Mitochondrial TSPO Deficiency Triggers Retrograde Signaling in MA-10 Mouse Tumor Leydig Cells
Abstract
1. Introduction
2. Results
2.1. RNA-seq Data Production and the Verification of Tspo Gene Indel Deletion Mutation after CRISPR Gene Editing
2.2. Transcriptome Changes after Tspo Mutation Affects the Extracellular Structure (ECS) and Extracellular Matrix (ECM)
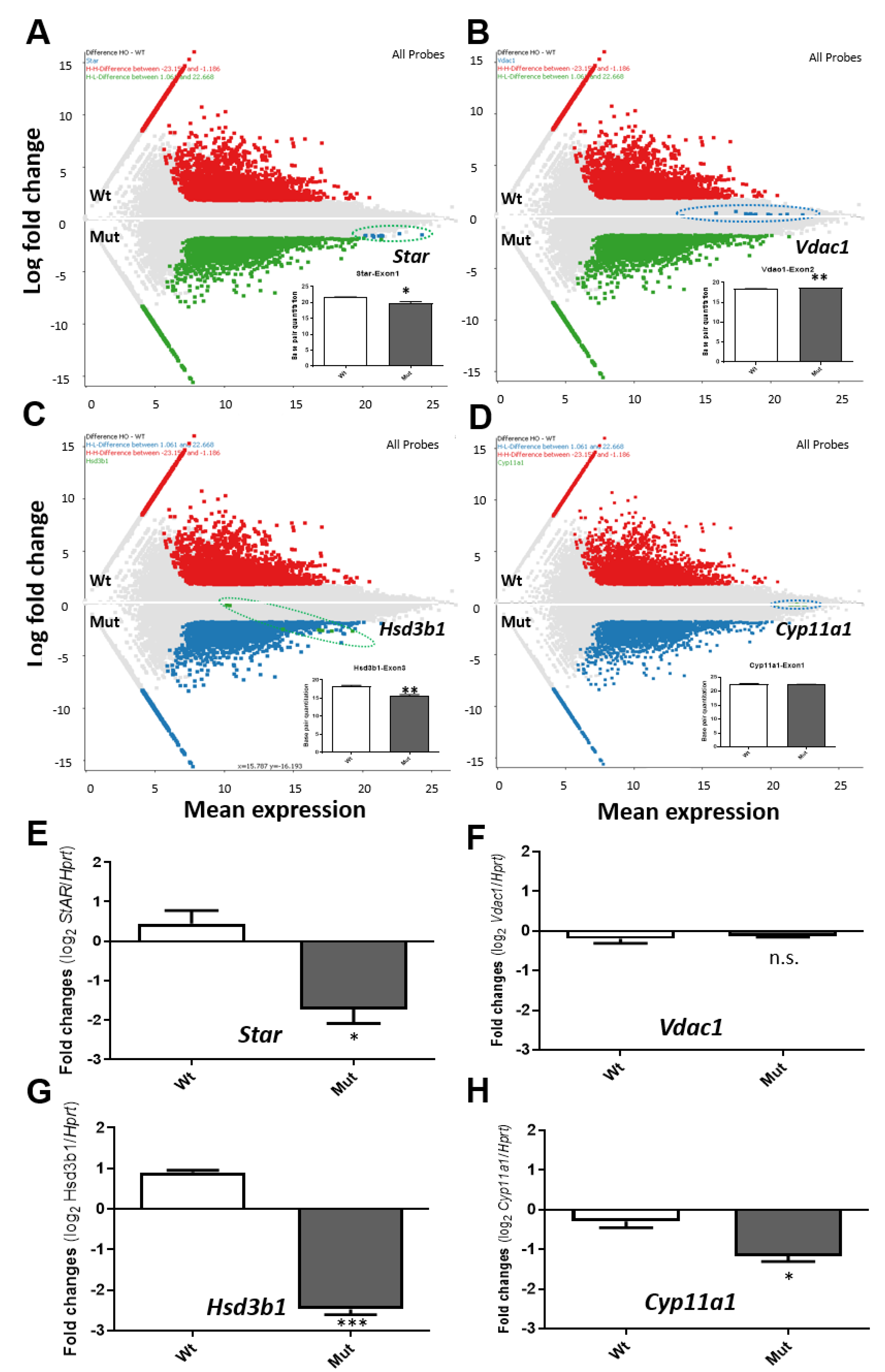
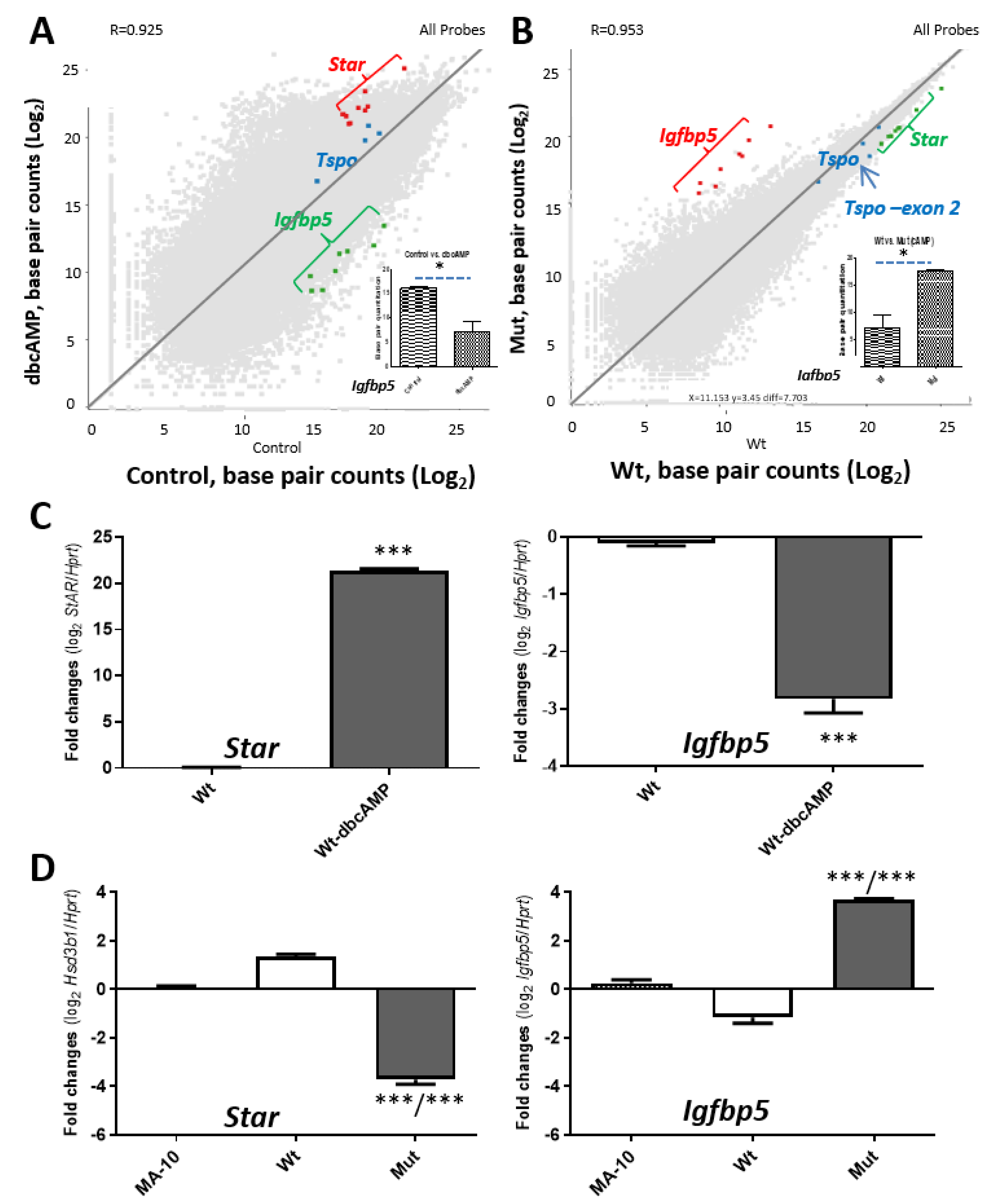
2.3. Tspo Deletion Affects Transcriptome Changes, including Star
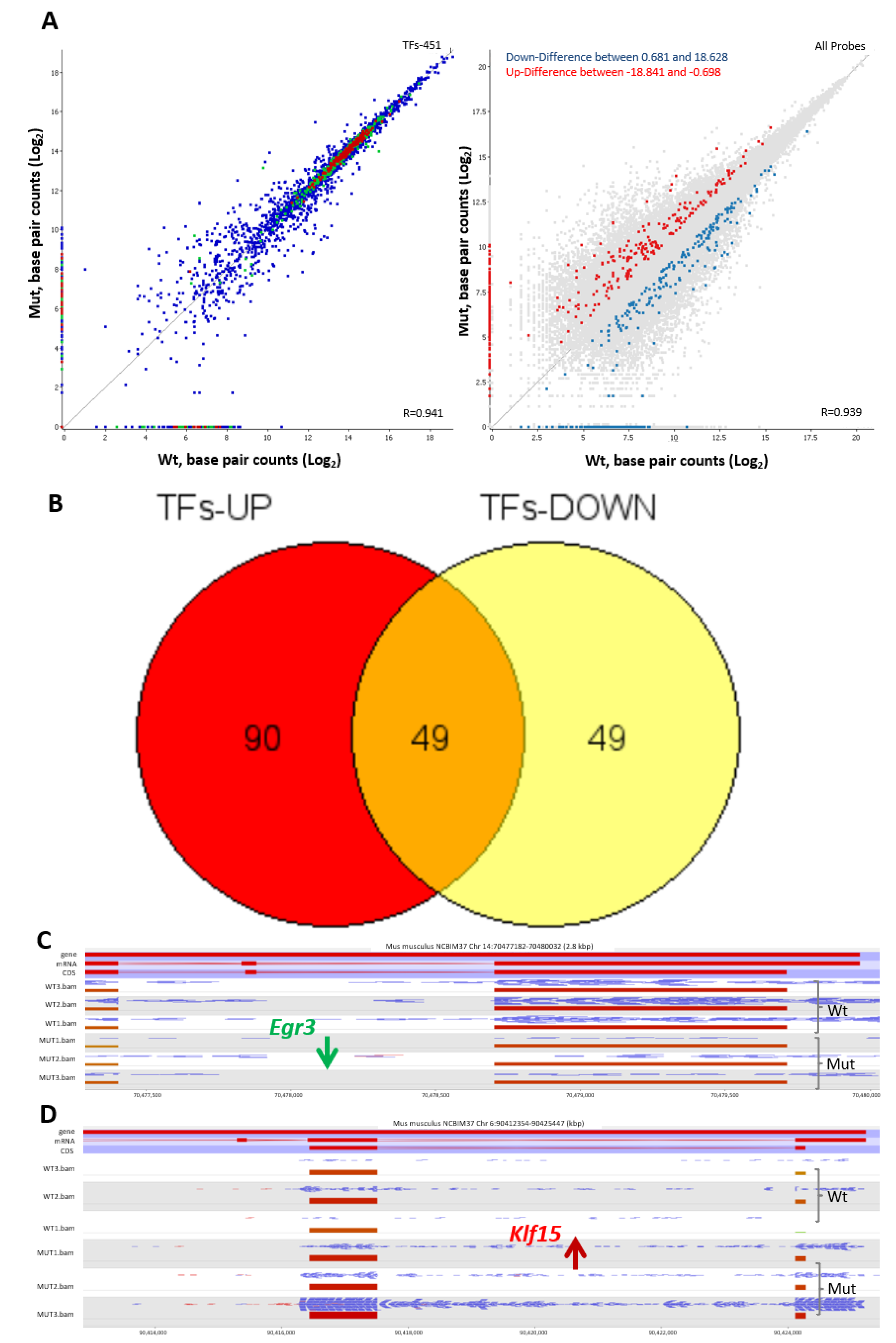
2.4. Tspo Mutation Leads to Differential Expresion of Multiple Transcription Factors
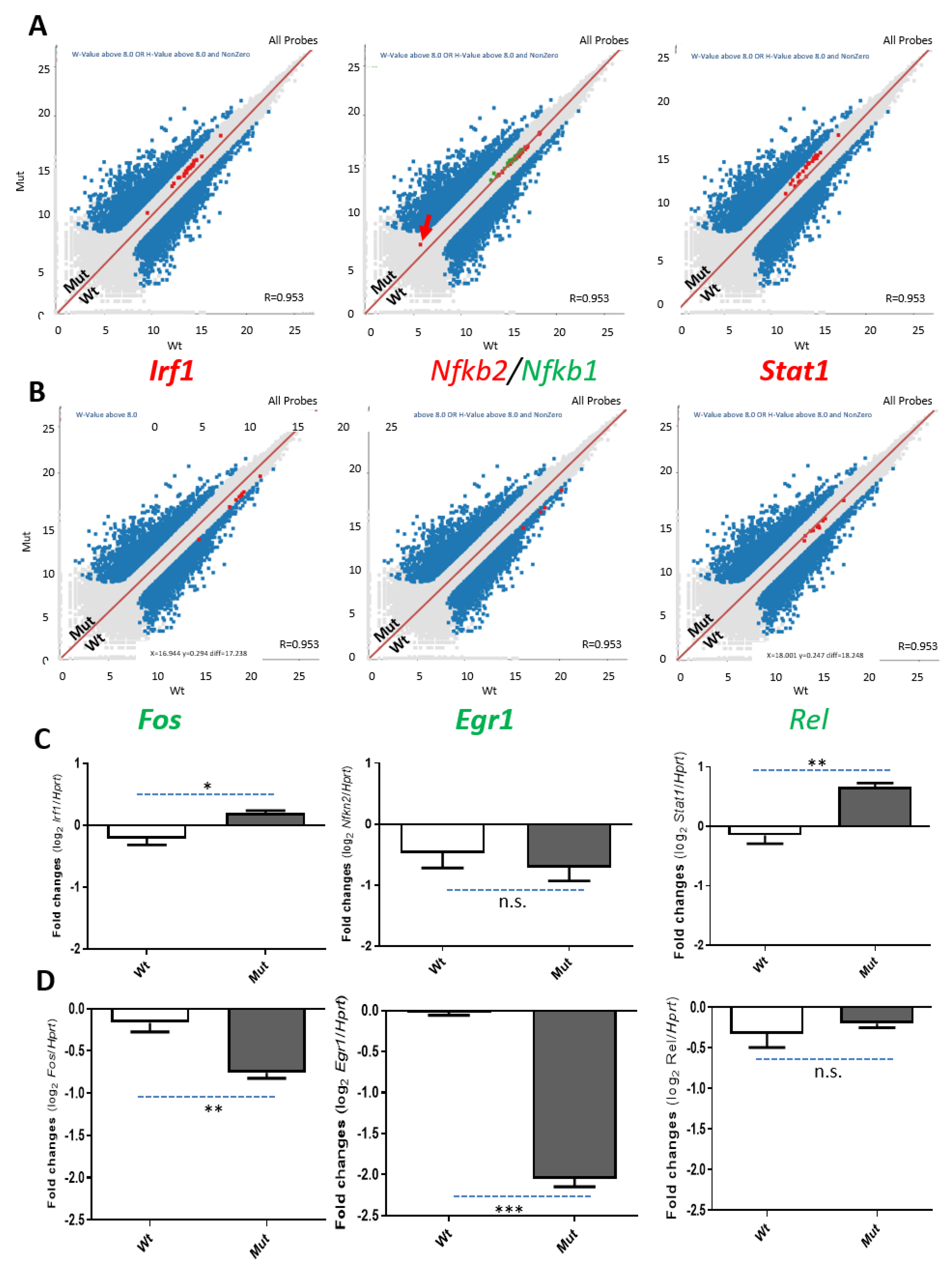
2.5. Transcriptome Changes Due to the Loss of TSPO Are Related to the NF-κB Pathway
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. RNA-seq Data Generation and Data Analysis
4.3. Real-Time PCR
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TSPO | translocator protein |
| CRISPR/Cas9 | clustered regularly interspaced short palindromic repeats/ CRISPR-associated protein 9 |
| dbcAMP | dibutyryl-cAMP |
| ΔΨm | mitochondrial membrane potential |
| Wt | wild-type |
| PCR | polymerase chain reaction |
| RNA-Seq | RNA sequencing |
| ECM | extracellular matrix |
| ECS | extracellular structure organization |
| EGTA | ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid |
| Ca2+ | Calcium ion |
References
- Papadopoulos, V.; Baraldi, M.; Guilarte, T.R.; Knudsen, T.B.; Lacapere, J.J.; Lindemann, P.; Norenberg, M.D.; Nutt, D.; Weizman, A.; Zhang, M.R.; et al. Translocator protein (18kDa): New nomenclature for the peripheral-type benzodiazepine receptor based on its structure and molecular function. Trends Pharmacol. Sci. 2006, 27, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Rupprecht, R.; Papadopoulos, V.; Rammes, G.; Baghai, T.C.; Fan, J.; Akula, N.; Groyer, G.; Adams, D.; Schumacher, M. Translocator protein (18 kDa) (TSPO) as a therapeutic target for neurological and psychiatric disorders. Nat. Rev. Drug Discov. 2010, 9, 971–988. [Google Scholar] [CrossRef] [PubMed]
- Rone, M.B.; Midzak, A.S.; Issop, L.; Rammouz, G.; Jagannathan, S.; Fan, J.; Ye, X.; Blonder, J.; Veenstra, T.; Papadopoulos, V. Identification of a dynamic mitochondrial protein complex driving cholesterol import, trafficking, and metabolism to steroid hormones. Mol. Endocrinol. 2012, 26, 1868–1882. [Google Scholar] [CrossRef] [PubMed]
- Owen, D.R.; Fan, J.; Campioli, E.; Venugopal, S.; Midzak, A.; Daly, E.; Harlay, A.; Issop, L.; Libri, V.; Kalogiannopoulou, D.; et al. TSPO mutations in rats and a human polymorphism impair the rate of steroid synthesis. Biochem. J. 2017, 474, 3985–3999. [Google Scholar] [CrossRef]
- Fan, J.; Campioli, E.; Midzak, A.; Culty, M.; Papadopoulos, V. Conditional steroidogenic cell-targeted deletion of TSPO unveils a crucial role in viability and hormone-dependent steroid formation. Proc. Natl. Acad. Sci. USA 2015, 112, 7261–7266. [Google Scholar] [CrossRef]
- Hauet, T.; Yao, Z.X.; Bose, H.S.; Wall, C.T.; Han, Z.; Li, W.; Hales, D.B.; Miller, W.L.; Culty, M.; Papadopoulos, V. Peripheral-type benzodiazepine receptor-mediated action of steroidogenic acute regulatory protein on cholesterol entry into leydig cell mitochondria. Mol. Endocrinol. 2005, 19, 540–554. [Google Scholar] [CrossRef]
- Fan, J.; Wang, K.; Zirkin, B.; Papadopoulos, V. CRISPR/Cas9-Mediated Tspo Gene Mutations Lead to Reduced Mitochondrial Membrane Potential and Steroid Formation in MA-10 Mouse Tumor Leydig Cells. Endocrinology 2018, 159, 1130–1146. [Google Scholar] [CrossRef]
- Zhao, A.H.; Tu, L.N.; Mukai, C.; Sirivelu, M.P.; Pillai, V.V.; Morohaku, K.; Cohen, R.; Selvaraj, V. Mitochondrial Translocator Protein (TSPO) Function Is Not Essential for Heme Biosynthesis. J. Biol. Chem. 2016, 291, 1591–1603. [Google Scholar] [CrossRef]
- Gatliff, J.; East, D.A.; Singh, A.; Alvarez, M.S.; Frison, M.; Matic, I.; Ferraina, C.; Sampson, N.; Turkheimer, F.; Campanella, M. A role for TSPO in mitochondrial Ca2+ homeostasis and redox stress signaling. Cell Death Dis. 2017, 8, e2896. [Google Scholar] [CrossRef]
- Meng, Y.; Tian, M.; Yin, S.; Lai, S.; Zhou, Y.; Chen, J.; He, M.; Liao, Z. Downregulation of TSPO expression inhibits oxidative stress and maintains mitochondrial homeostasis in cardiomyocytes subjected to anoxia/reoxygenation injury. Biomed. Pharmacother. 2020, 121, 109588. [Google Scholar] [CrossRef]
- Thai, P.N.; Daugherty, D.J.; Frederich, B.J.; Lu, X.; Deng, W.; Bers, D.M.; Dedkova, E.N.; Schaefer, S. Cardiac-specific Conditional Knockout of the 18-kDa Mitochondrial Translocator Protein Protects from Pressure Overload Induced Heart Failure. Sci. Rep. 2018, 8, 16213. [Google Scholar] [CrossRef] [PubMed]
- Tu, L.N.; Zhao, A.H.; Hussein, M.; Stocco, D.M.; Selvaraj, V. Translocator Protein (TSPO) Affects Mitochondrial Fatty Acid Oxidation in Steroidogenic Cells. Endocrinology 2016, 157, 1110–1121. [Google Scholar] [CrossRef] [PubMed]
- Krauss, S.; Zhang, C.Y.; Lowell, B.B. A significant portion of mitochondrial proton leak in intact thymocytes depends on expression of UCP2. Proc. Natl. Acad. Sci. USA 2002, 99, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Waldeck-Weiermair, M.; Duan, X.; Naghdi, S.; Khan, M.J.; Trenker, M.; Malli, R.; Graier, W.F. Uncoupling protein 3 adjusts mitochondrial Ca2+ uptake to high and low Ca2+ signals. Cell Calcium 2010, 48, 288–301. [Google Scholar] [CrossRef]
- Pierelli, G.; Stanzione, R.; Forte, M.; Migliarino, S.; Perelli, M.; Volpe, M.; Rubattu, S. Uncoupling Protein 2: A Key Player and a Potential Therapeutic Target in Vascular Diseases. Oxid. Med. Cell. Longev. 2017, 2017, 7348372. [Google Scholar] [CrossRef]
- Elmore, S.P.; Nishimura, Y.; Qian, T.; Herman, B.; Lemasters, J.J. Discrimination of depolarized from polarized mitochondria by confocal fluorescence resonance energy transfer. Arch. Biochem. Biophys. 2004, 422, 145–152. [Google Scholar] [CrossRef]
- Milenkovic, V.M.; Slim, D.; Bader, S.; Koch, V.; Heinl, E.S.; Alvarez-Carbonell, D.; Nothdurfter, C.; Rupprecht, R.; Wetzel, C.H. CRISPR-Cas9 Mediated TSPO Gene Knockout alters Respiration and Cellular Metabolism in Human Primary Microglia Cells. Int. J. Mol. Sci. 2019, 20, 3359. [Google Scholar] [CrossRef]
- Yao, R.; Pan, R.; Shang, C.; Li, X.; Cheng, J.; Xu, J.; Li, Y. Translocator Protein 18 kDa (TSPO) Deficiency Inhibits Microglial Activation and Impairs Mitochondrial Function. Front. Pharmacol. 2020, 11, 986. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Diaz, E.S.; Pellizzari, E.; Meroni, S.; Cigorraga, S.; Lustig, L.; Denduchis, B. Effect of extracellular matrix proteins on in vitro testosterone production by rat Leydig cells. Mol. Reprod. Dev. 2002, 61, 493–503. [Google Scholar] [CrossRef]
- Jauregui, E.J.; Mitchell, D.; Garza, S.M.; Topping, T.; Hogarth, C.A.; Griswold, M.D. Leydig cell genes change their expression and association with polysomes in a stage-specific manner in the adult mouse testis. Biol. Reprod. 2018, 98, 722–738. [Google Scholar] [CrossRef]
- Fan, J.; Traore, K.; Li, W.; Amri, H.; Huang, H.; Wu, C.; Chen, H.; Zirkin, B.; Papadopoulos, V. Molecular mechanisms mediating the effect of mono-(2-ethylhexyl) phthalate on hormone-stimulated steroidogenesis in MA-10 mouse tumor Leydig cells. Endocrinology 2010, 151, 3348–3362. [Google Scholar] [CrossRef] [PubMed]
- Midzak, A.; Papadopoulos, V. Adrenal Mitochondria and Steroidogenesis: From Individual Proteins to Functional Protein Assemblies. Front. Endocrinol. 2016, 7, 106. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.J.; Boucher, N.; El-Asmar, B.; Tremblay, J.J. cAMP-induced expression of the orphan nuclear receptor Nur77 in MA-10 Leydig cells involves a CaMKI pathway. J. Androl. 2009, 30, 134–145. [Google Scholar] [CrossRef] [PubMed]
- Kulakovskiy, I.V.; Vorontsov, I.E.; Yevshin, I.S.; Sharipov, R.N.; Fedorova, A.D.; Rumynskiy, E.I.; Medvedeva, Y.A.; Magana-Mora, A.; Bajic, V.B.; Papatsenko, D.A.; et al. HOCOMOCO: Towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res. 2018, 46, D252–D259. [Google Scholar] [CrossRef]
- Zhang, H.; Menzies, K.J.; Auwerx, J. The role of mitochondria in stem cell fate and aging. Development 2018, 145, 8. [Google Scholar] [CrossRef]
- Lilienbaum, A.; Israel, A. From calcium to NF-kappa B signaling pathways in neurons. Mol. Cell. Biol. 2003, 23, 2680–2698. [Google Scholar] [CrossRef]
- Tomic, M.; Dufau, M.L.; Catt, K.J.; Stojilkovic, S.S. Calcium signaling in single rat Leydig cells. Endocrinology 1995, 136, 3422–3429. [Google Scholar] [CrossRef]
- Barron, A.M.; Ji, B.; Kito, S.; Suhara, T.; Higuchi, M. Steroidogenic abnormalities in translocator protein knockout mice and significance in the aging male. Biochem. J. 2018, 475, 75–85. [Google Scholar] [CrossRef]
- Tu, L.N.; Morohaku, K.; Manna, P.R.; Pelton, S.H.; Butler, W.R.; Stocco, D.M.; Selvaraj, V. Peripheral benzodiazepine receptor/translocator protein global knock-out mice are viable with no effects on steroid hormone biosynthesis. J. Biol. Chem. 2014, 289, 27444–27454. [Google Scholar] [CrossRef]
- Wang, H.; Zhai, K.; Xue, Y.; Yang, J.; Yang, Q.; Fu, Y.; Hu, Y.; Liu, F.; Wang, W.; Cui, L.; et al. Global Deletion of TSPO Does Not Affect the Viability and Gene Expression Profile. PLoS ONE 2016, 11, e0167307. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Campioli, E.; Sottas, C.; Zirkin, B.; Papadopoulos, V. Amhr2-Cre-Mediated Global Tspo Knockout. J. Endocr. Soc. 2020, 4, bvaa001. [Google Scholar] [CrossRef] [PubMed]
- Cubbon, A.; Ivancic-Bace, I.; Bolt, E.L. CRISPR-Cas immunity, DNA repair and genome stability. Biosci. Rep. 2018, 38, 5. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, A.M.; Pan, Y.K.; Chandrapalan, T.; Kwong, R.W.M.; Perry, S.F. Loss-of-function approaches in comparative physiology: Is there a future for knockdown experiments in the era of genome editing? J. Exp. Biol. 2019, 222 Pt 7. [Google Scholar] [CrossRef] [PubMed]
- El-Brolosy, M.A.; Stainier, D.Y.R. Genetic compensation: A phenomenon in search of mechanisms. PLoS Genet. 2017, 13, e1006780. [Google Scholar] [CrossRef] [PubMed]
- Tuladhar, R.; Yeu, Y.; Tyler Piazza, J.; Tan, Z.; Rene Clemenceau, J.; Wu, X.; Barrett, Q.; Herbert, J.; Mathews, D.H.; Kim, J.; et al. CRISPR-Cas9-based mutagenesis frequently provokes on-target mRNA misregulation. Nat. Commun. 2019, 10, 4056. [Google Scholar] [CrossRef]
- Mou, H.; Smith, J.L.; Peng, L.; Yin, H.; Moore, J.; Zhang, X.O.; Song, C.Q.; Sheel, A.; Wu, Q.; Ozata, D.M.; et al. CRISPR/Cas9-mediated genome editing induces exon skipping by alternative splicing or exon deletion. Genome Biol. 2017, 18, 108. [Google Scholar] [CrossRef]
- Akbarinejad, V.; Tajik, P.; Movahedin, M.; Youssefi, R. Effect of extracellular matrix on testosterone production during in vitro culture of bovine testicular cells. Vet. Res. Forum 2017, 8, 7–13. [Google Scholar]
- Mazaud Guittot, S.; Verot, A.; Odet, F.; Chauvin, M.A.; le Magueresse-Battistoni, B. A comprehensive survey of the laminins and collagens type IV expressed in mouse Leydig cells and their regulation by LH/hCG. Reproduction 2008, 135, 479–488. [Google Scholar] [CrossRef]
- Vernon, R.B.; Lane, T.F.; Angello, J.C.; Sage, H. Adhesion, shape, proliferation, and gene expression of mouse Leydig cells are influenced by extracellular matrix in vitro. Biol. Reprod. 1991, 44, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Da Cunha, F.M.; Torelli, N.Q.; Kowaltowski, A.J. Mitochondrial Retrograde Signaling: Triggers, Pathways, and Outcomes. Oxid. Med. Cell. Longev. 2015, 2015, 482582. [Google Scholar] [CrossRef] [PubMed]
- Al-Anazi, A.; Parhar, R.; Saleh, S.; Al-Hijailan, R.; Inglis, A.; Al-Jufan, M.; Bazzi, M.; Hashmi, S.; Conca, W.; Collison, K.; et al. Intracellular calcium and NF-kB regulate hypoxia-induced leptin, VEGF, IL-6 and adiponectin secretion in human adipocytes. Life Sci. 2018, 212, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Altamirano, F.; Lopez, J.R.; Henriquez, C.; Molinski, T.; Allen, P.D.; Jaimovich, E. Increased resting intracellular calcium modulates NF-kappaB-dependent inducible nitric-oxide synthase gene expression in dystrophic mdx skeletal myotubes. J. Biol. Chem. 2012, 287, 20876–20887. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.J.; Kim, D.H.; Jung, H.; Chung, S.J.; Chi, S.W.; Cho, S.; Lee, S.C.; Park, B.C.; Park, S.G.; Bae, K.H. Annexin A4 interacts with the NF-kappaB p50 subunit and modulates NF-kappaB transcriptional activity in a Ca2+-dependent manner. Cell. Mol. Life Sci. 2010, 67, 2271–2281. [Google Scholar] [CrossRef]
- Jefferson, K.K.; Smith, M.F., Jr.; Bobak, D.A. Roles of intracellular calcium and NF-kappa B in the Clostridium difficile toxin A-induced up-regulation and secretion of IL-8 from human monocytes. J. Immunol. 1999, 163, 5183–5191. [Google Scholar]
- Takeda, K.; Yanagi, S. Mitochondrial retrograde signaling to the endoplasmic-reticulum regulates unfolded protein responses. Mol. Cell. Oncol. 2019, 6, e1659078. [Google Scholar] [CrossRef]
- Hunt, R.J.; Bateman, J.M. Mitochondrial retrograde signaling in the nervous system. FEBS Lett. 2018, 592, 663–678. [Google Scholar] [CrossRef]
- Biswas, G.; Adebanjo, O.A.; Freedman, B.D.; Anandatheerthavarada, H.K.; Vijayasarathy, C.; Zaidi, M.; Kotlikoff, M.; Avadhani, N.G. Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: A novel mode of inter-organelle crosstalk. EMBO J. 1999, 18, 522–533. [Google Scholar] [CrossRef]
- Papadopoulos, V.; Fan, J.; Zirkin, B. Translocator protein (18 kDa): An update on its function in steroidogenesis. J. Neuroendocrinol. 2017. [Google Scholar] [CrossRef]
- Bonsack, F.; Sukumari-Ramesh, S. TSPO: An Evolutionarily Conserved Protein with Elusive Functions. Int. J. Mol. Sci. 2018, 19, 1694. [Google Scholar] [CrossRef]
- Li, F.; Liu, J.; Liu, N.; Kuhn, L.A.; Garavito, R.M.; Ferguson-Miller, S. Translocator Protein 18 kDa (TSPO): An Old Protein with New Functions? Biochemistry 2016, 55, 2821–2831. [Google Scholar] [CrossRef] [PubMed]
- Hardwick, M.; Fertikh, D.; Culty, M.; Li, H.; Vidic, B.; Papadopoulos, V. Peripheral-type benzodiazepine receptor (PBR) in human breast cancer: Correlation of breast cancer cell aggressive phenotype with PBR expression, nuclear localization, and PBR-mediated cell proliferation and nuclear transport of cholesterol. Cancer Res. 1999, 59, 831–842. [Google Scholar] [PubMed]
- Desai, R.; East, D.A.; Hardy, L.; Faccenda, D.; Rigon, M.; Crosby, J.; Alvarez, M.S.; Singh, A.; Mainenti, M.; Hussey, L.K.; et al. Mitochondria form contact sites with the nucleus to couple prosurvival retrograde response. Sci. Adv. 2020, 6, eabc9955. [Google Scholar] [CrossRef]
- Casellas, P.; Galiegue, S.; Basile, A.S. Peripheral benzodiazepine receptors and mitochondrial function. Neurochem. Int. 2002, 40, 475–486. [Google Scholar] [CrossRef]
- Gong, J.; Szego, E.M.; Leonov, A.; Benito, E.; Becker, S.; Fischer, A.; Zweckstetter, M.; Outeiro, T.; Schneider, A. Translocator Protein Ligand Protects against Neurodegeneration in the MPTP Mouse Model of Parkinsonism. J. Neurosci. 2019, 39, 3752–3769. [Google Scholar] [CrossRef]
- Gorlach, A.; Bertram, K.; Hudecova, S.; Krizanova, O. Calcium and ROS: A mutual interplay. Redox Biol. 2015, 6, 260–271. [Google Scholar] [CrossRef]
- Yasin, N.; Veenman, L.; Singh, S.; Azrad, M.; Bode, J.; Vainshtein, A.; Caballero, B.; Marek, I.; Gavish, M. Classical and Novel TSPO Ligands for the Mitochondrial TSPO Can Modulate Nuclear Gene Expression: Implications for Mitochondrial Retrograde Signaling. Int. J. Mol. Sci. 2017, 18, 786. [Google Scholar] [CrossRef]
- Rosenberg, N.; Rosenberg, O.; Weizman, A.; Leschiner, S.; Sakoury, Y.; Fares, F.; Soudry, M.; Weisinger, G.; Veenman, L.; Gavish, M. In vitro mitochondrial effects of PK 11195, a synthetic translocator protein 18 kDa (TSPO) ligand, in human osteoblast-like cells. J. Bioenerg. Biomembr. 2011, 43, 739–746. [Google Scholar] [CrossRef]
- Ascoli, M. Characterization of several clonal lines of cultured Leydig tumor cells: Gonadotropin receptors and steroidogenic responses. Endocrinology 1981, 108, 88–95. [Google Scholar] [CrossRef]
- Fan, J.; Rone, M.B.; Papadopoulos, V. Translocator protein 2 is involved in cholesterol redistribution during erythropoiesis. J. Biol. Chem. 2009, 284, 30484–30497. [Google Scholar] [CrossRef]


| Primers | Sequence | Purpose |
|---|---|---|
| Cyp11a1-F | CGCCATCACCTCTTGGTTTA | Real-time PCR |
| Cyp11a1-R | GGTGGCCTATCACCAGTATTATC | Real-time PCR |
| Cyp11b1-F | CGGCAACATCACAGATACGA | Real-time PCR |
| Cyp11b1-R | CAGGAACAAGTGGCTGAAGA | Real-time PCR |
| Dnahc5-F | CTGAGAGCTGGGATGAAGATG | Real-time PCR |
| Dnahc5-R | AGCCATTACGCCTGAGAAC | Real-time PCR |
| Egr1-F | GAGTCGTTTGGCTGGGATAA | Real-time PCR |
| Egr1-R | AACAACCCTATGAGCACCTG | Real-time PCR |
| Fos-F | GCAACGCAGACTTCTCATCT | Real-time PCR |
| Fos-R | GAATCCGAAGGGAACGGAATAA | Real-time PCR |
| Fscn1-F | TCTCCTGATCGGTCTCTTCATC | Real-time PCR |
| Fscn1-R | TCTTCGCCCTGGAACAGA | Real-time PCR |
| Hprt-F | GAGCAAGTCTTTCAGTCCTGTCCA | Real-time PCR |
| Hprt-R | GCGTCGTGATTAGCGATGATGAAC | Real-time PCR |
| Hsd3b1-F | CTGTCACCTTGGTCTTTGTCT | Real-time PCR |
| Hsd3b1-R | TGGTGCAGGAGAAAGAACTG | Real-time PCR |
| Hspb8-F | CTTCTGCAGGGAGCTGTATTT | Real-time PCR |
| Hspb8-R | CCGGAAGAGCTGATGGTAAAG | Real-time PCR |
| Igfbp5-F | CAGCCTTCAGCTCGGAAAT | Real-time PCR |
| Igfbp5-R | GGCGAGCAAACCAAGATAGA | Real-time PCR |
| Irf1-F | TTAGTGTCTCGGCTGGACTT | Real-time PCR |
| Irf1-R | AGGATCAGAGTAGGAACAAGGG | Real-time PCR |
| Nfkb2-F | GATCCTTAGGCTCCACGATG | Real-time PCR |
| Nfkb2-R | GGGCCTAGCCCAGAGATA | Real-time PCR |
| Nrxn1-F | GTAGCCCGTTGTGGTAAGAA | Real-time PCR |
| Nrxn1-R | ATGTGAAAGTCACCAGGAATCT | Real-time PCR |
| Psd4-F | GGCAGTTAGGCTCCAGTAAA | Real-time PCR |
| Psd4-R | CAGGTCCCAGGAAACTCTTATC | Real-time PCR |
| Rel-F | AACACAGCCTCACCACATT | Real-time PCR |
| Rel-R | GATTAGTGCAGGAATCAATCCTTTC | Real-time PCR |
| Star-F | ACTCTATCTGGGTCTGCGATA | Real-time PCR |
| Star-R | GGAAGTCCCTCCAAGACTAAAC | Real-time PCR |
| Stat1-F | GGTCGCAAACGAGACATCATA | Real-time PCR |
| Stat1-R | CCCATGGAAATCAGACAGTACC | Real-time PCR |
| Thbs1-R | CTAGGTGTCCTGTTCCTGTTG | Real-time PCR |
| Thbs-F | CACCTCCAATGAGTTCAAAGATG | Real-time PCR |
| Tspo-wt-F | TGTGAAACCTCCCAGCTCTTTCCA | Real-time PCR-WT |
| Tspo-wt-R | TAGCTTGCAGAAACCCTCTTGGCA | Real-time PCR-WT |
| Tspo-mut-F | CCCATGGCTGAATACAGTGTG | Real-time PCR-Deletion |
| Tspo-mut-R | GGGAGCCTACTTTGTACGTG | Real-time PCR-Deletion |
| Vdac1-F | AGTCCATCTGTACTTGGTTTCC | Real-time PCR |
| Vdac1-R | ACGAAGTCAGAGAATGGATTGG | Real-time PCR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, J.; Papadopoulos, V. Mitochondrial TSPO Deficiency Triggers Retrograde Signaling in MA-10 Mouse Tumor Leydig Cells. Int. J. Mol. Sci. 2021, 22, 252. https://doi.org/10.3390/ijms22010252
Fan J, Papadopoulos V. Mitochondrial TSPO Deficiency Triggers Retrograde Signaling in MA-10 Mouse Tumor Leydig Cells. International Journal of Molecular Sciences. 2021; 22(1):252. https://doi.org/10.3390/ijms22010252
Chicago/Turabian StyleFan, Jinjiang, and Vassilios Papadopoulos. 2021. "Mitochondrial TSPO Deficiency Triggers Retrograde Signaling in MA-10 Mouse Tumor Leydig Cells" International Journal of Molecular Sciences 22, no. 1: 252. https://doi.org/10.3390/ijms22010252
APA StyleFan, J., & Papadopoulos, V. (2021). Mitochondrial TSPO Deficiency Triggers Retrograde Signaling in MA-10 Mouse Tumor Leydig Cells. International Journal of Molecular Sciences, 22(1), 252. https://doi.org/10.3390/ijms22010252







