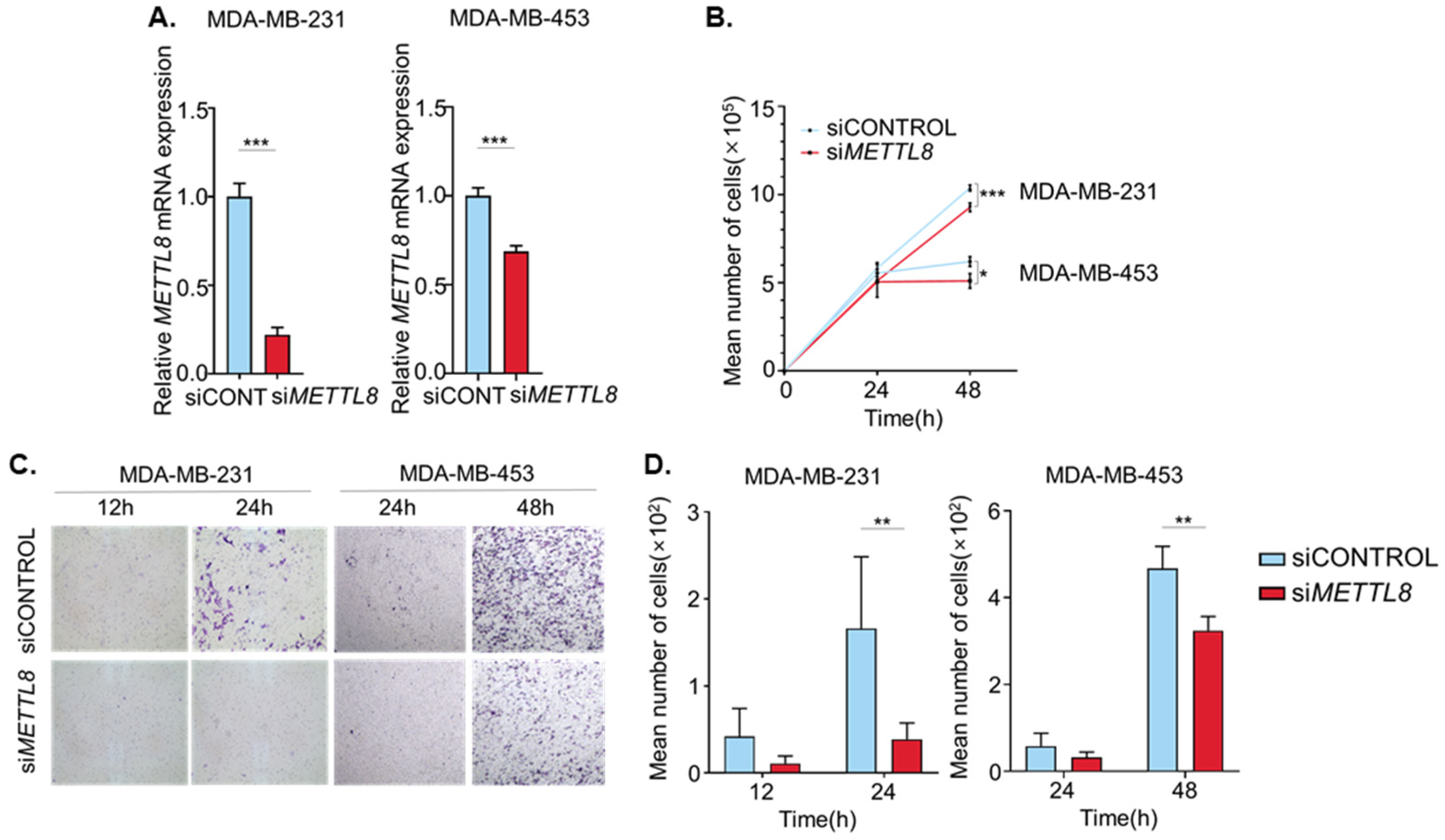
2.3. METTL8 Knockdown Decreased Proliferation and Migration of Breast Cancer Cell Line
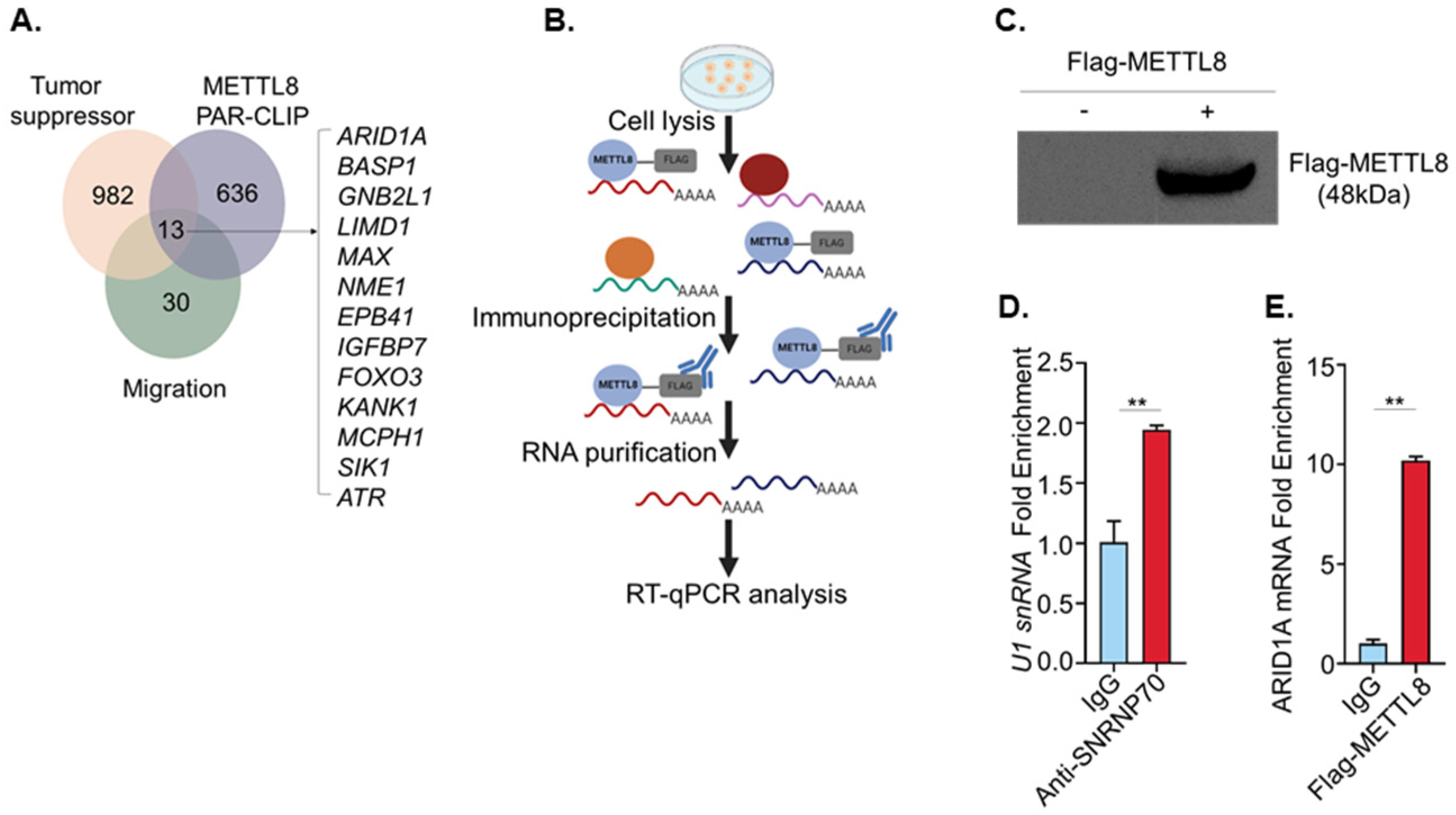
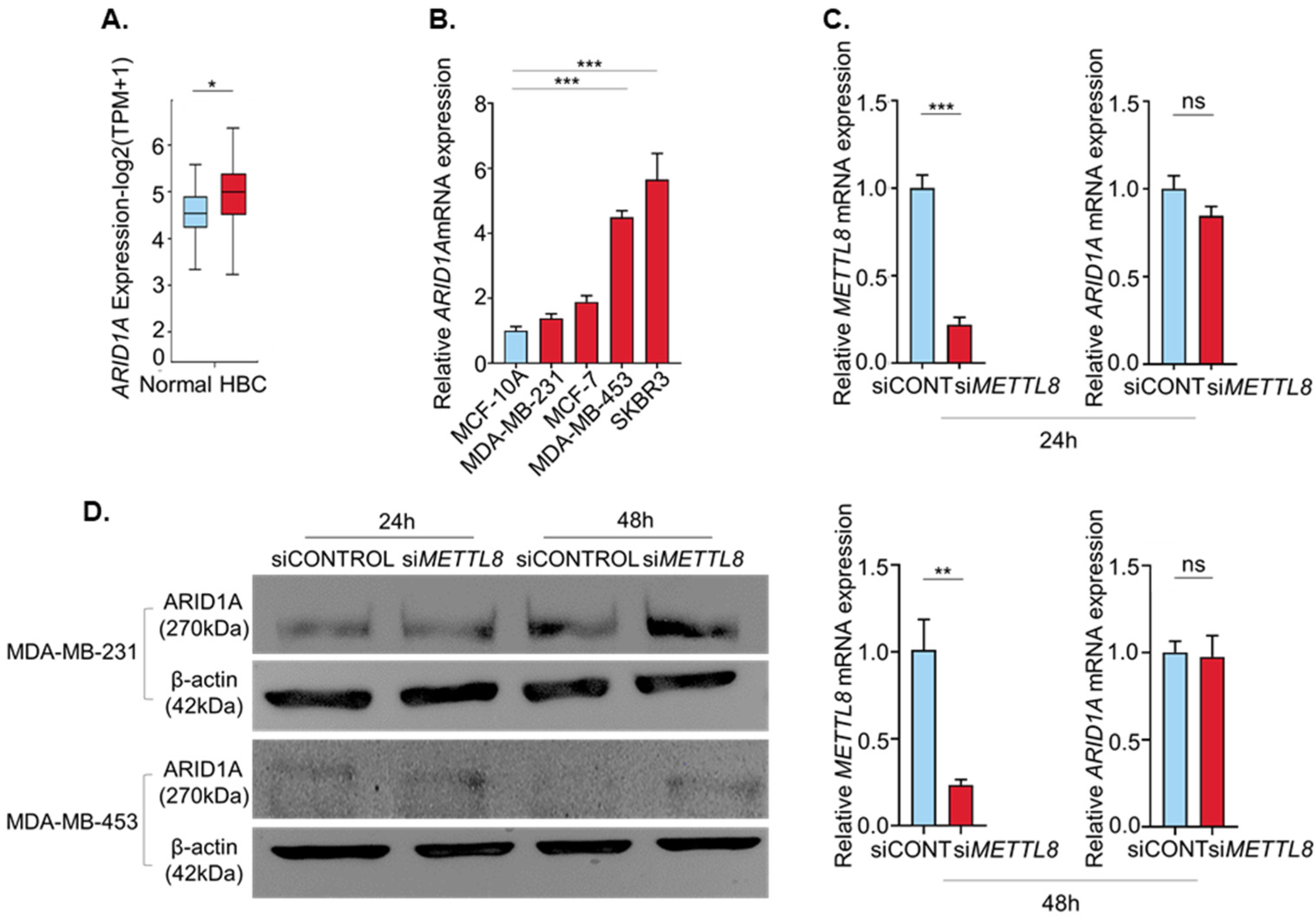
2.4. METTL8 Interacts with ARID1A mRNA
2.5. ARID1A mRNA Is a Candidate Transcript Associated with the Cell Migration Phenotype Regulated by METTL8
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Transfection
4.2. RNA Immunoprecipitation(RIP)
4.3. RNA Extraction and Quantitative RT-PCR
4.4. Protein Extraction and Western Blot
4.5. Cell Proliferation Assay
4.6. Cell Migration Assay
4.7. Public Data Acquisition
4.8. Gene Expression Database Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.A.; Caleffi, M.; Albert, U.-S.; Chen, T.H.H.; Duffy, S.W.; Franceschi, D.; Nyström, L. Global Summit Early Detection and Access to Care Panel Breast Cancer in Limited-Resource Countries: Early Detection and Access to Care. Breast J. 2006, 12, S16–S26. [Google Scholar] [CrossRef] [PubMed]
- Ghoncheh, M.; Pournamdar, Z.; Salehiniya, H. Incidence and Mortality and Epidemiology of Breast Cancer in the World. Asian Pac. J. Cancer Prev. 2016, 17, 43–46. [Google Scholar] [CrossRef]
- Nguyen, F.; Peña, L.; Ibisch, C.; Loussouarn, D.; Gama, A.; Rieder, N.; Belousov, A.; Campone, M.; Abadie, J. Canine invasive mammary carcinomas as models of human breast cancer. Part 1: Natural history and prognostic factors. Breast Cancer Res. Treat. 2018, 167, 635–648. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, S.K.; Saxena, S.; Lakshmanan, K.; Sangaiah, A.K.; Chauhan, H.; Shrivastava, S.; Singh, R.K. Deep feature learning for histopathological image classification of canine mammary tumors and human breast cancer. Inf. Sci. 2020, 508, 405–421. [Google Scholar] [CrossRef]
- Mohammed, S.I.; Meloni, G.B.; Parpaglia, M.P.; Marras, V.; Burrai, G.P.; Meloni, F.; Pirino, S.; Antuofermo, E. Mammography and Ultrasound Imaging of Preinvasive and Invasive Canine Spontaneous Mammary Cancer and Their Similarities to Human Breast Cancer. Cancer Prev. Res. 2011, 4, 1790–1798. [Google Scholar] [CrossRef][Green Version]
- Barbieri, I.; Kouzarides, T. Role of RNA modifications in cancer. Nat. Rev. Cancer 2020, 20, 303–322. [Google Scholar] [CrossRef] [PubMed]
- Han, S.H.; Choe, J. Diverse molecular functions of m6A mRNA modification in cancer. Exp. Mol. Med. 2020, 52, 738–749. [Google Scholar] [CrossRef]
- Lan, Q.; Liu, P.Y.; Haase, J.; Bell, J.L.; Hüttelmaier, S.; Liu, T. The Critical Role of RNA m6A Methylation in Cancer. Cancer Res. 2019, 79, 1285–1292. [Google Scholar] [CrossRef]
- Xu, L.; Liu, X.; Sheng, N.; Oo, K.S.; Liang, J.; Chionh, Y.H.; Xu, J.; Ye, F.; Gao, Y.-G.; Dedon, P.C.; et al. Three distinct 3-methylcytidine (m3C) methyltransferases modify tRNA and mRNA in mice and humans. J. Biol. Chem. 2017, 292, 14695–14703. [Google Scholar] [CrossRef]
- Gu, H.; Do, D.V.; Liu, X.; Xu, L.; Su, Y.; Nah, J.M.; Wong, Y.; Li, Y.; Sheng, N.; Tilaye, G.A.; et al. The STAT3 Target Mettl8 Regulates Mouse ESC Differentiation via Inhibiting the JNK Pathway. Stem Cell Rep. 2018, 10, 1807–1820. [Google Scholar] [CrossRef] [PubMed]
- Nair, N.U.; Das, A.; Rogkoti, V.-M.; Fokkelman, M.; Marcotte, R.; De Jong, C.G.; Koedoot, E.; Lee, J.S.; Meilijson, I.; Hannenhalli, S.; et al. Migration rather than proliferation transcriptomic signatures are strongly associated with breast cancer patient survival. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Zhang, L.-H.; Zhang, X.-Y.; Hu, T.; Chen, X.-Y.; Li, J.-J.; Raida, M.; Sun, N.; Luo, Y.; Gao, X. The SUMOylated METTL8 Induces R-loop and Tumorigenesis via m3C. iScience 2020, 23, 100968. [Google Scholar] [CrossRef] [PubMed]
- Yeon, S.Y.; Jo, Y.S.; Choi, E.J.; Kim, M.S.; Yoo, N.J.; Lee, S.H. Frameshift Mutations in Repeat Sequences of ANK3, HACD4, TCP10L, TP53BP1, MFN1, LCMT2, RNMT, TRMT6, METTL8 and METTL16 Genes in Colon Cancers. Pathol. Oncol. Res. 2018, 24, 617–622. [Google Scholar] [CrossRef]
- Friedl, P.; Alexander, S. Cancer Invasion and the Microenvironment: Plasticity and Reciprocity. Cell 2011, 147, 992–1009. [Google Scholar] [CrossRef]
- Xiao, Y.; Humphries, B.; Yang, C.; Wang, Z. MiR-205 Dysregulations in Breast Cancer: The Complexity and Opportunities. Non-Coding RNA 2019, 5, 53. [Google Scholar] [CrossRef]
- Lee, K.-H.; Park, H.-M.; Son, K.-H.; Shin, T.-J.; Cho, J.-Y. Transcriptome Signatures of Canine Mammary Gland Tumors and Its Comparison to Human Breast Cancers. Cancers 2018, 10, 317. [Google Scholar] [CrossRef]
- Zhao, M.; Sun, J.; Zhao, Z. TSGene: A web resource for tumor suppressor genes. Nucleic Acids Res. 2012, 41, D970–D976. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, X.; Yang, J.; Duan, J.; Wu, Z.; Yang, F.; Zhang, X.; Xiao, S. Loss of ARID1A promotes proliferation, migration and invasion via the Akt signaling pathway in NPC. Cancer Manag. Res. 2019, 11, 4931–4946. [Google Scholar] [CrossRef]
- Guo, R.-S.; Yu, Y.; Chen, J.; Chen, Y.-Y.; Shen, N.; Qiu, M. Restoration of Brain Acid Soluble Protein 1 Inhibits Proliferation and Migration of Thyroid Cancer Cells. Chin. Med. J. 2016, 129, 1439–1446. [Google Scholar] [CrossRef]
- Cheng, S.; Mao, Q.; Dong, Y.; Ren, J.; Su, L.; Liu, J.; Liu, Q.; Zhou, J.; Ye, X.; Zheng, S. GNB2L1 and its O-GlcNAcylation regulates metastasis via modulating epithelial-mesenchymal transition in the chemoresistance of gastric cancer. PLoS ONE 2017, 12, e0182696. [Google Scholar] [CrossRef]
- Ma, X.; Huang, J.; Tian, Y.; Chen, Y.; Yang, Y.; Zhang, X.; Zhang, F.; Xue, L. Myc suppresses tumor invasion and cell migration by inhibiting JNK signaling. Oncogene 2017, 36, 3159–3167. [Google Scholar] [CrossRef] [PubMed]
- Huggins, C.J.; Andrulis, I.L. Cell cycle regulated phosphorylation of LIMD1 in cell lines and expression in human breast cancers. Cancer Lett. 2008, 267, 55–66. [Google Scholar] [CrossRef]
- Novak, M.; Leonard, M.K.; Yang, X.H.; Kowluru, A.; Belkin, A.M.; Kaetzel, D.M. Metastasis suppressor NME 1 regulates melanoma cell morphology, self-adhesion and motility via induction of fibronectin expression. Exp. Dermatol. 2015, 24, 455–461. [Google Scholar] [CrossRef]
- Yang, X.; Yu, D.; Ren, Y.; Wei, J.; Pan, W.; Zhou, C.; Zhou, L.; Liu, Y.; Yang, M. Integrative Functional Genomics Implicates EPB41 Dysregulation in Hepatocellular Carcinoma Risk. Am. J. Hum. Genet. 2016, 99, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Amemiya, Y.; Yang, W.; Benatar, T.; Nofech-Mozes, S.; Yee, A.; Kahn, H.; Holloway, C.; Seth, A. Insulin like growth factor binding protein-7 reduces growth of human breast cancer cells and xenografted tumors. Breast Cancer Res. Treat. 2010, 126, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Sobolesky, P.M.; Halushka, P.V.; Garrett-Mayer, E.; Smith, M.T.; Moussa, O. Regulation of the Tumor Suppressor FOXO3 by the Thromboxane-A2 Receptors in Urothelial Cancer. PLoS ONE 2014, 9, e107530. [Google Scholar] [CrossRef]
- Gu, Y.; Zhang, M. Upregulation of the Kank1 gene inhibits human lung cancer progression in vitro and in vivo. Oncol. Rep. 2018, 40, 1243–1250. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Liu, W.; Liu, X.; Ai, Q.; Yu, J. Overexpression of MCPH1 inhibits the migration and invasion of lung cancer cells. OncoTargets Ther. 2018, ume 11, 3111–3117. [Google Scholar] [CrossRef]
- Yang, L.; Xie, N.; Huang, J.; Huang, H.; Xu, S.; Wang, Z.; Cai, J. SIK1-LNC represses the proliferative, migrative, and invasive abilities of lung cancer cells. OncoTargets Ther. 2018, 11, 4197–4206. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Li, G.; Zhou, J.; Han, N.; Liu, Z.; Yin, J. miR-107 Activates ATR/Chk1 Pathway and Suppress Cervical Cancer Invasion by Targeting MCL1. PLoS ONE 2014, 9, e111860. [Google Scholar] [CrossRef] [PubMed]
- Queiroga, F.L.; Raposo, T.; Carvalho, M.I.; Prada, J.; Pires, I. Canine mammary tumours as a model to study human breast cancer: Most recent findings. In Vivo 2011, 25, 455–465. [Google Scholar] [PubMed]
- Begon, D.Y.; Delacroix, L.; Vernimmen, D.; Jackers, P.; Winkler, R. Yin Yang 1 Cooperates with Activator Protein 2 to Stimulate ERBB2 Gene Expression in Mammary Cancer Cells. J. Biol. Chem. 2005, 280, 24428–24434. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Stovall, D.B.; Inoue, K.; Sui, G. The Oncogenic Role of Yin Yang 1. Crit. Rev. Oncog. 2011, 16, 163–197. [Google Scholar] [CrossRef] [PubMed]
- Arvey, A.; Agius, P.; Noble, W.S.; Leslie, C. Sequence and chromatin determinants of cell-type-specific transcription factor binding. Genome Res. 2012, 22, 1723–1734. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-Y.; Fong, Y.-C.; Lee, C.-Y.; Chen, M.-Y.; Tsai, H.-C.; Hsu, H.-C.; Tang, C.-H. CCL5 increases lung cancer migration via PI3K, Akt and NF-κB pathways. Biochem. Pharmacol. 2009, 77, 794–803. [Google Scholar] [CrossRef] [PubMed]
- Pavlidou, E.N.; Balis, V. Diagnostic significance and prognostic role of the ARID1A gene in cancer outcomes (Review). World Acad. Sci. J. 2020, 2, 49–64. [Google Scholar] [CrossRef][Green Version]
- Guan, B.; Wang, T.-L.; Shih, I.-M. ARID1A, a Factor That Promotes Formation of SWI/SNF-Mediated Chromatin Remodeling, Is a Tumor Suppressor in Gynecologic Cancers. Cancer Res. 2011, 71, 6718–6727. [Google Scholar] [CrossRef]
- Nottegar, A.; Veronese, N.; Solmi, M.; Luchini, C. Tumor suppressor gene ARID1A in cancer: Recent advances and future perspective. J. Carinog. Mutagen. 2016, 7, 2. [Google Scholar]
- Wu, J.N.; Roberts, C.W.M. ARID1A Mutations in Cancer: Another Epigenetic Tumor Suppressor? Cancer Discov. 2013, 3, 35–43. [Google Scholar] [CrossRef]
- Takeda, T.; Banno, K.; Okawa, R.; Yanokura, M.; Iijima, M.; Irie-Kunitomi, H.; Nakamura, K.; Iida, M.; Adachi, M.; Umene, K.; et al. ARID1A gene mutation in ovarian and endometrial cancers (Review). Oncol. Rep. 2015, 35, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.-A.; Lee, J.-H.; Kim, D.W.; Cho, J.-Y. E3-ubiquitin ligase NEDD4 enhances bone formation by removing TGFβ1-induced pSMAD1 in immature osteoblast. Bone 2018, 116, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.-A.; Kim, D.W.; Lee, D.-B.; Cho, J.-Y. NEDD4 Plays Roles in the Maintenance of Breast Cancer Stem Cell Characteristics. Front. Oncol. 2020, 10, 1680. [Google Scholar] [CrossRef] [PubMed]
| Gene | Direction | Sequence |
|---|---|---|
| METTL8 | Forward | 5′-CGAGGAGATGGTACCAGAGCATA-3′ |
| Reverse | 5′-AGCGGCGATCAACCAGATTT-3′ | |
| YY1 | Forward | 5′-AAAACATCTGCACACCCACG-3′ |
| Reverse | 5′-GTCTCCGGTATGGATTCGCA-3′ | |
| ARID1A | Forward | 5′-CTTCAACCTCAGTCAGCTCCCA-3′ |
| Reverse | 5′-GGTCACCCACCTCATACTCCTTT-3′ | |
| CCSER2 (control) | Forward | 5′-GACAGGAGCATTACCACCTCAG-3′ |
| Reverse | 5′-CTTCTGAGCCTGGAAAAAGGG-3′ | |
| 18S (control) | Forward | 5′-AACCCGTTGAACCCCATT-3′ |
| Reverse | 5′-CCATCCAATCGGTAGTAGCG-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.-A.; Lee, K.-H.; Kim, H.; Cho, J.-Y. METTL8 mRNA Methyltransferase Enhances Cancer Cell Migration via Direct Binding to ARID1A. Int. J. Mol. Sci. 2021, 22, 5432. https://doi.org/10.3390/ijms22115432
Lee S-A, Lee K-H, Kim H, Cho J-Y. METTL8 mRNA Methyltransferase Enhances Cancer Cell Migration via Direct Binding to ARID1A. International Journal of Molecular Sciences. 2021; 22(11):5432. https://doi.org/10.3390/ijms22115432
Chicago/Turabian StyleLee, Shin-Ae, Kang-Hoon Lee, Huisu Kim, and Je-Yoel Cho. 2021. "METTL8 mRNA Methyltransferase Enhances Cancer Cell Migration via Direct Binding to ARID1A" International Journal of Molecular Sciences 22, no. 11: 5432. https://doi.org/10.3390/ijms22115432
APA StyleLee, S.-A., Lee, K.-H., Kim, H., & Cho, J.-Y. (2021). METTL8 mRNA Methyltransferase Enhances Cancer Cell Migration via Direct Binding to ARID1A. International Journal of Molecular Sciences, 22(11), 5432. https://doi.org/10.3390/ijms22115432







