Insulin-Like Growth Factor Binding Protein-3 Binds to Histone 3
Abstract
1. Introduction
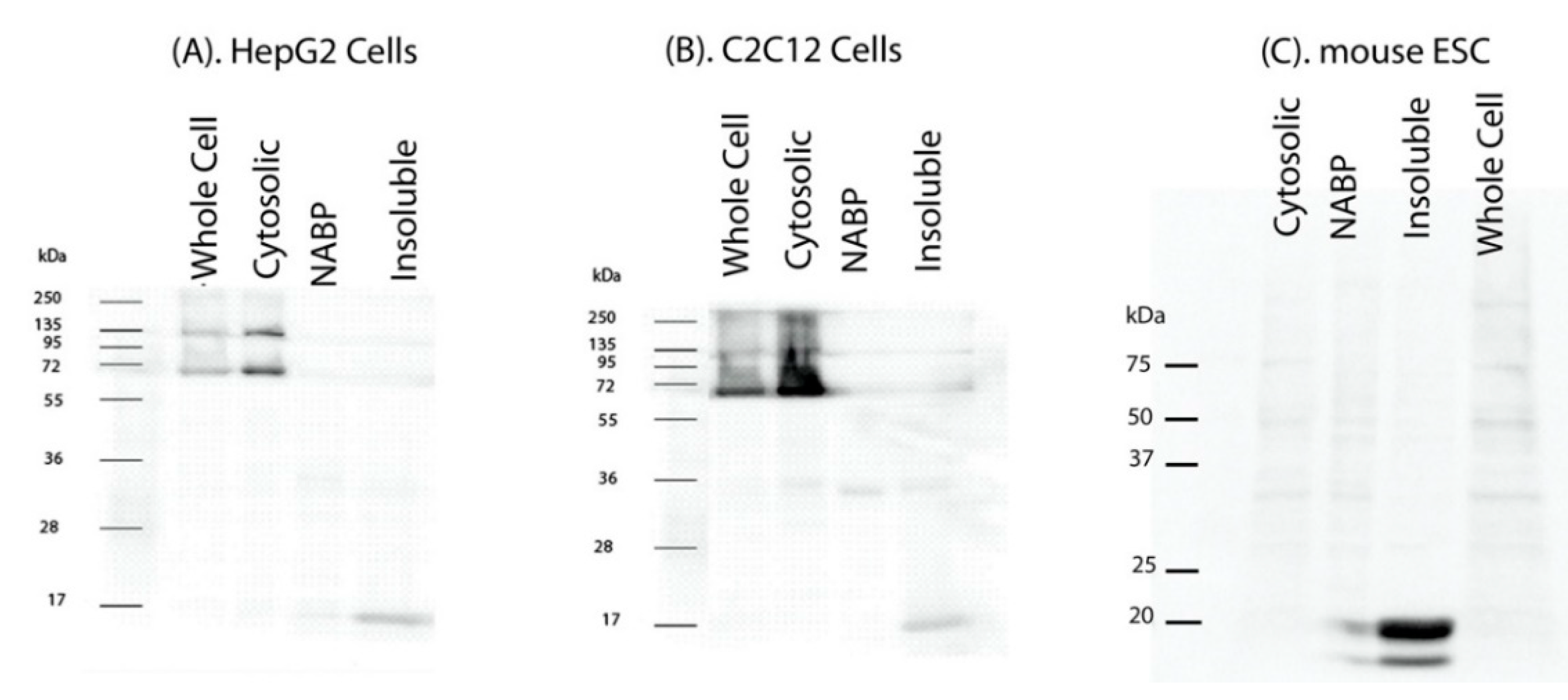
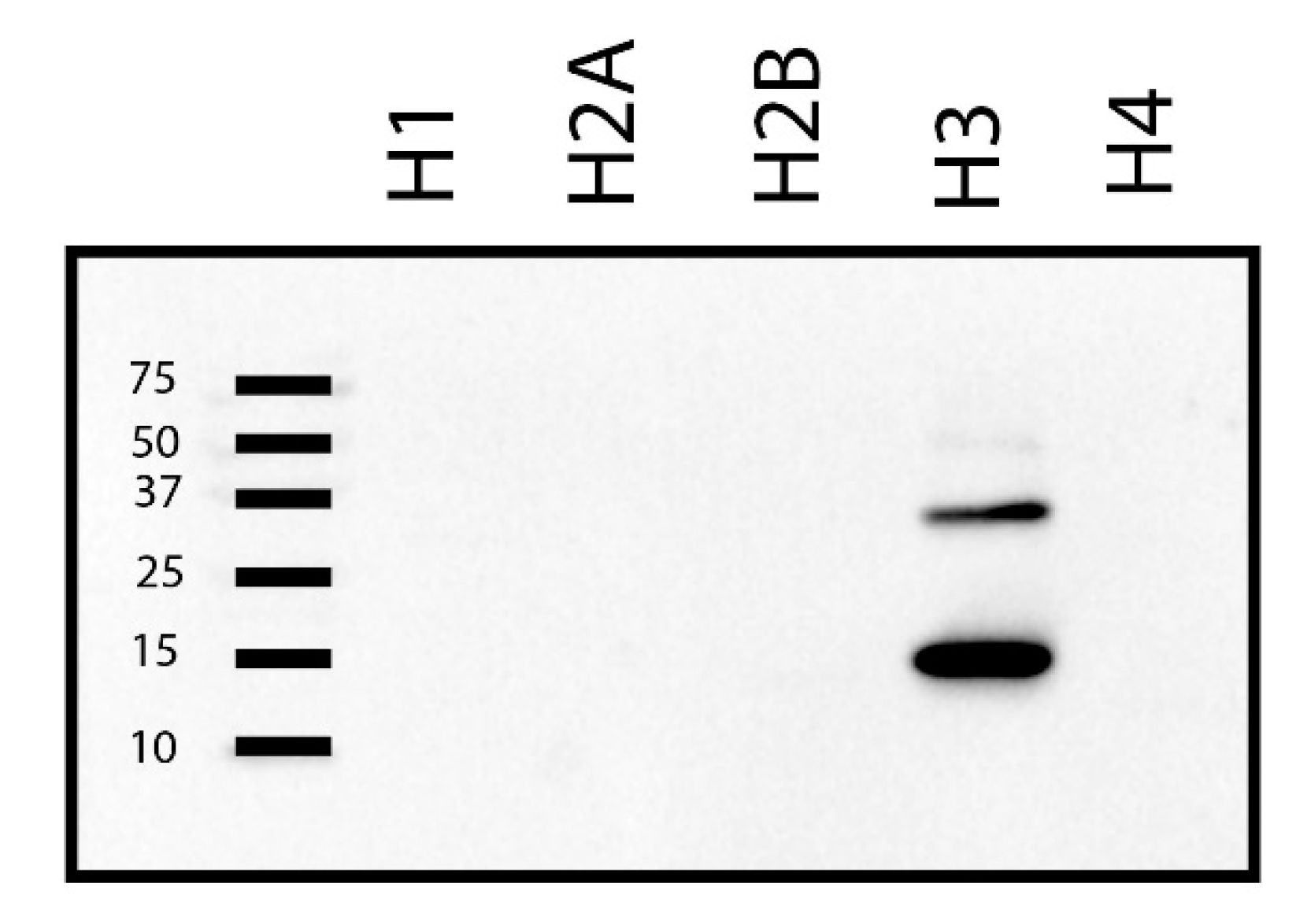
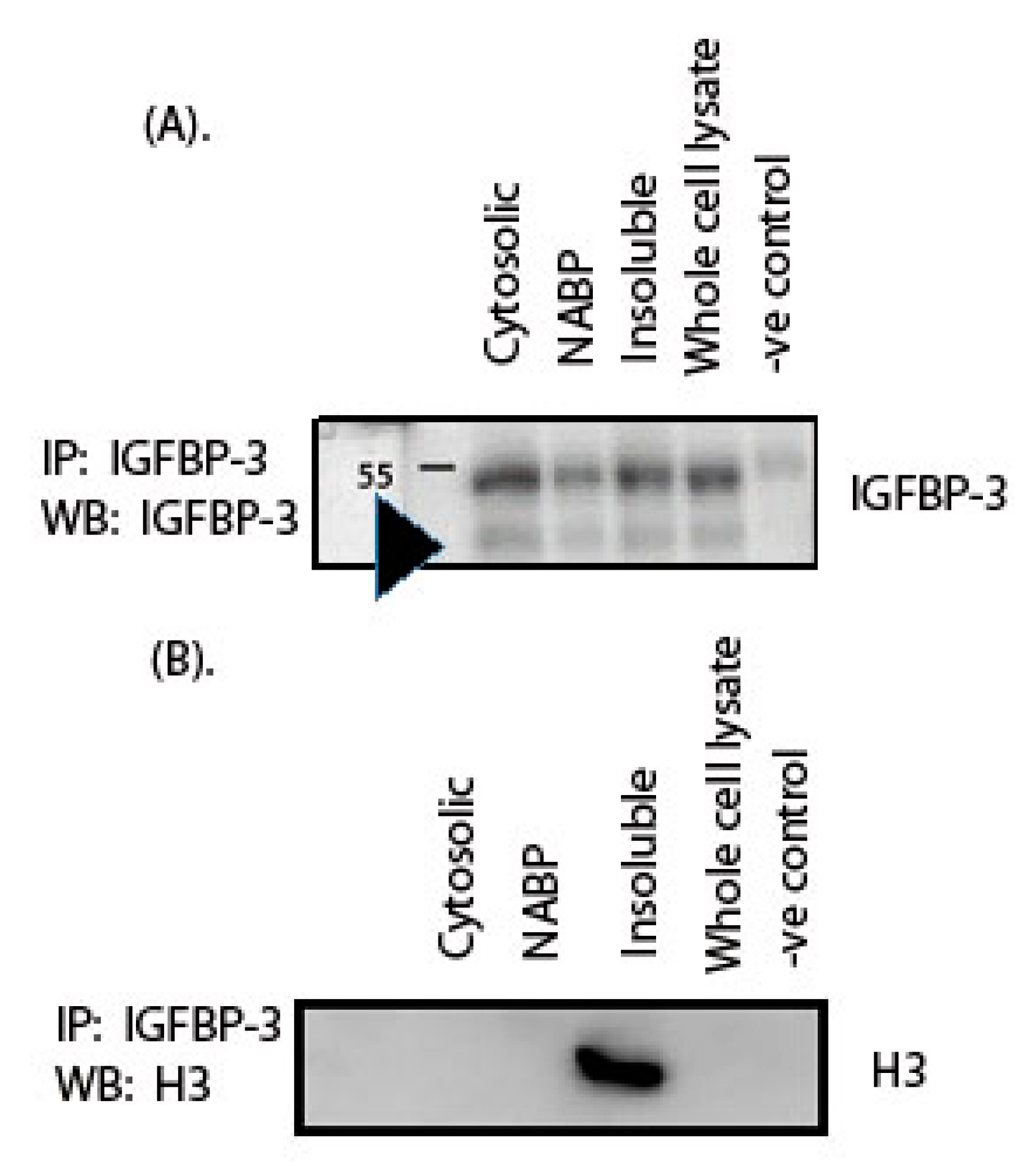
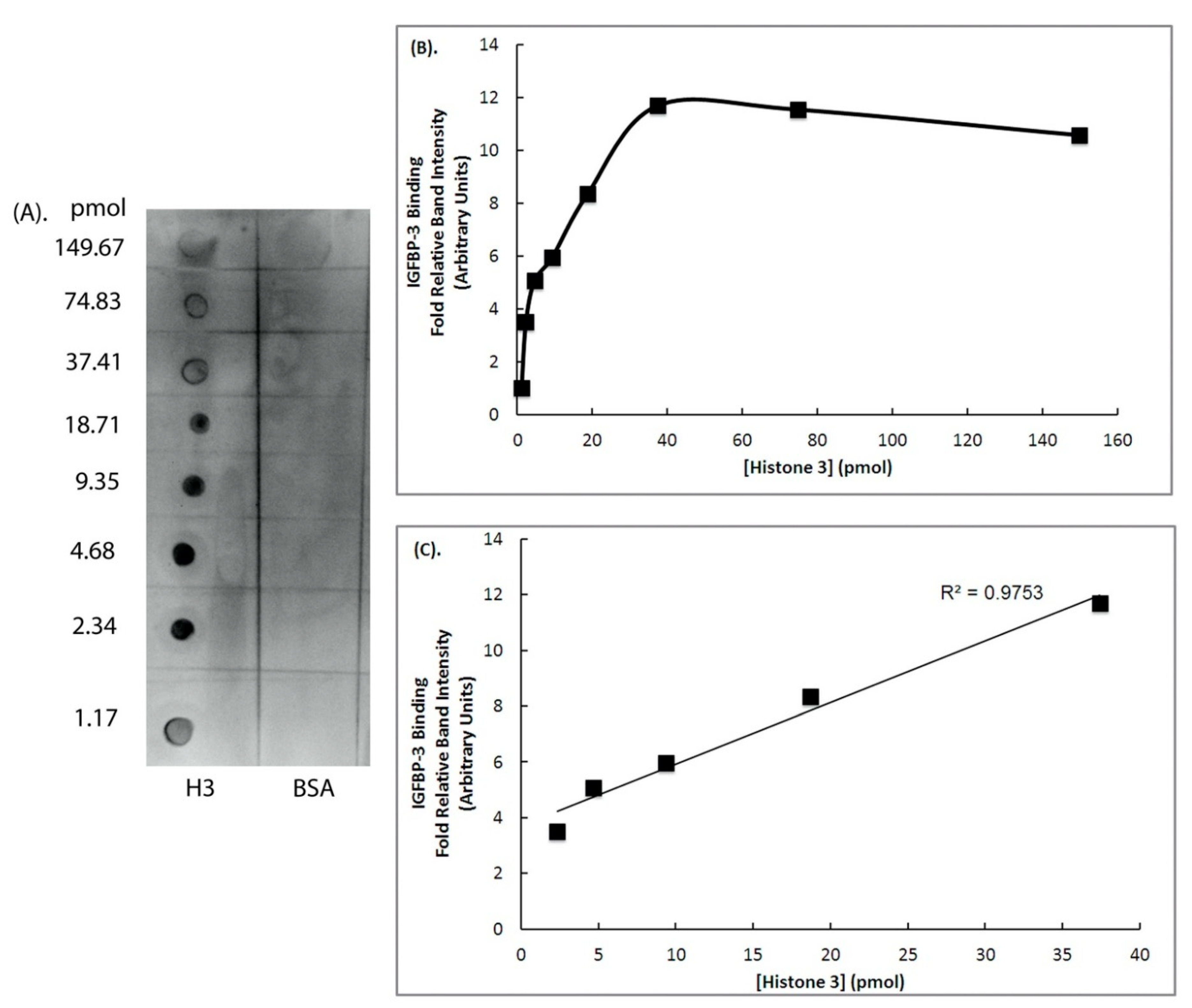
2. Results
3. Discussion
4. Material and Methods
4.1. Cell Lines
4.2. Cell Fractionation and Lysis
4.3. Whole Cell Lysis
4.4. Ligand Blotting
4.5. Ligand Dot Blot
4.6. Far-Western Blot Analyses
4.7. Co-Immunoprecipitation
4.8. Western Blot Analysis
4.9. Protein Estimation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Foulstone, E.J.; Savage, P.B.; Crown, A.L.; Holly, J.M.; Stewart, C.E. Role of insulin-like growth factor binding protein-3 (IGFBP-3) in the differentiation of primary human adult skeletal myoblasts. J. Cell. Physiol. 2003, 195, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Schedlich, L.J.; Le Page, S.L.; Firth, S.M.; Briggs, L.J.; Jans, D.A.; Baxter, R.C. Nuclear import of insulin-like growth factor-binding protein-3 and -5 is mediated by the importin beta subunit. J. Biol. Chem. 2000, 275, 23462–23470. [Google Scholar] [CrossRef] [PubMed]
- Micutkova, L.; Hermann, M.; Offterdinger, M.; Hess, M.W.; Matscheski, A.; Pircher, H.; Muck, C.; Ebner, H.L.; Laich, A.; Ferrando-May, E.; et al. Analysis of the cellular uptake and nuclear delivery of insulin-like growth factor binding protein-3 in human osteosarcoma cells. Int. J. Cancer 2012, 130, 1544–1557. [Google Scholar] [CrossRef] [PubMed]
- Granata, R.; Trovato, L.; Garbarino, G.; Taliano, M.; Ponti, R.; Sala, G.; Ghidoni, R.; Ghigo, E. Dual effects of IGFBP-3 on endothelial cell apoptosis and survival: Involvement of the sphingolipid signaling pathways. FASEB J. 2004, 18, 1456–1458. [Google Scholar] [CrossRef]
- Grkovic, S.; O’Reilly, V.C.; Han, S.; Hong, M.; Baxter, R.C.; Firth, S.M. IGFBP-3 binds GRP78, stimulates autophagy and promotes the survival of breast cancer cells exposed to adverse microenvironments. Oncogene 2013, 32, 2412–2420. [Google Scholar] [CrossRef]
- Lin, M.Z.; Marzec, K.A.; Martin, J.L.; Baxter, R.C. The role of insulin-like growth factor binding protein-3 in the breast cancer cell response to DNA-damaging agents. Oncogene 2014, 33, 85–96. [Google Scholar] [CrossRef]
- Chua, M.W.; Lin, M.Z.; Martin, J.L.; Baxter, R.C. Involvement of the insulin-like growth factor binding proteins in the cancer cell response to DNA damage. J. Cell Commun. Signal. 2015, 9, 167–176. [Google Scholar] [CrossRef]
- Shrivastav, S.V.; Bhardwaj, A.; Pathak, K.A.; Shrivastav, A. Insulin-Like Growth Factor Binding Protein-3 (IGFBP-3): Unraveling the Role in Mediating IGF-Independent Effects Within the Cell. Front. Cell Dev. Biol. 2020, 8, 286. [Google Scholar] [CrossRef]
- Butt, A.J.; Fraley, K.A.; Firth, S.M.; Baxter, R.C. IGF-binding protein-3-induced growth inhibition and apoptosis do not require cell surface binding and nuclear translocation in human breast cancer cells. Endocrinology 2002, 143, 2693–2699. [Google Scholar] [CrossRef][Green Version]
- Shang, Y.; Baumrucker, C.R.; Green, M.H. Signal relay by retinoic acid receptors alpha and beta in the retinoic acid-induced expression of insulin-like growth factor-binding protein-3 in breast cancer cells. J. Biol. Chem. 1999, 274, 18005–18010. [Google Scholar] [CrossRef]
- Gucev, Z.S.; Oh, Y.; Kelley, K.M.; Rosenfeld, R.G. Insulin-like growth factor binding protein 3 mediates retinoic acid- and transforming growth factor beta2-induced growth inhibition in human breast cancer cells. Cancer Res. 1996, 56, 1545–1550. [Google Scholar] [PubMed]
- Kansra, S.; Ewton, D.Z.; Wang, J.; Friedman, E. IGFBP-3 mediates TGF beta 1 proliferative response in colon cancer cells. Int. J. Cancer 2000, 87, 373–378. [Google Scholar] [CrossRef]
- Rajah, R.; Lee, K.W.; Cohen, P. Insulin-like growth factor binding protein-3 mediates tumor necrosis factor-alpha-induced apoptosis: Role of Bcl-2 phosphorylation. Cell Growth Differ. 2002, 13, 163–171. [Google Scholar] [PubMed]
- Lopez-Bermejo, A.; Buckway, C.K.; Devi, G.R.; Hwa, V.; Plymate, S.R.; Oh, Y.; Rosenfeld, R.G. Characterization of insulin-like growth factor-binding protein-related proteins (IGFBP-rPs) 1, 2, and 3 in human prostate epithelial cells: Potential roles for IGFBP-rP1 and 2 in senescence of the prostatic epithelium. Endocrinology 2000, 141, 4072–4080. [Google Scholar] [CrossRef]
- Goossens, K.; Esquenet, M.; Swinnen, J.V.; Manin, M.; Rombauts, W.; Verhoeven, G. Androgens decrease and retinoids increase the expression of insulin-like growth factor-binding protein-3 in LNcaP prostatic adenocarcinoma cells. Mol. Cell. Endocrinol. 1999, 155, 9–18. [Google Scholar] [CrossRef]
- Krishnan, A.V.; Peehl, D.M.; Feldman, D. Inhibition of prostate cancer growth by vitamin D: Regulation of target gene expression. J. Cell. Biochem. 2003, 88, 363–371. [Google Scholar] [CrossRef]
- Boyle, B.J.; Zhao, X.Y.; Cohen, P.; Feldman, D. Insulin-like growth factor binding protein-3 mediates 1 alpha,25-dihydroxyvitamin d(3) growth inhibition in the LNCaP prostate cancer cell line through p21/WAF1. J. Urol. 2001, 165, 1319–1324. [Google Scholar] [CrossRef]
- Ingermann, A.R.; Yang, Y.F.; Han, J.; Mikami, A.; Garza, A.E.; Mohanraj, L.; Fan, L.; Idowu, M.; Ware, J.L.; Kim, H.S.; et al. Identification of a novel cell death receptor mediating IGFBP-3-induced anti-tumor effects in breast and prostate cancer. J. Biol. Chem. 2010, 285, 30233–30246. [Google Scholar] [CrossRef]
- Burrows, C.; Holly, J.M.; Laurence, N.J.; Vernon, E.G.; Carter, J.V.; Clark, M.A.; McIntosh, J.; McCaig, C.; Winters, Z.E.; Perks, C.M. Insulin-like growth factor binding protein 3 has opposing actions on malignant and nonmalignant breast epithelial cells that are each reversible and dependent upon cholesterol-stabilized integrin receptor complexes. Endocrinology 2006, 147, 3484–3500. [Google Scholar] [CrossRef]
- Lee, K.W.; Liu, B.; Ma, L.; Li, H.; Bang, P.; Koeffler, H.P.; Cohen, P. Cellular internalization of insulin-like growth factor binding protein-3: Distinct endocytic pathways facilitate re-uptake and nuclear localization. J. Biol. Chem. 2004, 279, 469–476. [Google Scholar] [CrossRef]
- Weinzimer, S.A.; Gibson, T.B.; Collett-Solberg, P.F.; Khare, A.; Liu, B.; Cohen, P. Transferrin is an insulin-like growth factor-binding protein-3 binding protein. J. Clin. Endocrinol. Metab. 2001, 86, 1806–1813. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.S.; Ling, T.Y.; Tseng, W.F.; Huang, Y.H.; Tang, F.M.; Leal, S.M.; Huang, J.S. Cellular growth inhibition by IGFBP-3 and TGF-beta1 requires LRP-1. FASEB J. 2003, 17, 2068–2081. [Google Scholar] [CrossRef] [PubMed]
- Leal, S.M.; Liu, Q.; Huang, S.S.; Huang, J.S. The type V transforming growth factor beta receptor is the putative insulin-like growth factor-binding protein 3 receptor. J. Biol. Chem. 1997, 272, 20572–20576. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Raz, A.; Murphy, L.J. Insulin-like growth factor binding protein-3 interacts with autocrine motility factor/phosphoglucose isomerase (AMF/PGI) and inhibits the AMF/PGI function. Cancer Res. 2004, 64, 2516–2522. [Google Scholar] [CrossRef][Green Version]
- Li, C.; Harada, A.; Oh, Y. IGFBP-3 sensitizes antiestrogen-resistant breast cancer cells through interaction with GRP78. Cancer Lett. 2012, 325, 200–206. [Google Scholar] [CrossRef]
- Ikonen, M.; Liu, B.; Hashimoto, Y.; Ma, L.; Lee, K.W.; Niikura, T.; Nishimoto, I.; Cohen, P. Interaction between the Alzheimer’s survival peptide humanin and insulin-like growth factor-binding protein 3 regulates cell survival and apoptosis. Proc. Natl. Acad. Sci. USA 2003, 100, 13042–13047. [Google Scholar] [CrossRef]
- Wu, C.; Yao, G.; Zou, M.; Chen, G.; Wang, M.; Liu, J.; Wang, J.; Xu, D. N-Acetylgalactosaminyltransferase 14, a novel insulin-like growth factor binding protein-3 binding partner. Biochem. Biophys. Res. Commun. 2007, 357, 360–365. [Google Scholar] [CrossRef]
- Oufattole, M.; Lin, S.W.; Liu, B.; Mascarenhas, D.; Cohen, P.; Rodgers, B.D. Ribonucleic acid polymerase II binding subunit 3 (Rpb3), a potential nuclear target of insulin-like growth factor binding protein-3. Endocrinology 2006, 147, 2138–2146. [Google Scholar] [CrossRef]
- Liu, B.; Lee, H.Y.; Weinzimer, S.A.; Powell, D.R.; Clifford, J.L.; Kurie, J.M.; Cohen, P. Direct functional interactions between insulin-like growth factor-binding protein-3 and retinoid X receptor-alpha regulate transcriptional signaling and apoptosis. J. Biol. Chem. 2000, 275, 33607–33613. [Google Scholar] [CrossRef]
- Chan, S.S.; Schedlich, L.J.; Twigg, S.M.; Baxter, R.C. Inhibition of adipocyte differentiation by insulin-like growth factor-binding protein-3. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E654–E663. [Google Scholar] [CrossRef]
- Moreno-Santos, I.; Castellano-Castillo, D.; Lara, M.F.; Fernandez-Garcia, J.C.; Tinahones, F.J.; Macias-Gonzalez, M. IGFBP-3 Interacts with the Vitamin D Receptor in Insulin Signaling Associated with Obesity in Visceral Adipose Tissue. Int. J. Mol. Sci. 2017, 18, 2349. [Google Scholar] [CrossRef] [PubMed]
- Holden, P.; Horton, W.A. Crude subcellular fractionation of cultured mammalian cell lines. BMC Res. Notes 2009, 2, 243. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, C.; Avgousti, D.C.; Weitzman, M.D. Differential Salt Fractionation of Nuclei to Analyze Chromatin-associated Proteins from Cultured Mammalian Cells. Bio-Protocol 2017, 7, e2175. [Google Scholar] [CrossRef] [PubMed]
- Forcob, S.; Bulic, A.; Jonsson, F.; Lipps, H.J.; Postberg, J. Differential expression of histone H3 genes and selective association of the variant H3.7 with a specific sequence class in Stylonychia macronuclear development. Epigenetics Chromatin 2014, 7, 4. [Google Scholar] [CrossRef]
- Chakraborty, S.; Nguyen, P.K.; Ruyer-Quil, C.; Bontozoglou, V. Extreme solitary waves on falling liquid films. J. Fluid Mech. 2014, 745, 564–591. [Google Scholar] [CrossRef]
- Ruiz-Carrillo, A.; Jorcano, J.L. An octamer of core histones in solution: Central role of the H3-H4 tetramer in the self-assembly. Biochemistry 1979, 18, 760–768. [Google Scholar] [CrossRef]
- Thomas, J.O.; Butler, P.J. Characterization of the octamer of histones free in solution. J. Mol. Biol. 1977, 116, 769–781. [Google Scholar] [CrossRef]
- Eickbush, T.H.; Moudrianakis, E.N. The histone core complex: An octamer assembled by two sets of protein-protein interactions. Biochemistry 1978, 17, 4955–4964. [Google Scholar] [CrossRef]
- Chung, S.Y.; Hill, W.E.; Doty, P. Characterization of the histone core complex. Proc. Natl. Acad. Sci. USA 1978, 75, 1680–1684. [Google Scholar] [CrossRef]
- Annunziato, A.T. DNA Packaging: Nulceosomes and Chromatin. Nat. Educ. 2008, 1, 26. [Google Scholar]
- Forbes, B.E.; McCarthy, P.; Norton, R.S. Insulin-like growth factor binding proteins: A structural perspective. Front. Endocrinol. (Lausanne) 2012, 3, 38. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yin, P.; Bach, L.A.; Duan, C. Several acidic amino acids in the N-domain of insulin-like growth factor-binding protein-5 are important for its transactivation activity. J. Biol. Chem. 2006, 281, 14184–14191. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Duan, C. Lamprey IGF-Binding Protein-3 Has IGF-Dependent and -Independent Actions. Front. Endocrinol. (Lausanne) 2016, 7, 174. [Google Scholar] [CrossRef] [PubMed]
- Baxter, R.C. Insulin-like growth factor binding protein-3 (IGFBP-3): Novel ligands mediate unexpected functions. J. Cell Commun. Signal. 2013, 7, 179–189. [Google Scholar] [CrossRef]
- Kastner, P.; Mark, M.; Chambon, P. Nonsteroid nuclear receptors: What are genetic studies telling us about their role in real life? Cell 1995, 83, 859–869. [Google Scholar] [CrossRef]
- Ikezoe, T.; Tanosaki, S.; Krug, U.; Liu, B.; Cohen, P.; Taguchi, H.; Koeffler, H.P. Insulin-like growth factor binding protein-3 antagonizes the effects of retinoids in myeloid leukemia cells. Blood 2004, 104, 237–242. [Google Scholar] [CrossRef]
- Pon, C.K.; Firth, S.M.; Baxter, R.C. Involvement of insulin-like growth factor binding protein-3 in peroxisome proliferator-activated receptor gamma-mediated inhibition of breast cancer cell growth. Mol. Cell. Endocrinol. 2015, 399, 354–361. [Google Scholar] [CrossRef]
- Jia, Q.; Xiao-li, M.; Xin, W.; Hong, C.; Bing-Ren, H. Insulin-like growth factor binding protein-3 interacts with the thyroid hormone receptor alpha1 and modulates transcription of thyroid hormone responsive gene. Zhongguo Yi Xue Ke Xue Yuan Xue Bao 2011, 33, 156–161. [Google Scholar]
- Alessi, D.R.; James, S.R.; Downes, C.P.; Holmes, A.B.; Gaffney, P.R.; Reese, C.B.; Cohen, P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr. Biol. 1997, 7, 261–269. [Google Scholar] [CrossRef]
- Gui, Y.; Murphy, L.J. Insulin-like growth factor (IGF)-binding protein-3 (IGFBP-3) binds to fibronectin (FN): Demonstration of IGF-I/IGFBP-3/fn ternary complexes in human plasma. J. Clin. Endocrinol. Metab. 2001, 86, 2104–2110. [Google Scholar]
- Grulich-Henn, J.; Spiess, S.; Heinrich, U.; Schonberg, D.; Bettendorf, M. Ligand blot analysis of insulin-like growth factor-binding proteins using biotinylated insulin-like growth factor-I. Horm. Res. 1998, 49, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Towbin, H.; Staehelin, T.; Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhardwaj, A.; Pathak, K.A.; Shrivastav, A.; Varma Shrivastav, S. Insulin-Like Growth Factor Binding Protein-3 Binds to Histone 3. Int. J. Mol. Sci. 2021, 22, 407. https://doi.org/10.3390/ijms22010407
Bhardwaj A, Pathak KA, Shrivastav A, Varma Shrivastav S. Insulin-Like Growth Factor Binding Protein-3 Binds to Histone 3. International Journal of Molecular Sciences. 2021; 22(1):407. https://doi.org/10.3390/ijms22010407
Chicago/Turabian StyleBhardwaj, Apurva, Kumar Alok Pathak, Anuraag Shrivastav, and Shailly Varma Shrivastav. 2021. "Insulin-Like Growth Factor Binding Protein-3 Binds to Histone 3" International Journal of Molecular Sciences 22, no. 1: 407. https://doi.org/10.3390/ijms22010407
APA StyleBhardwaj, A., Pathak, K. A., Shrivastav, A., & Varma Shrivastav, S. (2021). Insulin-Like Growth Factor Binding Protein-3 Binds to Histone 3. International Journal of Molecular Sciences, 22(1), 407. https://doi.org/10.3390/ijms22010407




