First Report of CRISPR/Cas9 Mediated DNA-Free Editing of 4CL and RVE7 Genes in Chickpea Protoplasts
Abstract
1. Introduction
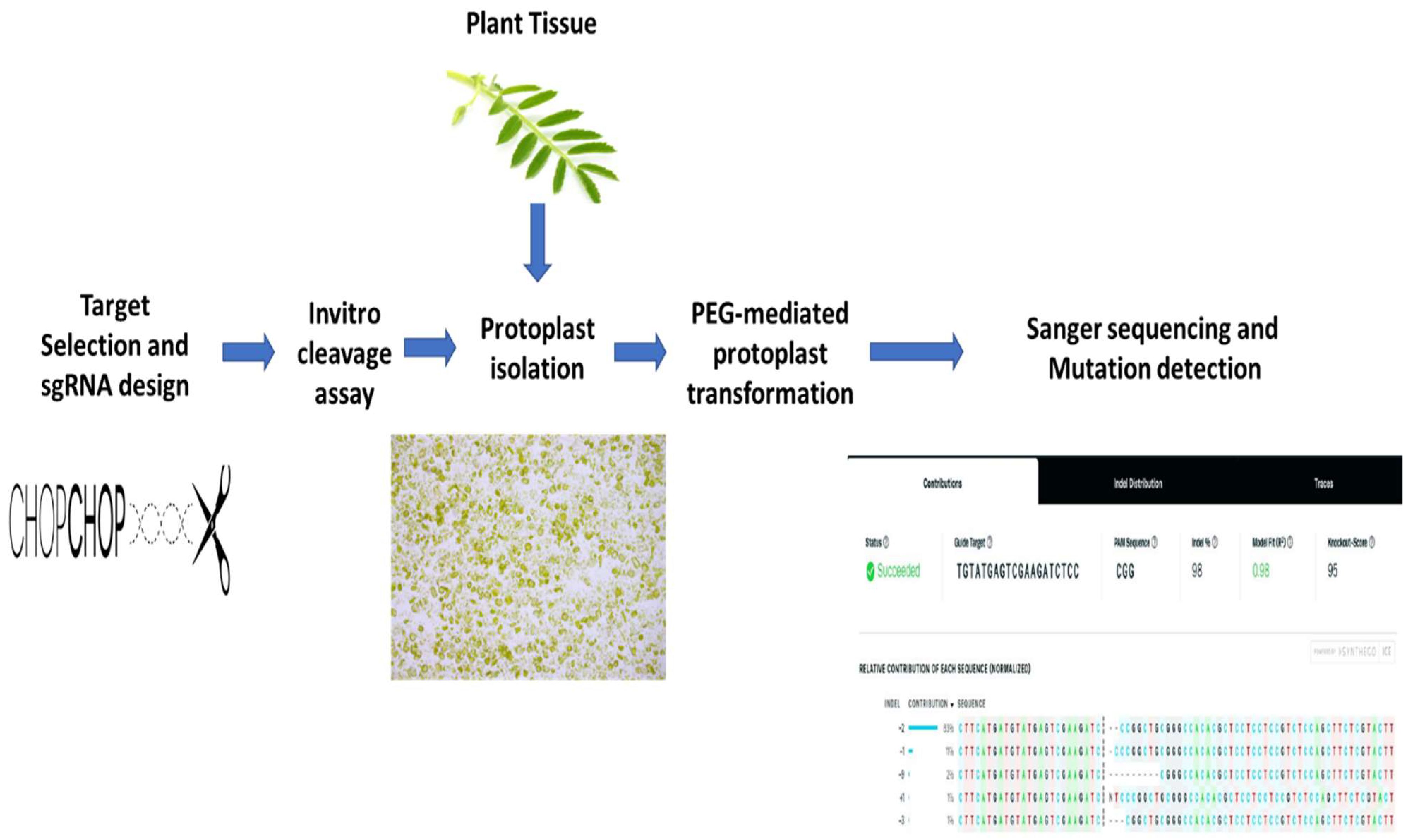
2. Results
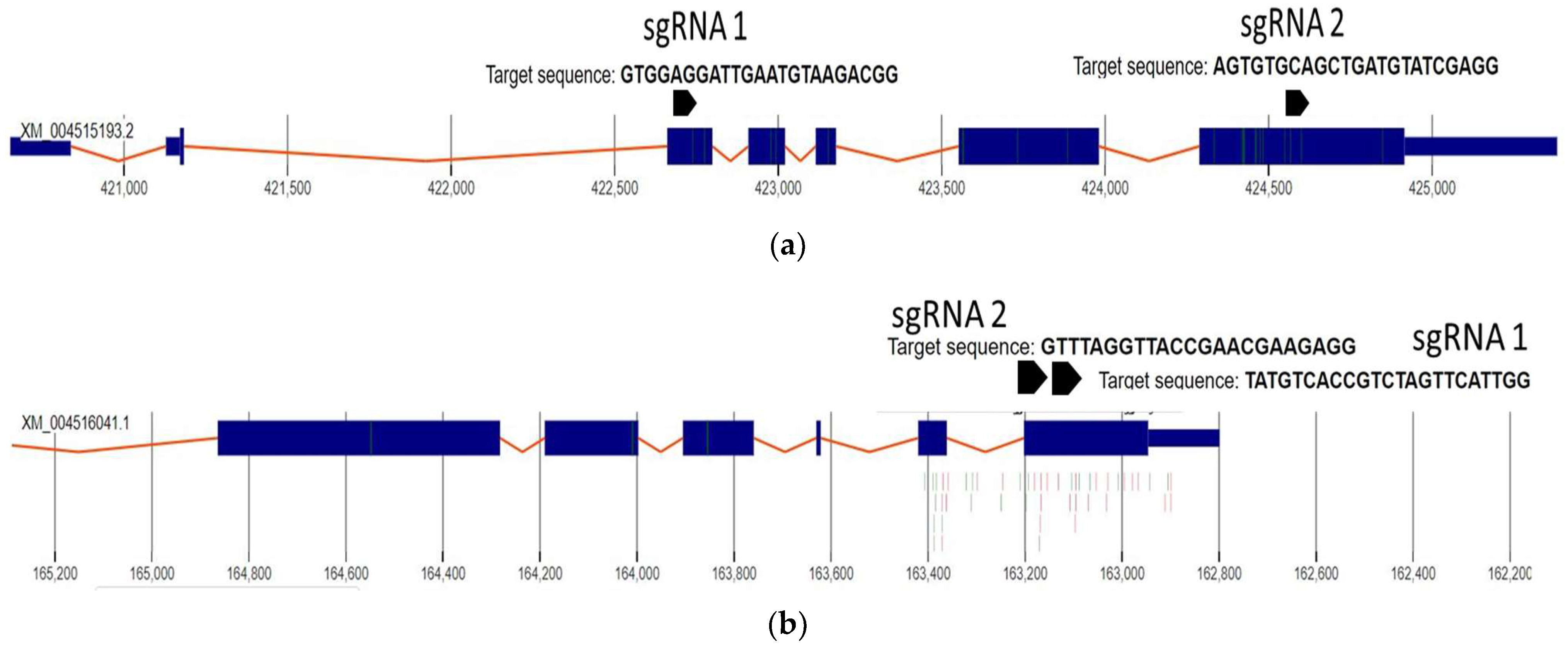
2.1. sgRNA Selection and Design
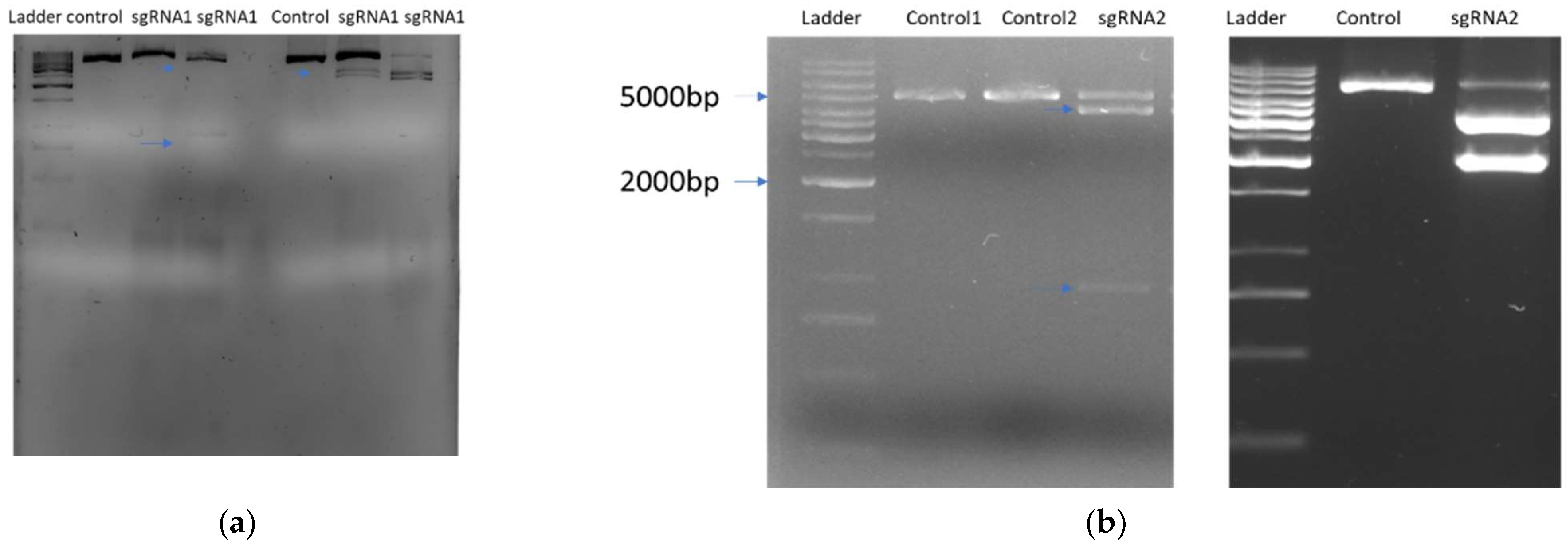
2.2. sgRNA Selection and Design In Vitro Digestion Assay
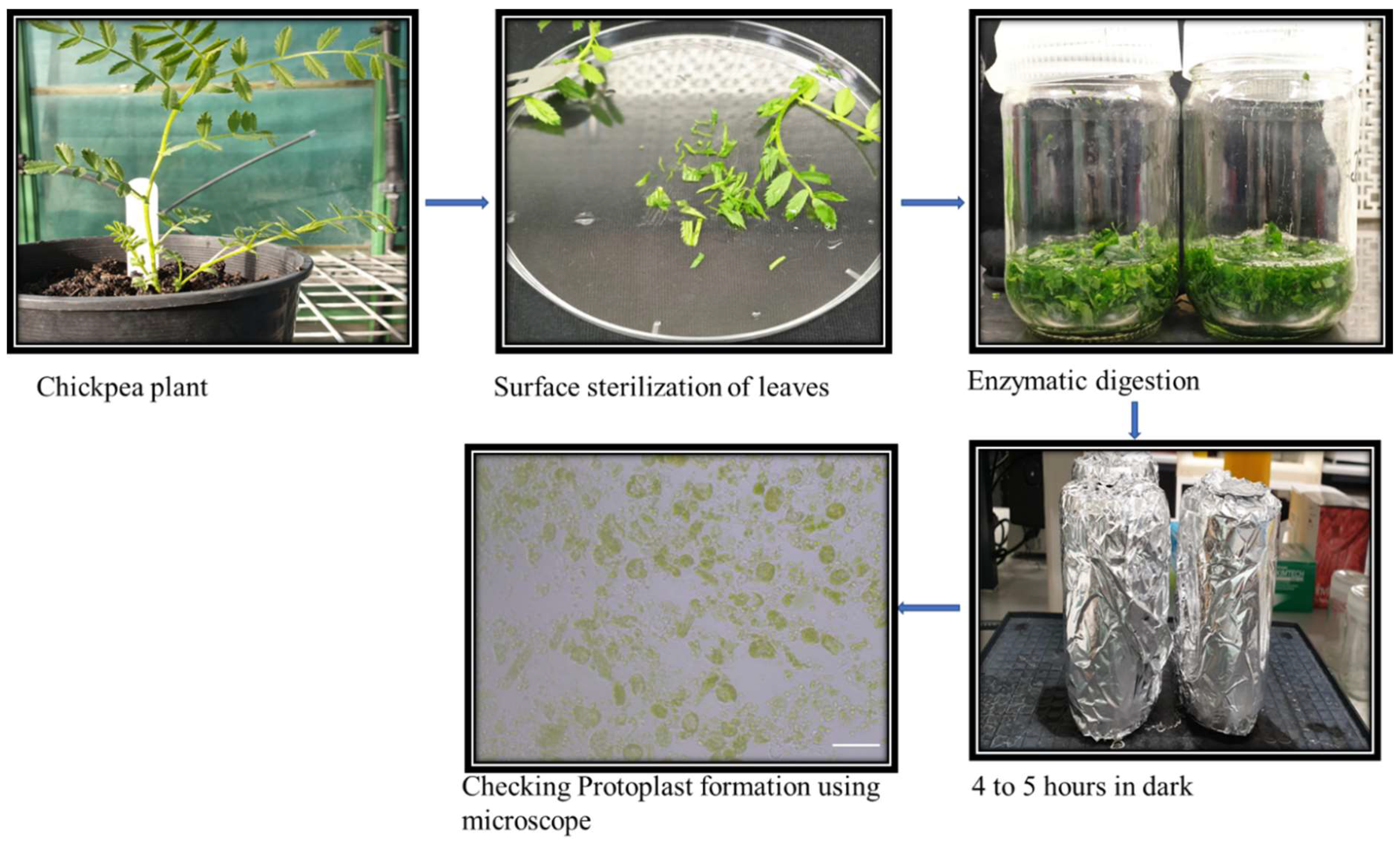
2.3. Protoplast Isolation and Transformation
2.4. Mutation Detection
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Chickpea Plant Material and Cas9 Protein
5.2. Target Site Selection and sgRNA Design
5.3. In Vitro Cleavage Assay
5.4. Protoplast Isolation
5.5. Protoplast Transformation with RNP Complex
5.6. DNA Extraction and PCR Amplification of Target Regions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| 4CL | 4-coumarate ligase |
| Cas9 | CRISPR associated protein 9 |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| GMO | Genetically modified organisms |
| gRNA | guide RNA |
| HDR | Homology-directed repair |
| NHEJ | Non-homologous end joining |
| NLS | Nuclear localization sequence |
| PAM | Protospacer adjacent motif |
| RNP | Ribonucleoprotein |
| sgRNA | Synthetic guide RNA |
| trcrRNA | tracer RNA |
References
- Baek, K.; Kim, D.H.; Jeong, J.; Sim, S.J.; Melis, A.; Kim, J.-S.; Jin, E.; Bae, S. DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins. Sci. Rep. 2016, 6, 30620. [Google Scholar] [CrossRef]
- Johansen, I.E.; Liu, Y.; Jørgensen, B.; Bennett, E.P.; Andreasson, E.; Nielsen, K.L.; Blennow, A.; Petersen, B.L. High efficacy full allelic CRISPR/Cas9 gene editing in tetraploid potato. Sci. Rep. 2019, 9, 17715. [Google Scholar] [CrossRef]
- Kim, D.; Alptekin, B.; Budak, H. CRISPR/Cas9 genome editing in wheat. Funct. Integr. Genom. 2018, 18, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Chen, K.; Li, T.; Zhang, Y.; Wang, Y.; Zhao, Q.; Liu, J.; Zhang, H.; Liu, C.; Ran, Y.; et al. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 2017, 8, 14261. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-S.; Hsu, C.-T.; Yang, L.-H.; Lee, L.-Y.; Fu, J.-Y.; Cheng, Q.-W.; Wu, F.-H.; Hsiao, H.C.-W.; Zhang, Y.; Zhang, R.; et al. Application of protoplast technology to CRISPR/Cas9 mutagenesis: From single-cell mutation detection to mutant plant regeneration. Plant Biotechnol. J. 2018, 16, 1295–1310. [Google Scholar] [CrossRef] [PubMed]
- Malnoy, M.; Viola, R.; Jung, M.-H.; Koo, O.-J.; Kim, S.; Kim, J.-S.; Velasco, R.; Nagamangala Kanchiswamy, C. DNA-Free Genetically Edited Grapevine and Apple Protoplast Using CRISPR/Cas9 Ribonucleoproteins. Front. Plant Sci. 2016, 7, 1904. [Google Scholar] [CrossRef] [PubMed]
- Murovec, J.; Guček, K.; Bohanec, B.; Avbelj, M.; Jerala, R. DNA-Free Genome Editing of Brassica oleracea and B. rapa Protoplasts Using CRISPR-Cas9 Ribonucleoprotein Complexes. Front. Plant Sci. 2018, 9, 1594. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Liang, Z.; Ren, C.; Nishitani, C.; Osakabe, K.; Wada, M.; Komori, S.; Malnoy, M.; Velasco, R.; Poli, M.; et al. CRISPR–Cas9-mediated genome editing in apple and grapevine. Nat. Protoc. 2018, 13, 2844–2863. [Google Scholar] [CrossRef]
- Petersen, B.L.; Möller, S.R.; Mravec, J.; Jørgensen, B.; Christensen, M.; Liu, Y.; Wandall, H.H.; Bennett, E.P.; Yang, Z. Improved CRISPR/Cas9 gene editing by fluorescence activated cell sorting of green fluorescence protein tagged protoplasts. BMC Biotechnol. 2019, 19, 36. [Google Scholar] [CrossRef]
- Woo, J.W.; Kim, J.; Kwon, S.I.; Corvalán, C.; Cho, S.W.; Kim, H.; Kim, S.-G.; Kim, S.-T.; Choe, S.; Kim, J.-S. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 2015, 33, 1162–1164. [Google Scholar] [CrossRef]
- Khatodia, S.; Bhatotia, K.; Passricha, N.; Khurana, S.M.P.; Tuteja, N. The CRISPR/Cas Genome-Editing Tool: Application in Improvement of Crops. Front. Plant Sci. 2016, 7, 506. [Google Scholar] [CrossRef] [PubMed]
- Noman, A.; Aqeel, M.; He, S. CRISPR-Cas9: Tool for Qualitative and Quantitative Plant Genome Editing. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Puchta, H. The repair of double-strand breaks in plants: Mechanisms and consequences for genome evolution. J. Exp. Bot. 2005, 56, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Puchta, H.; Dujon, B.; Hohn, B. Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc. Natl. Acad. Sci. USA 1996, 93, 5055. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Mao, Y.; Xu, N.; Zhang, B.; Wei, P.; Yang, D.-L.; Wang, Z.; Zhang, Z.; Zheng, R.; Yang, L.; et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 4632. [Google Scholar] [CrossRef] [PubMed]
- Klimek-Chodacka, M.; Oleszkiewicz, T.; Lowder, L.G.; Qi, Y.; Baranski, R. Efficient CRISPR/Cas9-based genome editing in carrot cells. Plant Cell Rep. 2018, 37, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, I.; Ban, Y.; Azuma, A.; Onoue, N.; Moriguchi, T.; Yamamoto, T.; Toki, S.; Endo, M. CRISPR/Cas9-mediated targeted mutagenesis in grape. PLoS ONE 2017, 12, e0177966. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, H.; Zhu, H. CRISPR technology is revolutionizing the improvement of tomato and other fruit crops. Hortic. Res. 2019, 6, 77. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, J.; Wei, P.; Zhang, B.; Gou, F.; Feng, Z.; Mao, Y.; Yang, L.; Zhang, H.; Xu, N.; et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 2014, 12, 797–807. [Google Scholar] [CrossRef]
- Liu, G.; Li, J.; Godwin, I.D. Genome Editing by CRISPR/Cas9 in Sorghum Through Biolistic Bombardment. In Sorghum: Methods and Protocols; Zhao, Z.-Y., Dahlberg, J., Eds.; Springer: New York, NY, USA, 2019; pp. 169–183. [Google Scholar] [CrossRef]
- Chilcoat, D.; Liu, Z.-B.; Sander, J. Chapter Two—Use of CRISPR/Cas9 for Crop Improvement in Maize and Soybean. In Progress in Molecular Biology and Translational Science; Weeks, D.P., Yang, B., Eds.; Academic Press: Cambridge, MA, USA, 2017; Volume 149, pp. 27–46. [Google Scholar]
- Zhang, Y.; Liang, Z.; Zong, Y.; Wang, Y.; Liu, J.; Chen, K.; Qiu, J.-L.; Gao, C. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 2016, 7, 12617. [Google Scholar] [CrossRef]
- Gao, X.; Chen, J.; Dai, X.; Zhang, D.; Zhao, Y. An Effective Strategy for Reliably Isolating Heritable and Cas9-Free Arabidopsis Mutants Generated by CRISPR/Cas9-Mediated Genome Editing. Plant Physiol. 2016, 171, 1794. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 2017, 15, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.W.; Lee, J.; Carroll, D.; Kim, J.-S.; Lee, J. Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9–sgRNA ribonucleoproteins. Genetics 2013, 195, 1177. [Google Scholar] [CrossRef] [PubMed]
- Croser, J.S.; Ahmad, F.; Clarke, H.J.; Siddique, K.H.M. Utilisation of wild Cicer in chickpea improvement—Progress, constraints, and prospects. Aust. J. Agric. Res. 2003, 54, 429–444. [Google Scholar] [CrossRef]
- Anwar, F.; Sharmila, P.; Saradhi, P.P. No more recalcitrant: Chickpea regeneration and genetic transformation. Afr. J. Biotechnol. 2010, 9, 782–797. [Google Scholar]
- Krishnamurthy, K.V.; Suhasini, K.; Sagare, A.P.; Meixner, M.; de Kathen, A.; Pickardt, T.; Schieder, O. Agrobacterium mediated transformation of chickpea (Cicer arietinum L.) embryo axes. Plant Cell Rep. 2000, 19, 235–240. [Google Scholar] [CrossRef]
- Acharjee, S.; Sarmah, B.K. Biotechnologically generating ’super chickpea’ for food and nutritional security. Plant Sci. Int. J. Exp. Plant Biol. 2013, 207, 108–116. [Google Scholar] [CrossRef]
- Bhatnagar-Mathur, P.; Vadez, V.; Devi, M.J.; Lavanya, M.; Vani, G.; Sharma, K.K. Genetic engineering of chickpea (Cicer arietinum L.) with the P5CSF129A gene for osmoregulation with implications on drought tolerance. Mol. Breed. 2009, 23, 591–606. [Google Scholar] [CrossRef]
- Rani, A.; Devi, P.; Jha, U.C.; Sharma, K.D.; Siddique, K.H.M.; Nayyar, H. Developing Climate-Resilient Chickpea Involving Physiological and Molecular Approaches With a Focus on Temperature and Drought Stresses. Front. Plant Sci. 2020, 10. [Google Scholar] [CrossRef]
- Movahedi, A.; Zhang, J.; Gao, P.; Yang, Y.; Wang, L.; Yin, T.; Kadkhodaei, S.; Ebrahimi, M.; Zhuge, Q. Expression of the chickpea CarNAC3 gene enhances salinity and drought tolerance in transgenic poplars. Plant Cell Tissue Organ Cult. (PCTOC) 2015, 120, 141–154. [Google Scholar] [CrossRef]
- Hajyzadeh, M.; Turktas, M.; Khawar, K.M.; Unver, T. miR408 overexpression causes increased drought tolerance in chickpea. Gene 2015, 555, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Anbazhagan, K.; Bhatnagar-Mathur, P.; Vadez, V.; Dumbala, S.R.; Kishor, P.B.K.; Sharma, K.K. DREB1A overexpression in transgenic chickpea alters key traits influencing plant water budget across water regimes. Plant Cell Rep. 2015, 34, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, I.; Singh, A.K.; Kaushik, M.; Amla, D.V. Agrobacterium-mediated transformation of chickpea (Cicer arietinum L.) with Bacillus thuringiensis cry1Ac gene for resistance against pod borer insect Helicoverpa armigera. Plant Sci. 2005, 168, 1135–1146. [Google Scholar] [CrossRef]
- Kumar, M.; Yusuf, M.A.; Nigam, M.; Kumar, M. An Update on Genetic Modification of Chickpea for Increased Yield and Stress Tolerance. Mol. Biotechnol. 2018, 60, 651–663. [Google Scholar] [CrossRef]
- Ehlting, J.; Büttner, D.; Wang, Q.; Douglas, C.J.; Somssich, I.E.; Kombrink, E. Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 1999, 19, 9–20. [Google Scholar] [CrossRef]
- Lee, D.; Meyer, K.; Chapple, C.; Douglas, C.J. Antisense suppression of 4-coumarate:coenzyme A ligase activity in Arabidopsis leads to altered lignin subunit composition. Plant Cell 1997, 9, 1985. [Google Scholar] [CrossRef]
- Liu, H.; Guo, Z.; Gu, F.; Ke, S.; Sun, D.; Dong, S.; Liu, W.; Huang, M.; Xiao, W.; Yang, G.; et al. 4-Coumarate-CoA Ligase-Like Gene OsAAE3 Negatively Mediates the Rice Blast Resistance, Floret Development and Lignin Biosynthesis. Front. Plant Sci. 2016, 7, 2041. [Google Scholar] [CrossRef]
- Yoshimura, K.; Masuda, A.; Kuwano, M.; Yokota, A.; Akashi, K. Programmed Proteome Response for Drought Avoidance/Tolerance in the Root of a C3 Xerophyte (Wild Watermelon) Under Water Deficits. Plant Cell Physiol. 2008, 49, 226–241. [Google Scholar] [CrossRef]
- Rawat, R.; Schwartz, J.; Jones, M.A.; Sairanen, I.; Cheng, Y.; Andersson, C.R.; Zhao, Y.; Ljung, K.; Harmer, S.L. REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc. Natl. Acad. Sci. USA 2009, 106, 16883–16888. [Google Scholar] [CrossRef]
- Kuno, N.; Moller, S.G.; Shinomura, T.; Xu, X.; Chua, N.H.; Furuya, M. The novel MYB protein EARLY-PHYTOCHROME-RESPONSIVE1 is a component of a slave circadian oscillator in Arabidopsis. Plant Cell 2003, 15, 2476–2488. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Lee, H. MYB-related transcription factors function as regulators of the circadian clock and anthocyanin biosynthesis in Arabidopsis. Plant Signal. Behav. 2016, 11, e1139278. [Google Scholar] [CrossRef] [PubMed]
- Farinas, B.; Mas, P. Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J. 2011, 66, 318–329. [Google Scholar] [CrossRef] [PubMed]
- Hilbeck, A.; Meier, M.; Römbke, J.; Jänsch, S.; Teichmann, H.; Tappeser, B. Environmental risk assessment of genetically modified plants—Concepts and controversies. Environ. Sci. Eur. 2011, 23, 13. [Google Scholar] [CrossRef]
- Carroll, D.; Charo, R.A. The societal opportunities and challenges of genome editing. Genome Biol. 2015, 16, 242. [Google Scholar] [CrossRef] [PubMed]
- Cheng, N.; Nakata, P.A. Development of a rapid and efficient protoplast isolation and transfection method for chickpea (Cicer arietinum). MethodsX 2020, 7, 101025. [Google Scholar] [CrossRef]
- Naim, F.; Shand, K.; Hayashi, S.; O’Brien, M.; McGree, J.; Johnson, A.A.T.; Dugdale, B.; Waterhouse, P.M. Are the current gRNA ranking prediction algorithms useful for genome editing in plants? PLoS ONE 2020, 15, e0227994. [Google Scholar] [CrossRef]
- Brandt, K.M.; Gunn, H.; Moretti, N.; Zemetra, R.S. A Streamlined Protocol for Wheat (Triticum aestivum) Protoplast Isolation and Transformation With CRISPR-Cas Ribonucleoprotein Complexes. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Wu, X.; Kriz, A.J.; Sharp, P.A. Target specificity of the CRISPR-Cas9 system. Quant Biol. 2014, 2, 59–70. [Google Scholar] [CrossRef]
- Ondřej, V.; Kitner, M.; Doležalová, I.; Nádvorník, P.; Navrátilová, B.; Lebeda, A. Chromatin structural rearrangement during dedifferentiation of protoplasts of Cucumis sativus L. Mol. Cells 2009, 27, 443–447. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Gao, C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 2014, 9, 2395–2410. [Google Scholar] [CrossRef]
- Li, H.; Yang, Y.; Hong, W.; Huang, M.; Wu, M.; Zhao, X. Applications of genome editing technology in the targeted therapy of human diseases: Mechanisms, advances and prospects. Signal Transduct. Target. Ther. 2020, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.T.; Fløe, L.; Petersen, T.S.; Huang, J.; Xu, F.; Bolund, L.; Luo, Y.; Lin, L. Chromatin accessibility and guide sequence secondary structure affect CRISPR-Cas9 gene editing efficiency. FEBS Lett. 2017, 591, 1892–1901. [Google Scholar] [CrossRef] [PubMed]
- Kocak, D.D.; Josephs, E.A.; Bhandarkar, V.; Adkar, S.S.; Kwon, J.B.; Gersbach, C.A. Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nat. Biotechnol. 2019, 37, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.M.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and classification of the CRISPR–Cas systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.Q.; Zheng, Z.; Nguyen, N.T.; Liebers, M.; Topkar, V.V.; Thapar, V.; Wyvekens, N.; Khayter, C.; Iafrate, A.J.; Le, L.P.; et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015, 33, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Cao, Y.; Han, Y.; Li, D.; Lin, Y.; Cai, Y. Systematic Analysis of the 4-Coumarate:Coenzyme A Ligase (4CL) Related Genes and Expression Profiling during Fruit Development in the Chinese Pear. Genes (Basel) 2016, 7, 89. [Google Scholar] [CrossRef]
- Dixon, R.A.; Paiva, N.L. Stress-Induced Phenylpropanoid Metabolism. Plant Cell 1995, 7, 1085–1097. [Google Scholar] [CrossRef]
- Hu, Y.; Li, W.C.; Xu, Y.; Li, G.; Liao, Y.; Fu, F.-L. Differential expression of candidate genes for lignin biosynthesis under drought stress in maize leaves. J. Appl. Genet. 2009, 50, 213–223. [Google Scholar] [CrossRef]
- Sun, S.-C.; Xiong, X.-P.; Zhang, X.-L.; Feng, H.-J.; Zhu, Q.-H.; Sun, J.; Li, Y.-J. Characterization of the Gh4CL gene family reveals a role of Gh4CL7 in drought tolerance. BMC Plant Biol. 2020, 20, 125. [Google Scholar] [CrossRef]
- Moenga, S.M.; Gai, Y.; Carrasquilla-Garcia, N.; Perilla-Henao, L.M.; Cook, D.R. Gene co-expression analysis reveals transcriptome divergence between wild and cultivated chickpea under drought stress. Plant J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.X.; Knowles, S.M.; Andronis, C.; Ong, M.S.; Tobin, E.M. Circadian clock associated1 and late elongated hypocotyl function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 2009, 150, 834–843. [Google Scholar] [CrossRef] [PubMed]
- Cavicchioli, R.; Ripple, W.J.; Timmis, K.N.; Azam, F.; Bakken, L.R.; Baylis, M.; Behrenfeld, M.J.; Boetius, A.; Boyd, P.W.; Classen, A.T.; et al. Scientists’ warning to humanity: Microorganisms and climate change. Nat. Rev. Microbiol. 2019, 17, 569–586. [Google Scholar] [CrossRef] [PubMed]
- Labun, K.; Montague, T.G.; Krause, M.; Torres Cleuren, Y.N.; Tjeldnes, H.; Valen, E. CHOPCHOP v3: Expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019, 47, W171–W174. [Google Scholar] [CrossRef] [PubMed]
- Subburaj, S.; Chung, S.J.; Lee, C.; Ryu, S.-M.; Kim, D.H.; Kim, J.-S.; Bae, S.; Lee, G.-J. Site-directed mutagenesis in Petunia × hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep. 2016, 35, 1535–1544. [Google Scholar] [CrossRef] [PubMed]
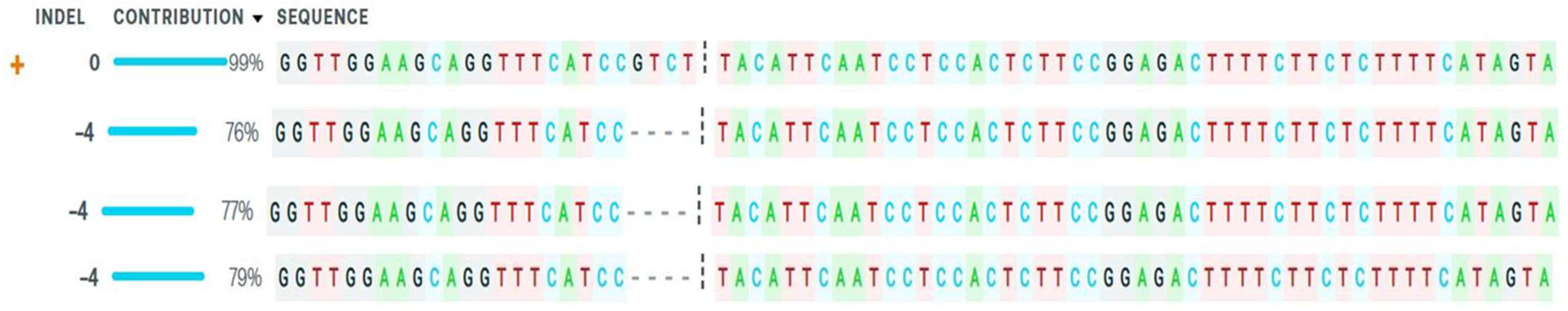
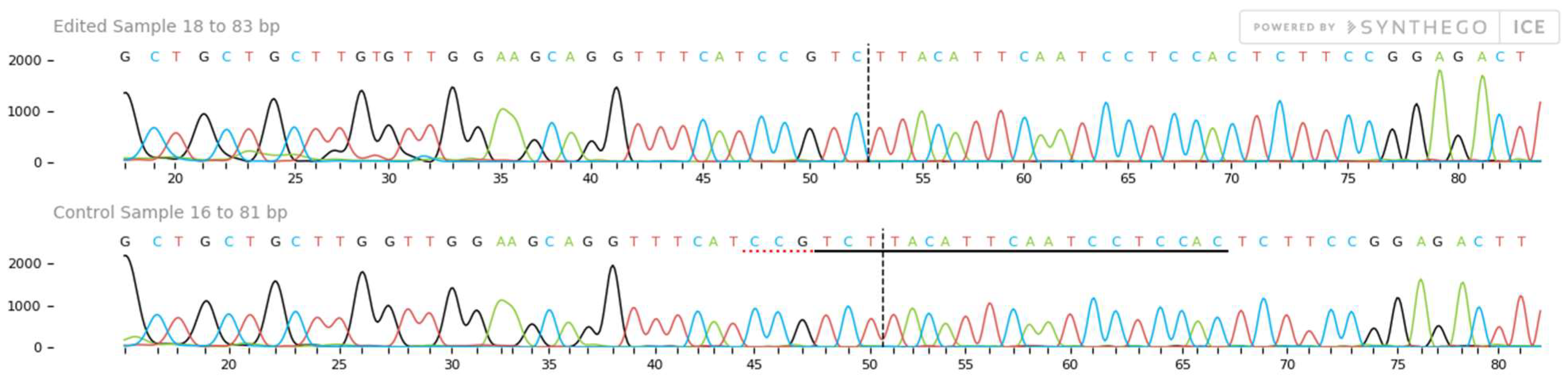
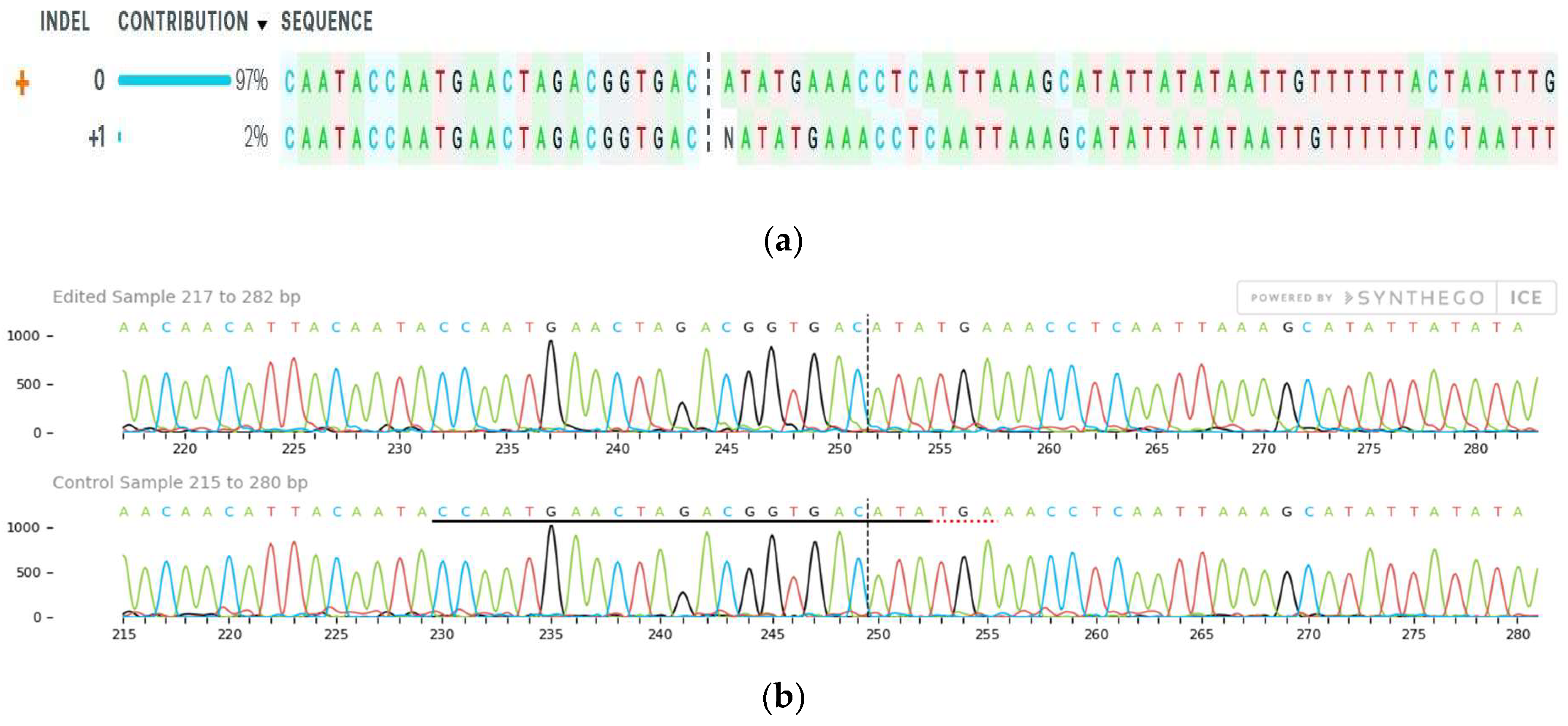
| Gene Name | ICC8261 (Drought Tolerant Genotype) Expression Levels | ICC283 (Sensitive Tolerant Genotype) Expression Levels e 3 |
|---|---|---|
| RVE7 | SAM 2.69, FB-3.4, POF0, FOF-1.29, YP1.9 | SAM 3.4, FB-3.02, POF0, FOF-5.9, YP 0 |
| 4CL | FB 10.39 | Not Differentially expressed data |
| Gene Name | sgRNA Name | sgRNA Sequence |
|---|---|---|
| 4-coumarate-CoA ligase-like 1 NW_004516753.1_162798-165892 Length 3094 | 4CLsgRNA1 4CLsgRNA2 | TATGTCACCGTCTAGTTCATTGG GTTTAGGTTACCGAACGAAGAGG |
| REVEILLE 7-like NW_004516329.1_420654-4253847 | RVE7sgRNA1 | GTGGAGGATTGAATGTAAGACGG |
| RVE7sgRNA2 | AGTGTGCAGCTGATGTATCGAGG |
| Primer Sets | Sequence | Expected Product Size |
|---|---|---|
| 4CL Primer Set1 | Forward: ACAATACCAATGAACTAGACGGTGA Reverse: TCCCTAACAAAATCCAACACATCT | 592 |
| 4CL Primer Set2 | Forward: ACAATACCAATGAACTAGACGGTG Reverse: TCCCTAACAAAATCCAACACATC | 590 |
| RVE7 Primer Set1 | Forward: AACATGCTGCTGCTTGGTTG Reverse: GACGAAGAGAGGGACTAATTTCA | 398 |
| RVE7 Primer Set2 | Forward: GGAAGCAGGTTTCATCCGTC Reverse: TGATGAAAGAAATTGATGCTCACTA | 420 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Badhan, S.; Ball, A.S.; Mantri, N. First Report of CRISPR/Cas9 Mediated DNA-Free Editing of 4CL and RVE7 Genes in Chickpea Protoplasts. Int. J. Mol. Sci. 2021, 22, 396. https://doi.org/10.3390/ijms22010396
Badhan S, Ball AS, Mantri N. First Report of CRISPR/Cas9 Mediated DNA-Free Editing of 4CL and RVE7 Genes in Chickpea Protoplasts. International Journal of Molecular Sciences. 2021; 22(1):396. https://doi.org/10.3390/ijms22010396
Chicago/Turabian StyleBadhan, Sapna, Andrew S. Ball, and Nitin Mantri. 2021. "First Report of CRISPR/Cas9 Mediated DNA-Free Editing of 4CL and RVE7 Genes in Chickpea Protoplasts" International Journal of Molecular Sciences 22, no. 1: 396. https://doi.org/10.3390/ijms22010396
APA StyleBadhan, S., Ball, A. S., & Mantri, N. (2021). First Report of CRISPR/Cas9 Mediated DNA-Free Editing of 4CL and RVE7 Genes in Chickpea Protoplasts. International Journal of Molecular Sciences, 22(1), 396. https://doi.org/10.3390/ijms22010396





