miR614 Expression Enhances Breast Cancer Cell Motility
Abstract
1. Introduction
2. Results
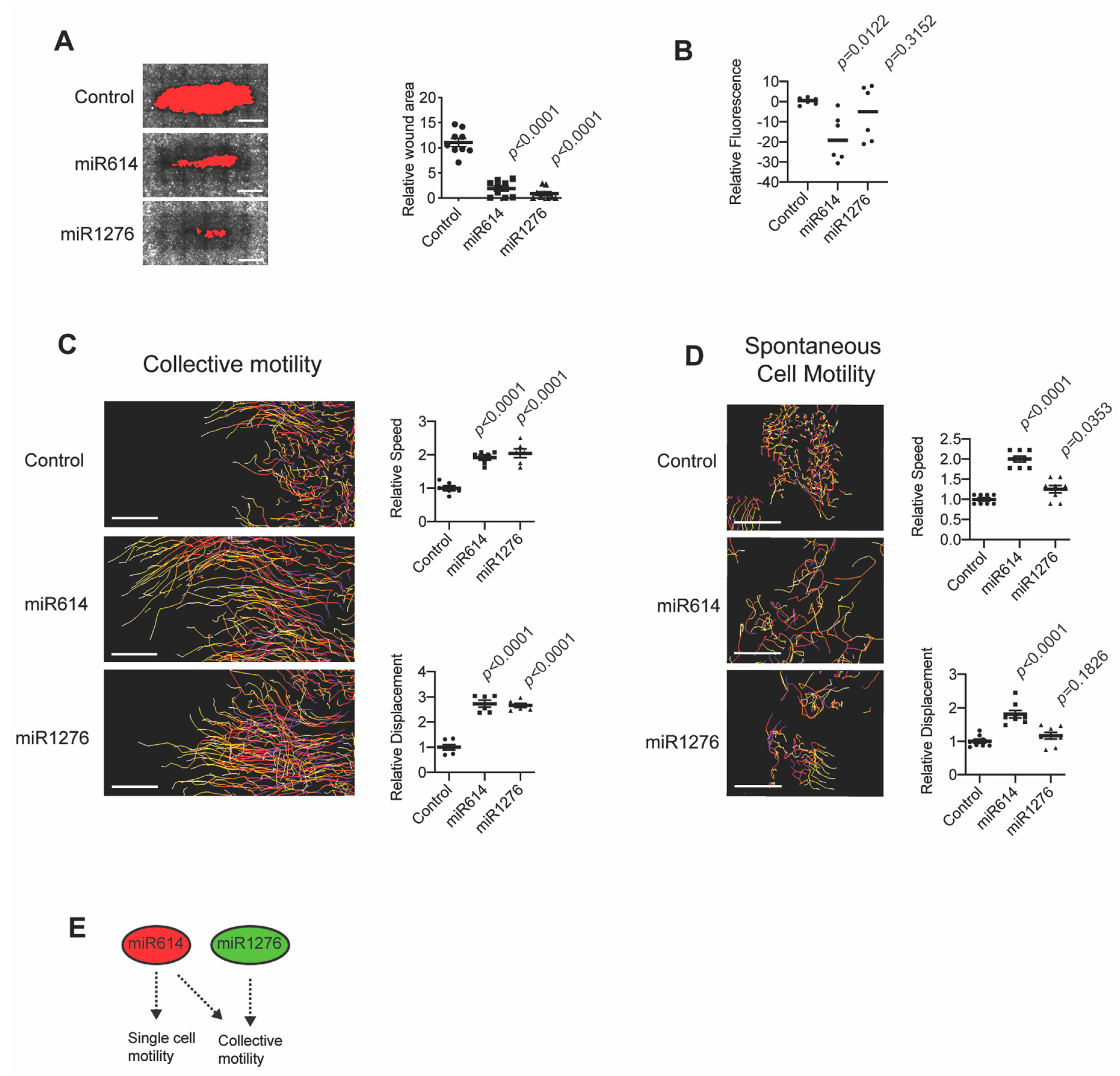
2.1. miR614 Enhances Cell Motility
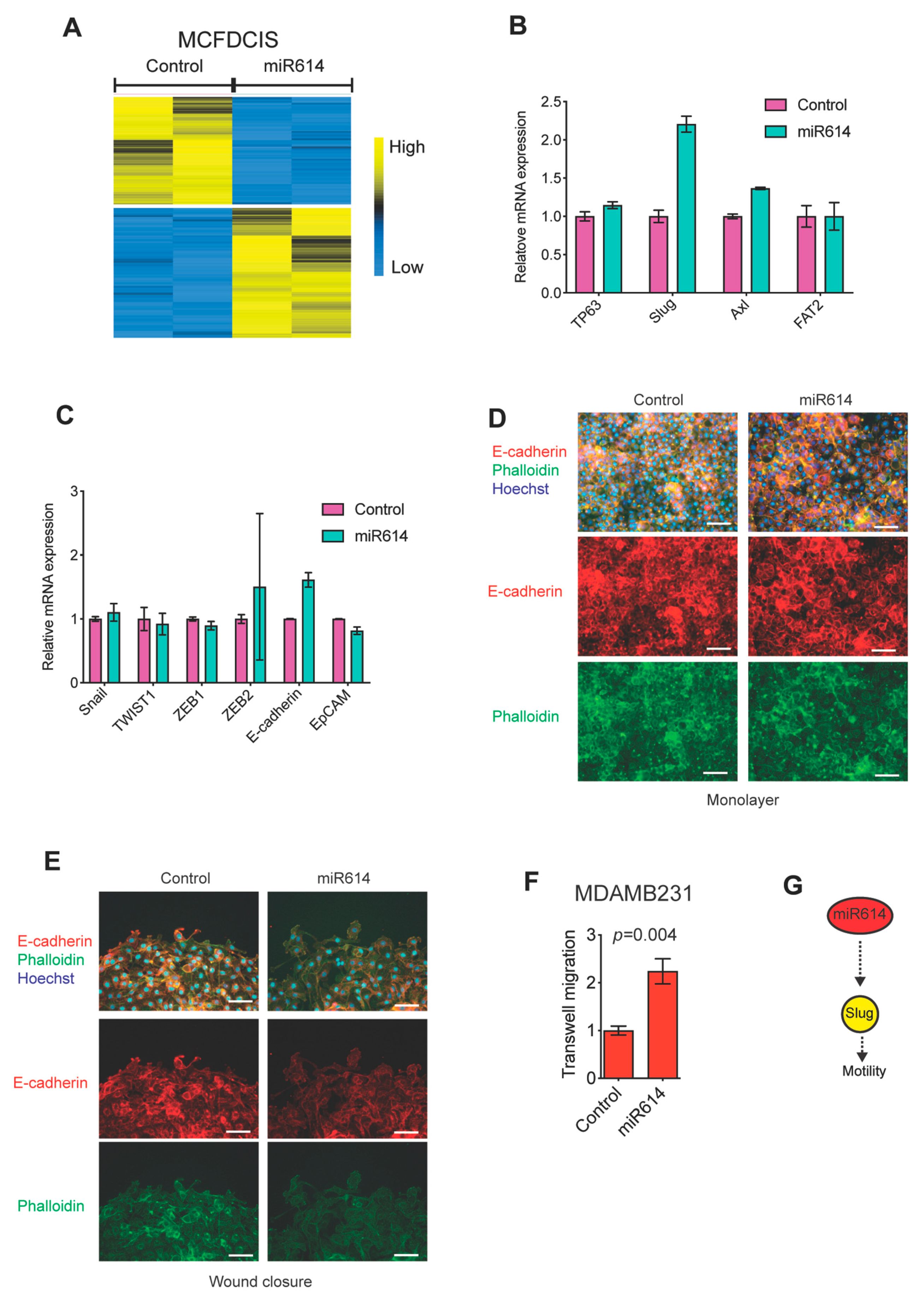
2.2. miR614 Increases Slug Expression but Does Not Suppress Epithelial Traits
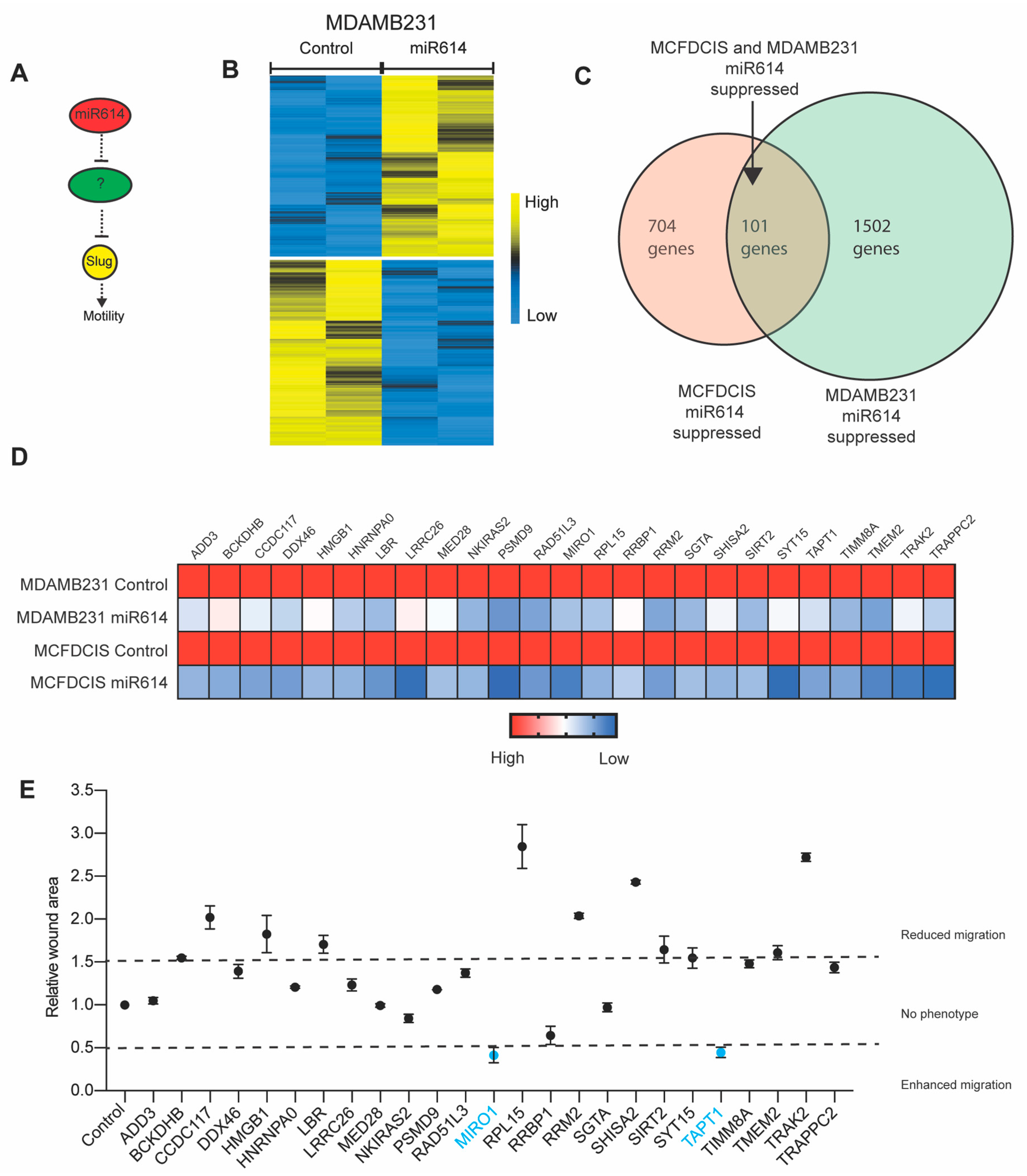
2.3. Evaluation of miR614 Target Genes as Regulators of Cell Motility
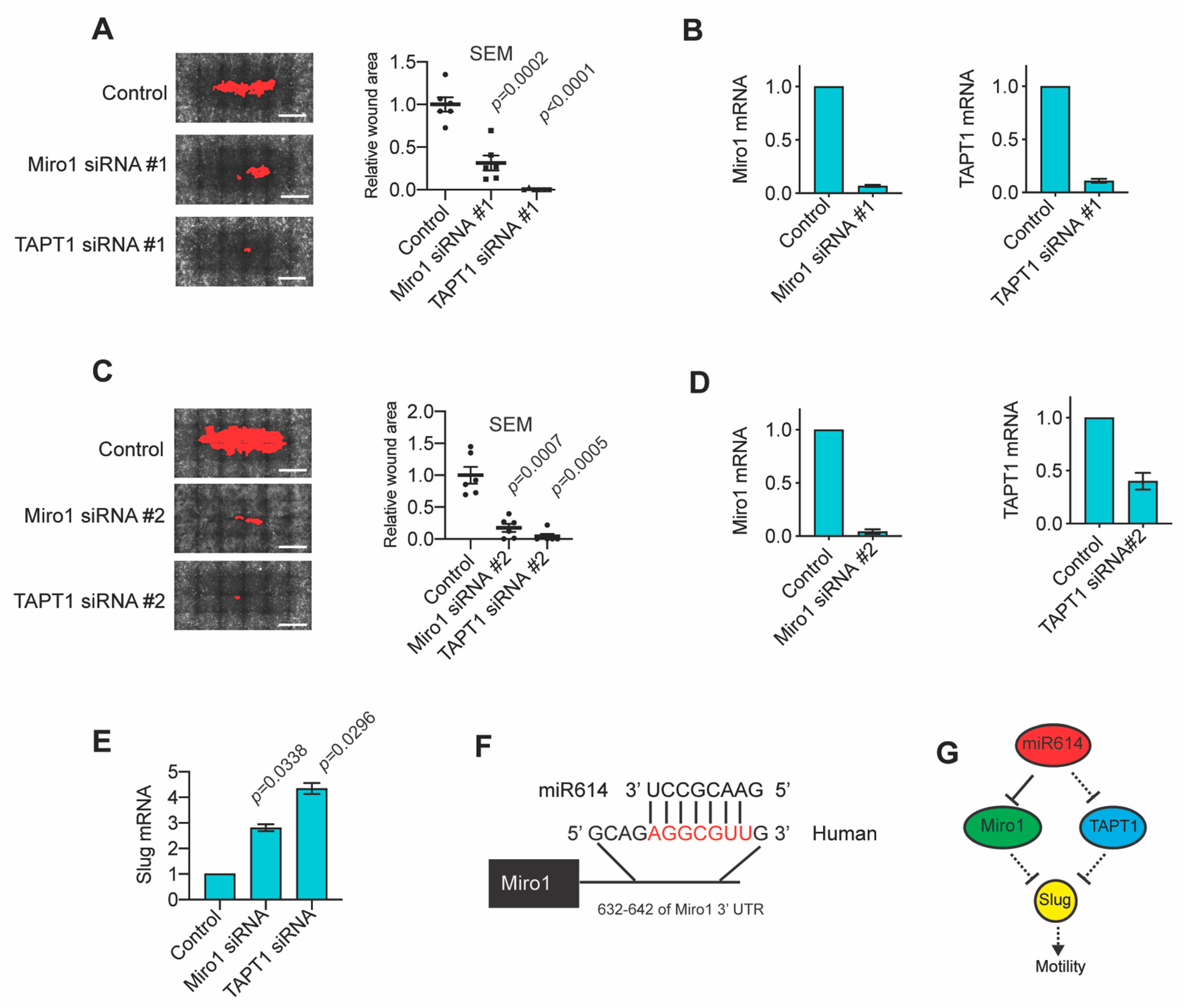
2.4. The miR614 Targets Miro1 and TAPT1 Suppress Migration and Slug Expression
2.5. High TAPT1 Expression Correlates with Improved Odds of Disease-Free Survival
3. Discussion
3.1. Use of Seed Sequences to Define Novel Regulatory Mechanisms
3.2. miR614
3.3. Miro1
3.4. TAPT1
4. Materials and Methods
4.1. Cell Culture
4.2. Immunofluorescence
4.3. Transfection of siRNAs and miRNAs
4.4. Wounding Assays
4.5. Time-Lapse Imaging
4.6. Transwell Migration Assays
4.7. Quantitative Real-Time PCR
4.8. Gene Expression Profiling
4.9. Patient Survival Analysis
4.10. Statistical Methods
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pearson, G.W. Control of Invasion by Epithelial-to-Mesenchymal Transition Programs during Metastasis. J. Clin. Med. 2019, 8, 646. [Google Scholar] [CrossRef] [PubMed]
- Fidler, I.J. Tumor heterogeneity and the biology of cancer invasion and metastasis. Cancer Res. 1978, 38, 2651–2660. [Google Scholar] [CrossRef] [PubMed]
- Vanharanta, S.; Massagué, J. Origins of Metastatic Traits. Cancer Cell 2013, 24, 410–421. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.A.; Cano, A. The epithelial–mesenchymal transition under control: Global programs to regulate epithelial plasticity. Semin. Cancer Biol. 2012, 22, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Westcott, J.M.; Prechtl, A.M.; Maine, E.A.; Dang, T.T.; Esparza, M.A.; Sun, H.; Zhou, Y.; Xie, Y.; Pearson, G.W. An epigenetically distinct breast cancer cell subpopulation promotes collective invasion. J. Clin. Investig. 2015, 125, 1927–1943. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.-M.; Li, A.; Olino, K.; Wolfgang, C.L.; Herman, J.M.; Schulick, R.D.; Iacobuzio-Donahue, C.; Hruban, R.H.; Goggins, M. Loss of E-cadherin expression and outcome among patients with resectable pancreatic adenocarcinomas. Mod. Pathol. 2011, 24, 1237–1247. [Google Scholar] [CrossRef]
- Horne, H.N.; Sherman, M.E.; Garcia-Closas, M.; Pharoah, P.D.; Blows, F.M.; Yang, X.R.; Hewitt, S.M.; Conway, C.M.; Lissowska, J.; Brinton, L.A.; et al. Breast cancer susceptibility risk associations and heterogeneity by E-cadherin tumor tissue expression. Breast Cancer Res. Treat. 2014, 143, 181–187. [Google Scholar] [CrossRef]
- Yang, Y.-L.; Chen, M.-W.; Xian, L. Prognostic and Clinicopathological Significance of Downregulated E-Cadherin Expression in Patients with Non-Small Cell Lung Cancer (NSCLC): A Meta-Analysis. PLoS ONE 2014, 9, e99763. [Google Scholar] [CrossRef]
- Christou, N.; Perraud, A.; Blondy, S.; Jauberteau, M.-O.; Battu, S.; Mathonnet, M. E-cadherin: A potential biomarker of colorectal cancer prognosis. Oncol. Lett. 2017, 13, 4571–4576. [Google Scholar] [CrossRef] [PubMed]
- Shamir, E.R.; Pappalardo, E.; Jorgens, D.M.; Coutinho, K.; Tsai, W.-T.; Aziz, K.; Auer, M.; Tran, P.T.; Bader, J.S.; Ewald, A.J. Twist1-induced dissemination preserves epithelial identity and requires E-cadherin. J. Cell Biol. 2014, 204, 839–856. [Google Scholar] [CrossRef] [PubMed]
- Dang, T.T.; Esparza, M.A.; Maine, E.A.; Westcott, J.M.; Pearson, G.W. ΔNp63alpha Promotes Breast Cancer Cell Motility through the Selective Activation of Components of the Epithelial-to-Mesenchymal Transition Program. Cancer Res. 2015, 75, 3925–3935. [Google Scholar] [CrossRef] [PubMed]
- Beerling, E.; Seinstra, D.; de Wit, E.; Kester, L.; van der Velden, D.; Maynard, C.; Schäfer, R.; van Diest, P.; Voest, E.; van Oudenaarden, A.; et al. Plasticity between Epithelial and Mesenchymal States Unlinks EMT from Metastasis-Enhancing Stem Cell Capacity. Cell Rep. 2016, 14, 2281–2288. [Google Scholar] [CrossRef]
- Westcott, J.M.; Camacho, S.; Nasir, A.; Huysman, M.E.; Rahhal, R.; Dang, T.T.; Riegel, A.T.; Brekken, R.A.; Pearson, G.W. ΔNp63 regulated epithelial-to-mesenchymal transition state heterogeneity confers a leader-follower relationship that drives collective invasion. Cancer Res. 2020, 80, 3933–3944. [Google Scholar] [CrossRef]
- Polyak, K. Molecular Markers for the Diagnosis and Management of Ductal Carcinoma in Situ. J. Natl. Cancer Inst. Monogr. 2010, 2010, 210–213. [Google Scholar] [CrossRef][Green Version]
- Lamouille, S.; Subramanyam, D.; Blelloch, R.; Derynck, R. Regulation of epithelial-mesenchymal and mesenchymal-epithelial transitions by microRNAs. Curr. Opin. Cell Biol. 2013, 25, 200–207. [Google Scholar] [CrossRef]
- Stinson, S.; Lackner, M.R.; Adai, A.T.; Yu, N.; Kim, H.J.; O’Brien, C.; Spoerke, J.; Jhunjhunwala, S.; Boyd, Z.; Januario, T.; et al. TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci. Signal. 2011, 4, ra41. [Google Scholar]
- Dang, T.T.; Westcott, J.M.; Maine, E.A.; Kanchwala, M.; Xing, C.; Pearson, G.W. ΔNp63alpha induces the expression of FAT2 and Slug to promote tumor invasion. Oncotarget 2016, 7, 28592. [Google Scholar] [CrossRef]
- Simpson, K.J.; Selfors, L.M.; Bui, J.; Reynolds, A.; Leake, D.; Khvorova, A.; Brugge, J.S. Identification of genes that regulate epithelial cell migration using an siRNA screening approach. Nat. Cell Biol. 2008, 10, 1027–1038. [Google Scholar] [CrossRef]
- Sugita, B.; Gill, M.; Mahajan, A.; Duttargi, A.; Kirolikar, S.; Almeida, R.; Regis, K.; Oluwasanmi, O.L.; Marchi, F.; Marian, C.; et al. Differentially expressed miRNAs in triple negative breast cancer between African-American and non-Hispanic white women. Oncotarget 2016, 7, 79274–79291. [Google Scholar] [CrossRef] [PubMed]
- Sturtz, L.A.; Melley, J.; Mamula, K.; Shriver, C.D.; Ellsworth, R.E. Outcome disparities in African American women with triple negative breast cancer: A comparison of epidemiological and molecular factors between African American and Caucasian women with triple negative breast cancer. BMC Cancer 2014, 14, 62. [Google Scholar] [CrossRef] [PubMed]
- Dietze, E.C.; Sistrunk, C.; Miranda-Carboni, G.; O’Regan, R.; Seewaldt, V.L. Triple-negative breast cancer in African-American women: Disparities versus biology. Nat. Rev. Cancer 2015, 15, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Hanna, J.; Hossain, G.S.; Kocerha, J. The Potential for microRNA Therapeutics and Clinical Research. Front. Genet. 2019, 10, 478. [Google Scholar] [CrossRef]
- Schultz, N.A.; Werner, J.; Willenbrock, H.; Roslind, A.; Giese, N.; Horn, T.; Wøjdemann, M.; Johansen, J.S. MicroRNA expression profiles associated with pancreatic adenocarcinoma and ampullary adenocarcinoma. Mod. Pathol. 2012, 25, 1609–1622. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Shen, J.; Lu, Y.; Lin, K.; Wang, H.; Li, Y.; Chang, P.; Walker, M.G.; Li, D. RNA sequencing analyses reveal novel differentially expressed genes and pathways in pancreatic cancer. Oncotarget 2017, 8, 42537–42547. [Google Scholar] [CrossRef]
- Zhang, J.; Gao, D.; Zhang, H. Upregulation of miR-614 promotes proliferation and inhibits apoptosis in ovarian cancer by suppressing PPP2R2A expression. Mol. Med. Rep. 2018, 17, 6285–6292. [Google Scholar] [CrossRef]
- Birsa, N.; Norkett, R.; Higgs, N.; Lopez-Domenech, G.; Kittler, J.T. Mitochondrial trafficking in neurons and the role of the Miro family of GTPase proteins. Biochem. Soc. Trans. 2013, 41, 1525–1531. [Google Scholar] [CrossRef]
- Eberhardt, E.L.; Ludlam, A.V.; Tan, Z.; Cianfrocco, M.A. Miro: A molecular switch at the center of mitochondrial regulation. Protein Sci. 2020, 29, 1269–1284. [Google Scholar] [CrossRef]
- López-Doménech, G.; Covill-Cooke, C.; Ivankovic, D.; Halff, E.F.; Sheehan, D.F.; Norkett, R.; Birsa, N.; Kittler, J.T. Miro proteins coordinate microtubule- and actin-dependent mitochondrial transport and distribution. EMBO J. 2018, 37, 321–336. [Google Scholar] [CrossRef]
- Kay, L.; Pienaar, I.S.; Cooray, R.; Black, G.; Soundararajan, M. Understanding Miro GTPases: Implications in the Treatment of Neurodegenerative Disorders. Mol. Neurobiol. 2018, 55, 7352–7365. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Sawada, T.; Lee, S.; Yu, W.; Silverio, G.; Alapatt, P.; Millan, I.; Shen, A.; Saxton, W.; Kanao, T.; et al. Parkinson’s Disease–Associated Kinase PINK1 Regulates Miro Protein Level and Axonal Transport of Mitochondria. PLoS Genet. 2012, 8, e1002537. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Kim, J. Mitochondrial Retrograde Signalling and Metabolic Alterations in the Tumour Microenvironment. Cells 2019, 8, 275. [Google Scholar] [CrossRef] [PubMed]
- Guha, M.; Srinivasan, S.P.; Ruthel, G.; Kashina, A.K.; Carstens, R.P.; Mendoza, A.; Khanna, C.; van Winkle, T.; Avadhani, N.G. Mitochondrial retrograde signaling induces epithelial–mesenchymal transition and generates breast cancer stem cells. Oncogene 2014, 33, 5238–5250. [Google Scholar] [CrossRef]
- Morlino, G.; Barreiro, O.; Baixauli, F.; Robles-Valero, J.; González-Granado, J.M.; Villa-Bellosta, R.; Cuenca-Alba, J.; Sorzano, C.; Óscar, S.; Veiga, E.; et al. Miro-1 Links Mitochondria and Microtubule Dynein Motors To Control Lymphocyte Migration and Polarity. Mol. Cell. Biol. 2014, 34, 1412–1426. [Google Scholar] [CrossRef]
- Schuler, M.-H.; Lewandowska, A.; Di Caprio, G.; Skillern, W.; Upadhyayula, S.; Kirchhausen, T.; Shaw, J.M.; Cunniff, B. Miro1-mediated mitochondrial positioning shapes intracellular energy gradients required for cell migration. Mol. Biol. Cell 2017, 28, 2159–2169. [Google Scholar] [CrossRef]
- Wang, X.; Winter, D.; Ashrafi, G.; Schlehe, J.; Wong, Y.L.; Selkoe, D.; Rice, S.; Steen, J.; Lavoie, M.J.; Schwarz, T.L. PINK1 and Parkin Target Miro for Phosphorylation and Degradation to Arrest Mitochondrial Motility. Cell 2011, 147, 893–906. [Google Scholar] [CrossRef]
- Grossmann, D.; Berenguer-Escuder, C.; Chemla, A.; Arena, G.; Krüger, R. The Emerging Role of RHOT1/Miro1 in the Pathogenesis of Parkinson’s Disease. Front. Neurol. 2020, 11, 587. [Google Scholar] [CrossRef]
- Bharat, V.; Wang, X. Precision Neurology for Parkinson’s Disease: Coupling Miro1-Based Diagnosis with Drug Discovery. Mov. Disord. 2020, 35, 1502–1508. [Google Scholar] [CrossRef]
- Howell, G.R.; Shindo, M.; Murray, S.; Gridley, T.; Wilson, L.A.; Schimenti, J.C. Mutation of a Ubiquitously Expressed Mouse Transmembrane Protein (Tapt1) Causes Specific Skeletal Homeotic Transformations. Genetics 2007, 175, 699–707. [Google Scholar] [CrossRef]
- Symoens, S.; Barnes, A.M.; Gistelinck, C.; Malfait, F.; Guillemyn, B.; Steyaert, W.; Syx, D.; D’Hondt, S.; Biervliet, M.; De Backer, J.; et al. Genetic Defects in TAPT1 Disrupt Ciliogenesis and Cause a Complex Lethal Osteochondrodysplasia. Am. J. Hum. Genet. 2015, 97, 521–534. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, L.; Bost, F.; Mazure, N.M. Primary Cilium in Cancer Hallmarks. Int. J. Mol. Sci. 2019, 20, 1336. [Google Scholar] [CrossRef] [PubMed]
- Kunitomi, H.; Kobayashi, Y.; Wu, R.-C.; Takeda, T.; Tominaga, E.; Banno, K.; Aoki, D. LAMC1 is a prognostic factor and a potential therapeutic target in endometrial cancer. J. Gynecol. Oncol. 2020, 31, e11. [Google Scholar] [CrossRef] [PubMed]
- Godfrey, L.; Crump, N.T.; O’Byrne, S.; Lau, I.J.; Rice, S.; Harman, J.R.; Jackson, T.; Elliott, N.; Buck, G.; Connor, C.; et al. H3K79me2/3 controls enhancer-promoter interactions and activation of the pan-cancer stem cell marker PROM1/CD133 in MLL-AF4 leukemia cells. Leukemia 2020, 1–17. [Google Scholar] [CrossRef]
- Dang, T.T.; Prechtl, A.M.; Pearson, G.W. Breast Cancer Subtype-Specific Interactions with the Microenvironment Dictate Mechanisms of Invasion. Cancer Res. 2011, 71, 6857–6866. [Google Scholar] [CrossRef]
- Maine, E.A.; Westcott, J.M.; Prechtl, A.M.; Dang, T.T.; Whitehurst, A.W.; Pearson, G.W. The cancer-testis antigens SPANX-A/C/D and CTAG2 promote breast cancer invasion. Oncotarget 2016, 7, 14708–14726. [Google Scholar] [CrossRef]
- Bookout, A.L.; Cummins, C.L.; Mangelsdorf, D.J.; Pesola, J.M.; Kramer, M.F. High-Throughput Real-Time Quantitative Reverse Transcription PCR. Curr. Protoc. Mol. Biol. 2006, 73, 15–18. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, X.; Story, M. Statistical methods of background correction for Illumina BeadArray data. Bioinformatics 2009, 25, 751–757. [Google Scholar] [CrossRef]
- Györffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res. Treat. 2010, 123, 725–731. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dang, T.T.; McIntosh, A.T.; Morales, J.C.; Pearson, G.W. miR614 Expression Enhances Breast Cancer Cell Motility. Int. J. Mol. Sci. 2021, 22, 112. https://doi.org/10.3390/ijms22010112
Dang TT, McIntosh AT, Morales JC, Pearson GW. miR614 Expression Enhances Breast Cancer Cell Motility. International Journal of Molecular Sciences. 2021; 22(1):112. https://doi.org/10.3390/ijms22010112
Chicago/Turabian StyleDang, Tuyen T., Alec T. McIntosh, Julio C. Morales, and Gray W. Pearson. 2021. "miR614 Expression Enhances Breast Cancer Cell Motility" International Journal of Molecular Sciences 22, no. 1: 112. https://doi.org/10.3390/ijms22010112
APA StyleDang, T. T., McIntosh, A. T., Morales, J. C., & Pearson, G. W. (2021). miR614 Expression Enhances Breast Cancer Cell Motility. International Journal of Molecular Sciences, 22(1), 112. https://doi.org/10.3390/ijms22010112





