Novel Insights into Selected Disease-Causing Mutations within the SLC35A1 Gene Encoding the CMP-Sialic Acid Transporter
Abstract
1. Introduction
2. Results
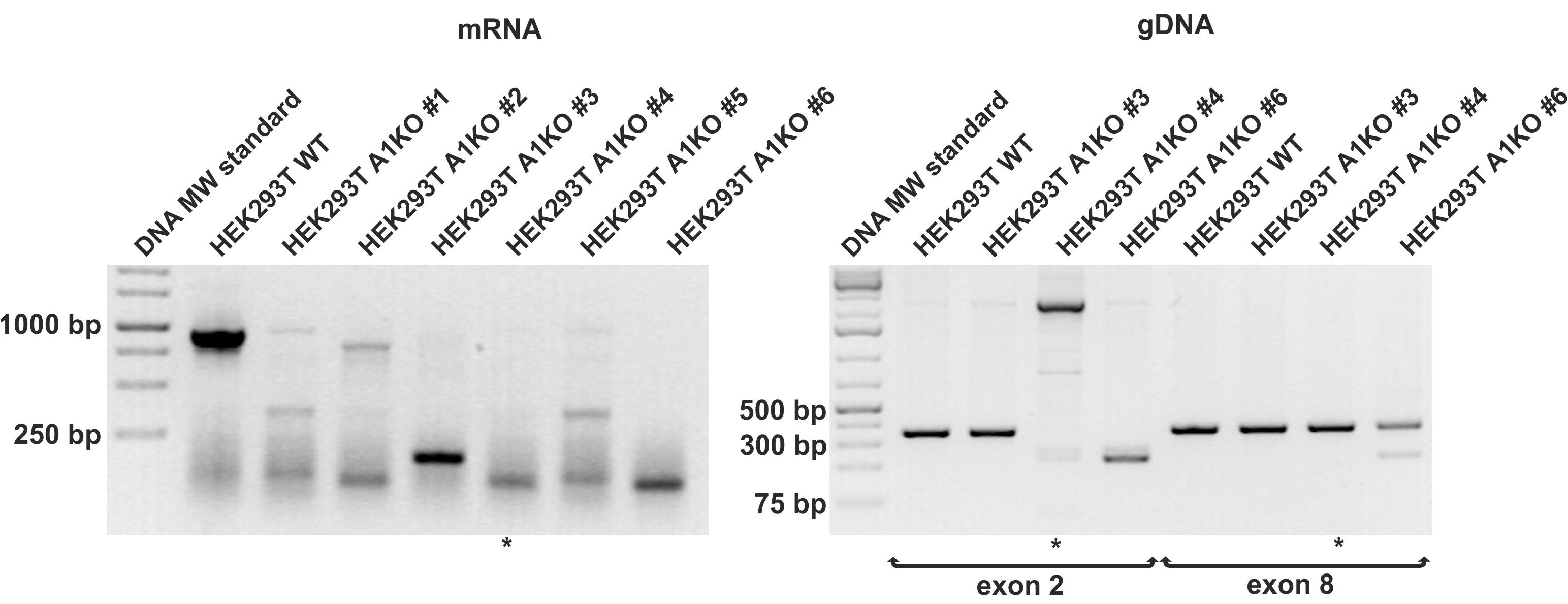
2.1. Inactivation of the SLC35A1 Gene in HEK293T Cells via CRISPR/Cas9 Strategy
2.2. Analysis of Subcellular Localization of CST Variants
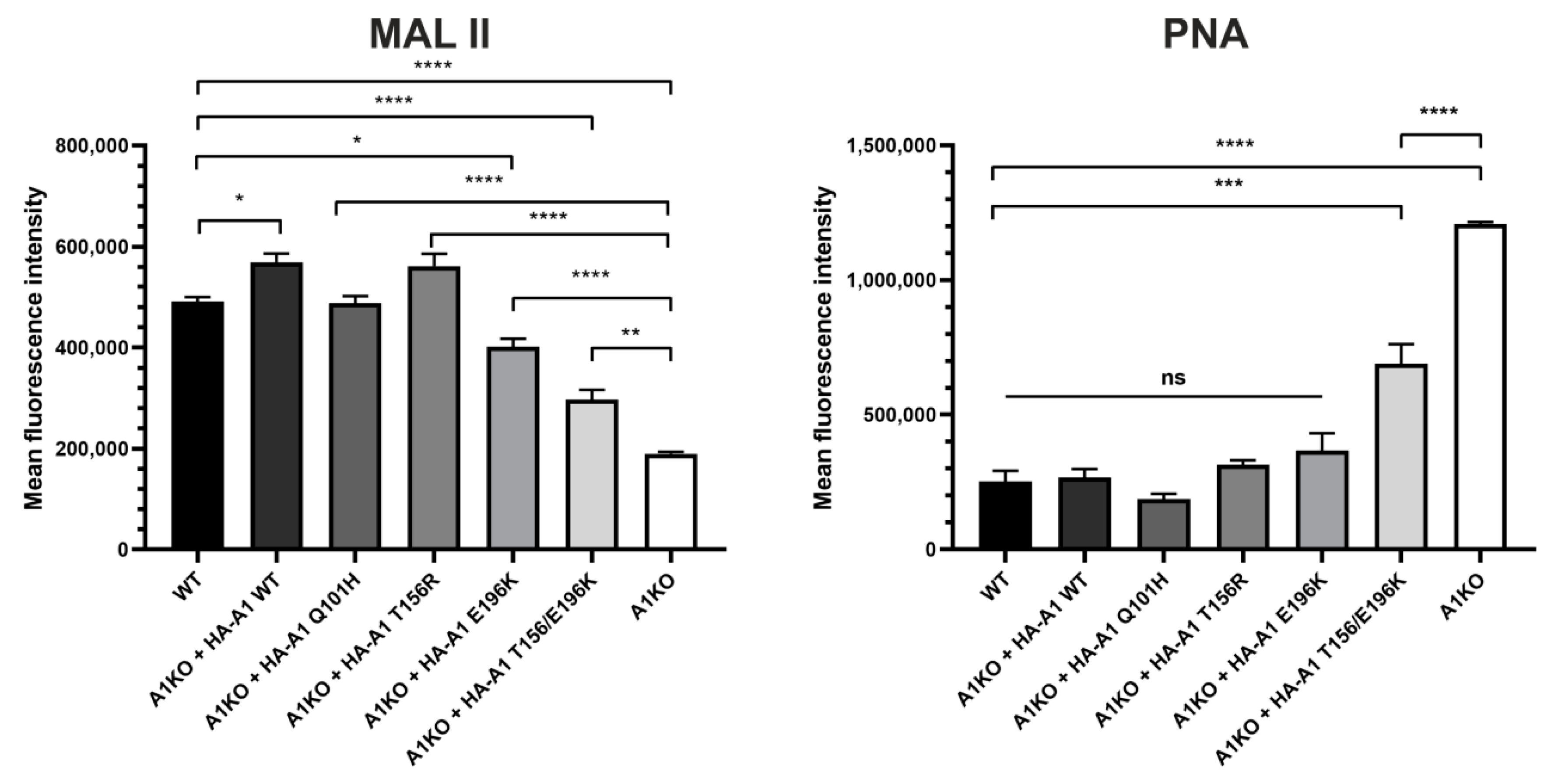
2.3. Analysis of Cell Surface Glycoconjugates Using Lectin-Based Flow Cytometry
2.4. Analysis of Cellular Glycoproteins Using Lectin Blotting
2.5. Structural Analysis of N-Glycans
2.6. Structural Analysis of O-Glycans
2.7. Analysis of Glycolipids Using Thin-Layer Chromatography
2.8. Analysis of the Ability of CST Variants to Form Dimers Using a Split Luciferase Complementation Assay
3. Discussion
4. Materials and Methods
4.1. Cell Maintenance
4.2. Generation of Knock-Out Cell Lines
4.3. Generation of Cells Lines Expressing Mutant Variants of the CMP-Sialic Acid Transporter
4.4. Immunofluorescence Staining
4.5. Lectin Blotting
4.6. Flow Cytometry Analysis of Cells Labeled with Lectins
4.7. Preparation of Cell-Derived N-Glycans for HPLC Analysis
4.8. Preparation of SEAP-Derived N-Glycans for HPLC Analysis
4.9. Exoglycosidase Digestion of SEAP-Derived N-Glycans
4.10. Analysis of O-Glycans
4.11. MALDI-TOF Analysis of N- and O-Glycans
4.12. Analysis of Cellular Glycolipids
4.13. Split Luciferase Complementation Assay
4.14. Bioluminescent Imaging
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AB | aminobenzamide |
| CDG | congenital disorder of glycosylation |
| CDP | cytidine diphosphate |
| CHO | Chinese hamster ovary |
| CMP | cytidine monophosphate |
| CORA | cellular O-glycome Reporter/amplification |
| CRISPR | clustered regularly-interspaced short palindromic repeats |
| CST | CMP-sialic acid transporter |
| DMEM | Dulbecco’s modified eagle medium |
| ECL | Erythrina cristagalli lectin |
| ER | endoplasmic reticulum |
| Gal | galactose |
| GalNAc | N-acetylgalactosamine |
| GAPDH | glyceraldehyde 3-phosphate dehydrogenase |
| GDP | guanosine diphosphate |
| GlcNAc | N-acetylglucosamine |
| GPI | glycosylphosphatidylinositol |
| HA | hemagglutinin |
| HEK | human embryonic kidney |
| HPLC | high performance liquid chromatography |
| LacCer | lactosylceramide |
| LgBiT | large NanoLuc subunit |
| MAG | myelin-associated glycoprotein |
| MAL/MAA | Maackia amurensis lectin/agglutinin |
| MALDI-TOF | matrix-assisted laser desorption/ionization-time-of-flight |
| Man | mannose |
| NanoBiT | NanoLuc Binary Technology |
| NCAM | neural cell adhesion molecule |
| Neu5Ac | 5-N-acetylneuraminic acid |
| NST | nucleotide sugar transporter |
| PML | progressive multifocal leukoencephalopathy |
| PNA | Arachis hypogea lectin |
| RCA | Ricinus communis agglutinin |
| SEAP | secreted alkaline phosphatase |
| SLC | solute carrier |
| SmBiT | small NanoLuc subunit |
| TMD | transmembrane domain |
| UDP | uridine diphosphate |
| UGT | UDP-galactose transporter |
| VSV | vesicular stomatitis virus |
| WGA | wheat germ agglutinin |
References
- Varki, A. Sialic acids in human health and disease. Trends Mol. Med. 2008, 14, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Fearon, D.T. Regulation by membrane sialic acid of β 1H-dependent decay-dissociation of amplification C3 convertase of the alternative complement pathway. Proc. Natl. Acad. Sci. USA 1978, 75, 1971–1975. [Google Scholar] [CrossRef] [PubMed]
- Varki, A. Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature 2007, 446, 1023–1029. [Google Scholar] [CrossRef] [PubMed]
- Rosen, S.D.; Singer, M.; Yednock, T.; Stoolman, L. Involvement of sialic acid on endothelial cells in organ-specific lymphocyte recirculation. Science 1985, 228, 1005–1007. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.L.; Varki, A.; McEver, R.P. GMP-140 binds to a glycoprotein receptor on human neutrophils: Evidence for a lectin-like interaction. J. Cell Biol. 1991, 112, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Rosen, S.D. Ligands for L-selectin: Homing, inflammation, and beyond. Annu. Rev. Immunol. 2004, 22, 129–156. [Google Scholar] [CrossRef]
- Collins, B.E.; Fralich, T.J.; Itonori, S.; Ichikawa, Y.; Schnaar, R.L. Conversion of cellular sialic acid expression from N-acetyl- to N-glycolylneuraminic acid using a synthetic precursor, N-glycolylmannosamine pentaacetate: Inhibition of myelin-associated glycoprotein binding to neural cells. Glycobiology 2000, 10, 11–20. [Google Scholar] [CrossRef][Green Version]
- Sun, J.; Shaper, N.L.; Itonori, S.; Heffer, M.; Sheikh, K.A.; Schnaar, R.L. Myelin-associated glycoprotein (Siglec-4) expression is progressively and selectively decreased in the brains of mice lacking complex gangliosides. Glycobiology 2004, 14, 851–857. [Google Scholar] [CrossRef]
- Varki, A.; Angata, T. Siglecs—The major subfamily of I-type lectins. Glycobiology 2006, 16, 1R–27R. [Google Scholar] [CrossRef]
- Schnaar, R.L.; Gerardy-Schahn, R.; Hildebrandt, H. Sialic acids in the brain: Gangliosides and polysialic acid in nervous system development, stability, disease, and regeneration. Physiol. Rev. 2014, 94, 461–518. [Google Scholar] [CrossRef]
- Sadoul, R.; Hirn, M.; Deagostini-Bazin, H.; Rougon, G.; Goridis, C. Adult and embryonic mouse neural cell adhesion molecules have different binding properties. Nature 1983, 304, 347–349. [Google Scholar] [CrossRef] [PubMed]
- Rutishauser, U.; Acheson, A.; Hall, A.K.; Mann, D.M.; Sunshine, J. The neural cell adhesion molecule (NCAM) as a regulator of cell-cell interactions. Science 1988, 240, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Capasso, J.M.; Hirschberg, C.B. Mechanisms of glycosylation and sulfation in the Golgi apparatus: Evidence for nucleotide sugar/nucleoside monophosphate and nucleotide sulfate/nucleoside monophosphate antiports in the Golgi apparatus membrane. Proc. Natl. Acad. Sci. USA 1984, 81, 7051–7055. [Google Scholar] [CrossRef] [PubMed]
- Hadley, B.; Maggioni, A.; Ashikov, A.; Day, C.J.; Haselhorst, T.; Tiralongo, J. Structure and function of nucleotide sugar transporters: Current progress. Comput. Struct. Biotechnol. J. 2014, 10, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Chintala, S.; Tan, J.; Gautam, R.; Rusiniak, M.E.; Guo, X.; Li, W.; Gahl, W.A.; Huizing, M.; Spritz, R.A.; Hutton, S.; et al. The Slc35d3 gene, encoding an orphan nucleotide sugar transporter, regulates platelet-dense granules. Blood 2007, 109, 1533–1540. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Yuan, Y.-F.; Jaouen, F.; Ma, M.-S.; Hao, C.-J.; Zhang, Z.; Chen, Q.; Yuan, Z.; Yu, L.; Li, W.; et al. SLC35D3 increases autophagic activity in midbrain dopaminergic neurons by enhancing BECN1-ATG14-PIK3C3 complex formation. Autophagy 2016, 12, 1168–1179. [Google Scholar] [CrossRef]
- Sosicka, P.; Maszczak-Seneczko, D.; Bazan, B.; Shauchuk, Y.; Kaczmarek, B.; Olczak, M. An insight into the orphan nucleotide sugar transporter SLC35A4. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 825–838. [Google Scholar] [CrossRef]
- Hadley, B.; Litfin, T.; Day, C.J.; Haselhorst, T.; Zhou, Y.; Tiralongo, J. Nucleotide sugar transporter SLC35 family structure and function. Comput. Struct. Biotechnol. J. 2019, 17, 1123–1134. [Google Scholar] [CrossRef]
- Puglielli, L.; Hirschberg, C.B. Reconstitution, identification, and purification of the rat liver Golgi membrane GDP-fucose transporter. J. Biol. Chem. 1999, 274, 35596–35600. [Google Scholar] [CrossRef]
- Puglielli, L.; Mandon, E.C.; Rancour, D.M.; Menon, A.K.; Hirschberg, C.B. Identification and purification of the rat liver Golgi membrane UDP-N-acetylgalactosamine transporter. J. Biol. Chem. 1999, 274, 4474–4479. [Google Scholar] [CrossRef]
- Gao, X.-D.; Dean, N. Distinct protein domains of the yeast Golgi GDP-mannose transporter mediate oligomer assembly and export from the endoplasmic reticulum. J. Biol. Chem. 2000, 275, 17718–17727. [Google Scholar] [CrossRef] [PubMed]
- Olczak, M.; Guillen, E. Characterization of a mutation and an alternative splicing of UDP-galactose transporter in MDCK-RCAr cell line. Biochim. Biophys. Acta 2006, 1763, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Maszczak-Seneczko, D.; Sosicka, P.; Majkowski, M.; Olczak, T.; Olczak, M. UDP-N-acetylglucosamine transporter and UDP-galactose transporter form heterologous complexes in the Golgi membrane. FEBS Lett. 2012, 586, 4082–4087. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.L.; Newstead, S. Structural basis of nucleotide sugar transport across the Golgi membrane. Nature 2017, 551, 521–524. [Google Scholar] [CrossRef]
- Wiertelak, W.; Sosicka, P.; Olczak, M.; Maszczak-Seneczko, D. Analysis of homologous and heterologous interactions between UDP-galactose transporter and beta-1,4-galactosyltransferase 1 using NanoBiT. Anal. Biochem. 2020, 593, 113599. [Google Scholar] [CrossRef]
- Maszczak-Seneczko, D.; Sosicka, P.; Kaczmarek, B.; Majkowski, M.; Luzarowski, M.; Olczak, T.; Olczak, M. UDP-galactose (SLC35A2) and UDP-N-acetylglucosamine (SLC35A3) transporters form glycosylation-related complexes with mannoside acetylglucosaminyltransferases (Mgats). J. Biol. Chem. 2015, 290, 15475–15486. [Google Scholar] [CrossRef]
- Khoder-Agha, F.; Sosicka, P.; Conde, M.E.; Hassinen, A.; Glumoff, T.; Olczak, M.; Kellokumpu, S. N-acetylglucosaminyltransferases and nucleotide sugar transporters form multi-enzyme–multi-transporter assemblies in golgi membranes in vivo. Cell. Mol. Life Sci. 2019, 76, 1821–1832. [Google Scholar] [CrossRef]
- Shauchuk, A.; Szulc, B.; Maszczak-Seneczko, D.; Wiertelak, W.; Skurska, E.; Olczak, M. N-glycosylation of the human β1,4-galactosyltransferase 4 is crucial for its activity and Golgi localization. Glycoconj. J. 2020, 37, 577–588. [Google Scholar] [CrossRef]
- Kellokumpu, S.; Hassinen, A.; Glumoff, T. Glycosyltransferase complexes in eukaryotes: Long-known, prevalent but still unrecognized. Cell. Mol. Life Sci. 2015, 73, 305–325. [Google Scholar] [CrossRef]
- Stanley, P.; Siminovitch, L. Complementation between mutants of CHO cells resistant to a variety of plant lectins. Somat. Cell Mol. Genet. 1977, 3, 391–405. [Google Scholar] [CrossRef]
- Briles, E.B.; Li, E.; Kornfeld, S. Isolation of wheat germ agglutinin-resistant clones of Chinese hamster ovary cells deficient in membrane sialic acid and galactose. J. Biol. Chem. 1977, 252, 1107–1116. [Google Scholar] [PubMed]
- Deutscher, S.L.; Nuwayhid, N.; Stanley, P.; Briles, E.I.B.; Hirschberg, C.B. Translocation across golgi vesicle membranes: A CHO glycosylation mutant deficient in CMP-sialic acid transport. Cell 1984, 39, 295–299. [Google Scholar] [CrossRef]
- Eckhardt, M.; Mühlenhoff, M.; Bethe, A.; Gerardy-Schahn, R. Expression cloning of the Golgi CMP-sialic acid transporter. Proc. Natl. Acad. Sci. USA 1996, 93, 7572–7576. [Google Scholar] [CrossRef] [PubMed]
- Eckhardt, M.; Gerardy-Schahn, R. Molecular cloning of the hamster CMP-sialic acid transporter. Eur. J. Biochem. 1997, 248, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Berninsone, P.; Eckhardt, M.; Gerardy-Schahn, R.; Hirschberg, C.B. Functional expression of the murine Golgi CMP-sialic acid transporter in Saccharomyces cerevisiae. J. Biol. Chem. 1997, 272, 12616–12619. [Google Scholar] [CrossRef]
- Ishida, N.; Ito, M.; Yoshioka, S.; Sun-Wada, G.-H.; Kawakita, M. Functional expression of human golgi CMP-sialic acid transporter in the Golgi complex of a transporter-deficient Chinese hamster ovary cell mutant. J. Biochem. 1998, 124, 171–178. [Google Scholar] [CrossRef]
- Lim, S.F.; Lee, M.M.; Zhang, P.; Song, Z. The Golgi CMP-sialic acid transporter: A new CHO mutant provides functional insights [published correction appears in Glycobiology 2009, 19, 192]. Glycobiology 2008, 18, 851–860. [Google Scholar] [CrossRef]
- Eckhardt, M.; Gotza, B.; Gerardy-Schahn, R. Membrane topology of the mammalian CMP-sialic acid transporter. J. Biol. Chem. 1999, 274, 8779–8787. [Google Scholar] [CrossRef]
- Aoki, K.; Sun-Wada, G.-H.; Segawa, H.; Yoshioka, S.; Ishida, N.; Kawakita, M. Expression and activity of chimeric molecules between human UDP-galactose transporter and CMP-sialic acid transporter. J. Biochem. 1999, 126, 940–950. [Google Scholar] [CrossRef]
- Aoki, K.; Ishida, N.; Kawakita, M. Substrate recognition by UDP-galactose and CMP-sialic acid transporters. Different sets of transmembrane helices are utilized for the specific recognition of UDP-galactose and CMP-sialic acid. J. Biol. Chem. 2001, 276, 21555–21561. [Google Scholar] [CrossRef]
- Nji, E.; Gulati, A.; Qureshi, A.A.; Coincon, M.; Drew, D. Structural basis for the delivery of activated sialic acid into Golgi for sialyation. Nat. Struct. Mol. Biol. 2019, 26, 415–423. [Google Scholar] [CrossRef]
- Ahuja, S.; Whorton, M.R. Structural basis for mammalian nucleotide sugar transport. Elife 2019, 8, e45221. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.L.; Corey, R.A.; Stansfeld, P.J.; Newstead, S. Structural basis for substrate specificity and regulation of nucleotide sugar transporters in the lipid bilayer. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Ströh, L.J.; Stehle, T. Glycan engagement by viruses: Receptor switches and specificity. Annu. Rev. Virol. 2014, 1, 285–306. [Google Scholar] [CrossRef] [PubMed]
- Urbanek, K.; Sutherland, D.M.; Orchard, R.C.; Wilen, C.B.; Knowlton, J.J.; Aravamudhan, P.; Taylor, G.M.; Virgin, H.W.; Dermody, T.S. Cytidine monophosphate N-acetylneuraminic acid synthetase and solute carrier family 35 member A1 are required for reovirus binding and infection. J. Virol. 2020, Oct 21, 01571-20. [Google Scholar] [CrossRef]
- Han, J.; Perez, J.T.; Chen, C.; Li, Y.; Benitez, A.; Kandasamy, M.; Lee, Y.; Andrade, J.; Tenoever, B.; Manicassamy, B. Genome-wide CRISPR/Cas9 screen identifies host factors essential for influenza virus replication. Cell Rep. 2018, 23, 596–607. [Google Scholar] [CrossRef] [PubMed]
- Mascarenhas, J.X.; Korokhov, N.; Burger, L.; Kassim, A.; Tuter, J.; Miller, D.; Borgschulte, T.; George, H.J.; Chang, A.; Pintel, D.J.; et al. Genetic engineering of CHO cells for viral resistance to minute virus of mice. Biotechnol. Bioeng. 2017, 114, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Moskovskich, A.; Goldmann, U.; Kartnig, F.; Lindinger, S.; Konecka, J.; Fiume, G.; Girardi, E.; Superti-Furga, G. The transporters SLC35A1 and SLC30A1 play opposite roles in cell survival upon VSV virus infection. Sci. Rep. 2019, 9, 10471. [Google Scholar] [CrossRef]
- Geoghegan, E.M.; Pastrana, D.V.; Schowalter, R.M.; Ray, U.; Gao, W.; Ho, M.; Pauly, G.T.; Sigano, D.M.; Kaynor, C.; Cahir-McFarland, E.; et al. Infectious entry and neutralization of pathogenic JC polyomaviruses. Cell Rep. 2017, 21, 1169–1179. [Google Scholar] [CrossRef]
- Martínez-Duncker, I.; Dupré, T.; Piller, V.; Piller, F.; Candelier, J.-J.; Trichet, C.; Tchernia, G.; Oriol, R.; Mollicone, R. Genetic complementation reveals a novel human congenital disorder of glycosylation of type II, due to inactivation of the Golgi CMP-sialic acid transporter. Blood 2005, 105, 2671–2676. [Google Scholar] [CrossRef]
- Mohamed, M.; Ashikov, A.; Guillard, M.; Robben, J.H.; Schmidt, S.; Heuvel, B.V.D.; De Brouwer, A.P.; Gerardy-Schahn, R.; Deen, P.M.; Wevers, R.A.; et al. Intellectual disability and bleeding diathesis due to deficient CMP-sialic acid transport. Neurology 2013, 81, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Riemersma, M.; Sandrock, J.; Boltje, T.J.; Büll, C.; Heise, T.; Ashikov, A.; Adema, G.J.; van Bokhoven, H.; Lefeber, D.J. Disease mutations in CMP-sialic acid transporter SLC35A1 result in ab-normal alpha-dystroglycan O-mannosylation, independent from sialic acid. Hum. Mol. Genet. 2015, 24, 2241–2246. [Google Scholar] [CrossRef] [PubMed]
- Van Tol, W.; Alsady, M.; van Hove, H.; van Scherpenzeel, M.; Willemsen, M.A.; Ashikov, A.; Lefeber, D.J. Ribitol Supplementation Restores the O-Mannosyl Glycans of Alpha-Dystroglycan in SLC35A1 Deficiency (Chapter 5). Ph.D. Thesis, Radboud Universiteit, Nijmegen, The Netherlands, 23 January 2020. [Google Scholar]
- Ng, B.G.; Asteggiano, C.G.; Kircher, M.; Buckingham, K.J.; Raymond, K.; Nickerson, D.A.; Shendure, J.; Bamshad, M.J.; Ensslen, M.; Freeze, H.H. Encephalopathy caused by novel mutations in the CMP-sialic acid transporter, SLC35A1. Am. J. Med. Genet. A 2017, 173, 2906–2911. [Google Scholar] [CrossRef] [PubMed]
- Kauskot, A.; Pascreau, T.; Adam, F.; Bruneel, A.; Reperant, C.; Lourenco-Rodrigues, M.-D.; Rosa, J.-P.; Petermann, R.; Maurey, H.; Auditeau, C.; et al. A mutation in the gene coding for the sialic acid transporter SLC35A1 is required for platelet life span but not proplatelet formation. Haematologica 2018, 103, e613–e617. [Google Scholar] [CrossRef]
- Ma, X.; Li, Y.; Kondo, Y.; Shi, H.; Han, J.; Jiang, Y.; Bai, X.; Archer-Hartmann, S.A.; Azadi, P.; Ruan, C.; et al. Slc35a1 deficiency causes thrombocytopenia due to impaired megakaryocytopoiesis and excessive platelet clearance in the liver. Haematologica 2020. [Google Scholar] [CrossRef]
- Kudelka, M.R.; Antonopoulos, A.; Wang, Y.; Duong, D.M.; Song, X.; Seyfried, N.T.; Dell, A.; Haslam, S.M.; Cummings, R.D.; Ju, T. Cellular O-glycome reporter/amplification to explore O-glycans of living cells. Nat. Methods 2016, 13, 81–86. [Google Scholar] [CrossRef]
- Stanley, P.; Sudo, T.; Carver, J.P. Differential involvement of cell surface sialic acid residues in wheat germ agglutinin binding to parental and wheat germ agglutinin-resistant Chinese hamster ovary cells. J. Cell Biol. 1980, 85, 60–69. [Google Scholar] [CrossRef]
- Sosicka, P.; Bazan, B.; Maszczak-Seneczko, D.; Shauchuk, Y.; Olczak, T.; Olczak, M. SLC35A5 Protein—A Golgi complex member with putative nucleotide sugar transport activity. Int. J. Mol. Sci. 2019, 20, 276. [Google Scholar] [CrossRef]
- Lee, E.U.; Roth, J.; Paulson, J.C. Alteration of terminal glycosylation sequences on N-Linked oligosaccharides of Chinese hamster ovary cells by expression of β-galactoside α2,6-sialyltransferase. J. Biol. Chem. 1989, 264, 13848–13855. [Google Scholar]
- Szulc, B.; Sosicka, P.; Maszczak-Seneczko, D.; Skurska, E.; Shauchuk, A.; Olczak, T.; Freeze, H.H.; Olczak, M. Biosynthesis of GlcNAc-rich N- and O-glycans in the Golgi apparatus does not require the nucleotide sugar transporter SLC35A3. J. Biol. Chem. 2020, 295, 16445–16463. [Google Scholar] [CrossRef]
- Shaw, G.; Morse, S.; Ararat, M.; Graham, F.L. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 2002, 16, 869–871. [Google Scholar] [CrossRef] [PubMed]
- Marquardt, T.; Bzduch, V.; Hogrebe, M.; Rust, S.; Reunert, J.; Grüneberg, M.; Park, J.; Callewaert, N.; Lachmann, R.; Wada, Y.; et al. SLC37A4-CDG: Mislocalization of the glucose-6-phosphate transporter to the Golgi causes a new congenital disorder of glycosylation. Mol. Genet. Metab. Rep. 2020, 25, 100636. [Google Scholar] [CrossRef] [PubMed]
- Ederveen, A.L.H.; de Haan, N.; Baerenfaenger, M.; Lefeber, D.J.; Wuhrer, M. Dissecting total plasma and protein-specific glycosylation profiles in congenital disorders of glycosylation. Int. J. Mol. Sci. 2020, 21, 7635. [Google Scholar] [CrossRef]
- Maszczak-Seneczko, D.; Olczak, T.; Wunderlich, L.; Olczak, M. Comparative analysis of involvement of UGT1 and UGT2 splice variants of UDP-galactose transporter in glycosylation of macromolecules in MDCK and CHO cell lines. Glycoconj. J. 2011, 28, 481–492. [Google Scholar] [CrossRef] [PubMed]
- Eckhardt, M.; Gotza, B.; Gerardy-Schahn, R. Mutants of the CMP-sialic acid transporter causing the Lec2 phenotype. J. Biol. Chem. 1998, 273, 20189–20195. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Chen, T.-L.L.; Vertel, B.M.; Colley, K.J. The CMP-sialic acid transporter is localized in the medial-trans Golgi and possesses two specific endoplasmic reticulum export motifs in its carboxyl-terminal cytoplasmic tail. J. Biol. Chem. 2006, 281, 31106–31118. [Google Scholar] [CrossRef] [PubMed]
- Bazan, B.; Wiktor, M.; Maszczak-Seneczko, D.; Olczak, T.; Kaczmarek, B.; Olczak, M. Lysine at position 329 within a C-terminal dilysine motif is crucial for the ER localization of human SLC35B4. PLoS ONE 2018, 13, e0207521. [Google Scholar] [CrossRef]
- Kaczmarek, R.; Mikolajewicz, K.; Szymczak, K.; Duk, M.; Majorczyk, E.; Krop-Watorek, A.; Buczkowska, A.; Czerwinski, M. Evaluation of an amino acid residue critical for the specificity and activity of human Gb3/CD77 synthase. Glycoconj. J. 2016, 33, 963–973. [Google Scholar] [CrossRef][Green Version]




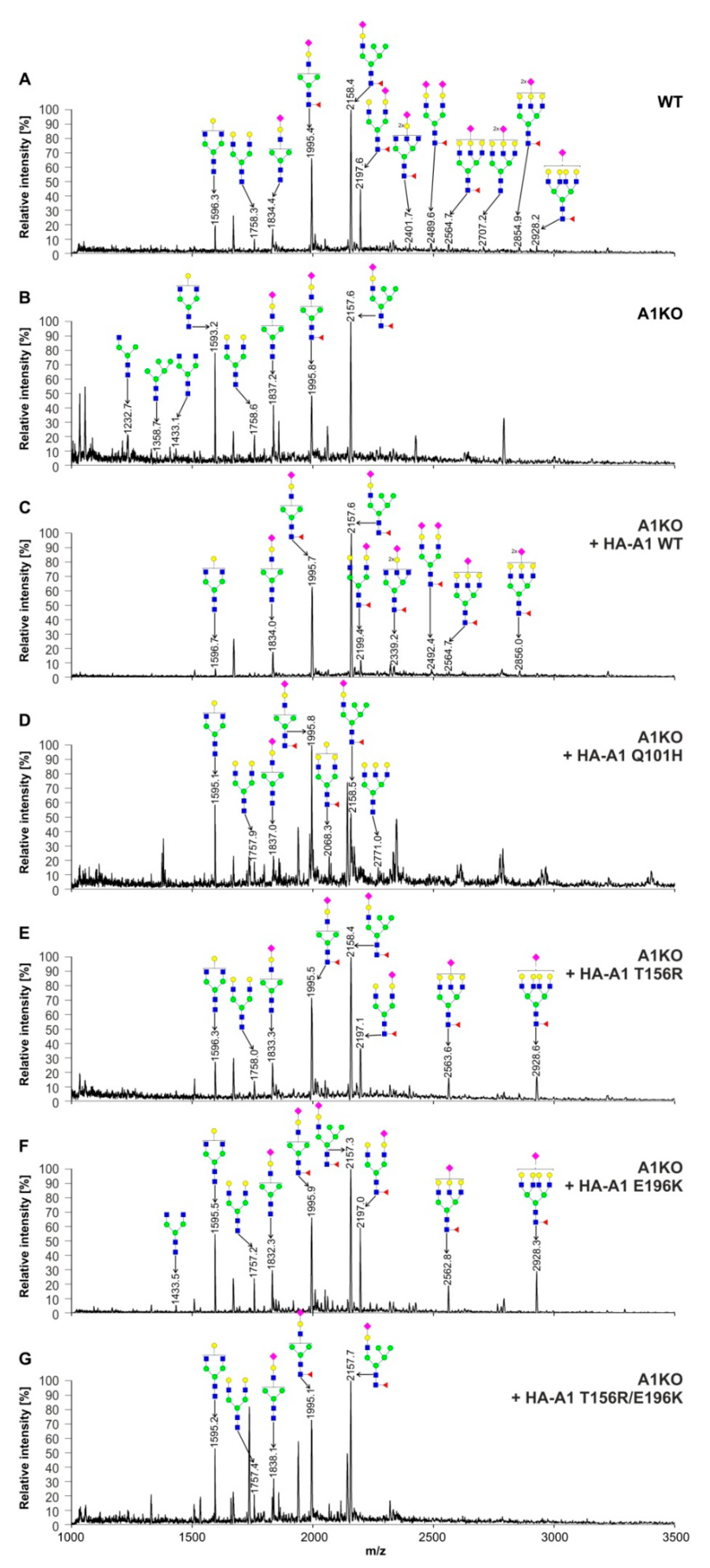



| CST Variant | N-Glycans | O-Glycans |
|---|---|---|
| Wild-type | +++ | +++ |
| Q101H | - | +++ |
| T156R | ++ | +++ |
| E196K | ++ | + |
| T156R/E196K | - | + |
| Antibody | Origin | Dilution | Manufacturer |
|---|---|---|---|
| anti-TGN46 | rabbit | 1:500 | Proteintech (Rosemont, IL, USA) |
| anti-calnexin | rabbit | 1:200 | Abcam (Cambridge, UK) |
| anti-HA | mouse | 1:100 | Thermo Fisher Scientific (Breda, The Netherlands) |
| anti-rabbit Alexa Fluor 488 | donkey | 1:200 | Life Technologies (Carlsbad, CA, USA) |
| anti-mouse Alexa Fluor 568 | donkey | 1:200 | Life Technologies (Carlsbad, CA, USA) |
| Lectin | Dilution |
|---|---|
| PNA | 1:200 |
| RCA I | 1:200 |
| ECL | 1:300 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szulc, B.; Zadorozhna, Y.; Olczak, M.; Wiertelak, W.; Maszczak-Seneczko, D. Novel Insights into Selected Disease-Causing Mutations within the SLC35A1 Gene Encoding the CMP-Sialic Acid Transporter. Int. J. Mol. Sci. 2021, 22, 304. https://doi.org/10.3390/ijms22010304
Szulc B, Zadorozhna Y, Olczak M, Wiertelak W, Maszczak-Seneczko D. Novel Insights into Selected Disease-Causing Mutations within the SLC35A1 Gene Encoding the CMP-Sialic Acid Transporter. International Journal of Molecular Sciences. 2021; 22(1):304. https://doi.org/10.3390/ijms22010304
Chicago/Turabian StyleSzulc, Bożena, Yelyzaveta Zadorozhna, Mariusz Olczak, Wojciech Wiertelak, and Dorota Maszczak-Seneczko. 2021. "Novel Insights into Selected Disease-Causing Mutations within the SLC35A1 Gene Encoding the CMP-Sialic Acid Transporter" International Journal of Molecular Sciences 22, no. 1: 304. https://doi.org/10.3390/ijms22010304
APA StyleSzulc, B., Zadorozhna, Y., Olczak, M., Wiertelak, W., & Maszczak-Seneczko, D. (2021). Novel Insights into Selected Disease-Causing Mutations within the SLC35A1 Gene Encoding the CMP-Sialic Acid Transporter. International Journal of Molecular Sciences, 22(1), 304. https://doi.org/10.3390/ijms22010304





