Proteomic Profiling of the First Human Dental Pulp Mesenchymal Stem/Stromal Cells from Carbonic Anhydrase II Deficiency Osteopetrosis Patients
Abstract
1. Introduction
2. Results
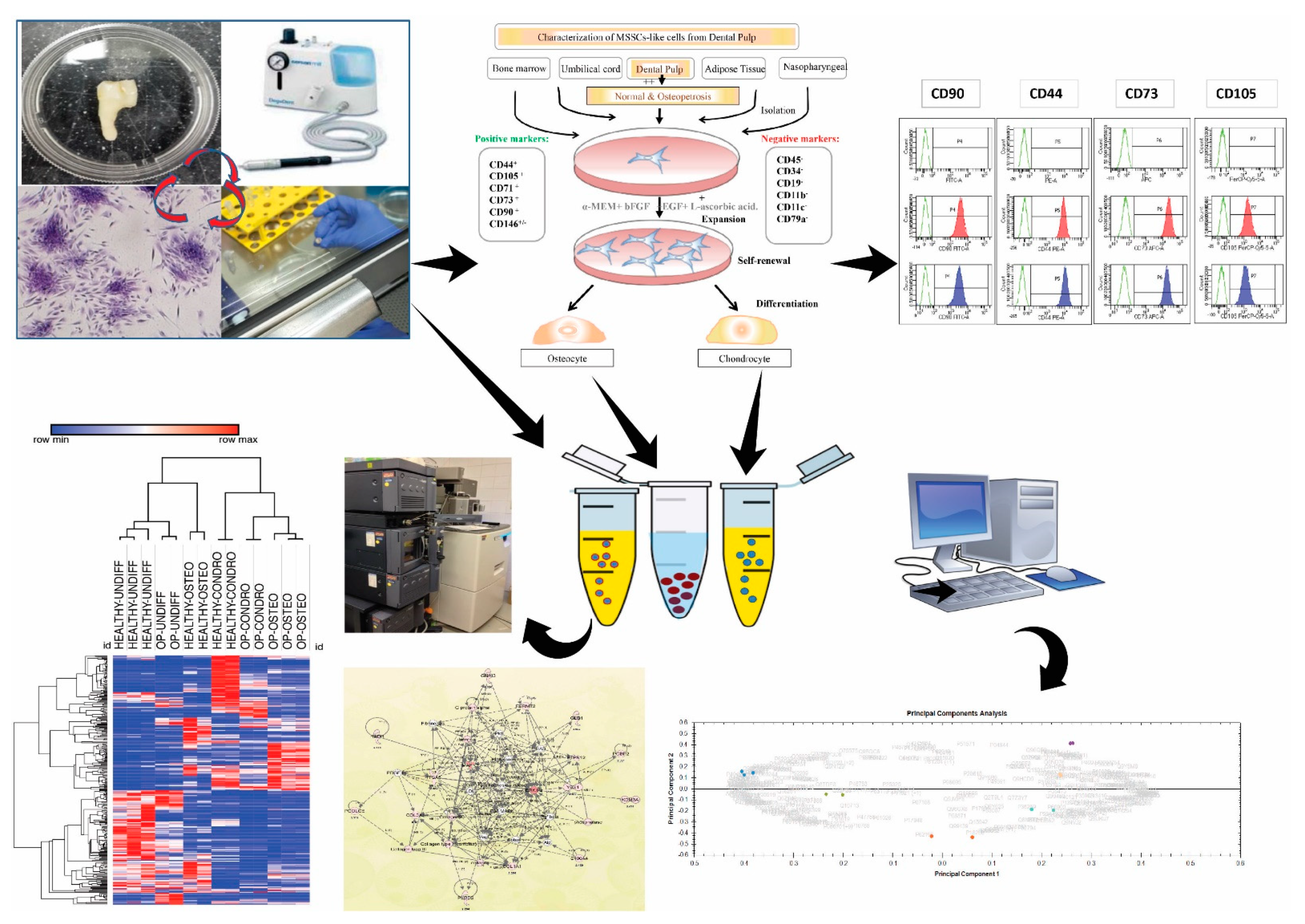
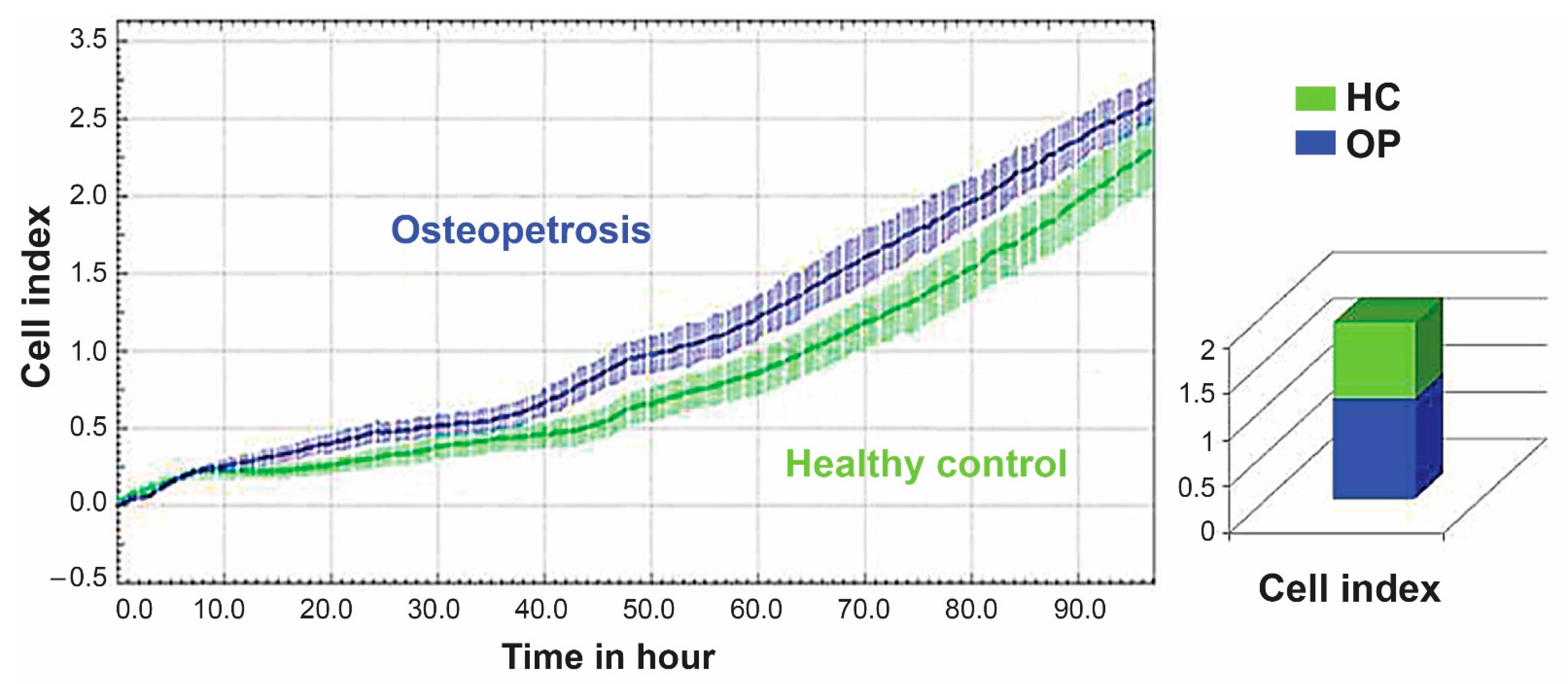
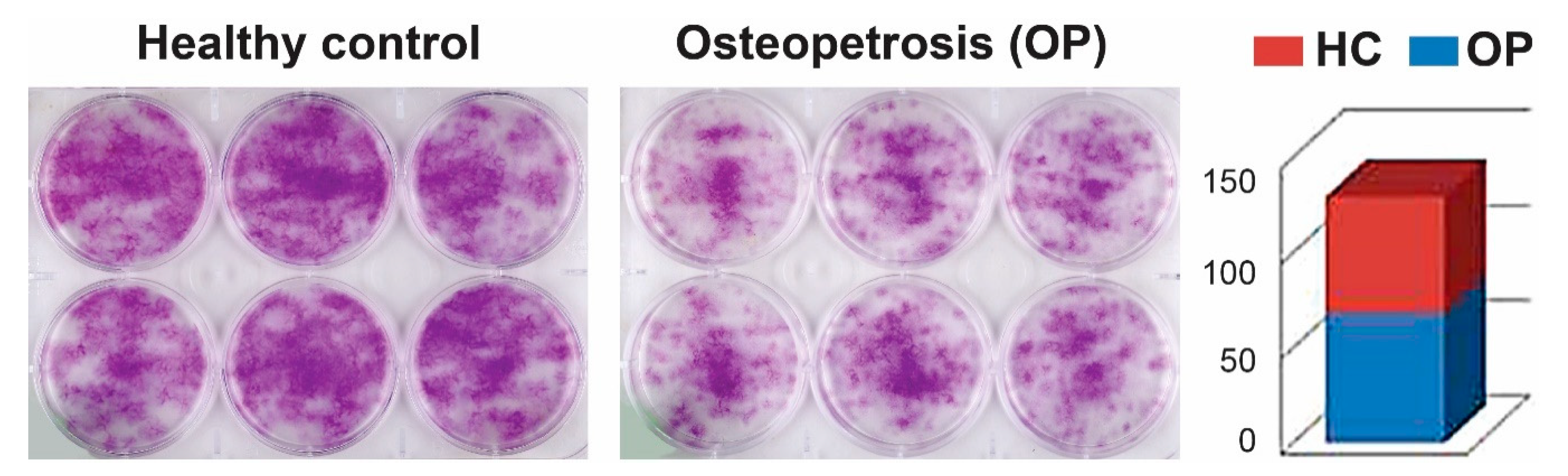
2.1. Cell Proliferation and Colony-Forming Capacity of Mesenchymal Cells from Dental Pulp
2.2. Characterization of MSC-Like Cells from Dental Pulp
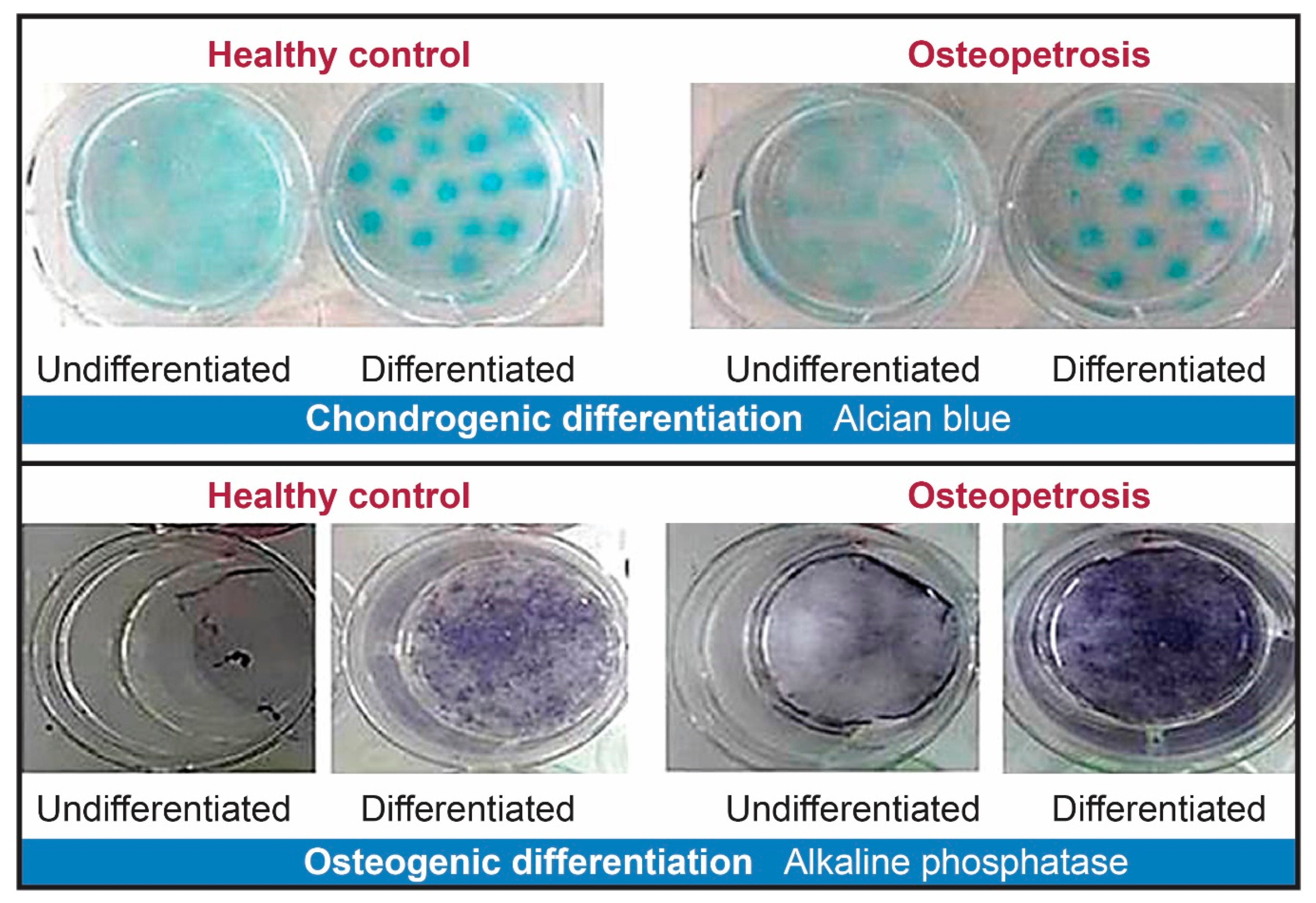
2.3. Cell Differentiation of Dental Pulp Mesenchymal Cells
2.4. Protein Expression Signatures of Undifferentiated DP MSCs
2.5. Protein Signatures Related to Osteogenesis in HC and OP DP-MSSCs
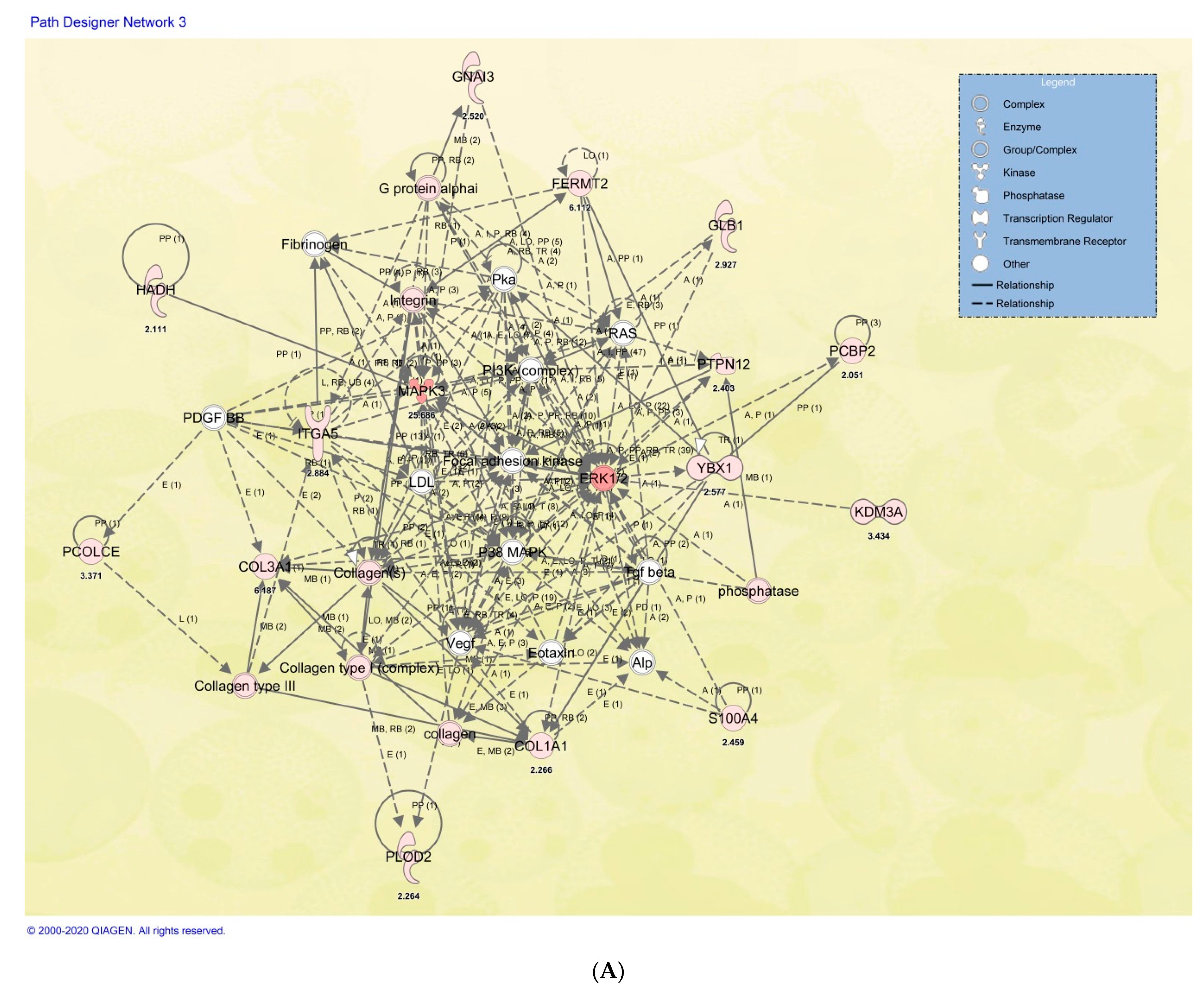
2.6. Cellular and Functional Annotations of the Identified DP-MSSCs
3. Discussion
4. Materials and Methods
4.1. Patient Materials
4.2. Cell Isolation, Culture, and Differentiation of Dental Pulp Mesenchymal Cells
4.3. Cell Proliferation Assay
4.4. Dental Pulp Mesenchymal Stem Cell Differentiation
4.5. Flow Cytometry
4.6. Cell Differentiation, Expression Proteomics, and Biomarker Discovery for SHED
4.7. Colony Forming Potential of Isolated Dental Pulp Mesenchymal Cells
4.8. Protein in-Solution Digestion
4.9. Protein Identification by Liquid Chromatography Mass Spectrometry (LC–MSE) Analysis
4.10. Data Analysis and Informatics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SHED | stem cells from human exfoliated deciduous teeth |
| MSCs | mesenchymal stem cells |
| LC–MS/MS | liquid chromatography–tandem mass spectrometry |
| CA-II | carbonic anhydrase II |
| HC | healthy control |
References
- Penna, S.; Capo, V.; Palagano, E.; Sobacchi, C.; Villa, A. One Disease, Many Genes: Implications for the Treatment of Osteopetroses. Front. Endocrinol. 2019, 10, 85. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.C.; Econs, M.J.; DiMeglio, L.A.; Insogna, K.L.; Levine, M.A.; Orchard, P.J.; Miller, W.P.; Petryk, A.; Rush, E.T.; Shoback, D.M.; et al. Diagnosis and Management of Osteopetrosis: Consensus Guidelines From the Osteopetrosis Working Group. J.Clin. Endocrinol. Metab. 2017, 102, 3111–3123. [Google Scholar] [CrossRef] [PubMed]
- Stark, Z.; Savarirayan, R. Osteopetrosis. Orphanet J. Rare Dis. 2009, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Bedi, R.S.; Goel, P.; Pasricha, N.; Sachin; Goel, A. Osteopetrosis-A rare entity with osteomyelitis. Ann. Maxillofac. Surg. 2011, 1, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Nagai, R.; Kooh, S.W.; Balfe, J.W.; Fenton, T.; Halperin, M.L. Renal tubular acidosis and osteopetrosis with carbonic anhydrase II deficiency: Pathogenesis of impaired acidification. Pediatric Nephrol. 1997, 11, 633–636. [Google Scholar] [CrossRef] [PubMed]
- Ohlsson, A.; Stark, G.; Sakati, N. Marble brain disease: Recessive osteopetrosis, renal tubular acidosis and cerebral calcification in three Saudi Arabian families. Dev. Med. Child Neurol. 1980, 22, 72–84. [Google Scholar] [CrossRef]
- Shah, G.N.; Bonapace, G.; Hu, P.Y.; Strisciuglio, P.; Sly, W.S. Carbonic anhydrase II deficiency syndrome (osteopetrosis with renal tubular acidosis and brain calcification): Novel mutations in CA2 identified by direct sequencing expand the opportunity for genotype-phenotype correlation. Hum. Mutat. 2004, 24, 272. [Google Scholar] [CrossRef]
- Sly, W.S.; Hewett-Emmett, D.; Whyte, M.P.; Yu, Y.S.; Tashian, R.E. Carbonic anhydrase II deficiency identified as the primary defect in the autosomal recessive syndrome of osteopetrosis with renal tubular acidosis and cerebral calcification. Proc. Natl. Acad. Sci. USA 1983, 80, 2752–2756. [Google Scholar] [CrossRef]
- Del Fattore, A.; Peruzzi, B.; Rucci, N.; Recchia, I.; Cappariello, A.; Longo, M.; Fortunati, D.; Ballanti, P.; Iacobini, M.; Luciani, M.; et al. Clinical, genetic, and cellular analysis of 49 osteopetrotic patients: Implications for diagnosis and treatment. J. Med. Genet. 2006, 43, 315–325. [Google Scholar] [CrossRef][Green Version]
- Sharpe, P.T. Dental mesenchymal stem cells. Development 2016, 143, 2273–2280. [Google Scholar] [CrossRef]
- Miura, M.; Gronthos, S.; Zhao, M.; Lu, B.; Fisher, L.W.; Robey, P.G.; Shi, S. SHED: Stem cells from human exfoliated deciduous teeth. Proc. Natl. Acad. Sci. USA 2003, 100, 5807–5812. [Google Scholar] [CrossRef] [PubMed]
- Yasui, T.; Mabuchi, Y.; Morikawa, S.; Onizawa, K.; Akazawa, C.; Nakagawa, T.; Okano, H.; Matsuzaki, Y. Isolation of dental pulp stem cells with high osteogenic potential. Inflamm. Regen. 2017, 37, 8. [Google Scholar] [CrossRef] [PubMed]
- Yasui, T.; Mabuchi, Y.; Toriumi, H.; Ebine, T.; Niibe, K.; Houlihan, D.D.; Morikawa, S.; Onizawa, K.; Kawana, H.; Akazawa, C.; et al. Purified Human Dental Pulp Stem Cells Promote Osteogenic Regeneration. J. Dent. Res. 2016, 95, 206–214. [Google Scholar] [CrossRef]
- Yasui, T.; Uezono, N.; Nakashima, H.; Noguchi, H.; Matsuda, T.; Noda-Andoh, T.; Okano, H.; Nakashima, K. Hypoxia Epigenetically Confers Astrocytic Differentiation Potential on Human Pluripotent Cell-Derived Neural Precursor Cells. Stem Cell Rep. 2017, 8, 1743–1756. [Google Scholar] [CrossRef] [PubMed]
- Arora, V.; Arora, P.; Munshi, A.K. Banking stem cells from human exfoliated deciduous teeth (SHED): Saving for the future. J. Clin. Pediatr. Dent. 2009, 33, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Tang, X.; Cehreli, Z.C.; Dai, X.; Xu, J.; Zhu, H. Autologous transplantation of deciduous tooth pulp into necrotic young permanent teeth for pulp regeneration in a dog model. J. Int. Med. Res. 2019, 47, 5094–5105. [Google Scholar] [CrossRef]
- Ma, L.; Makino, Y.; Yamaza, H.; Akiyama, K.; Hoshino, Y.; Song, G.; Kukita, T.; Nonaka, K.; Shi, S.; Yamaza, T. Cryopreserved dental pulp tissues of exfoliated deciduous teeth is a feasible stem cell resource for regenerative medicine. PLoS ONE 2012, 7, e51777. [Google Scholar] [CrossRef]
- Taguchi, T.; Yanagi, Y.; Yoshimaru, K.; Zhang, X.Y.; Matsuura, T.; Nakayama, K.; Kobayashi, E.; Yamaza, H.; Nonaka, K.; Ohga, S.; et al. Regenerative medicine using stem cells from human exfoliated deciduous teeth (SHED): A promising new treatment in pediatric surgery. Surg. Today 2019, 49, 316–322. [Google Scholar] [CrossRef]
- Andrukhov, O.; Behm, C.; Blufstein, A.; Rausch-Fan, X. Immunomodulatory properties of dental tissue-derived mesenchymal stem cells: Implication in disease and tissue regeneration. World J. Stem Cells 2019, 11, 604–617. [Google Scholar] [CrossRef]
- Yamaza, T.; Kentaro, A.; Chen, C.; Liu, Y.; Shi, Y.; Gronthos, S.; Wang, S.; Shi, S. Immunomodulatory properties of stem cells from human exfoliated deciduous teeth. Stem Cell Res. Ther. 2010, 1, 5. [Google Scholar] [CrossRef]
- Bento, L.W.; Zhang, Z.; Imai, A.; Nor, F.; Dong, Z.; Shi, S.; Araujo, F.B.; Nor, J.E. Endothelial differentiation of SHED requires MEK1/ERK signaling. J. Dent. Res. 2013, 92, 51–57. [Google Scholar] [CrossRef]
- Fu, X.; Jin, L.; Ma, P.; Fan, Z.; Wang, S. Allogeneic stem cells from deciduous teeth in treatment for periodontitis in miniature swine. J. Periodontol. 2014, 85, 845–851. [Google Scholar] [CrossRef] [PubMed]
- Urraca, N.; Memon, R.; El-Iyachi, I.; Goorha, S.; Valdez, C.; Tran, Q.T.; Scroggs, R.; Miranda-Carboni, G.A.; Donaldson, M.; Bridges, D.; et al. Characterization of neurons from immortalized dental pulp stem cells for the study of neurogenetic disorders. Stem Cell Res. 2015, 15, 722–730. [Google Scholar] [CrossRef] [PubMed]
- Verma, K.; Bains, R.; Bains, V.K.; Rawtiya, M.; Loomba, K.; Srivastava, S.C. Therapeutic potential of dental pulp stem cells in regenerative medicine: An overview. Dent. Res. J. 2014, 11, 302–308. [Google Scholar]
- Yang, J.W.; Shin, Y.Y.; Seo, Y.; Kim, H.S. Therapeutic Functions of Stem Cells from Oral Cavity: An Update. Int. J. Mol. Sci. 2020, 21, 4389. [Google Scholar] [CrossRef]
- Cao, H.; Yan, Q.; Wang, D.; Lai, Y.; Zhou, B.; Zhang, Q.; Jin, W.; Lin, S.; Lei, Y.; Ma, L.; et al. Focal adhesion protein Kindlin-2 regulates bone homeostasis in mice. Bone Res. 2020, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; van den Hil, F.E.; Mummery, C.L.; Orlova, V.V. Generation and Functional Characterization of Monocytes and Macrophages Derived from Human Induced Pluripotent Stem Cells. Curr. Protoc. Stem Cell Biol. 2020, 52, e108. [Google Scholar] [CrossRef]
- Kerkis, I.; Caplan, A.I. Stem cells in dental pulp of deciduous teeth. Tissue Eng. Part B Rev. 2012, 18, 129–138. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Gronthos, S.; Brahim, J.; Li, W.; Fisher, L.W.; Cherman, N.; Boyde, A.; DenBesten, P.; Robey, P.G.; Shi, S. Stem cell properties of human dental pulp stem cells. J. Dent. Res. 2002, 81, 531–535. [Google Scholar] [CrossRef]
- Okur, F.V.; Cevher, I.; Ozdemir, C.; Kocaefe, C.; Cetinkaya, D.U. Osteopetrotic induced pluripotent stem cells derived from patients with different disease-associated mutations by non-integrating reprogramming methods. Stem Cell Res. Ther. 2019, 10, 211. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro, M.M.; Dong, Z.; Kaneko, T.; Zhang, Z.; Miyazawa, M.; Shi, S.; Smith, A.J.; Nor, J.E. Dental pulp tissue engineering with stem cells from exfoliated deciduous teeth. J. Endod. 2008, 34, 962–969. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.T.; Yamaza, T.; Shea, L.D.; Djouad, F.; Kuhn, N.Z.; Tuan, R.S.; Shi, S. Stem/progenitor cell-mediated de novo regeneration of dental pulp with newly deposited continuous layer of dentin in an in vivo model. Tissue Eng. Part A 2010, 16, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, M.; Iohara, K. Mobilized dental pulp stem cells for pulp regeneration: Initiation of clinical trial. J. Endod. 2014, 40, S26–S32. [Google Scholar] [CrossRef] [PubMed]
- Parra-Torres, A.Y.; Valdés-Flores, M.; Orozco, L.; Velázquez-Cruz, R. Molecular Aspects of Bone Remodeling. Top. Osteoporos. 2013. [Google Scholar] [CrossRef]
- Jilka, R.L. Molecular and cellular mechanisms of the anabolic effect of intermittent PTH. Bone 2007, 40, 1434–1446. [Google Scholar] [CrossRef] [PubMed]
- Arany, P.R.; Cho, A.; Hunt, T.D.; Sidhu, G.; Shin, K.; Hahm, E.; Huang, G.X.; Weaver, J.; Chen, A.C.; Padwa, B.L.; et al. Photoactivation of endogenous latent transforming growth factor-beta1 directs dental stem cell differentiation for regeneration. Sci. Transl. Med. 2014, 6, 238ra269. [Google Scholar] [CrossRef]
- Choi, S.C.; Kim, S.J.; Choi, J.H.; Park, C.Y.; Shim, W.J.; Lim, D.S. Fibroblast growth factor-2 and -4 promote the proliferation of bone marrow mesenchymal stem cells by the activation of the PI3K-Akt and ERK1/2 signaling pathways. Stem Cells Dev. 2008, 17, 725–736. [Google Scholar] [CrossRef]
- Schneider, M.R.; Sibilia, M.; Erben, R.G. The EGFR network in bone biology and pathology. Trends Endocrinol. Metab. 2009, 20, 517–524. [Google Scholar] [CrossRef]
- Lee, D.Y.; Yeh, C.R.; Chang, S.F.; Lee, P.L.; Chien, S.; Cheng, C.K.; Chiu, J.J. Integrin-mediated expression of bone formation-related genes in osteoblast-like cells in response to fluid shear stress: Roles of extracellular matrix, Shc, and mitogen-activated protein kinase. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2008, 23, 1140–1149. [Google Scholar] [CrossRef]
- Healy, J.A.; Nugent, A.; Rempel, R.E.; Moffitt, A.B.; Davis, N.S.; Jiang, X.; Shingleton, J.R.; Zhang, J.; Love, C.; Datta, J.; et al. GNA13 loss in germinal center B cells leads to impaired apoptosis and promotes lymphoma in vivo. Blood 2016, 127, 2723–2731. [Google Scholar] [CrossRef] [PubMed]
- Love, C.; Sun, Z.; Jima, D.; Li, G.; Zhang, J.; Miles, R.; Richards, K.L.; Dunphy, C.H.; Choi, W.W.; Srivastava, G.; et al. The genetic landscape of mutations in Burkitt lymphoma. Nat. Genet. 2012, 44, 1321–1325. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.X.; Yun, M.; Xu, Y.; Chen, J.W.; Weng, H.W.; Zheng, Z.S.; Chen, C.; Xie, D.; Ye, S. GNA13 as a prognostic factor and mediator of gastric cancer progression. Oncotarget 2016, 7, 4414–4427. [Google Scholar] [CrossRef] [PubMed]
- Volk, S.W.; Shah, S.R.; Cohen, A.J.; Wang, Y.; Brisson, B.K.; Vogel, L.K.; Hankenson, K.D.; Adams, S.L. Type III collagen regulates osteoblastogenesis and the quantity of trabecular bone. Calcif. Tissue Int. 2014, 94, 621–631. [Google Scholar] [CrossRef]
- Alaiya, A.A.; Aljurf, M.; Shinwari, Z.; Almohareb, F.; Malhan, H.; Alzahrani, H.; Owaidah, T.; Fox, J.; Alsharif, F.; Mohamed, S.Y.; et al. Protein signatures as potential surrogate biomarkers for stratification and prediction of treatment response in chronic myeloid leukemia patients. Int. J. Oncol. 2016, 49, 913–933. [Google Scholar] [CrossRef]
- Kassem, D.H.; Kamal, M.M.; El-Kholy Ael, L.; El-Mesallamy, H.O. Exendin-4 enhances the differentiation of Wharton’s jelly mesenchymal stem cells into insulin-producing cells through activation of various beta-cell markers. Stem Cell Res. Ther. 2016, 7, 108. [Google Scholar] [CrossRef]
- Kassem, D.H.; Kamal, M.M.; El-Kholy Ael, L.; El-Mesallamy, H.O. Association of expression levels of pluripotency/stem cell markers with the differentiation outcome of Wharton’s jelly mesenchymal stem cells into insulin producing cells. Biochimie 2016, 127, 187–195. [Google Scholar] [CrossRef]
- Kassem, M.; Kristiansen, M.; Abdallah, B.M. Mesenchymal stem cells: Cell biology and potential use in therapy. Basic Clin. Pharmacol. Toxicol. 2004, 95, 209–214. [Google Scholar] [CrossRef]
- Murakami, M.; Horibe, H.; Iohara, K.; Hayashi, Y.; Osako, Y.; Takei, Y.; Nakata, K.; Motoyama, N.; Kurita, K.; Nakashima, M. The use of granulocyte-colony stimulating factor induced mobilization for isolation of dental pulp stem cells with high regenerative potential. Biomaterials 2013, 34, 9036–9047. [Google Scholar] [CrossRef]
- Basmaeil, Y.S.; Algudiri, D.; Alenzi, R.; Al Subayyil, A.; Alaiya, A.; Khatlani, T. HMOX1 is partly responsible for phenotypic and functional abnormalities in mesenchymal stem cells/stromal cells from placenta of preeclampsia (PE) patients. Stem Cell Res. Ther. 2020, 11, 30. [Google Scholar] [CrossRef]


| Patient | Age (Years) | Gender | Mode/Gene | Developmental | Cranial/Bone | Renal |
|---|---|---|---|---|---|---|
| DP#42-5242 | 9 | M | CAII (IVS2 + 1G > A) | Delayed | Fracture ++ | RTA |
| DP#47-6342 | 8 | F | AR-CAII c.232 + 1G > A, IVS2 + 1G > A | Short Stature | Calcification | RTA |
| DP#52-4774 | 14 | F | CAII | ADHD, Delayed | Calcification | RTA |
| DP#53-3492 | 13 | M | CAII | Delayed | Calcification, Fractures | RTA |
| DP#54-5544 | 7 | M | AD-CAII c.232 + 1G > A; IVS2 + 1G > A | Delayed | Calcification, Fractures | RTA |
| DP#31-7530 | 9 | F | Healthy | Normal | No Abnormalities | N/A |
| DP#41-1082 | 7 | F | Healthy | Normal | No Abnormalities | N/A |
| DP#46-7880 | 8 | M | Healthy | Normal | No Abnormalities | N/A |
| DP#50-4300 | 14 | M | Healthy | Normal | No Abnormalities | N/A |
| DP#51-5231 | 7 | M | Healthy | Normal | No Abnormalities | N/A |
| DP#57-4600 | 11 | F | Healthy | Normal | No Abnormalities | N/A |
| Symbol | Entrez Gene Name | GenPept/ UniProt/Swiss-Prot Accession | Expr Fold Change OP:HC | Location | Family | Bone Involvement |
|---|---|---|---|---|---|---|
| COL1A1 | Collagen type I alpha 1 chain | P02452 | −2.26587705 | Extracellular Space | Other | Bone matrix/Osteoblast |
| COL3A1 | Collagen type III alpha 1 chain | P02461 | −6.1868361 | Extracellular Space | Other | Bone matrix/Osteoblast |
| FERMT2 | Fermitin family member 2 | Q96AC1 | −6.11159085 | Cytoplasm | Other | Bone homeostasis regulation |
| GLB1 | Galactosidase beta 1 | P16278 | 2.92715725 | Cytoplasm | Enzyme | |
| GNAI3 | G protein subunit alpha i3 | P08754 | −2.52033996 | Cytoplasm | Enzyme | Impaired activation of integrin and ERK1/2. |
| HADH | Hydroxyacyl-CoA dehydrogenase | Q16836 | 2.1108754 | Cytoplasm | Enzyme | In OP gene panel |
| ITGA5 | Integrin subunit alpha 5 | P08648 | 2.88383695 | Plasma Membrane | Transmembrane Receptor | osteoclastogenesis |
| KDM3A | Lysine demethylase 3A | Q9Y4C1 | 3.43441641 | Nucleus | Transcription regulator | OP associated |
| MAPK3 | Mitogen-activated protein kinase 3 | P27361 | 25.6856293 | Cytoplasm | Kinase | Bone Diseases |
| PCBP2 | Poly(rC) binding protein 2 | Q15366 | 2.05142965 | Nucleus | Other | |
| PCOLCE | Procollagen C-endopeptidase enhancer | Q15113 | 3.37138722 | Extracellular Space | Other | osteoclast–osteoblast bone modeling |
| PLOD2 | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 | O00469 | 2.26356027 | Cytoplasm | Enzyme | bone mass syndromes |
| PTPN12 | Protein tyrosine phosphatase non-receptor type 12 | Q05209 | 2.40278726 | Cytoplasm | Phosphatase | Bone resorbing |
| S100A4 | S100 calcium binding protein A4 | P26447 | 2.45896457 | Cytoplasm | Other | Bone resorption osteoclast function |
| YBX1 | Y-box binding protein 1 | P67809 | 2.57704761 | Nucleus | Transcription regulator |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alkhayal, Z.; Shinwari, Z.; Gaafar, A.; Alaiya, A. Proteomic Profiling of the First Human Dental Pulp Mesenchymal Stem/Stromal Cells from Carbonic Anhydrase II Deficiency Osteopetrosis Patients. Int. J. Mol. Sci. 2021, 22, 380. https://doi.org/10.3390/ijms22010380
Alkhayal Z, Shinwari Z, Gaafar A, Alaiya A. Proteomic Profiling of the First Human Dental Pulp Mesenchymal Stem/Stromal Cells from Carbonic Anhydrase II Deficiency Osteopetrosis Patients. International Journal of Molecular Sciences. 2021; 22(1):380. https://doi.org/10.3390/ijms22010380
Chicago/Turabian StyleAlkhayal, Zikra, Zakia Shinwari, Ameera Gaafar, and Ayodele Alaiya. 2021. "Proteomic Profiling of the First Human Dental Pulp Mesenchymal Stem/Stromal Cells from Carbonic Anhydrase II Deficiency Osteopetrosis Patients" International Journal of Molecular Sciences 22, no. 1: 380. https://doi.org/10.3390/ijms22010380
APA StyleAlkhayal, Z., Shinwari, Z., Gaafar, A., & Alaiya, A. (2021). Proteomic Profiling of the First Human Dental Pulp Mesenchymal Stem/Stromal Cells from Carbonic Anhydrase II Deficiency Osteopetrosis Patients. International Journal of Molecular Sciences, 22(1), 380. https://doi.org/10.3390/ijms22010380





