Tuning G-Quadruplex Nanostructures with Lipids. Towards Designing Hybrid Scaffolds for Oligonucleotide Delivery †
Abstract
1. Introduction
2. Results and Discussion
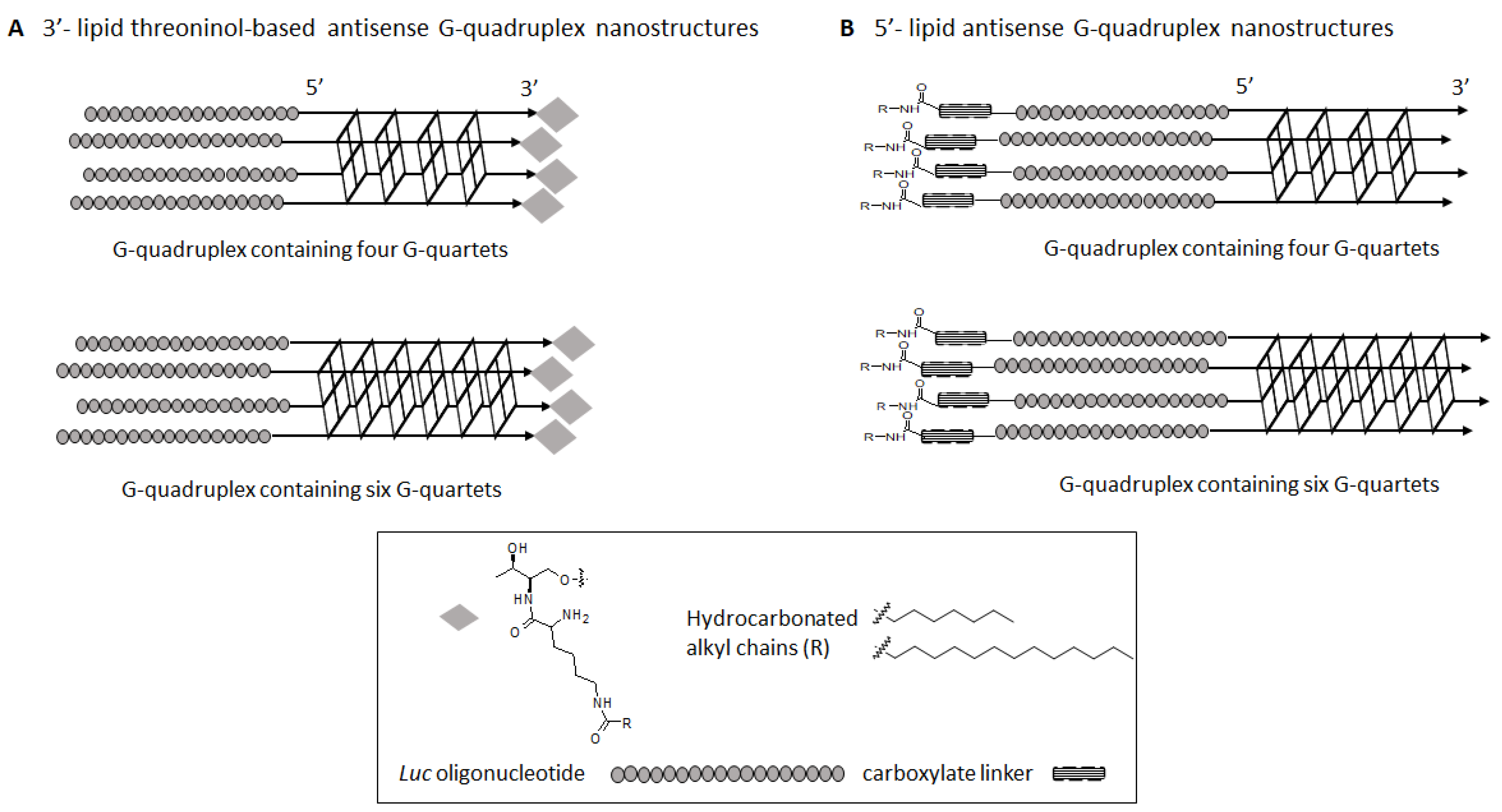
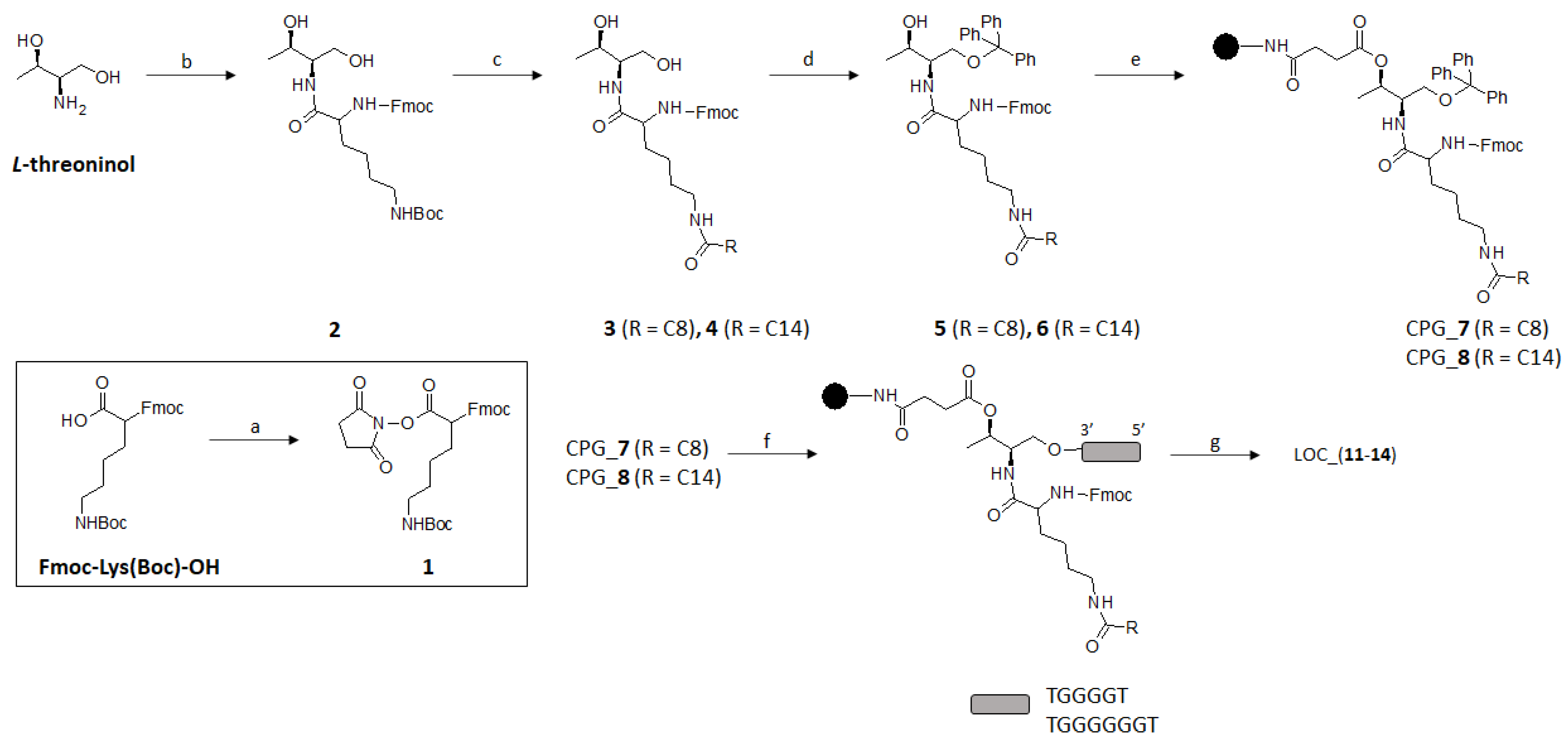
2.1. Synthesis of Lipid Threoninol Derivatives and Preparation of Lipid Oligonucleotide Conjugates (LOCs) with G-Rich Sequences
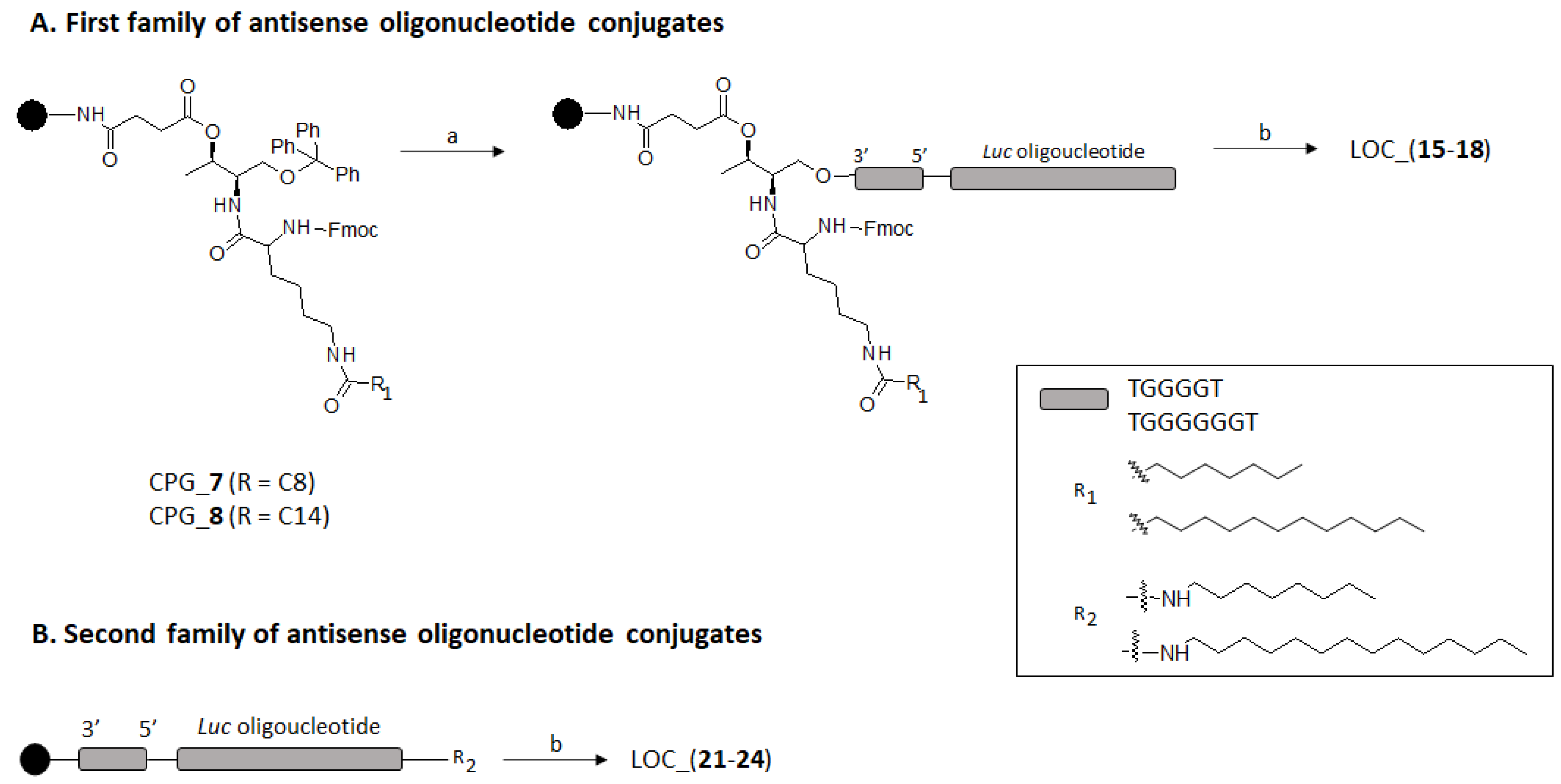
2.2. Preparation of Lipid Oligonucleotide Conjugates (LOC) Hybrids Containing the Luc Sequence
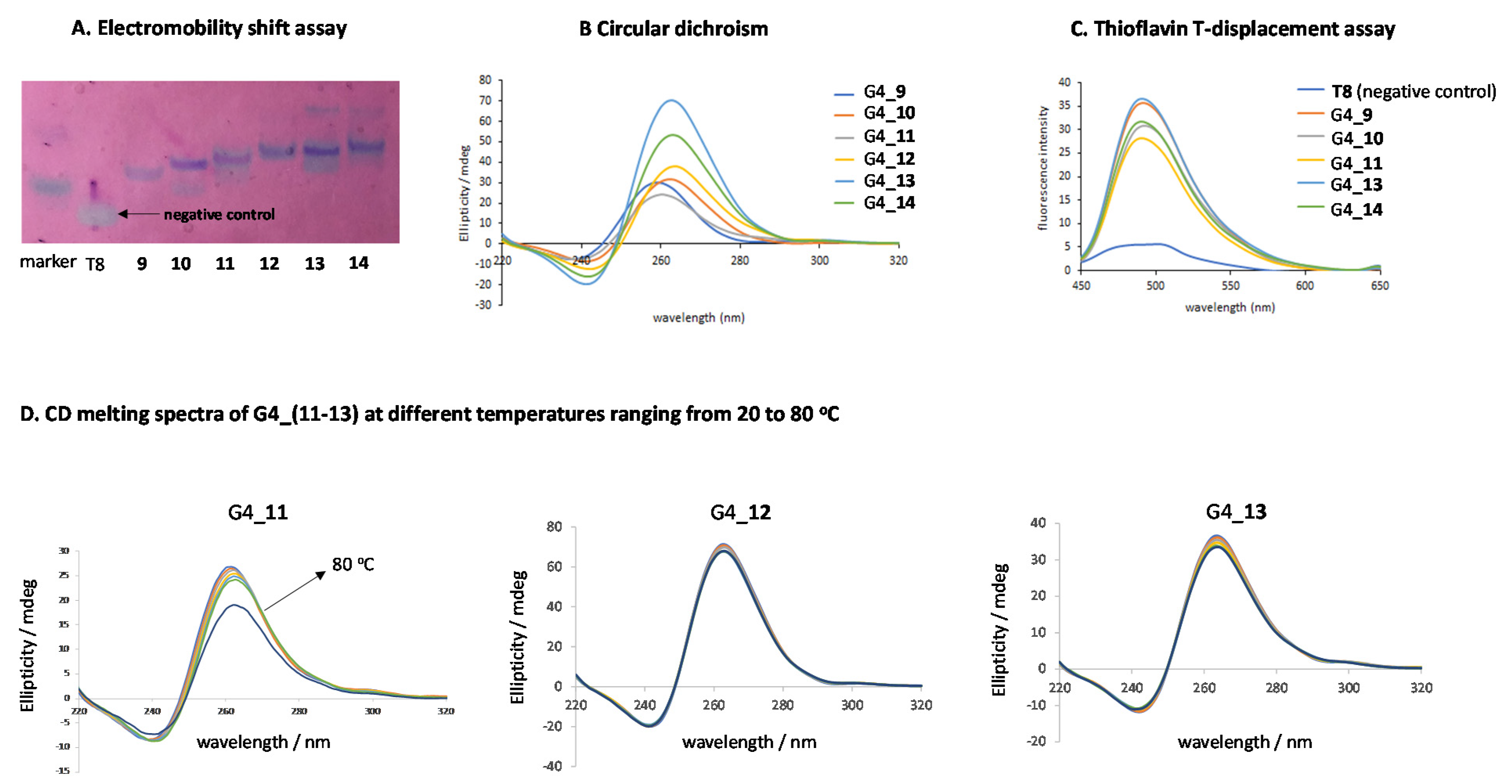
2.3. G-Quadruplex Formation and Biophysical Characterization
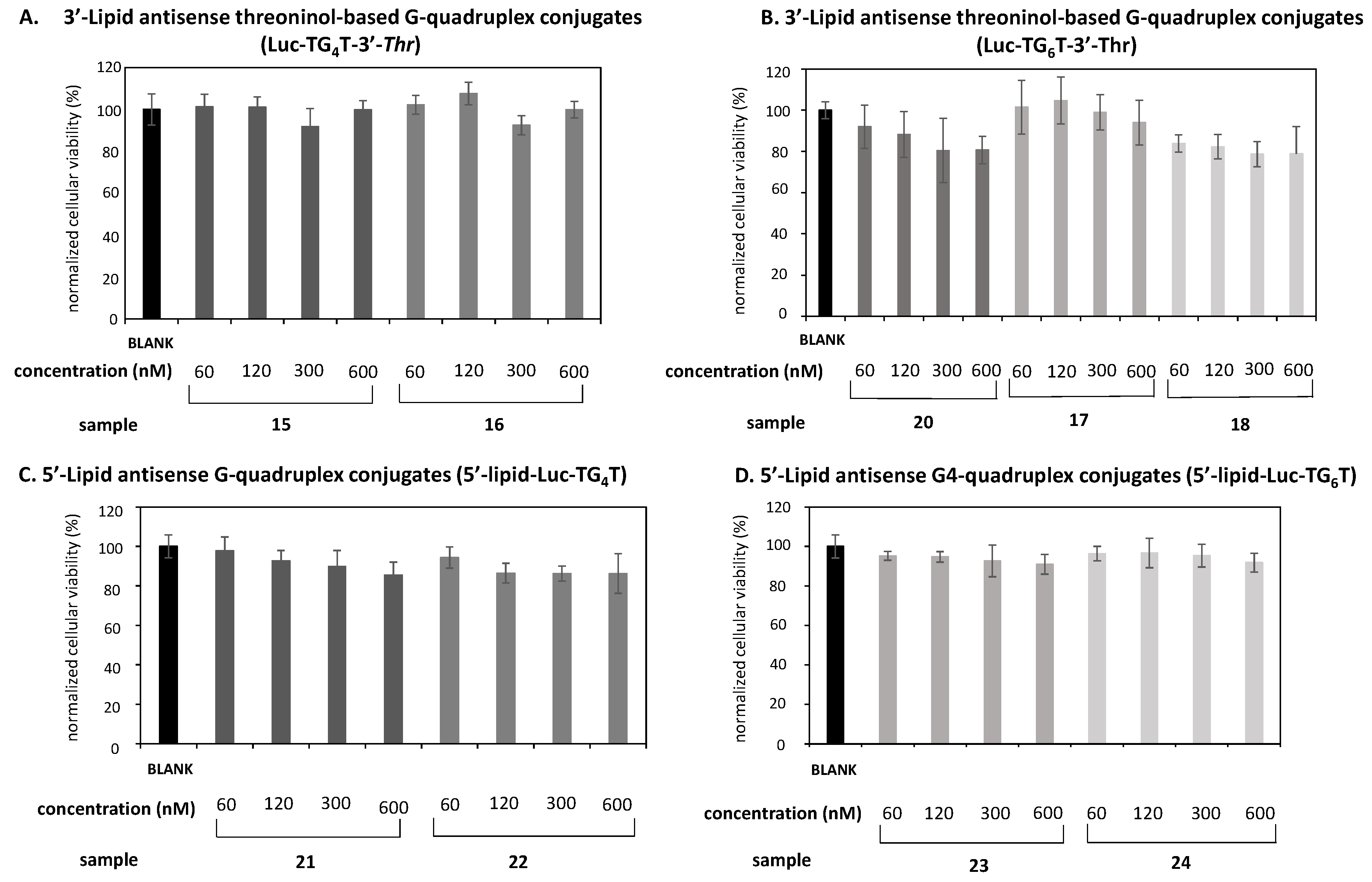
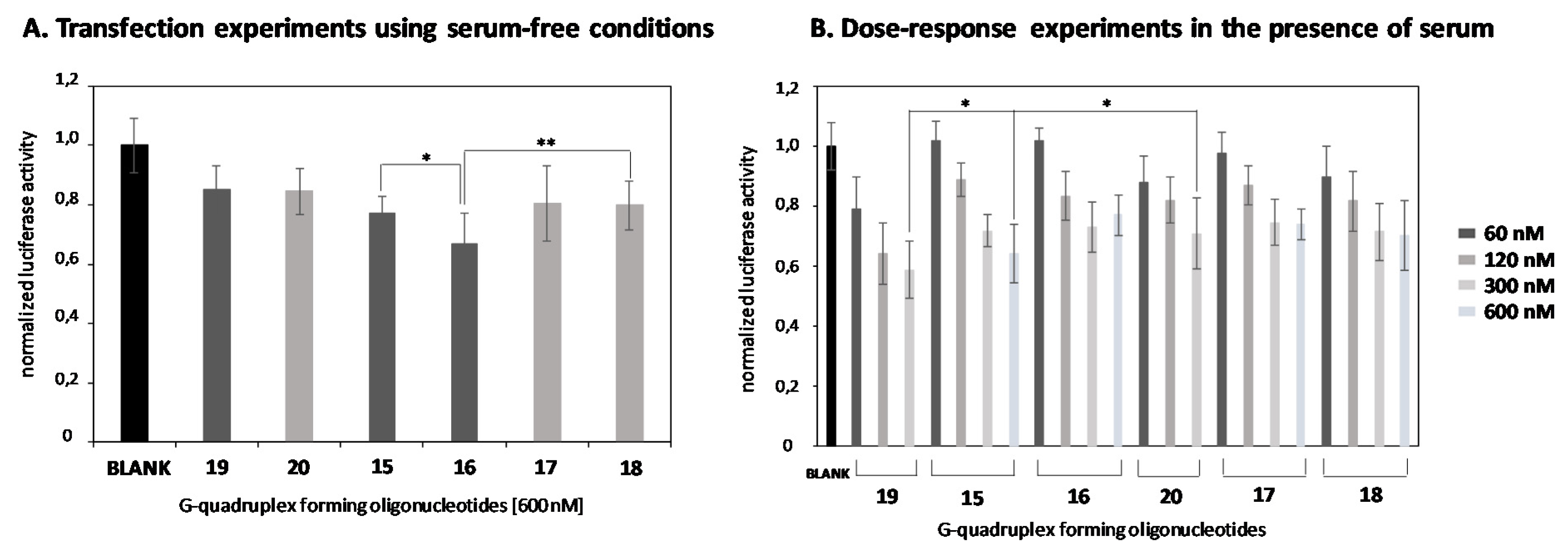
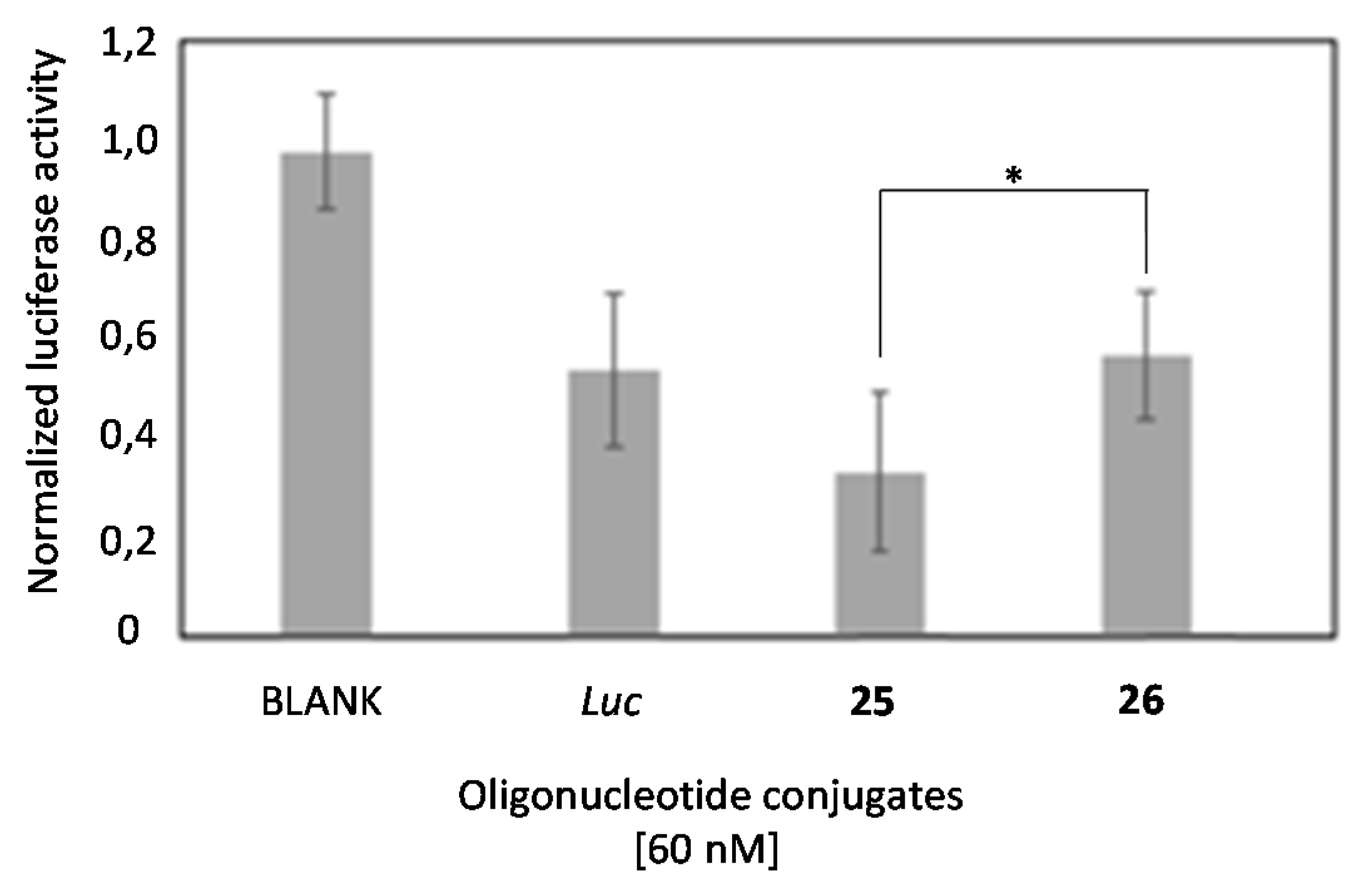
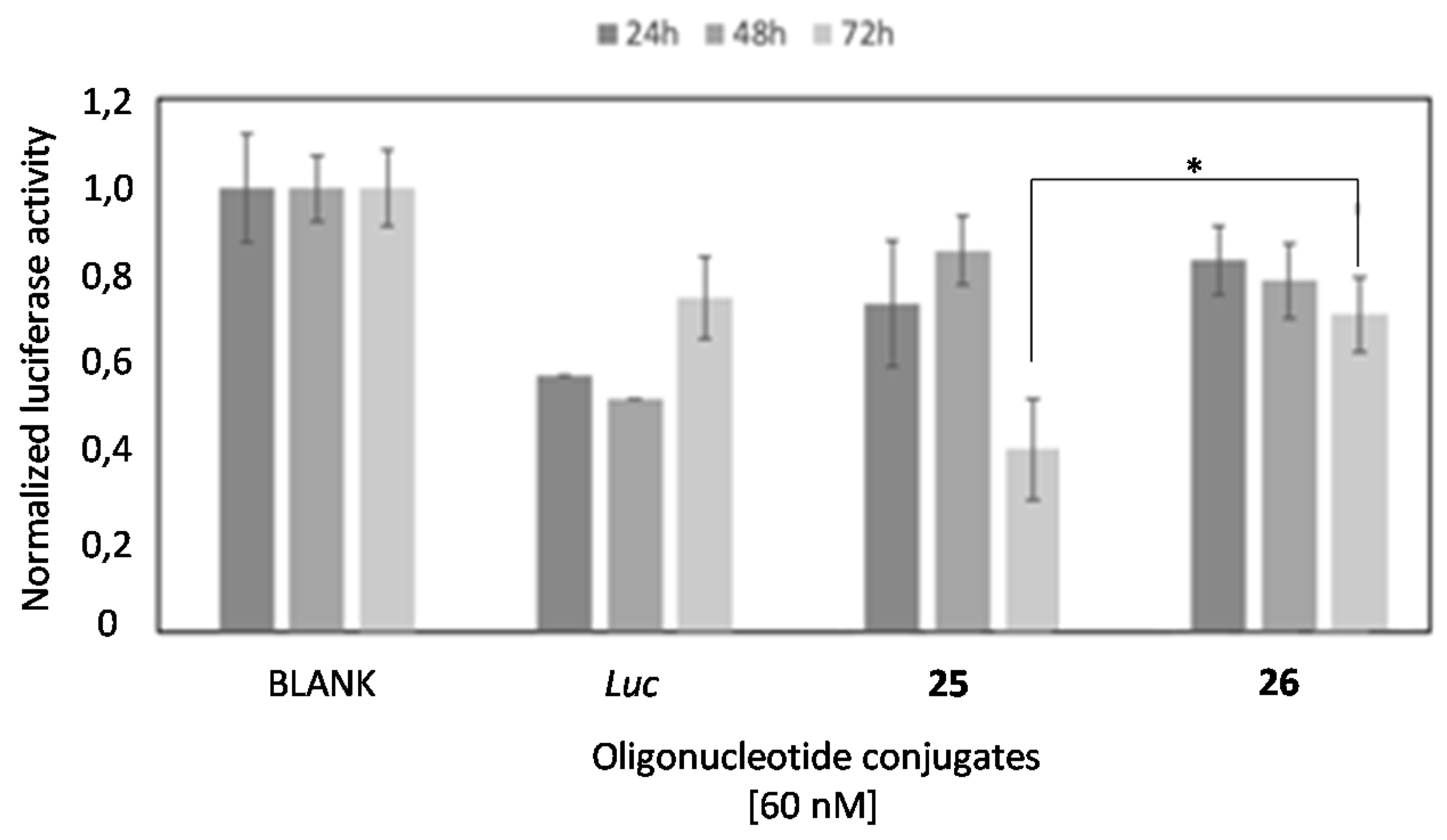
2.4. In Vitro Transfection Studies
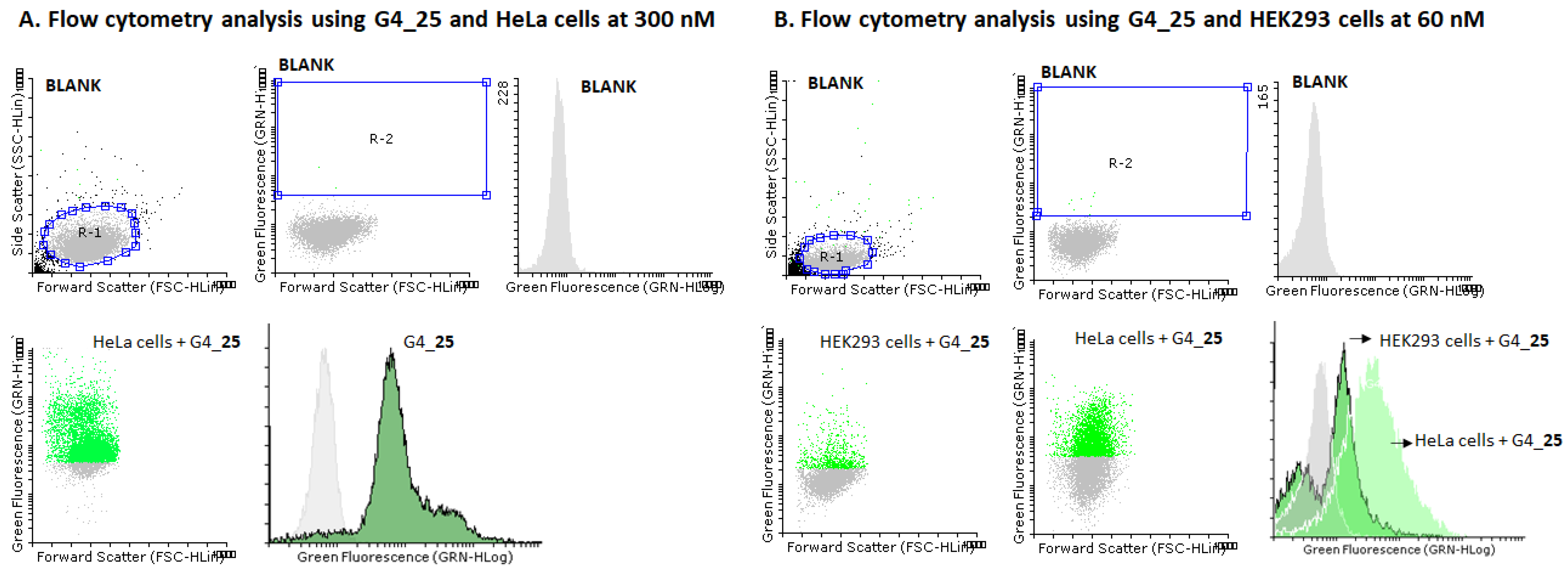
2.5. Cellular Uptake Studies
2.6. Comparison of the Luciferase Inhibitory Properties of Antisense Oligonucleotides Carrying TG4T or T6 Sequences
3. Materials and Methods
3.1. General Methods and Materials
3.2. General Protocol for Derivatizing L-Threoninol with Fmoc-Lys(Boc)-OH
3.3. General Protocol for Derivatizing 1 with Alkyl Residues of Different Length (C8 and C14)
3.4. General Protocol for a Selective Primary Alcohol Protection Using a Trityl Unit as a Protecting Group
3.5. General Protocol for CPG Functionalization and DNA Synthesis
3.6. General Protocol for Introducing Aminolipids on Solid-Phase
3.7. General Protocol for the G-Quadruplex Formation
3.8. CD Spectroscopy and Melting Experiments
3.9. Electromobility Shift Assay
3.10. Fluorescence Spectroscopy Experiments
3.11. MTT Assay
3.12. Gene Transfection Studies
3.12.1. Transfection Studies in the Presence of Lipofectamine
3.12.2. Transfection Studies in the Abscene of Lipofectamine
3.13. Flow Cytometry
3.14. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lane, A.N.; Chaires, J.B.; Gray, R.D.; Trent, J.O. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008, 36, 5482–5515. [Google Scholar] [CrossRef] [PubMed]
- Gellert, M.; Lipsett, M.N.; Davies, D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. USA 1962, 48, 2013–2018. [Google Scholar] [CrossRef] [PubMed]
- Maiti, S.; Saha, P.; Das, T.; Bessi, I.; Schwalbe, H.; Dash, J. Human Telomeric G-Quadruplex Selective Fluoro-Isoquinolines Induce Apoptosis in Cancer Cells. Bioconjug. Chem. 2018, 29, 1141–1154. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Brosh, R.M., Jr. G-quadruplex nucleic acids and human disease. FEBS J. 2010, 277, 3470–3488. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Hurley, L.H. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie 2008, 90, 1149–1171. [Google Scholar] [CrossRef] [PubMed]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef]
- Neidle, S.; Harrison, R.; Reszka, A.P.; A Read, M. Structure-activity relationships among guanine-quadruplex telomerase inhibitors. Pharmacol. Ther. 2000, 85, 133–139. [Google Scholar] [CrossRef]
- Read, M.A.; Wood, A.A.; Harrison, J.R.; Gowan, S.M.; Kelland, L.R.; Dosanjh, H.S.; Neidle, S. Molecular Modeling Studies on G-Quadruplex Complexes of Telomerase Inhibitors: Structure−Activity Relationships. J. Med. Chem. 1999, 42, 4538–4546. [Google Scholar] [CrossRef]
- Seeman, N.C.; Sleiman, H.F. DNA nanotechnology. Nat. Rev. Mater. 2018, 3, 17068. [Google Scholar] [CrossRef]
- Han, J.; Zhao, D.; Li, D.; Wang, X.; Jin, Z.; Zhao, K. Polymer-Based Nanomaterials and Applications for Vaccines and Drugs. Polymers 2018, 10, 31. [Google Scholar] [CrossRef]
- Lai, W.; Wong, W.-T. Design of Polymeric Gene Carriers for Effective Intracellular Delivery. Trends Biotechnol. 2018, 36, 713–728. [Google Scholar] [CrossRef] [PubMed]
- Grijalvo, S.; Puras, G.; Zárate, J.; Sainz-Ramos, M.; Qtaish, A.N.; López, T.; Mashal, M.; Attia, N.; Díaz Díaz, D.; Pons, R.; et al. Cationic Niosomes as Non-Viral Vehicles for Nucleic Acids: Challenges and Opportunities in Gene Delivery. Pharmaceutics 2019, 11, 50. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, P.P.; Biswas, S.; Torchilin, V.P. Current trends in the use of liposomes for tumor targeting. Nanomedicine 2013, 8, 1509–1528. [Google Scholar] [CrossRef] [PubMed]
- Lehner, R.; Wang, X.; Marsch, S.; Hunziker, P.R. Intelligent nanomaterials for medicine: Carrier platforms and targeting strategies in the context of clinical application. Nanomed. Nanotechnol. Biol. Med. 2013, 9, 742–757. [Google Scholar] [CrossRef]
- Angell, C.; Xie, S.; Zhang, L.; Chen, Y. DNA Nanotechnology for Precise Control over Drug Delivery and Gene Therapy. Small 2016, 12, 1117–1132. [Google Scholar] [CrossRef]
- Zadegan, R.; Norton, M.L. Structural DNA Nanotechnology: From Design to Applications. Int. J. Mol. Sci. 2012, 13, 7149–7162. [Google Scholar] [CrossRef]
- Tintoré, M.; Eritja, R.; Fàbrega, C. DNA Nanoarchitectures: Steps towards Biological Applications. ChemBioChem 2014, 15, 1374–1390. [Google Scholar] [CrossRef]
- Jorge, A.F.; Aviñó, A.; Pais, A.A.C.C.; Eritja, R.; Fàbrega, C. DNA-based nanoscaffolds as vehicles for 5-fluoro-2′-deoxyuridine oligomers in colorectal cancer therapy. Nanoscale 2018, 10, 7238–7249. [Google Scholar] [CrossRef]
- Bujold, K.E.; Lacroix, A.; Sleiman, H.F. DNA Nanostructures at the Interface with Biology. Chem 2018, 4, 495–521. [Google Scholar] [CrossRef]
- Bujold, K.E.; Hsu, J.C.C.; Sleiman, H.F. Optimized DNA “Nanosuitcases” for Encapsulation and Conditional Release of siRNA. J. Am. Chem. Soc. 2016, 138, 14030–14038. [Google Scholar] [CrossRef]
- Yan, J.; Chen, J.; Zhang, N.; Yang, Y.; Zhu, W.; Li, L.; He, B. Mitochondria-targeted tetrahedral DNA nanostructures for doxorubicin delivery and enhancement of apoptosis. J. Mater. Chem. B 2020, 8, 492–503. [Google Scholar] [CrossRef]
- Li, J.; Pei, H.; Zhu, B.; Liang, L.; Wei, M.; He, Y.; Chen, N.; Li, D.; Huang, Q.; Fan, C. Self-Assembled Multivalent DNA Nanostructures for Noninvasive Intracellular Delivery of Immunostimulatory CpG Oligonucleotides. ACS Nano 2011, 5, 8783–8789. [Google Scholar] [CrossRef] [PubMed]
- Platella, C.; Riccardi, C.; Montesarchio, D.; Roviello, G.N.; Musumeci, D. G-quadruplex-based aptamers against protein targets in therapy and diagnostics. Biochim. Biophys. Acta (BBA)—Gen. Subj. 2017, 1861, 1429–1447. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.; Yang, C.-S.; Huang, D.-M. Aptamer-Conjugated DNA Icosahedral Nanoparticles as a Carrier of Doxorubicin for Cancer Therapy. ACS Nano 2011, 5, 6156–6163. [Google Scholar] [CrossRef] [PubMed]
- Cosconati, S.; Rizzo, A.; Trotta, R.; Pagano, B.; Iachettini, S.; De Tito, S.; Lauri, I.; Fotticchia, I.; Giustiniano, M.; Marinelli, L.; et al. Shooting for Selective Druglike G-Quadruplex Binders: Evidence for Telomeric DNA Damage and Tumor Cell Death. J. Med. Chem. 2012, 55, 9785–9792. [Google Scholar] [CrossRef] [PubMed]
- Sando, S.; Matsui, K.; Niinomi, Y.; Sato, N.; Aoyama, Y. Facile preparation of DNA-tagged carbohydrates. Bioorg. Med. Chem. Lett. 2003, 13, 2633–2636. [Google Scholar] [CrossRef]
- Grijalvo, S.; Alagia, A.; Gargallo, R.; Eritja, R. Cellular uptake studies of antisense oligonucleotides using G-quadruplex-nanostructures. The effect of cationic residue on the biophysical and biological properties. RSC Adv. 2016, 6, 76099–76109. [Google Scholar] [CrossRef]
- Lyonnais, S.; Grijalvo, S.; Alvarez-Fernández, C.; Fleta-Soriano, E.; Martinez, J.P.; Meyerhans, A.; Sánchez-Palomino, S.; Mirambeau, G.; Eritja, R. Lipid-Oligonucleotide Conjugates Forming G-Quadruplexes (Lipoquads) as Potent Inhibitors of HIV Entry. Proceedings 2017, 1, 670. [Google Scholar] [CrossRef]
- Koutsoudakis, G.; De León, A.P.; Herrera, C.; Dorner, M.; Pérez-Vilaró, G.; Lyonnais, S.; Grijalvo, S.; Eritja, R.; Meyerhans, A.; Mirambeau, G.; et al. Oligonucleotide-Lipid Conjugates Forming G-Quadruplex Structures Are Potent and Pangenotypic Hepatitis C Virus Entry Inhibitors In Vitro and Ex Vivo. Antimicrob. Agents Chemother. 2017, 61, e02354-16. [Google Scholar] [CrossRef]
- Patwa, A.N.; Gissot, A.; Bestel, I.; Philippe, B. Hybrid lipid oligonucleotide conjugates: Synthesis, self-assemblies and biomedical applications. Chem. Soc. Rev. 2011, 40, 5844–5854. [Google Scholar] [CrossRef]
- Liu, H.; Li, Y.; Mozhi, A.; Zhang, L.; Liu, Y.; Xu, X.; Xing, J.; Liang, X.; Ma, G.; Yang, J.; et al. SiRNA-phospholipid conjugates for gene and drug delivery in cancer treatment. Biomaterials 2014, 35, 6519–6533. [Google Scholar] [CrossRef]
- Grijalvo, S.; Ocampo, S.M.; Perales, J.C.; Eritja, R. Synthesis of Oligonucleotides Carrying Amino Lipid Groups at the 3′-End for RNA Interference Studies. J. Org. Chem. 2010, 75, 6806–6813. [Google Scholar] [CrossRef]
- Raouane, M.; Desmaële, D.; Urbinati, G.; Massaad-Massade, L.; Couvreur, P. Lipid Conjugated Oligonucleotides: A Useful Strategy for Delivery. Bioconjug. Chem. 2012, 23, 1091–1104. [Google Scholar] [CrossRef]
- Grijalvo, S.; Ocampo, S.M.; Perales, J.C.; Eritja, R. Synthesis of Lipid-Oligonucleotide Conjugates for RNA Interference Studies. Chem. Biodivers. 2011, 8, 287–299. [Google Scholar] [CrossRef]
- Prakash, T.P.; Mullick, A.E.; Lee, R.G.; Yu, J.; Yeh, S.T.; Low, A.; Chappell, A.E.; Østergaard, M.E.; Murray, S.; Gaus, H.J.; et al. Fatty acid conjugation enhances potency of antisense oligonucleotides in muscle. Nucleic Acids Res. 2019, 47, 6029–6044. [Google Scholar] [CrossRef]
- Lennox, K.A.; Behlke, M.A. Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther. 2011, 18, 1111–1120. [Google Scholar] [CrossRef]
- Soutschek, J.; Akinc, A.; Bramlage, B.; Charisse, K.; Constien, R.; Donoghue, M.; Elbashir, S.; Geick, A.; Hadwiger, P.; Harborth, J.; et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nat. Cell Biol. 2004, 432, 173–178. [Google Scholar] [CrossRef]
- Wilner, S.E.; Sparks, S.E.; Cowburn, D.; Girvin, M.E.; Levy, M. Controlling Lipid Micelle Stability Using Oligonucleotide Headgroups. J. Am. Chem. Soc. 2015, 137, 2171–2174. [Google Scholar] [CrossRef]
- Cozzoli, L.; Gjonaj, L.; Stuart, M.C.A.; Poolman, B.; Roelfes, G. Responsive DNA G-quadruplex micelles. Chem. Commun. 2018, 54, 260–263. [Google Scholar] [CrossRef]
- Vialet, B.; Gissot, A.; Delzor, R.; Philippe, B.; Brune, V. Controlling G-quadruplex formation via lipid modification of oligonucleotide sequences. Chem. Commun. 2017, 53, 11560–11563. [Google Scholar] [CrossRef]
- Pérez-Rentero, S.; Grijalvo, S.; Peñuelas, G.; Fàbrega, C.; Eritja, R. Thioctic Acid Derivatives as Building Blocks to Incorporate DNA Oligonucleotides onto Gold Nanoparticles. Molecules 2014, 19, 10495–10523. [Google Scholar] [CrossRef]
- Villorbina, G.; Canals, D.; Carde, L.; Grijalvo, S.; Pascual, R.; Rabal, O.; Teixidó, J.; Fabrias, G.; Llebaria, A.; Casas, J.; et al. Solid-phase synthesis of a combinatorial library of dihydroceramide analogues and its activity in human alveolar epithelial cells. Bioorg. Med. Chem. 2007, 15, 50–62. [Google Scholar] [CrossRef]
- Gupta, K.C.; Kumar, P.; Bhatia, D.; Sharma, A.K. A Rapid Method for the Functionalisation of Polymer Supports for Solid Phase Oligonucleotide Synthesis. Nucleosides Nucleotides Nucleic Acids 1995, 14, 829–832. [Google Scholar] [CrossRef]
- Zhang, H.; Mao, J.; Zhou, D.; Xu, Y.; Thonberg, H.; Liang, Z.; Wahlestedt, C. mRNA accessible site tagging (MAST): A novel high throughput method for selecting effective antisense oligonucleotides. Nucleic Acids Res. 2003, 31, e72. [Google Scholar] [CrossRef]
- Kachalova, A.V.; Stetsenko, D.A.; Romanova, E.A.; Tashlitsky, V.N.; Gait, M.J.; Oretskaya, T.S. A New and Efficient Method for Synthesis of 5′-Conjugates of Oligonucleotides through Amide-Bond Formation on Solid Phase. Helv. Chim. Acta 2002, 85, 2409–2416. [Google Scholar] [CrossRef]
- Saccà, B. The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides. Nucleic Acids Res. 2005, 33, 1182–1192. [Google Scholar] [CrossRef]
- Petraccone, L.; Erra, E.; Duro, I.; Esposito, V.; Randazzo, A.; Mayol, L.; Mattia, C.A.; Barone, G.; Giancola, C. Relative stability of quadruplexes containing different number of G-tetrads. Nucleosides Nucleotides Nucleic Acids 2005, 24, 757–760. [Google Scholar] [CrossRef]
- Guan, A.-J.; Zhang, X.-F.; Sun, X.; Li, Q.; Xiang, J.-F.; Wang, L.-X.; Lan, L.; Yang, F.-M.; Xu, S.-J.; Guo, X.-M.; et al. Ethyl-substitutive Thioflavin T as a highly-specific fluorescence probe for detecting G-quadruplex structure. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Mohanty, J.; Barooah, N.; Dhamodharan, V.; Harikrishna, S.; Pradeepkumar, P.I.; Bhasikuttan, A.C. Thioflavin T as an Efficient Inducer and Selective Fluorescent Sensor for the Human Telomeric G-Quadruplex DNA. J. Am. Chem. Soc. 2013, 135, 367–376. [Google Scholar] [CrossRef]
- Mergny, J.-L. Kinetics of tetramolecular quadruplexes. Nucleic Acids Res. 2005, 33, 81–94. [Google Scholar] [CrossRef]
- Dapić, V. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 2003, 31, 2097–2107. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Manoharan, M. Oligonucleotide Conjugates as Potential Antisense Drugs with Improved Uptake, Biodistribution, Targeted Delivery, and Mechanism of Action. Antisense Nucleic Acid Drug Dev. 2002, 12, 103–128. [Google Scholar] [CrossRef]
- Yessine, M.-A.; Meier, C.; Petereit, H.-U.; Leroux, J.-C. On the role of methacrylic acid copolymers in the intracellular delivery of antisense oligonucleotides. Eur. J. Pharm. Biopharm. 2006, 63, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sarett, S.M.; Werfel, T.A.; Lee, L.; Jackson, M.A.; Kilchrist, K.V.; Brantley-Sieders, D.; Duvall, C.L. Lipophilic siRNA targets albumin in situ and promotes bioavailability, tumor penetration, and carrier-free gene silencing. Proc. Natl. Acad. Sci. USA 2017, 114, E6490–E6497. [Google Scholar] [CrossRef]
- Wolfrum, C.; Shi, S.; Jayaprakash, K.N.; Jayaraman, M.; Wang, G.; Pandey, R.K.; Rajeev, K.G.; Nakayama, T.; Charrise, K.; Ndungo, E.M.; et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat. Biotechnol. 2007, 25, 1149–1157. [Google Scholar] [CrossRef]
- Choi, J.-K.; Ho, J.; Curry, S.; Qin, D.; Bittman, R.; Hamilton, J. Interactions of very long-chain saturated fatty acids with serum albumin. J. Lipid Res. 2002, 43, 1000–1010. [Google Scholar] [CrossRef]
- Brown, D.A.; Kang, S.H.; Gryaznov, S.M.; DeDionisio, L.; Heidenreich, O.; Sullivan, S.; Xu, X.; Nerenberg, M.I. Effect of phosphorothioate modification of oligodeoxynucleotides on specific protein binding. J. Biol. Chem. 1994, 269, 26801–26805. [Google Scholar]
- Liang, X.-H.; Sun, H.; Shen, W.; Crooke, S.T. Identification and characterization of intracellular proteins that bind oligonucleotides with phosphorothioate linkages. Nucleic Acids Res. 2015, 43, 2927–2945. [Google Scholar] [CrossRef]
- Weidner, D.A.; Valdez, B.C.; Henning, D.; Greenberg, S.; Busch, H. Phosphorothioate oligonucleotides bind in a non sequence-specific manner to the nucleolar protein C23/nucleolin. FEBS Lett. 1995, 366, 146–150. [Google Scholar] [CrossRef]
- Ly, S.; Echeverria, D.; Sousa, J.; Khvorova, A. Single-Stranded Phosphorothioated Regions Enhance Cellular Uptake of Cholesterol-Conjugated siRNA but Not Silencing Efficacy. Mol. Ther. Nucleic Acids 2020, 21, 991–1005. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.K.; Shen, W.; Liang, X.-H.; Crooke, S.T. Nucleic acid binding proteins affect the subcellular distribution of phosphorothioate antisense oligonucleotides. Nucleic Acids Res. 2017, 45, 10649–10671. [Google Scholar] [CrossRef] [PubMed]

| Name | Sequence | Backbone | Modification (Mod) | Mass (Calcd) | Mass (Found) | T1/2 Na+ (°C) d |
|---|---|---|---|---|---|---|
| 9 | TG4T | PO | unmod. | 1863 | 1860 | 59.2 e |
| 10 | TG6T | PO | unmod. | 2521 | 2519 | >80 |
| 11 | TG4T_C8 | PO | 3′_Thr_C8 a | 2284 | 2281 | >80 |
| 12 | TG4T_C14 | PO | 3′_Thr_C14 a | 2368 | 2366 | >80 |
| 13 | TG6T_C8 | PO | 3′_Thr_C8 a | 2942 | 2940 | >80 |
| 14 | TG6T_C14 | PO | 3′_Thr_C14 a | 3026 | 3025 | >80 |
| 15 | Luc-TG4T_C8 | PS/PO | 3′_Thr_C8 b | 8095 | 8081 | - |
| 16 | Luc-TG4T_C14 | PS/PO | 3′_Thr_C14 b | 8179 | 8168 | - |
| 17 | Luc-TG6T_C8 | PS/PO | 3′_Thr_C8 b | 8751 | 8749 | |
| 18 | Luc-TG6T_C14 | PS/PO | 3′_Thr_C14 b | 8837 | 8838 | - |
| 19 | Luc-TG4T | PS/PO | unmod. | 7672 | 7365 f | - |
| 20 | Luc-TG6T | PS/PO | unmod. | 8330 | 8328 | - |
| 21 | C8_Luc-TG4T | PS/PO | 5′_C8_NH c | 7980 | 8040 g | - |
| 22 | C14_Luc-TG4T | PS/PO | 5′_C14_NH c | 8063 | 8100 g | - |
| 23 | C8_Luc-TG6T | PS/PO | 5′_C8_NH c | 8637 | 8635 | - |
| 24 | C14_Luc-TG6T | PS/PO | 5′_C14_NH c | 8722 | 8573 h | - |
| 25 | F_Luc_TG4T_C8 | PS/PO | 3′_Thr_C8 a | 8646 | 8648 | - |
| 26 | F_Luc_T6_C8 | PS/PO | 3′_Thr_C8 a | 8546 | 8548 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grijalvo, S.; Clua, A.; Eres, M.; Gargallo, R.; Eritja, R. Tuning G-Quadruplex Nanostructures with Lipids. Towards Designing Hybrid Scaffolds for Oligonucleotide Delivery. Int. J. Mol. Sci. 2021, 22, 121. https://doi.org/10.3390/ijms22010121
Grijalvo S, Clua A, Eres M, Gargallo R, Eritja R. Tuning G-Quadruplex Nanostructures with Lipids. Towards Designing Hybrid Scaffolds for Oligonucleotide Delivery. International Journal of Molecular Sciences. 2021; 22(1):121. https://doi.org/10.3390/ijms22010121
Chicago/Turabian StyleGrijalvo, Santiago, Anna Clua, Marc Eres, Raimundo Gargallo, and Ramon Eritja. 2021. "Tuning G-Quadruplex Nanostructures with Lipids. Towards Designing Hybrid Scaffolds for Oligonucleotide Delivery" International Journal of Molecular Sciences 22, no. 1: 121. https://doi.org/10.3390/ijms22010121
APA StyleGrijalvo, S., Clua, A., Eres, M., Gargallo, R., & Eritja, R. (2021). Tuning G-Quadruplex Nanostructures with Lipids. Towards Designing Hybrid Scaffolds for Oligonucleotide Delivery. International Journal of Molecular Sciences, 22(1), 121. https://doi.org/10.3390/ijms22010121






