Exploring the Potential of Drug Response Assays for Precision Medicine in Ovarian Cancer
Abstract
1. Introduction
2. Commercially Tested Drug-Response Assays in Ovarian Cancer
Current Clinical Use of CRAs
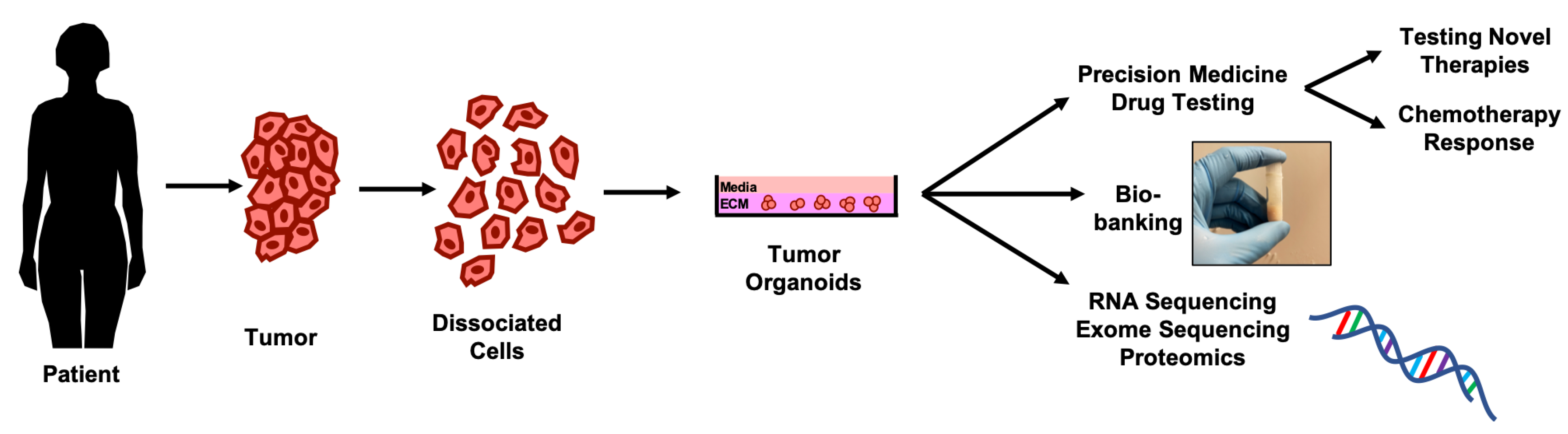
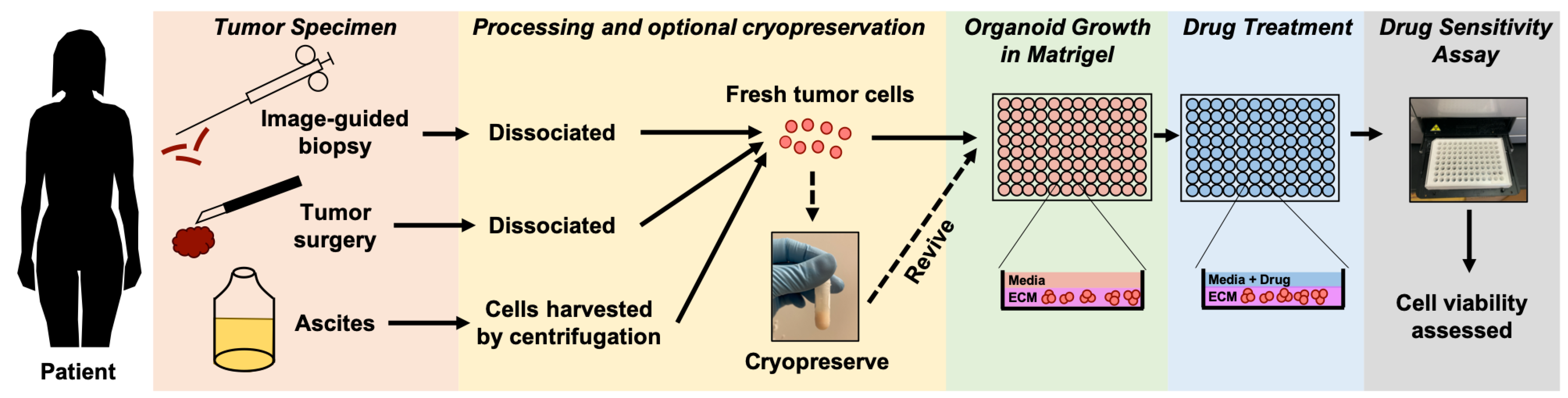
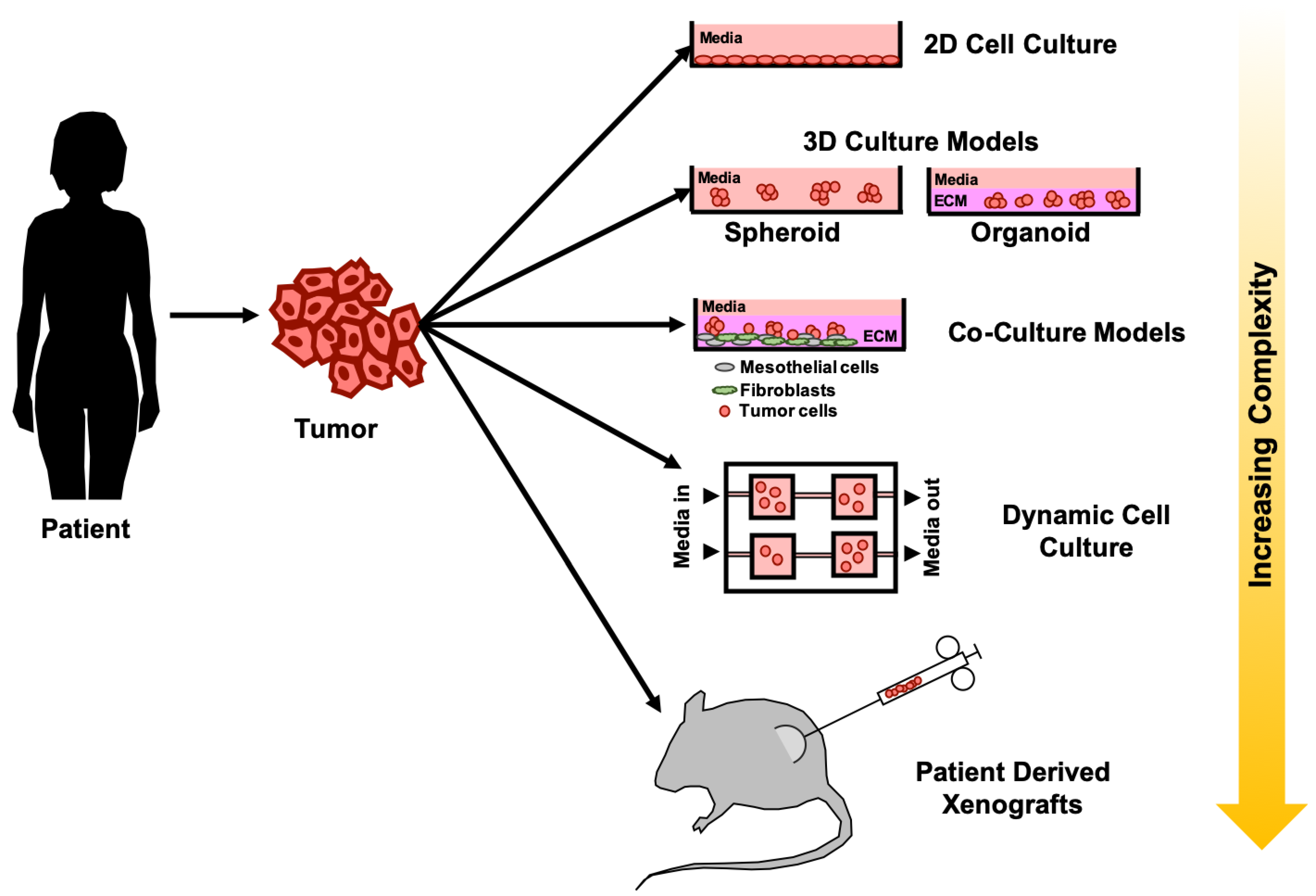
3. Emergence of 3D Cell Culture Models for Drug-Testing in Ovarian Cancer
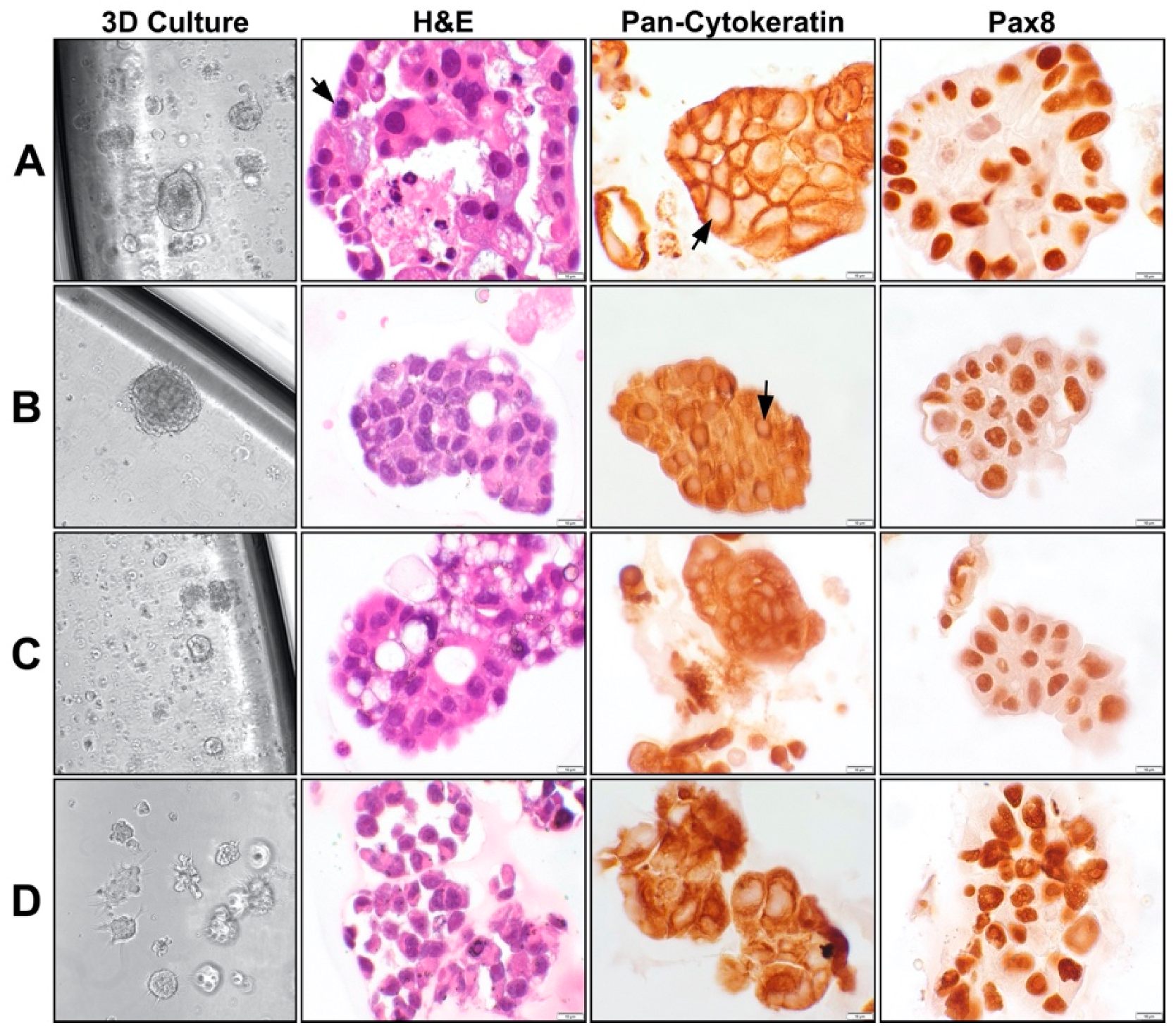
Applications of Organoid Culture Models in Ovarian Cancer
4. Advanced 3D Cell Culture Models as Preclinical Drug Screening Platforms
5. Concluding Remarks and Future Directions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 2D | Two dimensional |
| 3D | Three dimensional |
| ASCO | American Society of Clinical Oncology |
| BRCA | Breast cancer susceptibility gene |
| CDx | Companion diagnostic |
| CRA | Chemo-response assay |
| CSCs | Cancer Stem Cells |
| DAPI | 4′,6-diamidino-2-phenylindole |
| ECM | Extracellular matrix |
| FDA | United States Food and Drug Administration |
| H&E | Hematoxylin and eosin |
| HER2 | HER2/neu, receptor tyrosine kinase erbB-2 |
| IHC | Immunohistochemistry |
| IRB | Institutional review board |
| mm3 | Cubic millimeters |
| NSCLC | Non-small cell lung cancer |
| OD | Optical Density |
| OS | Overall survival |
| PARP | Poly (ADP-ribose) polymerase |
| Pax8 | Paired box gene 8 |
| PD-L1 | Programmed death ligand 1 |
| PDX | Patient derived xenograft |
| PFS | Progression free survival |
| TAM | Tumor-associated macrophage |
| TME | Tumor microenvironment |
| US | United States |
| WHO | World Health Organization |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.-P.; Li, J.; Kros, J.M. Breakthroughs in modern cancer therapy and elusive cardiotoxicity: Critical research-practice gaps, challenges, and insights. Med. Res. Rev. 2017, 38, 325–376. [Google Scholar] [CrossRef] [PubMed]
- Zugazagoitia, J.; Guedes, C.; Ponce, S.; Ferrer, I.; Molina-Pinelo, S.; Paz-Ares, L. Current Challenges in cancer treatment. Clin. Ther. 2016, 38, 1551–1566. [Google Scholar] [CrossRef] [PubMed]
- Sambi, M.; Bagheri, L.; Szewczuk, M.R. Current challenges in cancer immunotherapy: Multimodal approaches to improve efficacy and patient response rates. J. Oncol. 2019, 2019, 1–12. [Google Scholar] [CrossRef]
- Krzyszczyk, P.; Acevedo, A.; Davidoff, E.J.; Timmins, L.M.; Marrero-Berrios, I.; Patel, M.; White, C.; Lowe, C.; Sherba, J.J.; Hartmanshenn, C.; et al. The growing role of precision and personalized medicine for cancer treatment. Technology 2018, 6, 79–100. [Google Scholar] [CrossRef]
- Scheerens, H.; Malong, A.; Bassett, K.; Boyd, Z.; Gupta, V.; Harris, J.; Mesick, C.; Simnett, S.; Stevens, H.; Gilbert, H.; et al. Current status of companion and complementary diagnostics: Strategic considerations for development and launch. Clin. Transl. Sci. 2017, 10, 84–92. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. FDA Approved Olaparib (LYNPARZA, AstraZeneca Pharmaceuticals LP) for the Maintenance Treatment of Adult Patients with Deleterious or Suspected Deleterious Germline or Somatic BRCA-Mutated (gBRCAm or sBRCAm) Advanced Epithelial Ovarian, Fallopian Tube or Primary Peritoneal Cancer Who Are in Complete or Partial Response to First-Line Platinum-Based; FDA: Silver Spring, MD, USA, 2018. [Google Scholar]
- U.S. Food and Drug Administration. List of Cleared or Approved Companion Diagnostic Devices (In Vitro and Imaging Tools); FDA: Silver Spring, MD, USA, 2020. [Google Scholar]
- Davis, A.; Tinker, A.V.; Friedlander, M. “Platinum resistant” ovarian cancer: What is it, who to treat and how to measure benefit? Gynecol. Oncol. 2014, 133, 624–631. [Google Scholar] [CrossRef]
- Guan, L.-Y.; Lu, Y. New developments in molecular targeted therapy of ovarian cancer. Discov. Med. 2018, 26, 219–229. [Google Scholar]
- Hoffman, R.M. In vitro assays for chemotherapy sensitivity. Crit. Rev. Oncol. 1993, 15, 99–111. [Google Scholar] [CrossRef]
- Samson, D.J.; Seidenfeld, J.; Ziegler, K.; Aronson, N. Chemotherapy sensitivity and resistance assays: A systematic review. J. Clin. Oncol. 2004, 22, 3618–3630. [Google Scholar] [CrossRef]
- Bosserman, L.; Prendergast, F.; Herbst, R.; Fleisher, M.; Salom, E.; Strickland, S.A.; Raptis, A.; Hallquist, A.; Perree, M.; Rajurkar, S.; et al. The microculture-kinetic (MiCK) assay: The role of a drug-induced apoptosis assay in drug development and clinical care. Cancer Res. 2012, 72, 3901–3905. [Google Scholar] [CrossRef][Green Version]
- Bosserman, L.; Rogers, K.; Willis, C.; Davidson, D.; Whitworth, P.; Karimi, M.; Upadhyaya, G.; Rutledge, J.; Hallquist, A.; Perree, M.; et al. Application of a drug-induced apoptosis assay to identify treatment strategies in recurrent or metastatic breast cancer. PLoS ONE 2015, 10, e0122609. [Google Scholar] [CrossRef] [PubMed]
- Bosserman, L.D.; Rajurkar, S.P.; Rogers, K.; Davidson, D.C.; Chernick, M.; Hallquist, A.; Malouf, D.; Presant, C.A. Correlation of drug-induced apoptosis assay results with oncologist treatment decisions and patient response and survival. Cancer 2012, 118, 4877–4883. [Google Scholar] [CrossRef]
- Ballard, K.S.; Homesley, H.D.; Hodson, C.; Presant, C.A.; Rutledge, J.; Hallquist, A.; Perree, M. Endometrial carcinoma in vitro chemosensitivity testing of single and combination chemotherapy regimens using the novel microculture kinetic apoptosis assay: Implications for endometrial cancer treatment. J. Gynecol. Oncol. 2010, 21, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Salom, E.; Penalver, M.; Homesley, H.; Burrell, M.; Garrett, A.; Presant, C.A.; Rutledge, J.; Chernick, M.; Hallquist, A.; Perree, M. Correlation of pretreatment drug induced apoptosis in ovarian cancer cells with patient survival and clinical response. J. Transl. Med. 2012, 10, 162. [Google Scholar] [CrossRef][Green Version]
- Herzog, T.J.; Krivak, T.C.; Fader, A.N.; Coleman, R.L. Chemosensitivity testing with ChemoFx and overall survival in primary ovarian cancer. Am. J. Obstet. Gynecol. 2010, 203, 68.e1–68.e6. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, T.; Orr, J.; Grendys, E.; Edwards, R.; Krivak, T.C.; Holloway, R.; Moore, R.G.; Puls, L.; Tillmanns, T.; Schink, J.C.; et al. A prospective study evaluating the clinical relevance of a chemoresponse assay for treatment of patients with persistent or recurrent ovarian cancer. Gynecol. Oncol. 2013, 131, 362–367. [Google Scholar] [CrossRef]
- Richard, S.; Wells, A.; Connor, J.; Price, F. Use of ChemoFx® for identification of effective treatments in epithelial ovarian cancer. PLoS Curr. 2015, 7, 7. [Google Scholar] [CrossRef]
- Mi, Z.; Holmes, F.A.; Hellerstedt, B.; Pippen, J.; Collea, R.; Backner, A.; Bush, J.E.; Gallion, H.H.; Wells, A.; O’Shaughnessy, J.A. Feasibility assessment of a chemoresponse assay to predict pathologic response in neoadjuvant chemotherapy for breast cancer patients. Anticancer Res. 2008, 28, 1733–1740. [Google Scholar]
- Konecny, G.; Crohns, C.; Pegram, M.; Felber, M.; Lude, S.; Kurbacher, C.; Cree, I.A.; Hepp, H.; Untch, M. Correlation of drug response with the ATP tumorchemosensitivity assay in primary FIGO stage III ovarian cancer. Gynecol. Oncol. 2000, 77, 258–263. [Google Scholar] [CrossRef]
- Sharma, S.; Neale, M.H.; Di Nicolantonio, F.; Knight, L.A.; Whitehouse, P.A.; Mercer, S.; Higgins, B.R.; Lamont, A.; Osborne, R.; Hindley, A.C.; et al. Outcome of ATP-based tumor chemosensitivity assay directed chemotherapy in heavily pre-treated recurrent ovarian carcinoma. BMC Cancer 2003, 3, 19. [Google Scholar] [CrossRef]
- Loizzi, V.; Chan, J.K.; Osann, K.; Cappuccini, F.; DiSaia, P.J.; Berman, M.L. Survival outcomes in patients with recurrent ovarian cancer who were treated with chemoresistance assay-guided chemotherapy. Am. J. Obstet. Gynecol. 2003, 189, 1301–1307. [Google Scholar] [CrossRef]
- Cree, I.A.; Kurbacher, C.M.; Lamont, A.; Hindley, A.C.; Love, S. A prospective randomized controlled trial of tumour chemosensitivity assay directed chemotherapy versus physician’s choice in patients with recurrent platinum-resistant ovarian cancer. Anticancer Drugs 2007, 18, 1093–1101. [Google Scholar] [CrossRef] [PubMed]
- Burstein, H.J.; Mangu, P.B.; Somerfield, M.R.; Schrag, D.; Samson, D.; Holt, L.; Zelman, D.; Ajani, J.A. American Society of Clinical Oncology clinical practice guideline update on the use of chemotherapy sensitivity and resistance assays. J. Clin. Oncol. 2011, 29, 3328–3330. [Google Scholar] [CrossRef] [PubMed]
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, L.; Lamperska, K. 2D and 3D cell cultures—A comparison of different types of cancer cell cultures. Arch. Med. Sci. 2018, 14, 910–919. [Google Scholar] [CrossRef] [PubMed]
- Bissell, M.J.; Hall, H.; Parry, G. How does the extracellular matrix direct gene expression? J. Theor. Biol. 1982, 99, 31–68. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef]
- Riedl, A.; Schlederer, M.; Pudelko, K.; Stadler, M.; Walter, S.; Unterleuthner, D.; Unger, C.; Kramer, N.; Hengstschläger, M.; Kenner, L.; et al. Comparison of cancer cells in 2D vs 3D culture reveals differences in AKT-mTOR-S6K signaling and drug responses. J. Cell Sci. 2017, 130, 203–218. [Google Scholar] [CrossRef]
- Reynolds, D.S.; Tevis, K.M.; Blessing, W.A.; Colson, Y.L.; Zaman, M.; Grinstaff, M.W. Breast cancer spheroids reveal a differential cancer stem cell response to chemotherapeutic treatment. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Imamura, Y.; Mukohara, T.; Shimono, Y.; Funakoshi, Y.; Chayahara, N.; Toyoda, M.; Kiyota, N.; Takao, S.; Kono, S.; Nakatsura, T.; et al. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol. Rep. 2015, 33, 1837–1843. [Google Scholar] [CrossRef]
- Clevers, H. Modeling development and disease with organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef] [PubMed]
- Kenny, P.A.; Lee, G.Y.; Myers, C.A.; Neve, R.M.; Semeiks, J.R.; Spellman, P.T.; Lorenz, K.; Lee, E.H.; Barcellos-Hoff, M.H.; Petersen, O.W.; et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol. Oncol. 2007, 1, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Fatehullah, A.; Tan, S.H.; Barker, N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016, 18, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Kopper, O.; de Witte, C.J.; Lõhmussaar, K.; Valle-Inclan, J.E.; Hami, N.; Kester, L.; Balgobind, A.V.; Korving, J.; Proost, N.; Begthel, H.; et al. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat. Med. 2019, 25, 838–849. [Google Scholar] [CrossRef]
- Maru, Y.; Tanaka, N.; Itami, M.; Hippo, Y. Efficient use of patient-derived organoids as a preclinical model for gynecologic tumors. Gynecol. Oncol. 2019, 154, 189–198. [Google Scholar] [CrossRef]
- Tibbitt, M.W.; Anseth, K.S. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol. Bioeng. 2009, 103, 655–663. [Google Scholar] [CrossRef]
- Maru, Y.; Hippo, Y. Current status of patient-derived ovarian cancer models. Cells 2019, 8, 505. [Google Scholar] [CrossRef]
- Lee, J.; Kotliarova, S.; Kotliarov, Y.; Li, A.; Su, Q.; Donin, N.M.; Pastorino, S.; Purow, B.W.; Christopher, N.; Zhang, W.; et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 2006, 9, 391–403. [Google Scholar] [CrossRef]
- Krohn, A.; Song, Y.-H.; Muehlberg, F.; Droll, L.; Beckmann, C.; Alt, E. CXCR4 receptor positive spheroid forming cells are responsible for tumor invasion in vitro. Cancer Lett. 2009, 280, 65–71. [Google Scholar] [CrossRef]
- Cui, X.; Hartanto, Y.; Zhang, H. Advances in multicellular spheroids formation. J. R. Soc. Interface 2017, 14, 20160877. [Google Scholar] [CrossRef]
- Herheliuk, T.; Perepelytsina, O.; Ugnivenko, A.; Ostapchenko, L.; Sydorenko, M. Investigation of multicellular tumor spheroids enriched for a cancer stem cell phenotype. Stem Cell Investig. 2019, 6, 21. [Google Scholar] [CrossRef]
- Al Habyan, S.; Kalos, C.; Szymborski, J.; McCaffrey, L. Multicellular detachment generates metastatic spheroids during intra-abdominal dissemination in epithelial ovarian cancer. Oncogene 2018, 37, 5127–5135. [Google Scholar] [CrossRef] [PubMed]
- Shieldartin, K.L.; Ackland, M.L.; Ahmed, N.; Rice, G.E. Multicellular spheroids in ovarian cancer metastases: Biology and pathology. Gynecol. Oncol. 2009, 113, 143–148. [Google Scholar] [CrossRef]
- Liao, J.; Qian, F.; Tchabo, N.; Mhawech-Fauceglia, P.; Beck, A.; Qian, Z.; Wang, X.; Huss, W.J.; Lele, S.B.; Morrison, C.D.; et al. Ovarian cancer spheroid cells with stem cell-like properties contribute to tumor generation, metastasis and chemotherapy resistance through hypoxia-resistant metabolism. PLoS ONE 2014, 9, e84941. [Google Scholar] [CrossRef] [PubMed]
- Frankel, A.; Buckman, R.; Kerbel, R.S. Abrogation of taxol-induced G2-M arrest and apoptosis in human ovarian cancer cells grown as multicellular tumor spheroids. Cancer Res. 1997, 57, 2388–2393. [Google Scholar] [PubMed]
- Xing, H.; Wang, S.; Hu, K.; Tao, W.; Li, J.; Gao, Q.; Yang, X.; Weng, D.; Lu, Y.; Ma, D. Effect of the cyclin-dependent kinases inhibitor p27 on resistance of ovarian cancer multicellular spheroids to anticancer chemotherapy. J. Cancer Res. Clin. Oncol. 2005, 131, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Wang, S.; Weng, D.; Chen, G.; Yang, X.; Zhou, J.; Xu, G.; Lu, Y.; Ma, D. Knock-down of P-glycoprotein reverses taxol resistance in ovarian cancer multicellular spheroids. Oncol. Rep. 2007, 17, 117–122. [Google Scholar] [CrossRef][Green Version]
- Kryczek, I.; Liu, S.; Roh, M.; Vatan, L.; Szeliga, W.; Wei, S.; Banerjee, M.; Mao, Y.; Kotarski, J.; Wicha, M.S.; et al. Expression of aldehyde dehydrogenase and CD133 defines ovarian cancer stem cells. Int. J. Cancer 2011, 130, 29–39. [Google Scholar] [CrossRef]
- Silva, I.A.; Bai, S.; McLean, K.; Yang, K.; A Griffith, K.; Thomas, D.; Ginestier, C.; Johnston, C.; Kueck, A.; Reynolds, R.K.; et al. Aldehyde dehydrogenase in combination with CD133 defines angiogenic ovarian cancer stem cells that portend poor patient survival. Cancer Res. 2011, 71, 3991–4001. [Google Scholar] [CrossRef]
- Xu, S.; Yang, Y.; Dong, L.; Qiu, W.; Yang, L.; Wang, X.; Liu, L. Construction and characteristics of an E-cadherin-related three-dimensional suspension growth model of ovarian cancer. Sci. Rep. 2015, 4, srep05646. [Google Scholar] [CrossRef]
- Ishiguro, T.; Sato, A.; Ohata, H.; Ikarashi, Y.; Takahashi, R.-U.; Ochiya, T.; Yoshida, M.; Tsuda, H.; Onda, T.; Kato, T.; et al. Establishment and characterization of an in vitro model of ovarian cancer stem-like cells with an enhanced proliferative capacity. Cancer Res. 2015, 76, 150–160. [Google Scholar] [CrossRef]
- Raghavan, S.; Ward, M.R.; Rowley, K.R.; Wold, R.M.; Takayama, S.; Buckanovich, R.J.; Mehta, G. Formation of stable small cell number three-dimensional ovarian cancer spheroids using hanging drop arrays for preclinical drug sensitivity assays. Gynecol. Oncol. 2015, 138, 181–189. [Google Scholar] [CrossRef]
- Aihara, A.; Abe, N.; Saruhashi, K.; Kanaki, T.; Nishino, T. Novel 3-D cell culture system for in vitro evaluation of anticancer drugs under anchorage-independent conditions. Cancer Sci. 2016, 107, 1858–1866. [Google Scholar] [CrossRef]
- Raghavan, S.; Mehta, P.; Ward, M.R.; Bregenzer, M.E.; Fleck, E.M.A.; Tan, L.; McLean, K.; Buckanovich, R.J.; Mehta, G. Personalized medicine–based approach to model patterns of chemoresistance and tumor recurrence using ovarian cancer stem cell spheroids. Clin. Cancer Res. 2017, 23, 6934–6945. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-W.; Yang, S.-T.; Chien, M.-H.; Hua, K.-T.; Wu, C.-J.; Hsiao, S.; Lin, H.; Hsiao, M.; Su, J.-L.; Wei, L.-H. The STAT3-miRNA-92-Wnt signaling pathway regulates spheroid formation and malignant progression in ovarian cancer. Cancer Res. 2017, 77, 1955–1967. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Bankhead, A.; Ljungman, M.; Neamati, N. Multi-omics profiling reveals key signaling pathways in ovarian cancer controlled by STAT3. Theranostics 2019, 9, 5478–5496. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, S.; Sun, Y.; Zhang, D.; Zhao, Z.; Liu, L. Reversing platinum resistance in ovarian cancer multicellular spheroids by targeting Bcl-2. OncoTargets Ther. 2019, 12, 897–906. [Google Scholar] [CrossRef]
- Rashidi, M.R.W.; Mehta, P.; Bregenzer, M.; Raghavan, S.; Fleck, E.M.; Horst, E.N.; Harissa, Z.; Ravikumar, V.; Brady, S.; Bild, A.; et al. Engineered 3D model of cancer stem cell enrichment and chemoresistance. Neoplasia 2019, 21, 822–836. [Google Scholar] [CrossRef]
- Shuford, S.; Wilhelm, C.; Rayner, M.; Elrod, A.; Millard, M.; Mattingly, C.; Lotstein, A.; Smith, R.; Guo, Q.J.; O’Donnell, L.; et al. Prospective validation of an ex vivo, patient-derived 3D spheroid model for response predictions in newly diagnosed ovarian cancer. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Boylan, K.L.; Buchanan, P.C.; Manion, R.D.; Shukla, D.M.; Braumberger, K.; Bruggemeyer, C.; Skubitz, A.P. The expression of Nectin-4 on the surface of ovarian cancer cells alters their ability to adhere, migrate, aggregate, and proliferate. Oncotarget 2017, 8, 9717–9738. [Google Scholar] [CrossRef]
- Boylan, K.L.M.; Manion, R.D.; Shah, H.; Skubitz, K.M.; Skubitz, A.P.N. Inhibition of ovarian cancer cell spheroid formation by synthetic peptides derived from Nectin-4. Int. J. Mol. Sci. 2020, 21, 4637. [Google Scholar] [CrossRef]
- Gilazieva, Z.; Ponomarev, A.; Rutland, C.; Rizvanov, A.A.; Solovyeva, V. Promising applications of tumor spheroids and organoids for personalized medicine. Cancers 2020, 12, 2727. [Google Scholar] [CrossRef] [PubMed]
- Dumont, S.; Jan, Z.; Heremans, R.; Van Gorp, T.; Vergote, I.; Timmerman, D. Organoids of epithelial ovarian cancer as an emerging preclinical in vitro tool: A review. J. Ovarian Res. 2019, 12, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bleijs, M.; van de Wetering, M.; Clevers, H.; Drost, J. Xenograft and organoid model systems in cancer research. EMBO J. 2019, 38, e101654. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Bockorny, B.; Paul, I.; Akshinthala, D.; Frappart, P.-O.; Gandarilla, O.; Bose, A.; Sanchez-Gonzalez, V.; Rouse, E.E.; Lehoux, S.D.; et al. PDX-Derived Organoids Model In Vivo Drug Response and Secrete Biomarkers. Available online: https://insight.jci.org/articles/view/135544/pdf (accessed on 9 December 2020).
- Weeber, F.; van de Wetering, M.; Hoogstraat, M.; Dijkstra, K.K.; Krijgsman, O.; Kuilman, T.; Gadellaa-van Hooijdonk, C.G.M.; van der Velden, D.L.; Peeper, D.S.; Cuppen, E.P.J.G.; et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc. Natl. Acad. Sci. USA 2015, 112, 13308–13311. [Google Scholar] [CrossRef] [PubMed]
- Van de Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; van Houdt, W.; van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015, 161, 933–945. [Google Scholar] [CrossRef] [PubMed]
- Beshiri, M.L.; Tice, C.M.; Tran, C.; Nguyen, H.M.; Sowalsky, A.G.; Agarwal, S.; Jansson, K.H.; Yang, Q.; McGowen, K.M.; Yin, J.J.; et al. A PDX/organoid biobank of advanced prostate cancers captures genomic and phenotypic heterogeneity for disease modeling and therapeutic screening. Clin. Cancer Res. 2018, 24, 4332–4345. [Google Scholar] [CrossRef]
- Sachs, N.; de Ligt, J.; Kopper, O.; Gogola, E.; Bounova, G.; Weeber, F.; Balgobind, A.V.; Wind, K.; Gracanin, A.; Begthel, H.; et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell 2018, 172, 373–386.e10. [Google Scholar] [CrossRef]
- Hill, S.J.; Decker, B.; Roberts, E.A.; Horowitz, N.S.; Muto, M.G.; Worley, J.M.J.; Feltmate, C.M.; Nucci, M.R.; Swisher, E.M.; Nguyen, H.; et al. Prediction of DNA repair inhibitor response in short-term patient-derived ovarian cancer organoids. Cancer Discov. 2018, 8, 1404–1421. [Google Scholar] [CrossRef]
- Maenhoudt, N.; Defraye, C.; Boretto, M.; Jan, Z.; Heremans, R.; Boeckx, B.; Hermans, F.; Arijs, I.; Cox, B.; Van Nieuwenhuysen, E.; et al. Developing organoids from ovarian cancer as experimental and preclinical models. Stem Cell Rep. 2020, 14, 717–729. [Google Scholar] [CrossRef]
- Hoffmann, K.; Berger, H.; Kulbe, H.; Thillainadarasan, S.; Mollenkopf, H.; Zemojtel, T.; Taube, E.; Darb-Esfahani, S.; Mangler, M.; Sehouli, J.; et al. Stable expansion of high-grade serous ovarian cancer organoids requires a low-Wnt environment. EMBO J. 2020, 39, e104013. [Google Scholar] [CrossRef] [PubMed]
- Loessner, D.; Stok, K.S.; Lutolf, M.P.; Hutmacher, D.W.; Clements, J.A.; Rizzi, S.C. Bioengineered 3D platform to explore cell–ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials 2010, 31, 8494–8506. [Google Scholar] [CrossRef]
- Yang, Z.; Zhao, X. A 3D model of ovarian cancer cell lines on peptide nanofiber scaffold to explore the cell–scaffold interaction and chemotherapeutic resistance of anticancer drugs. Int. J. Nanomed. 2011, 6, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Phan, N.; Hong, J.J.; Tofig, B.; Mapua, M.; Elashoff, D.; Moatamed, N.A.; Huang, J.; Memarzadeh, S.; Damoiseaux, R.; Soragni, A. A simple high-throughput approach identifies actionable drug sensitivities in patient-derived tumor organoids. Commun. Biol. 2019, 2, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Gotimer, K.; De Souza, C.; Tepper, C.G.; Karnezis, A.N.; Leiserowitz, G.S.; Chien, J.; Smith, L.H. Short-term organoid culture for drug sensitivity testing of high-grade serous carcinoma. Gynecol. Oncol. 2020, 157, 783–792. [Google Scholar] [CrossRef] [PubMed]
- De Witte, C.J.; Valle-Inclan, J.E.; Hami, N.; Lõhmussaar, K.; Kopper, O.; Vreuls, C.P.H.; Jonges, G.N.; van Diest, P.; Nguyen, L.; Clevers, H.; et al. Patient-derived ovarian cancer organoids mimic clinical response and exhibit heterogeneous inter- and intrapatient drug responses. Cell Rep. 2020, 31, 107762. [Google Scholar] [CrossRef]
- Nanki, Y.; Chiyoda, T.; Hirasawa, A.; Ookubo, A.; Itoh, M.; Ueno, M.; Akahane, T.; Kameyama, K.; Yamagami, W.; Kataoka, F.; et al. Patient-derived ovarian cancer organoids capture the genomic profiles of primary tumours applicable for drug sensitivity and resistance testing. Sci. Rep. 2020, 10, 12581. [Google Scholar] [CrossRef]
- Zhang, S.; Iyer, S.; Ran, H.; Dolgalev, I.; Gu, S.; Wei, W.; Foster, C.J.; Loomis, C.A.; Olvera, N.; Dao, F.; et al. Genetically defined, syngeneic organoid platform for developing combination therapies for ovarian cancer. Cancer Discov. 2020. [Google Scholar] [CrossRef]
- Nguyen, H.T.L.; Soragni, A. Patient-derived tumor organoid rings for histologic characterization and high-throughput screening. STAR Protoc. 2020, 1, 100056. [Google Scholar] [CrossRef]
- Yuan, Y.; Jiang, Y.-C.; Sun, C.-K.; Chen, Q. Role of the tumor microenvironment in tumor progression and the clinical applications (review). Oncol. Rep. 2016, 35, 2499–2515. [Google Scholar] [CrossRef]
- Lengyel, E.; Burdette, J.E.; Kenny, H.A.; Matei, D.; Pilrose, J.; Haluska, P.; Nephew, K.P.; Hales, D.B.; Stack, M.S. Epithelial ovarian cancer experimental models. Oncogene 2013, 33, 3619–3633. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.; Yang, H.; Zhang, F.; Yang, S. 3D cell coculture tumor model: A promising approach for future cancer drug discovery. Process Biochem. 2019, 78, 148–160. [Google Scholar] [CrossRef]
- Paschos, N.K.; Brown, W.E.; Eswaramoorthy, R.; Hu, J.; Athanasiou, K.A. Advances in tissue engineering through stem cell-based co-culture. J. Tissue Eng. Regen. Med. 2015, 9, 488–503. [Google Scholar] [CrossRef] [PubMed]
- Hoarau-Véchot, J.; Rafii, A.; Touboul, C.; Pasquier, J. Halfway between 2D and animal models: Are 3D cultures the ideal tool to study cancer-microenvironment interactions? Int. J. Mol. Sci. 2018, 19, 181. [Google Scholar] [CrossRef] [PubMed]
- Kenny, H.A.; Krausz, T.; Yamada, S.D.; Lengyel, E. Use of a novel 3D culture model to elucidate the role of mesothelial cells, fibroblasts and extra-cellular matrices on adhesion and invasion of ovarian cancer cells to the omentum. Int. J. Cancer 2007, 121, 1463–1472. [Google Scholar] [CrossRef] [PubMed]
- Kenny, H.A.; Lal-Nag, M.; White, E.A.; Shen, M.; Chiang, C.-Y.; Mitra, A.K.; Zhang, Y.; Curtis, M.W.; Schryver, E.M.; Bettis, S.; et al. Quantitative high throughput screening using a primary human three-dimensional organotypic culture predicts in vivo efficacy. Nat. Commun. 2015, 6, 1–11. [Google Scholar] [CrossRef]
- Wan, X.; Bovornchutichai, P.; Cui, Z.; O’Neill, E.; Ye, H. Morphological analysis of human umbilical vein endothelial cells co-cultured with ovarian cancer cells in 3D: An oncogenic angiogenesis assay. PLoS ONE 2017, 12, e0180296. [Google Scholar] [CrossRef]
- Long, L.; Yin, M.; Min, W. 3D co-culture system of tumor-associated macrophages and ovarian cancer cells. Bio Protoc. 2018, 8, e2815. [Google Scholar] [CrossRef]
- Loessner, D.; Rockstroh, A.; Shokoohmand, A.; Holzapfel, B.M.; Wagner, F.; Baldwin, J.; Boxberg, M.; Schmalfeldt, B.; Lengyel, E.; Clements, J.A.; et al. A 3D tumor microenvironment regulates cell proliferation, peritoneal growth and expression patterns. Biomaterials 2019, 190–191, 63–75. [Google Scholar] [CrossRef]
- Hedegaard, C.L.; Redondo-Gómez, C.; Tan, B.Y.; Ng, K.W.; Loessner, D.; Mata, A. Peptide-protein coassembling matrices as a biomimetic 3D model of ovarian cancer. Sci. Adv. 2020, 6, eabb3298. [Google Scholar] [CrossRef]
- Li, S.-S.; Ip, C.K.M.; Tang, M.Y.H.; Sy, S.K.H.; Yung, S.; Chan, T.M.; Yang, M.; Shum, H.C.; Wong, A.S.T. Modeling ovarian cancer multicellular spheroid behavior in a dynamic 3D peritoneal microdevice. J. Vis. Exp. 2017. [Google Scholar] [CrossRef] [PubMed]
- Arellano, J.A.; Howell, T.A.; Gammon, J.; Cho, S.; Janát-Amsbury, M.M.; Gale, B. Use of a highly parallel microfluidic flow cell array to determine therapeutic drug dose response curves. Biomed. Microdevices 2017, 19, 25. [Google Scholar] [CrossRef] [PubMed]
- Masiello, T.; Dhall, A.; Hemachandra, L.P.M.; Tokranova, N.; Melendez, J.A.; Castracane, J. A Dynamic culture method to produce ovarian cancer spheroids under physiologically-relevant shear stress. Cells 2018, 7, 277. [Google Scholar] [CrossRef] [PubMed]
- Flont, M.; Jastrzębska, E.; Brzózka, Z. A multilayered cancer-on-a-chip model to analyze the effectiveness of new-generation photosensitizers. Analyst 2020, 145, 6937–6947. [Google Scholar] [CrossRef] [PubMed]
- Coluccio, M.L.; Perozziello, G.; Malara, N.; Parrotta, E.; Zhang, P.; Gentile, F.; Limongi, T.; Raj, P.M.; Cuda, G.; Candeloro, P.; et al. Microfluidic platforms for cell cultures and investigations. Microelectron. Eng. 2019, 208, 14–28. [Google Scholar] [CrossRef]
- Tehranirokh, M.; Kouzani, A.Z.; Francis, P.S.; Kanwar, J.R. Microfluidic devices for cell cultivation and proliferation. Biomicrofluidics 2013, 7, 051502. [Google Scholar] [CrossRef]
- Roberts, C.M.; Cardenas, C.; Tedja, R. The role of intra-tumoral heterogeneity and its clinical relevance in epithelial ovarian cancer recurrence and metastasis. Cancers 2019, 11, 1083. [Google Scholar] [CrossRef]
| MiCK Assay | ||||
|---|---|---|---|---|
| Study | Study Type | Patient Population | Results 1 | Reference |
| Salom et al. (2012) | Prospective | n = 104; epithelial ovarian cancer patients | OS of chemonaive patients with stage III or IV disease was longer when treated with assay-predicted chemotherapy | [17] |
| Bosserman et al. (2012) | Prospective and non-blinded | n = 44; breast cancer, non-small cell lung cancer, ovarian cancer, and others (n = 2 ovarian cancers) | Patients receiving assay-predicted chemotherapy demonstrated improved median OS compared to patients who were treated empirically | [15] |
| ChemoFX Assay | ||||
| Study | Study Type | Patient Population | Results 1 | Reference |
| Herzog et al. (2010) | Retrospective | n = 192; ovarian cancer patients | A trend towards increased OS was seen in patients who received a treatment also found efficacious in vitro | [18] |
| Rutherford et al. (2013) | Prospective | n = 262; recurrent or persistent ovarian cancer | Improved PFS and OS for patients with assay-sensitive tumors compared to resistant or intermediate response based on in vitro assay. | [19] |
| Study | Study Type | Patient Population | Cell Viability Assessment | Results 1 | Reference |
|---|---|---|---|---|---|
| Konecny et al. (2000) | Non-randomized | n = 38; FIGO stage III ovarian cancer patients | ATP-based assay | Patients predicted to be sensitive showed a trend for increased PFS and OS compared to assay-predicted resistant patients | [22] |
| Sharma et al. (2003) | Prospective and non-randomized | n = 44; chemotherapy-treated recurrent epithelial ovarian cancer patients | ATP-based assay | Patients were treated according to the assay-predicted results. The overall response rate was 61% for evaluable patients | [23] |
| Loizzi et al. (2003) | Retrospective and non-randomized | n = 100; recurrent ovarian cancer | Tritiated thymidine uptake drug resistance assay | For patients with platinum-sensitive disease, 1-year OS and PFS were increased when treated with assay-directed chemotherapy compared to the control group. | [24] |
| Cree et al. (2007) | Prospective, randomized | n = 180; platinum-resistant recurrent ovarian cancer patients | ATP-based assay | Patients treated with assay-predicted chemotherapy regimens demonstrated a small trend toward improved PFS and response rate compared to physician’s choice treated patients (not statistically significant). | [25] |
| Natural ECM Gels | Synthetic ECM Gels |
|---|---|
| Formed from extracellular matrix components or materials derived from biological sources such as chitosan and alginate. | Made of non-natural molecules |
| Biocompatible and bioactive | Inert molecules |
| Complex composition with presence of undefined endogenous components | Defined components |
| Altering properties of these gels can be difficult | Easily manufactured in a cost-effective manner, highly reproducible |
| Restricted clinical applications due to presence unknown endogenous factors | Used for clinical applications as well as fundamental studies of cell physiology |
| Examples: alginate, collagen, Matrigel, fibrin, hyaluronic acid | Examples: Poly(2-hydroxy ethyl methacrylate), Polyethylene glycol, Poly(vinyl alcohol) |
| Study | Outcomes of the Study | Reference |
|---|---|---|
| Xing et al. (2005) Xing et al. (2007) | Analysis of spheroid culture from ovarian cancer cell lines revealed higher expression of p27 protein in spheroids associated with drug resistance to taxol | [48,49] |
| Kryczek et al.(2012) Silva et al. (2011) | CD133 and ALDH are identified as CSC markers of ovarian cancer using spheroid culture models | [50,51] |
| Xu et al. (2014) | E-cadherin plays an important role in spheroid formation and drug resistance to cisplatin. | [52] |
| Ishiguro et al. (2016) | ROCK inhibition of ovarian cancer spheroids may promote CSC phenotype | [53] |
| Raghavan et al. (2015) | Developed a novel 384-well hanging drop tumor spheroid platform used to test sensitivity to cisplatin chemotherapy. | [54] |
| Aihara et al. (2016) | Developed a novel 3D cell culture technique using FP001 polymer for screening of anticancer agents. FP001 facilitated homogenous spheroid culture. | [55] |
| Raghavan et al. (2017) | Developed a patient-derived 3D hanging drop spheroid platform with ALDH+ CD133+ ovarian cancer cells to screen the effects of chemotherapy drugs | [56] |
| Chen et al. (2017) Lu et al. (2019) | Activation of STAT3 plays an important role in formation of epithelial ovarian cancer spheroids and regulating putative stem-like cell markers | [57,58] |
| Yang et al. (2019) | Role of bcl-2 was explored in ovarian cancer spheroids in response to platinum-drugs | [59] |
| Rashidi et al. (2019) | Developed an in vitro 3D model to study stemness and chemoresistance in ovarian cancer. Serial passaging of spheroids using this technique demonstrated enrichment of cells with stem cell markers and emergence of platinum-resistance phenotype. | [60] |
| Shuford et al. (2019) | Developed an ex vivo patient derived 3D spheroid model for drug testing. A correlation between clinical response to therapy and in vitro response was seen in some patients. | [61] |
| Boylan et al.(2016) Boylan et al. (2020) | Cell adhesion molecule Nectin-4 may be involved in ovarian cancer spheroid formation | [62,63] |
| Study | Type of ECM | Results | Reference |
|---|---|---|---|
| Loessner et al. (2010) | Bioengineered PEG-based hydrogel matrix | Developed a 3D cell culture platform using a biomimetic synthetic hydrogel. The resulting multicellular structures were tested for sensitivity to paclitaxel compared to 2D monolayer culture. | [75] |
| Yang and Zhao (2011) | Nanofiber scaffold based 3D cell culture | Ovarian cancer cells grown in 3D on nanofiber scaffold were found to exhibit higher therapeutic resistance to anti-cancer drugs, like 5-FU, paclitaxel, and curcumin compared to conventional 2D cell culture. | [76] |
| Hill et al. (2018) | Matrigel-based | Developed short-term patient-derived ovarian cancer organoids for drug screening analyses. | [72] |
| Phan et al. (2019) | Matrigel-based | Developed a high throughput drug screening platform to evaluate the therapeutic response of tumor organoids derived from clinical samples and cell lines. | [77] |
| Kopper et al. (2019) | Basement membrane extract-based | Developed patient-derived organoid lines from main subtypes of ovarian cancer as a platform for drug screening assays. | [36] |
| Maenhoudt et al. (2020) | Matrigel-based | Developed patient-derived organoid lines from predominately high grade serous ovarian cancers and performed mutational analyses and chemosensitivity assays. | [73] |
| Chen et al. (2020) | Basement membrane extract-based | Developed a short duration organoid culture platform derived from HGSOC malignant effusion specimens and utilized it for drug sensitivity testing | [78] |
| de Witte et al. (2020) | Basement membrane extract-based | Utilized a patient-derived organoid (PDO) platform to assess the chemotherapy response to various drugs. The in vitro drug response of 7 PDOs treated with carboplatin and paclitaxel correlated with clinical outcomes seen in those patients. | [79] |
| Nanki et al. (2020) | Matrigel-based | Developed a PDO platform that recapitulated the in vivo architecture and genetic signature of original ovarian cancer tumor. This was utilized for drug sensitivity testing using 23 FDA-approved drugs | [80] |
| Zhang et al. (2020) | Matrigel-based | Modelled HGSOC by genetically manipulating mouse fallopian tube epithelium. Sensitivity of derived organoids was tested in drug assays. | [81] |
| Co-Culture 3D Models | ||
|---|---|---|
| Study | Purpose of Model | Reference |
| Kenny et al. (2007) | Developed a 3D co-culture model to study ovarian cancer metastasis to omentum. The model demonstrated that attachment and invasion of ovarian cancer cells is promoted by fibroblasts but may be inhibited by mesothelial cells. | [88] |
| Kenny et al. (2015) | Developed a high throughput drug screening assay using primary human 3D co-culture incorporating fibroblasts, mesothelial cells, and ovarian cancer cells. | [89] |
| Wan et al. (2017) | Developed a 3D co-culture model using human endothelial cells and ovarian cancer cells to mimic the interaction between the two types of cells in cancer. Utilized this model for anti-cancer drug testing using paclitaxel and cisplatin. | [90] |
| Long et al. (2018) | Developed a 3D co-culture platform using tumor-associated macrophages (TAMs) and cancer cells to form ovarian cancer organoids. | [91] |
| Loessner et al. (2019) | Developed a 3D co-culture model using ovarian cancer and mesothelial cells. The model demonstrated a potential increase in cancer cell proliferation in vitro and in vivo compared to monoculture. | [92] |
| Hedegaard et al. (2020) | Developed a 3D co-culture model with human umbilical vein endothelial, human mesenchymal, and ovarian cancer cells using a novel peptide-derived hydrogel as alternative to Matrigel | [93] |
| Dynamic 3D Cell Culture Models | ||
| Study | Purpose of Model | Reference |
| Li et al. (2017) | Developed a dynamic 3D co-culture system that mimics the interaction between ovarian cancer cells and mesothelium in a microfluidic device. | [94] |
| Arellano et al. (2017) | Utilized a high-throughput, microfluidic-based platform for in vitro drug testing of ovarian cancer cells | [95] |
| Masiello et al. (2018) | Developed a novel dynamic cell culture platform using an orbital shaker for generating ovarian cancer spheroids by mimicking in vivo dynamic tumor microenvironment using fluid shear stress. | [96] |
| Flont et al. (2020) | Developed a microfluidic based co-culture model with fibroblasts and ovarian cancer cells. This platform was utilized to evaluate the potential cytotoxic effects of photosensitisers. | [97] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, T.; Neal, A.S.; Moatamed, N.A.; Memarzadeh, S. Exploring the Potential of Drug Response Assays for Precision Medicine in Ovarian Cancer. Int. J. Mol. Sci. 2021, 22, 305. https://doi.org/10.3390/ijms22010305
Singh T, Neal AS, Moatamed NA, Memarzadeh S. Exploring the Potential of Drug Response Assays for Precision Medicine in Ovarian Cancer. International Journal of Molecular Sciences. 2021; 22(1):305. https://doi.org/10.3390/ijms22010305
Chicago/Turabian StyleSingh, Tanya, Adam S. Neal, Neda A. Moatamed, and Sanaz Memarzadeh. 2021. "Exploring the Potential of Drug Response Assays for Precision Medicine in Ovarian Cancer" International Journal of Molecular Sciences 22, no. 1: 305. https://doi.org/10.3390/ijms22010305
APA StyleSingh, T., Neal, A. S., Moatamed, N. A., & Memarzadeh, S. (2021). Exploring the Potential of Drug Response Assays for Precision Medicine in Ovarian Cancer. International Journal of Molecular Sciences, 22(1), 305. https://doi.org/10.3390/ijms22010305




