Similarities in the General Chemical Composition of Colon Cancer Cells and Their Microvesicles Investigated by Spectroscopic Methods-Potential Clinical Relevance
Abstract
1. Introduction
2. Results
2.1. Tumor-Derived Microvesicles (TMVs) Characterization
2.2. General Chemical Composition of the Colon Cancer Cell Lines and the Corresponding TMVs
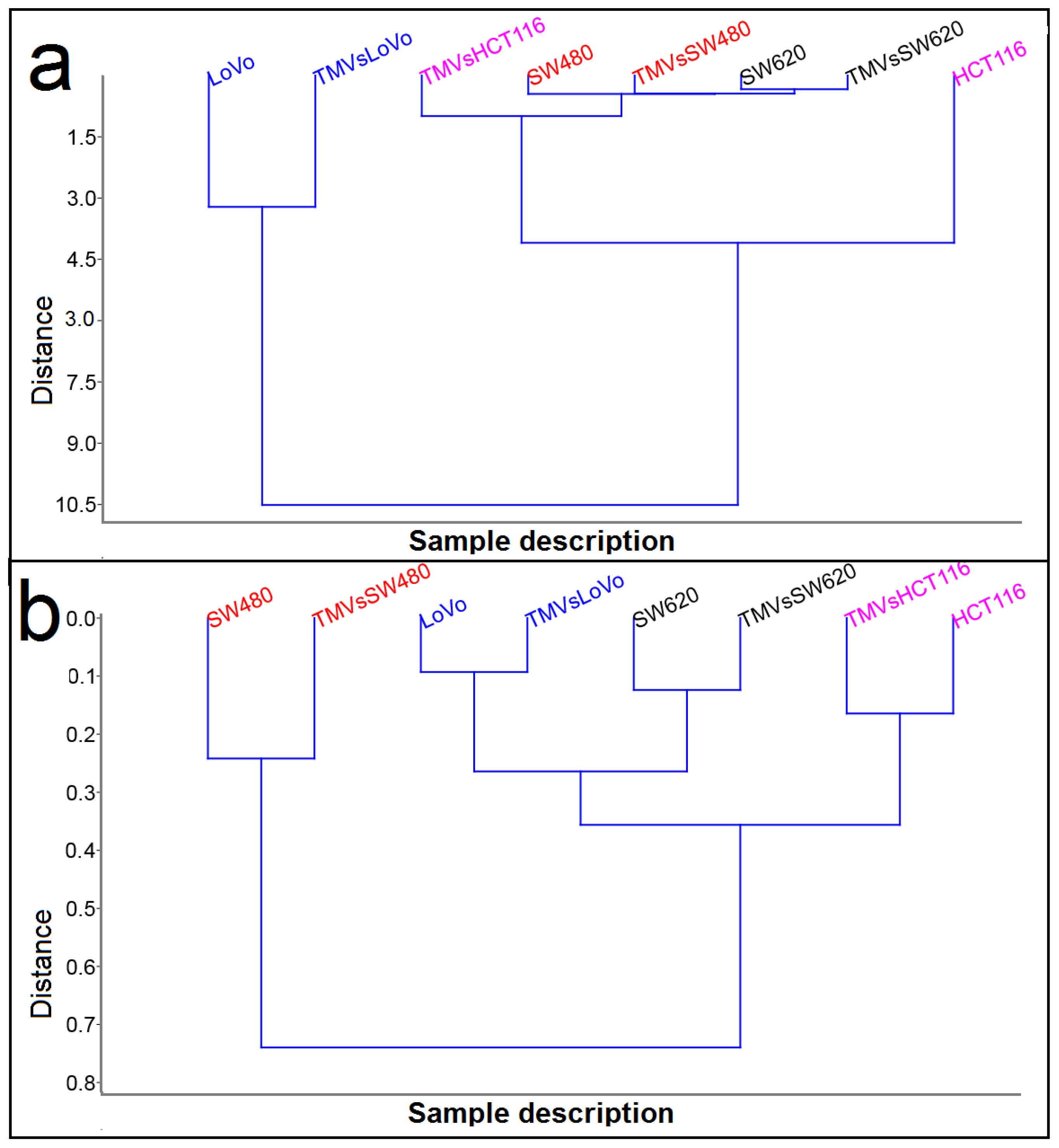
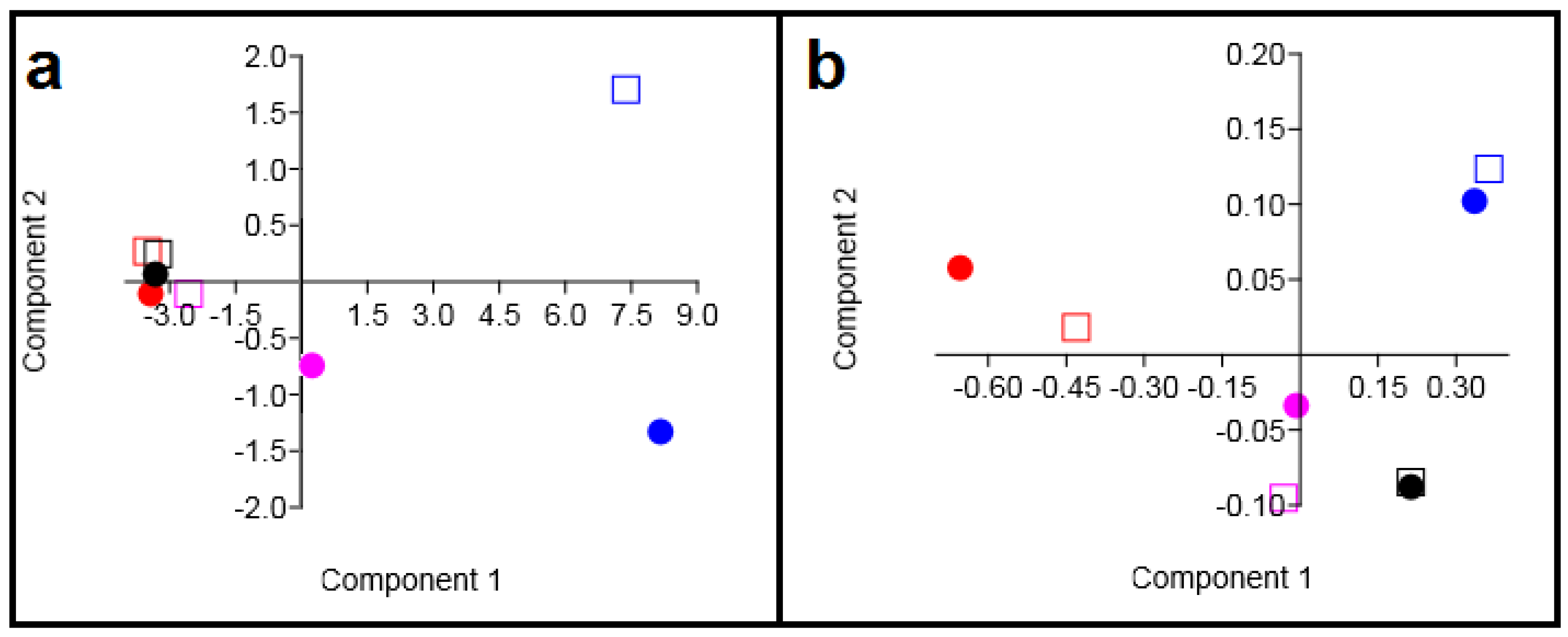
2.3. Determination of Similarities Between TMVs and Cell Lines
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Isolation of TMVs
4.3. Western Blotting
4.4. Nanoparticle Tracking Analysis (NTA)
4.5. Cell and TMVs Preparation for Fourier Transrorm IntraRed (FTIR) and-Raman Measurements
4.6. FTIR Spectroscopy
4.7. FT-Raman Spectroscopy
4.8. Deconvolution of Amide I Region (1600–1700 cm−1)
4.9. Data Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MDPI | Multidisciplinary Digital Publishing Institute |
| DOAJ | Directory of open access journals |
| TLA | Three letter acronym |
| LD | linear dichroism |
References
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Prz. Gastroenterol. 2019, 14, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Ewing, I.; Hurley, J.J.; Josephides, E.; Millar, A. The molecular genetics of colorectal cancer. Frontline Gastroenterol. 2014, 5, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Freddie, B.; Jacques, F.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar]
- Singh, A.B.; Harris, R.C. Autocrine, paracrine an juxtacrine signalling by EGFR ligands. Cell Signal. 2005, 17, 1183–1193. [Google Scholar] [CrossRef] [PubMed]
- Al-Nedawi, K.; Meehan, B.; Rak, J. Microvesicles: Messengers and mediators of tumor progression. Cell Cycle 2009, 8, 2014–2018. [Google Scholar] [CrossRef]
- Cocucci, E.; Racchetti, G.; Meldolesi, J. Shedding microveiscles: Artefacts no more. Trends Cell Biol. 2009, 19, 43–51. [Google Scholar] [CrossRef]
- Gould, S.J.; Raposo, G. As we wait: Coping with an imperfect nomenclature for extracellular vesicles. J. Extracell. Vesicles 2013, 2, 20389. [Google Scholar] [CrossRef]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intracellular communication. J. Proteomics 2010, 73, 1907–1920. [Google Scholar] [CrossRef]
- Tetta, C.; Ghigo, E.; Silengo, L.; Deregibus, M.C.; Camussi, G. Extracellular vesicles as an emerging mechanism of cell-to-cell communication. Endocrine 2013, 44, 11–19. [Google Scholar] [CrossRef]
- Turturici, G.; Tinnirello, R.; Sconzo, G.; Geraci, F. Extracellular membrane vesicles as a mechanism of cell-to-cell communication: Advantages and disadvantages. Am. J. Physiol. Cell Physiol. 2014, 306, C621–C633. [Google Scholar] [CrossRef]
- Peinado, H.; Aleckowic, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Herqueta-Redono, M.; Williams, C.; Garcia-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Bobrie, A.; Colombo, M.; Raposo, G.; Thery, C. Exosome secretion: Molecular mechanisms and roles in immune responses. Traffic 2011, 12, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Delabranche, X.; Berger, A.; Boisrame-Helms, J.; Meziani, F. Microparticles and infectious diseases. Med. Mal. Infect. 2013, 42, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Schneider, A.; Simons, M. Exosomes: Vesicular carriers for intracellular communication in neurodegenerative disorders. Cell Tissue Res. 2013, 352, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Shiromizu, T.; Kume, H.; Ishida, M.; Adachi, J.; Kano, M.; Matsubara, H.; Tomonaga, T. Quantitation of putative colorectal cancer biomarker candidates in serum extracellular vesicles by targeted proteomics. Sci. Rep. 2017, 7, 12782. [Google Scholar]
- Gallart-Palau, X.; Serra, A.; Wong, A.S.; Sandin, S.; Lai, M.K.; Chen, C.P.; Kon, O.L.; Sze, S.K. Extracellular vesicles are rapidly purified from human plasma by Protein Organic Solvent PRecipitation (PROSPR). Sci. Rep. 2015, 30, 14664. [Google Scholar] [CrossRef] [PubMed]
- Baran, J.; Baj-Krzyworzeka, M.; Węglarczyk, K.; Szatanek, R.; Zembala, M.; Barbasz, J.; Czupryna, A.; Szczepanik, A.; Zembala, M. Circulating tumour-derived microvesicles in plasma of gastric cancer patients. Cancer Immunol. Immunother. 2010, 59, 841–850. [Google Scholar] [CrossRef]
- Hon, K.W.; Abu, N.; Ab Mutalib, N.S.; Jamal, R. Exosomes as potential biomarkers and targeted therapy in colorectal cancer: A mini-review. Front. Pharmacol. 2017, 8, 583. [Google Scholar] [CrossRef]
- Lee, B.R.; Kim, J.H.; Choi, E.S.; Cho, J.H.; Kim, E. Effect of young exosomes injected in aged mice. Int. J. Nanomed. 2018, 13, 5335–5345. [Google Scholar] [CrossRef]
- Suchorska, W.M.; Lach, M.S. The role of exosomes in tumor progression and metastasis (review). Oncol. Rep. 2016, 35, 1237–1244. [Google Scholar] [CrossRef]
- Luo, D.; Zhan, S.; Xia, W.; Huang, L.; Ge, W.; Wang, T. Proteomics study of serum exosomes from papillary thyroid cancer patients. Endocr. Relat. Cancer. 2018, 25, 879–891. [Google Scholar] [CrossRef] [PubMed]
- Skotland, T.; Sandvig, K.; Llorente, A. Lipids in exosomes: Current knowledge and the way forward. Prog. Lipid Res. 2017, 66, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Herzberg, G. Molecular Spectra and Molecular Structure. Part II. Infrared and Raman Spectra of Polyatomic Molecules; D. Van Nanostrand Company: New York, NY, USA, 1945. [Google Scholar]
- Woodward, L.A. Introduction to the Theory of Molecular Vibrations & Vibrational Spectroscopy; Oxford University Press: Oxford, UK, 1972. [Google Scholar]
- Baj-Krzyworzeka, M.; Mytar, B.; Szatanek, R.; Surmiak, M.; Węglarczyk, K.; Baran, J.; Siedlar, M. Colorectal cancer-derived microvesicles modulate differentiation of human monocytes to macrophages. J. Trans. Med. 2016, 14, 36. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Thery, C. Biogenesis, secretion, and intracellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Maziak, D.E.; Do, M.T.; Shamji, F.M.; Sundaresan, S.R.; Perkins, D.G.; Wong, P.T. Fourier-transform infrare spectroscopic study of characteristic molecular structure in cancer cells of esophagus: An explaratory study. Cancer Detect Prev. 2007, 31, 244–253. [Google Scholar] [CrossRef]
- Krishna, C.M.; Sockalingum, G.D.; Bhat, R.A.; Venteo, L.; Kushtagi, P.; Pluot, M.; Manfait, M. FTIR and Raman microspectroscopy of normal, benign, and malignant formalin-fixed ovarian tissues. Anal. Bioanal. Chem. 2007, 387, 1649–1656. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; Rehman, I. Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2008, 43, 134–179. [Google Scholar] [CrossRef]
- Elmi, F.; Movaghar, A.F.; Alinezhad, H.; Nikbakhsh, N. Application of FT-IR spectroscopy on breast cancer serum analysis. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2017, 187, 87–91. [Google Scholar] [CrossRef]
- Yano, K.; Okoshima, S.; Gotou, Y.; Kumaido, K.; Moriguchi, T.; Katayama, H. Direct measurement of human lung cancerous and noncancerous tissues by fourier transform infrared microscopy: Can an infrared microscope be used as clinical tool? Anal. Biochem. 2000, 287, 218–225. [Google Scholar] [CrossRef]
- Barth, A.; Zscherp, C. What vibrations tellu s about proteins. Q. Rev. Biophys. 2002, 35, 369–430. [Google Scholar] [CrossRef] [PubMed]
- Wrobel, T.P.; Mateuszuk, L.; Chlopicki, S.; Malek, K.; Baranska, M. Imaging of lipids in artherosclerotic lesion in aorta from ApoE/LDLR-/- mice by FT-IR spectroscopy an hierarchical cluster analysis. Analyst 2011, 136, 5247–5255. [Google Scholar] [CrossRef] [PubMed]
- Barth, A. Infrated spectroscopy of proteins. Biochim. Biophys. Acta 2007, 1767, 1073–1101. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.W.; Taylor, D.S.; Zwerdling, T.; Lane, S.M.; Ihara, K.; Huser, T. Micro-Raman spectroscopy detects indiviual neoplastic and normal hematopoietic cells. Biophys. J. 2006, 90, 648–656. [Google Scholar] [CrossRef]
- Krafft, C.; Neudert, L.; Simat, T.; Salzer, R. Near infrared Raman spectra of human brain lipids. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2005, 61, 1529–1535. [Google Scholar] [CrossRef]
- Silveira, L., Jr.; Sathaiah, S.; Zangaro, R.A.; Pacheco, M.T.; Chavantes, M.C.; Pasqualucci, C.A. Correlation between near-infrared Raman spectroscopy and the histopathological analysis of artherosclerosis in human coronary arteries. Lasers Surg. Med. 2002, 30, 290–297. [Google Scholar] [CrossRef]
- Stone, N.; Kendall, C.; Smith, J.; Crow, P.; Barr, H. Raman spectroscopy for identification of epithelial cancers. Faraday Discuss. 2004, 126, 141–157. [Google Scholar] [CrossRef]
- Huang, Z.; McWilliams, A.; Lui, H.; McLean, D.I.; Lam, S.; Zeng, H. Near-infrared Raman spectroscopy for optical diagnosis of lung cancer. Int. J. Cancer. 2003, 107, 1047–1052. [Google Scholar] [CrossRef]
- Huang, Z.; McWilliams, A.; Lam, S.; English, J.; McLean, D.; Lui, H.; Zeng, H. Effect of formalin fixation on the near-infrare Raman spectroscopy of normal and cancerous human bronchial tissues. Int. J. Oncol. 2003, 23, 649–655. [Google Scholar]
- Stremersch, S.; Marro, M.; Pinchasik, B.E.; Hendrix, A.; De Smedt, S.C.; Loza-Alvarez, P.; Skirtach, A.G.; Raemdonck, K.; Braeckmans, K. Identification of individual exosome-like vesicles by surface enhanced Raman spectroscopy. Small 2016, 12, 3292–3301. [Google Scholar] [CrossRef]
- Malini, R.; Venkatakrishna, K.; Kurien, J.; Pai, K.M.; Rao, L.; Kartha, V.B.; Krishna, C.M. Discrimination of normal, inflammatory, premalignant, and malignant oral tissue: A Raman spectroscopy study. Biopolymers 2006, 81, 179–193. [Google Scholar] [CrossRef] [PubMed]
- Skornyakov, I.V.; Tolstorozhev, G.B.; Butra, V.A. Method for Diagnosis of Malignant Neoplasm of the Breast. Belarussian Patent No. 13242, 30 April 2010. [Google Scholar]
- Tolstorozhev, G.B.; Skornyakov, I.V.; Butra, V.A. IR spectroscopic signs of malignant neoplasms in the thyroid gland. J. Appl. Spectrosc. 2012, 79, 143–147. [Google Scholar] [CrossRef]
- Fraser, R.D.B.; MacRae, T.P. Conformations in Fibrous Proteins and Related Synthetic Polypeptides; Academic Press: New York, NY, USA, 1973. [Google Scholar]
- Yang, H.; Yang, S.; Kong, J.; Dong, A.; Yu, S. Obtaining information about protein secondary structures in aqueous solution using Fourier transform IR spectroscopy. Nat. Protoc. 2015, 10, 382–396. [Google Scholar] [CrossRef]
- Tuschel, D. Practical group theory and Raman spectroscopy, part I: Normal vibrational modes. Spectroscopy 2014, 29, 14–23. [Google Scholar]
- Steele, D. Infrared Spectroscopy: Theory, Handbook of Vibrational Spectroscopy; John Wiley & Sons, Ltd.: Weinheim, Germany, 2016. [Google Scholar]
- Wartewing, S. IR and Raman Spectroscopy: Fundamental Processing; Wiley-vch Verlag GmbH & Co. KGaA: Weinheim, Germany, 2003. [Google Scholar]
- Lau, W.S. Infrared Characterization for Microelectronics; Horiba Ltd.: London, UK, 1999. [Google Scholar]
- Leibovitz, A.; Stinson, J.C.; McCombs, W.B.; McCoy, C.E.; Mazur, K.C.; Mabry, N.D. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 1976, 36, 4562–4569. [Google Scholar] [PubMed]
- Ji, H.; Greening, D.W.; Barnes, T.W.; Lim, J.W.; Tauro, B.J.; Rai, A.; Xu, R.; Adda, C.; Mathivanan, S.; Zhao, W.; et al. Proteome profiling of exosomes derived from human primary and metastatic colorectal cancer cells reveal differentia expression of key metastatic factors and signal transduction components. Proteomics 2013, 13, 1672–1686. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.S.; Choi, D.Y.; Hong, B.S.; Jang, S.C.; Kim, D.K.; Lee, J.; Kim, Y.K.; Kim, K.P.; Gho, Y.S. Quantitative proteomics of extracellular vesicles derived from human primary and metastatic colorectal cancer cells. J. Extracell. Vesicles 2012, 1, 18704. [Google Scholar] [CrossRef]
- Lobb, R.J.; Lima, L.G.; Moller, A. Exosomes: Key mediators of metastasis and pre-metastatic niche formation. Semin. Cell Dev. Biol. 2017, 67, 3–10. [Google Scholar] [CrossRef]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reynies, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Berg, K.C.G.; Eide, P.W.; Eilersten, I.A.; Johannessen, B.; Bruun, J.; Danielsen, S.A.; Bjornslett, M.; Meza-Zepeda, L.A.; Eknaes, M.; Lind, G.E.; et al. Multi-omics of 34 colorectal cancer cell lines—A resource for biomedical studies. Mol. Cancer 2017, 16, 116. [Google Scholar] [CrossRef]
- Quinn, L.A.; Moore, G.E.; Morgan, R.T.; Woods, L.K. Cell lines from human colon carcinoma with unusual cell products, double minutes, and homogeneously staning regions. Cancer Res. 1976, 39, 4914–4924. [Google Scholar]
- Pablo, J.G.; Armistead, F.J.; Peyman, S.A.; Bonthron, D.; Lones, M.; Smith, S.; Evans, S.D. Biochemical fingerprint of colorectal cancer cell lines using label-free live single-cell Raman spectroscopy, J. Raman Spectrosc. 2018, 49, 1323–1332. [Google Scholar] [CrossRef] [PubMed]
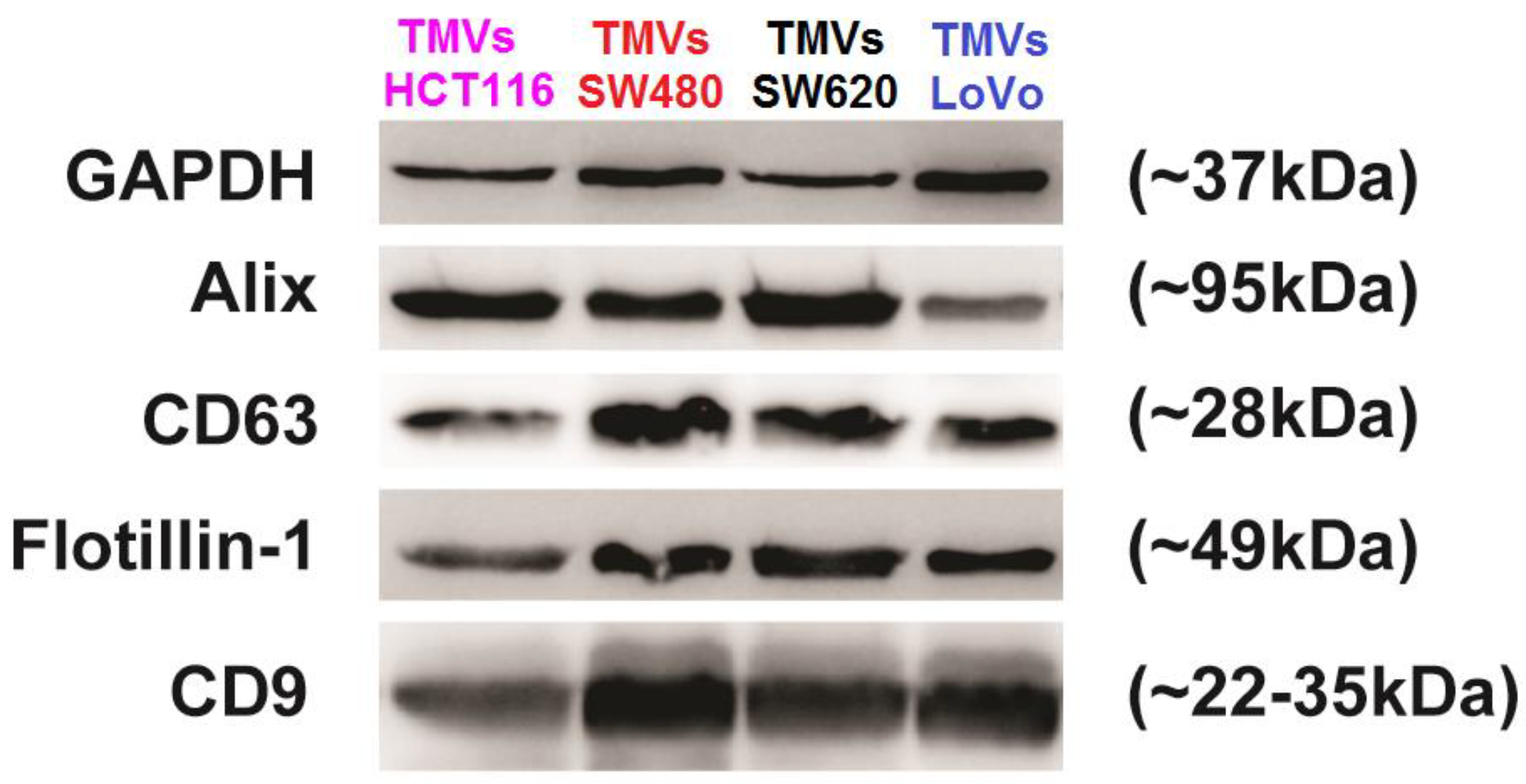


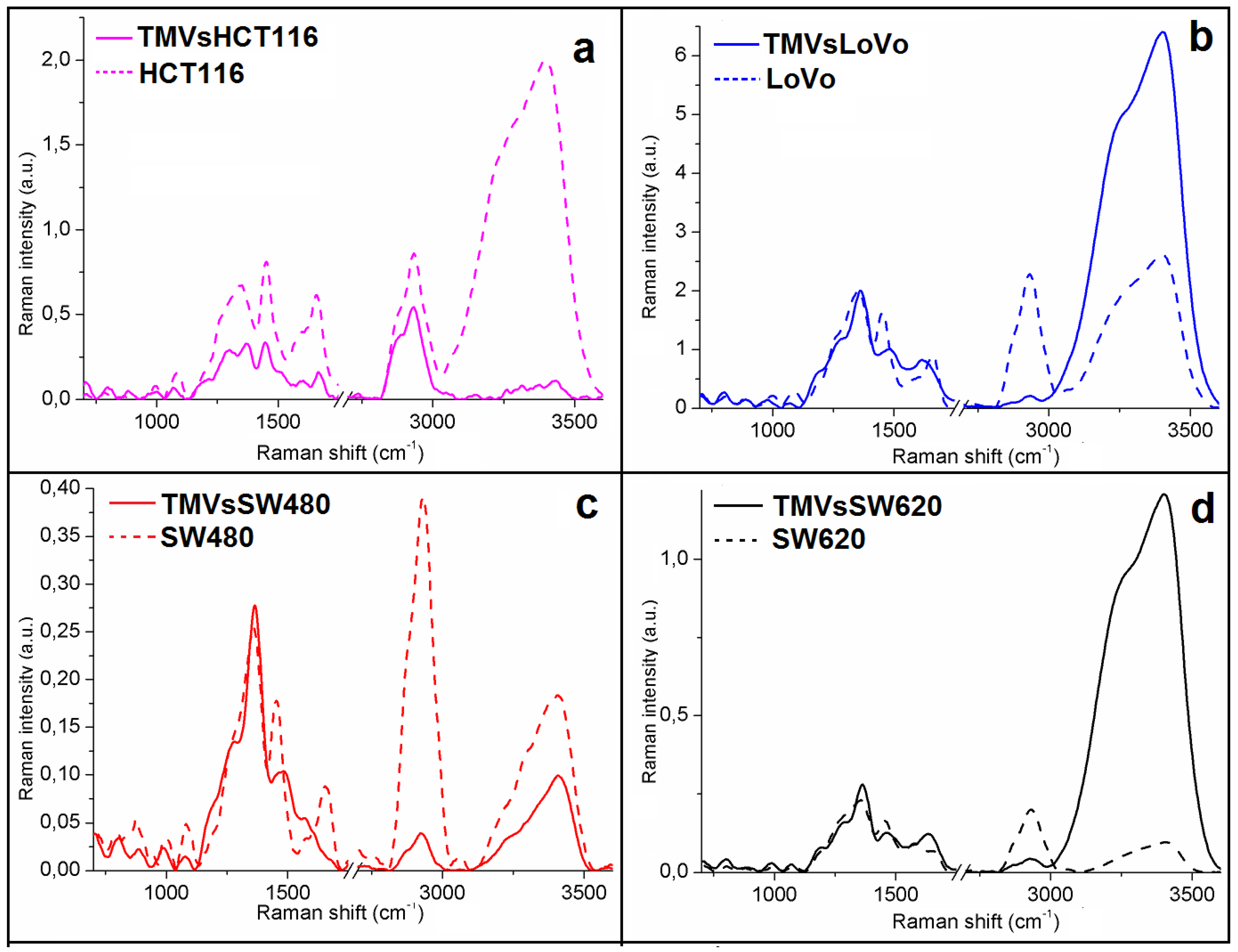
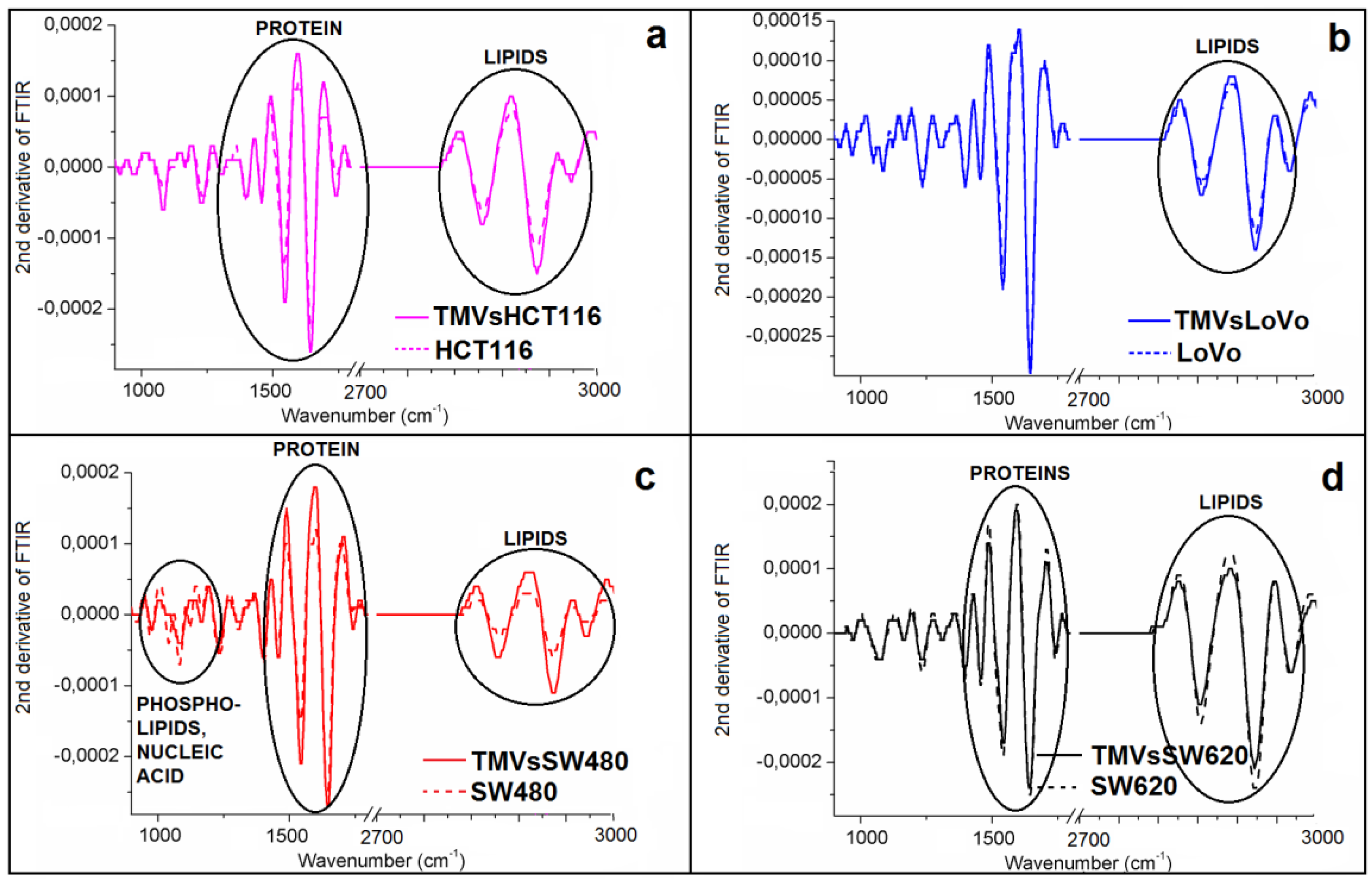
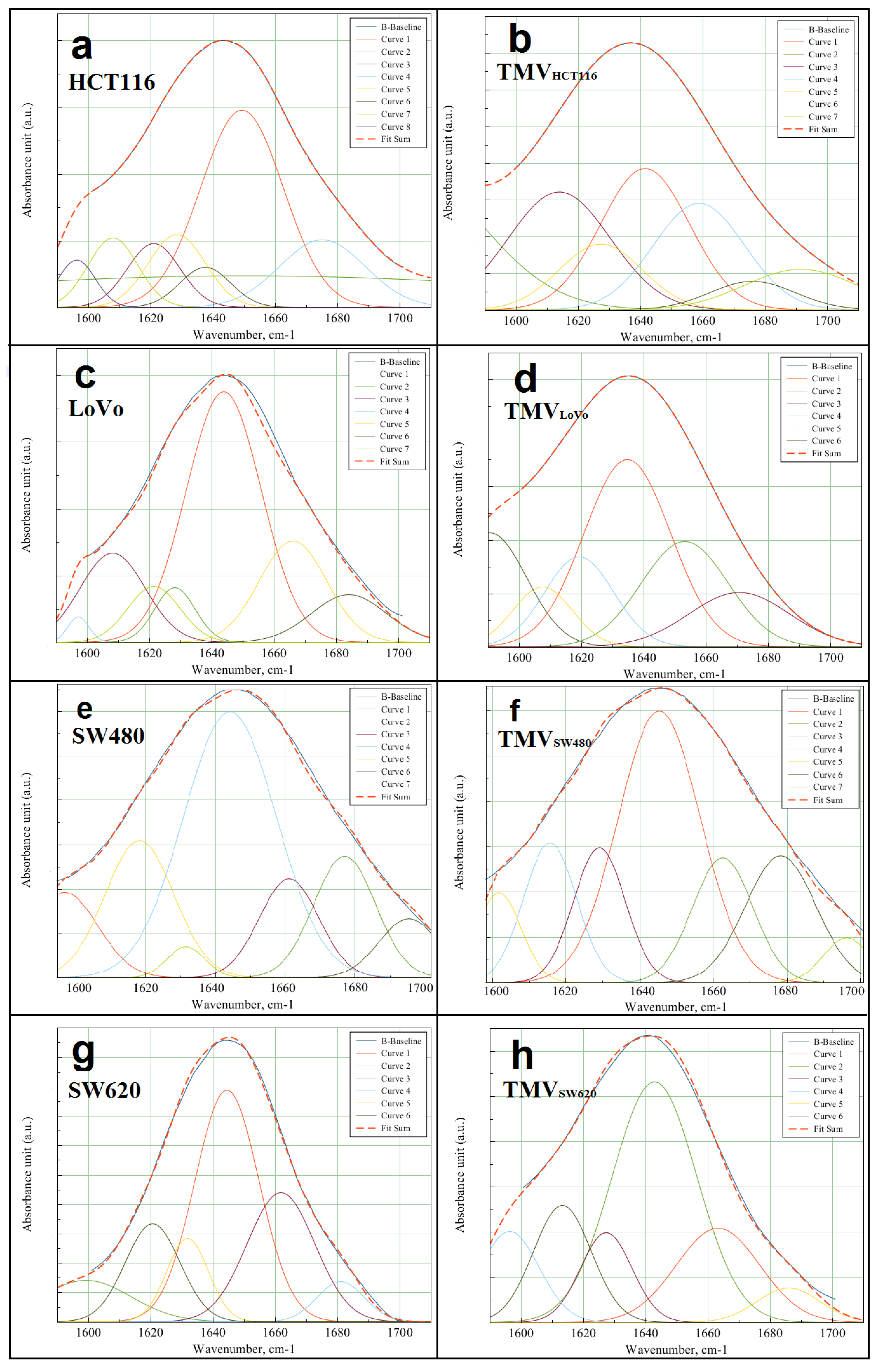
| FTIR Spectroscopy Peaks (cm−1) | ||||||||
| HCT116 | TMVHCT116 | LoVo | TMVLoVo | SW480 | TMVSW480 | SW620 | TMVSW620 | Vibrations |
| 972 | 970 | 973 | 976 | 981 | 983 | 976 | 975 | PO3−2 group from DNA, RNA and phospholipids [28] |
| 1083 | 1083 | 1085 | 1081 | 1081 | 1080 | 1072 | 1067 | C–O group from glycogen [29] |
| 1232 | 1237 | 1239 | 1238 | 1237 | 1236 | 1234 | 1238 | Amide III [30] |
| 1302 | 1306 | 1307 | 1308 | 1309 | 1309 | 1309 | 1307 | CH2 group from protein [31,32] |
| 1400 | 1403 | 1402 | 1404 | 1400 | 1402 | 1402 | 1401 | CH2 group from protein and lipids [33,34] |
| 1456 | 1457 | 1458 | 1457 | 1459 | 1458 | 1458 | 1458 | CH2 group from cholesterol [35] |
| 1546 | 1544 | 1543 | 1547 | 1544 | 1544 | 1541 | 1542 | Amide II [32] |
| 1643 | 1644 | 1646 | 1646 | 1646 | 1645 | 1645 | 1643 | Amide I [32] |
| 1738 | Absent | Absent | Absent | Absent | 1731 | 1733 | 1741 | C–O group from lipids [34] |
| 2858 | 2862 | 2861 | 2861 | 2860 | 2860 | 2860 | 2858 | Symmetric stretching vibrations of CH2 [34] |
| 2926 | 2923 | 2927 | 2928 | 2927 | 2926 | 2924 | 2925 | Asymmetric stretching vibrations of CH2 [34] |
| 3064 | Absent | 3075 | Absent | 2963 | Absent | Absent | Absent | CH3 asymmetric stretching [34] |
| 3284 | 3290 | 3287 | 3294 | 3301 | 3284 | 3287 | 3284 | Amide A and OH group from water [35] |
| Raman Spectroscopy Peaks (cm−1) | ||||||||
| HCT116 | TMVHCT116 | LoVo | TMVLoVo | SW480 | TMVSW480 | SW620 | TMVSW620 | Vibrations |
| 1001 | 999 | 981 | 1000 | 988 | 987 | 986 | 984 | C–H in-plane bending mode of phenylalanine [36] |
| 1072 | 1080 | 1068 | 1083 | 1077 | 1080 | 1072 | 1072 | Glucose triglycerides, C–C (lipid) [37,38] |
| Absent | Absent | Absent | Absent | 1280 | Absent | 1281 | 1284 | Amide III [39] |
| 1301 | 1351 | 1291 | 1360 | 1362 | 1355 | 1362 | 1362 | CH3/CH2 twisting or bending mode of lipid/collagen [40,41] |
| 1450 | 1451 | 1481 | 1454 | 1479 | 1453 | 1461 | 1466 | Fatty acids, CH2 (lipids and proteins) [40,42] |
| 1668 | 1658 | 1620 | 1657 | 1647 | 1654 | 1637 | 1635 | Amide I [43] |
| 1784 | 1771 | 1761 | 1764 | 1786 | 1776 | Absent | Absent | C55O ester (lipids) [42] |
| Absent | Absent | Absent | 2700 | 2722 | 2728 | Absent | Absent | Stretching vibrations of CH, NH, and OH groups [40] |
| 2931 | 2933 | 2932 | 2931 | 2926 | 2929 | 2929 | 2926 | CH band of lipids [42] |
| 3145 | Absent | Absent | Absent | Absent | Absent | 3124 | Absent | CH from cholesterol and cholesterol ester [42] |
| Sample | Peak Areas | Percentage [%] | α-Helix/β-Harmonica Ratio | ||
|---|---|---|---|---|---|
| α-Helix | β-Harmonica | α-Helix | β-Harmonica | ||
| HCT116 | 55.36 | 59.91 | 48.03 | 51.97 | 0.92 |
| TMVHCT116 | 12.24 | 13.47 | 47.61 | 52.39 | 0.91 |
| LoVo | 63.34 | 59.40 | 51.61 | 48.39 | 1.07 |
| TMVLoVo | 19.70 | 17.80 | 52.53 | 47.47 | 1.11 |
| SW480 | 33.35 | 35.53 | 48.42 | 51.58 | 0.94 |
| TMVSW480 | 33.99 | 34.45 | 49.67 | 50.33 | 0.99 |
| SW620 | 40.78 | 63.42 | 39.14 | 60.86 | 0.64 |
| TMVSW620 | 22.25 | 19.75 | 52.98 | 47.02 | 1.13 |
| Sensitivity | HCT116 | TMVHCT116 | LoVo | TMVLoVo | SW480 | TMVSW480 | SW620 | TMVSW620 |
|---|---|---|---|---|---|---|---|---|
| Specificity | ||||||||
| HCT116 | 100% | 85% | 94% | 98% | 96% | 98% | 95% | 95% |
| 100% | 87% | 98% | 94% | 96% | 99% | 90% | 64% | |
| TMVHCT116 | 85% | 100% | 93% | 92% | 87% | 89% | 90% | 96% |
| 87% | 100% | 94% | 96% | 95% | 85% | 98% | 63% | |
| LoVo | 94% | 93% | 100% | 89% | 92% | 93% | 93% | 98% |
| 98% | 94% | 100% | 98% | 99% | 98% | 97% | 51% | |
| TMVLoVo | 98% | 92% | 89% | 100% | 96% | 97% | 85% | 99% |
| 94% | 96% | 98% | 100% | 99% | 95% | 99% | 57% | |
| SW480 | 96% | 87% | 92% | 96% | 100% | 96% | 96% | 98% |
| 96% | 95% | 99% | 99% | 100% | 96% | 98% | 55% | |
| TMVSW480 | 98% | 89% | 93% | 97% | 96% | 100% | 91% | 99% |
| 99% | 85% | 98% | 95% | 96% | 100% | 90% | 65% | |
| SW620 | 95% | 90% | 93% | 85% | 96% | 91% | 100% | 97% |
| 90% | 98% | 97% | 99% | 98% | 90% | 100% | 68% | |
| TMVSW620 | 95% | 96% | 98% | 99% | 98% | 99% | 97% | 100% |
| 64% | 63% | 51% | 57% | 55% | 65% | 68% | 100% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Depciuch, J.; Klębowski, B.; Stec, M.; Szatanek, R.; Węglarczyk, K.; Baj-Krzyworzeka, M.; Parlińska-Wojtan, M.; Baran, J. Similarities in the General Chemical Composition of Colon Cancer Cells and Their Microvesicles Investigated by Spectroscopic Methods-Potential Clinical Relevance. Int. J. Mol. Sci. 2020, 21, 1826. https://doi.org/10.3390/ijms21051826
Depciuch J, Klębowski B, Stec M, Szatanek R, Węglarczyk K, Baj-Krzyworzeka M, Parlińska-Wojtan M, Baran J. Similarities in the General Chemical Composition of Colon Cancer Cells and Their Microvesicles Investigated by Spectroscopic Methods-Potential Clinical Relevance. International Journal of Molecular Sciences. 2020; 21(5):1826. https://doi.org/10.3390/ijms21051826
Chicago/Turabian StyleDepciuch, Joanna, Bartosz Klębowski, Małgorzata Stec, Rafał Szatanek, Kazimierz Węglarczyk, Monika Baj-Krzyworzeka, Magdalena Parlińska-Wojtan, and Jarek Baran. 2020. "Similarities in the General Chemical Composition of Colon Cancer Cells and Their Microvesicles Investigated by Spectroscopic Methods-Potential Clinical Relevance" International Journal of Molecular Sciences 21, no. 5: 1826. https://doi.org/10.3390/ijms21051826
APA StyleDepciuch, J., Klębowski, B., Stec, M., Szatanek, R., Węglarczyk, K., Baj-Krzyworzeka, M., Parlińska-Wojtan, M., & Baran, J. (2020). Similarities in the General Chemical Composition of Colon Cancer Cells and Their Microvesicles Investigated by Spectroscopic Methods-Potential Clinical Relevance. International Journal of Molecular Sciences, 21(5), 1826. https://doi.org/10.3390/ijms21051826







