New Perspectives Related to the Bioluminescent System in Dinoflagellates: Pyrocystis lunula, a Case Study
Abstract
1. Introduction
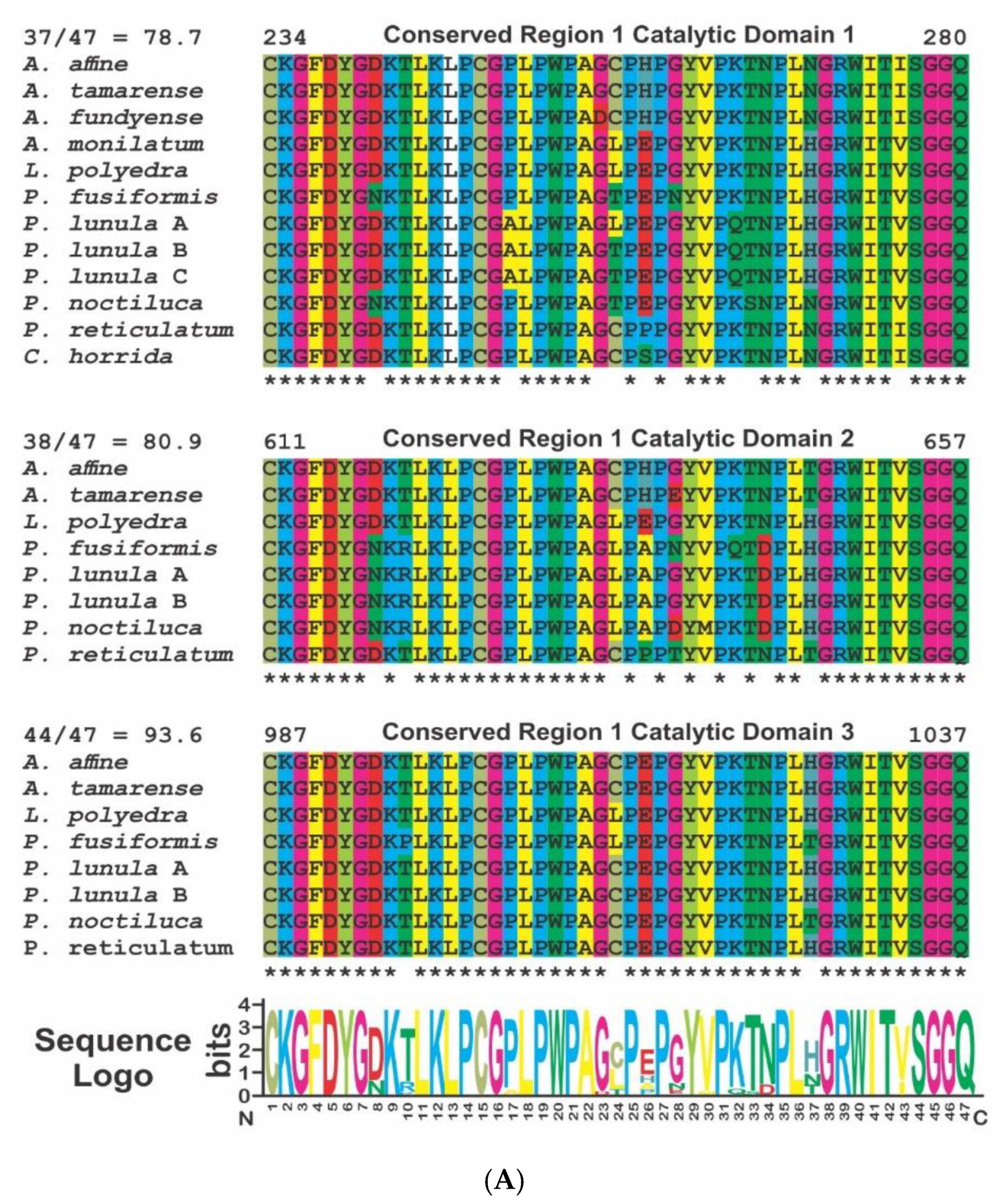
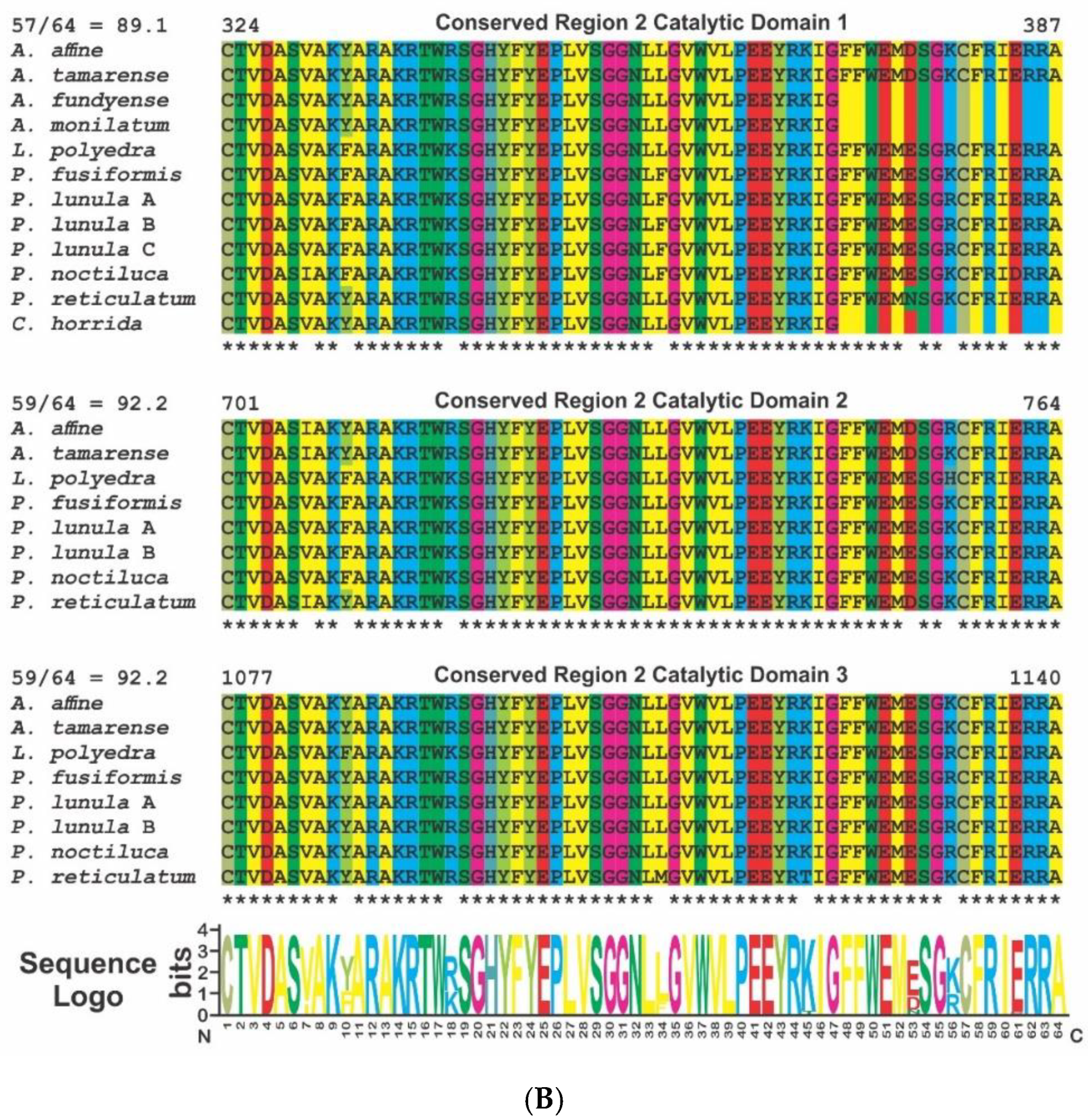
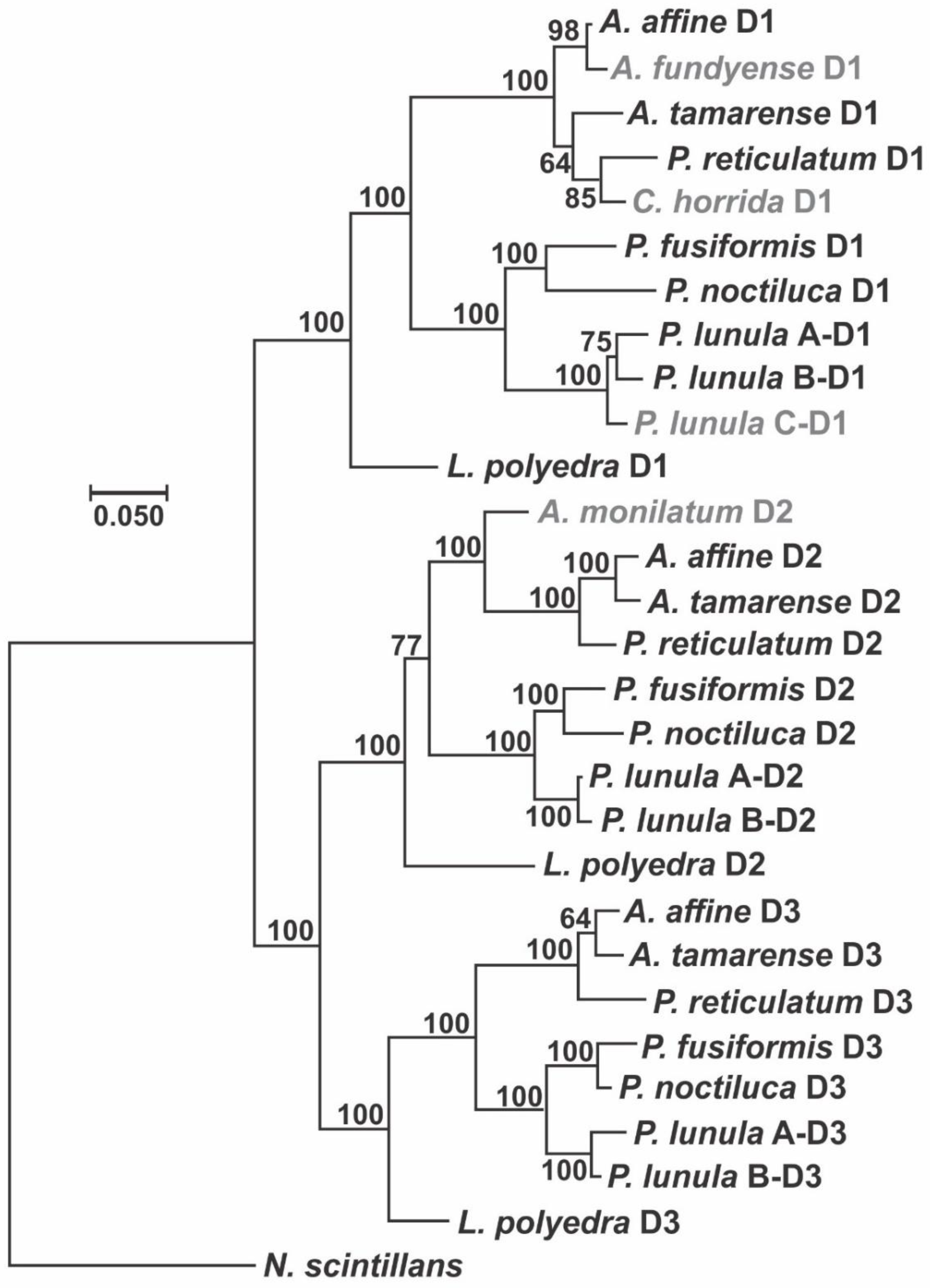
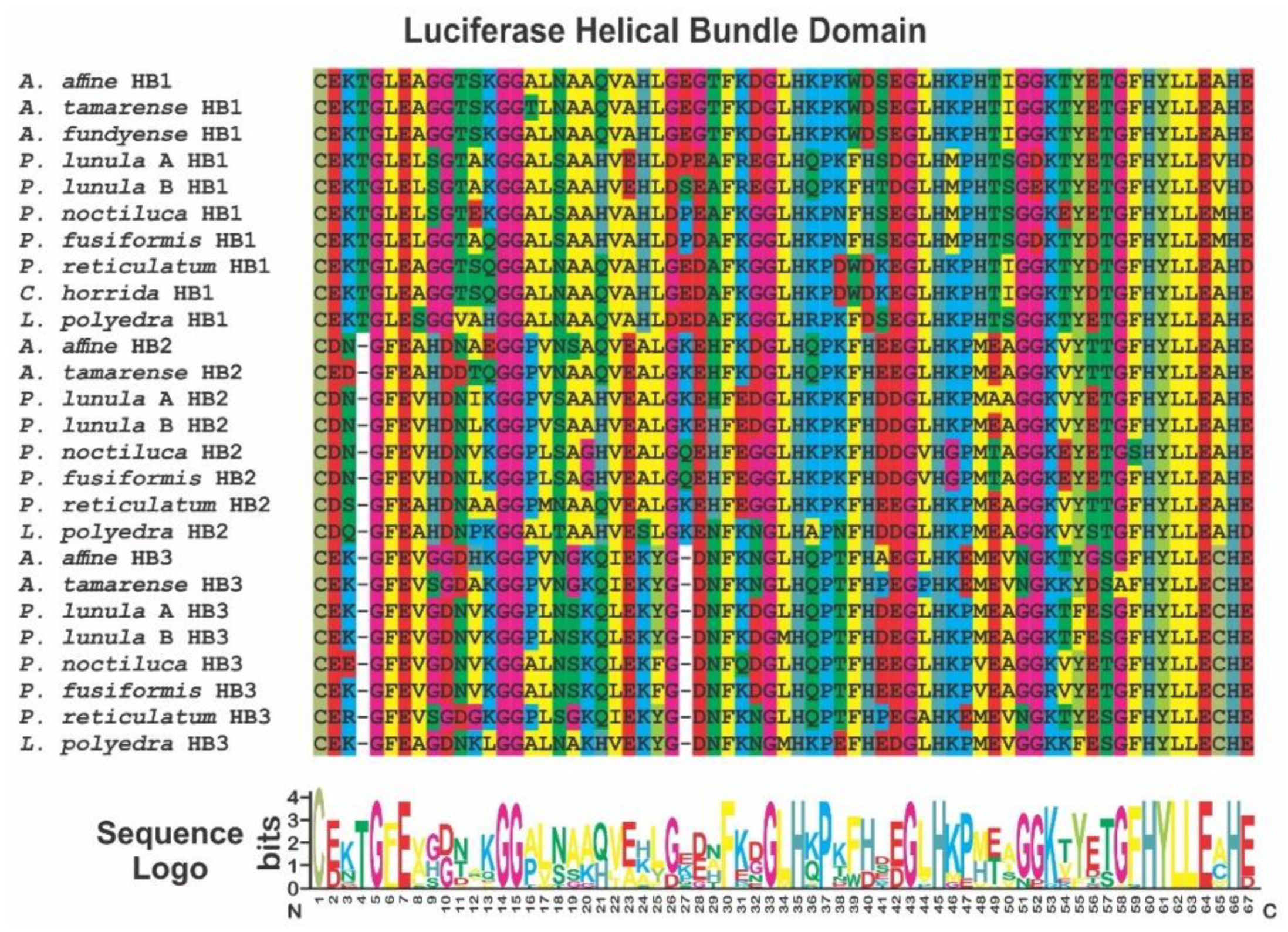
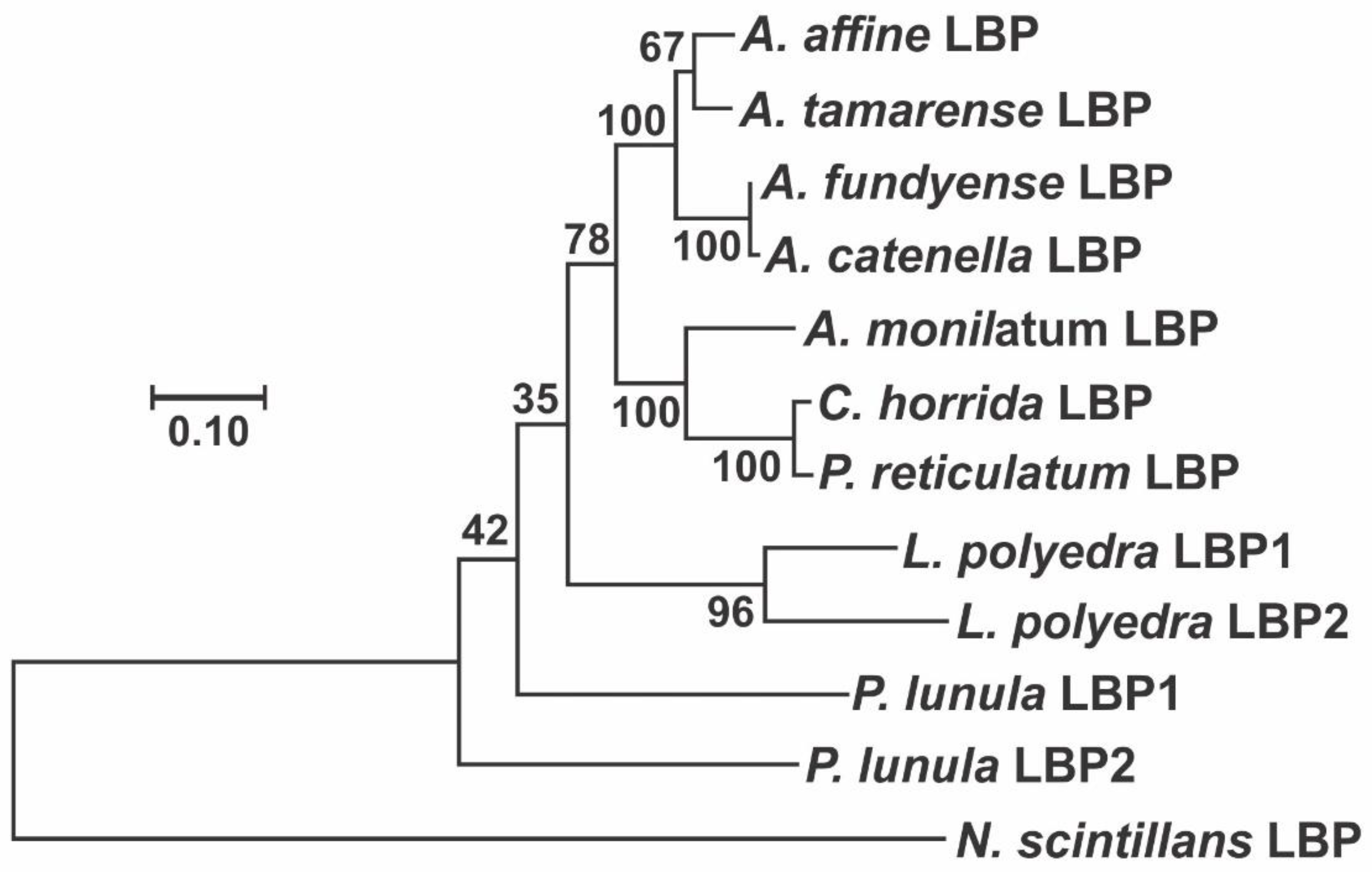
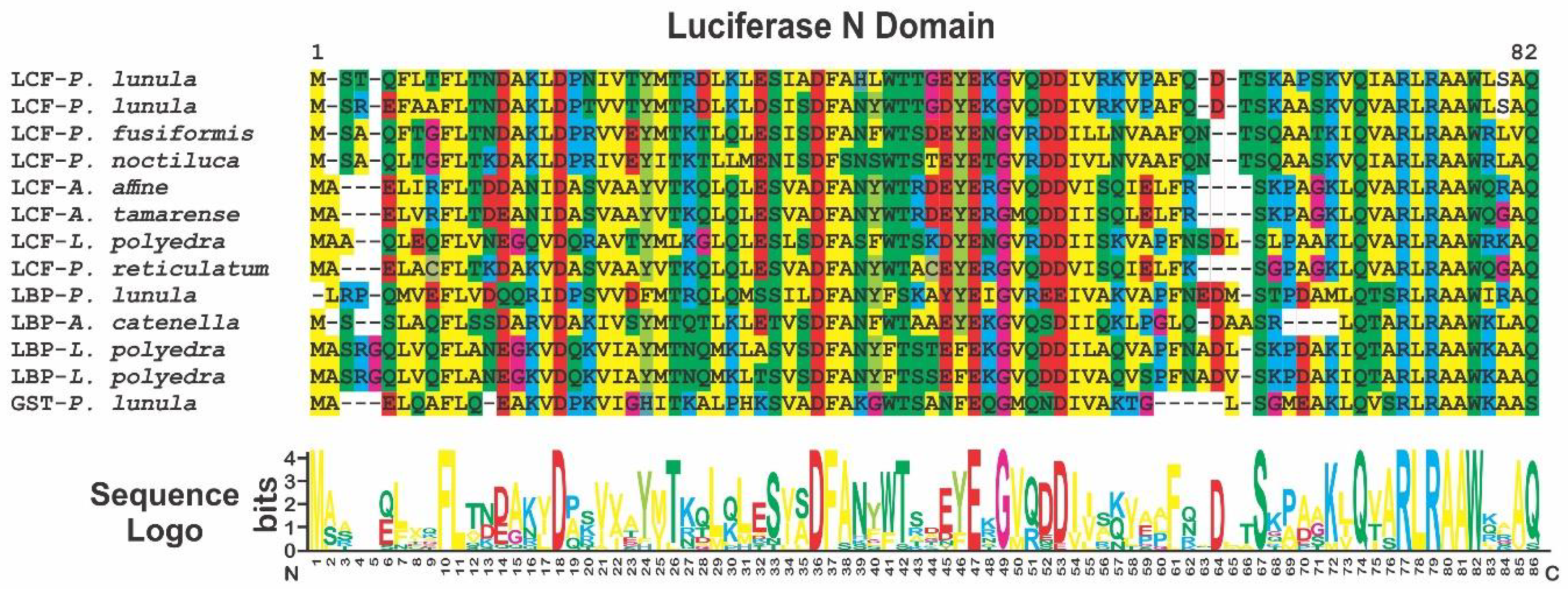
2. Phylogenetic and Structural Analyses
3. Dinoflagellate Bioluminescent System
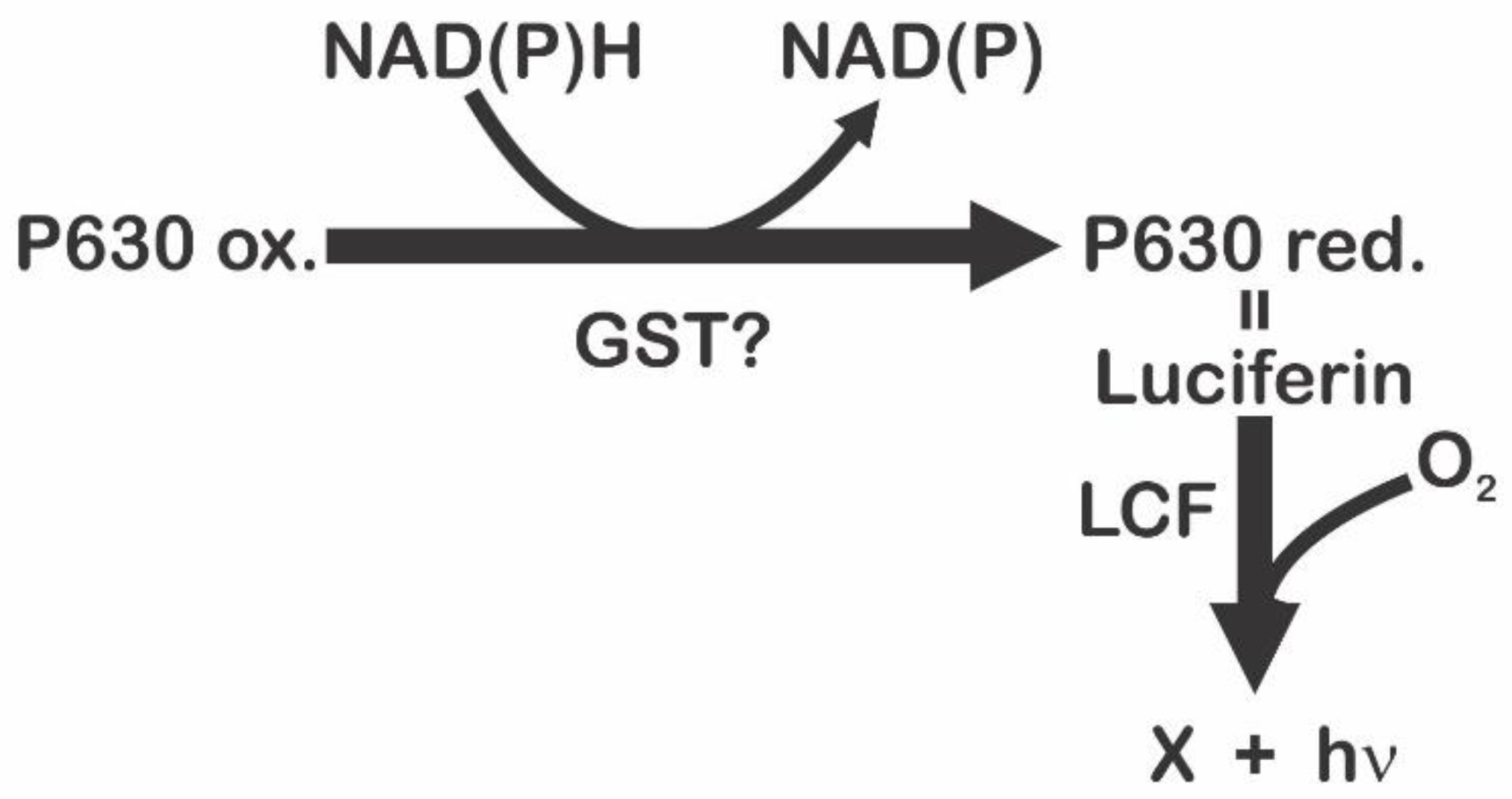
3.1. Luciferase (LCF)
3.2. Luciferin-Binding Protein (LBP)
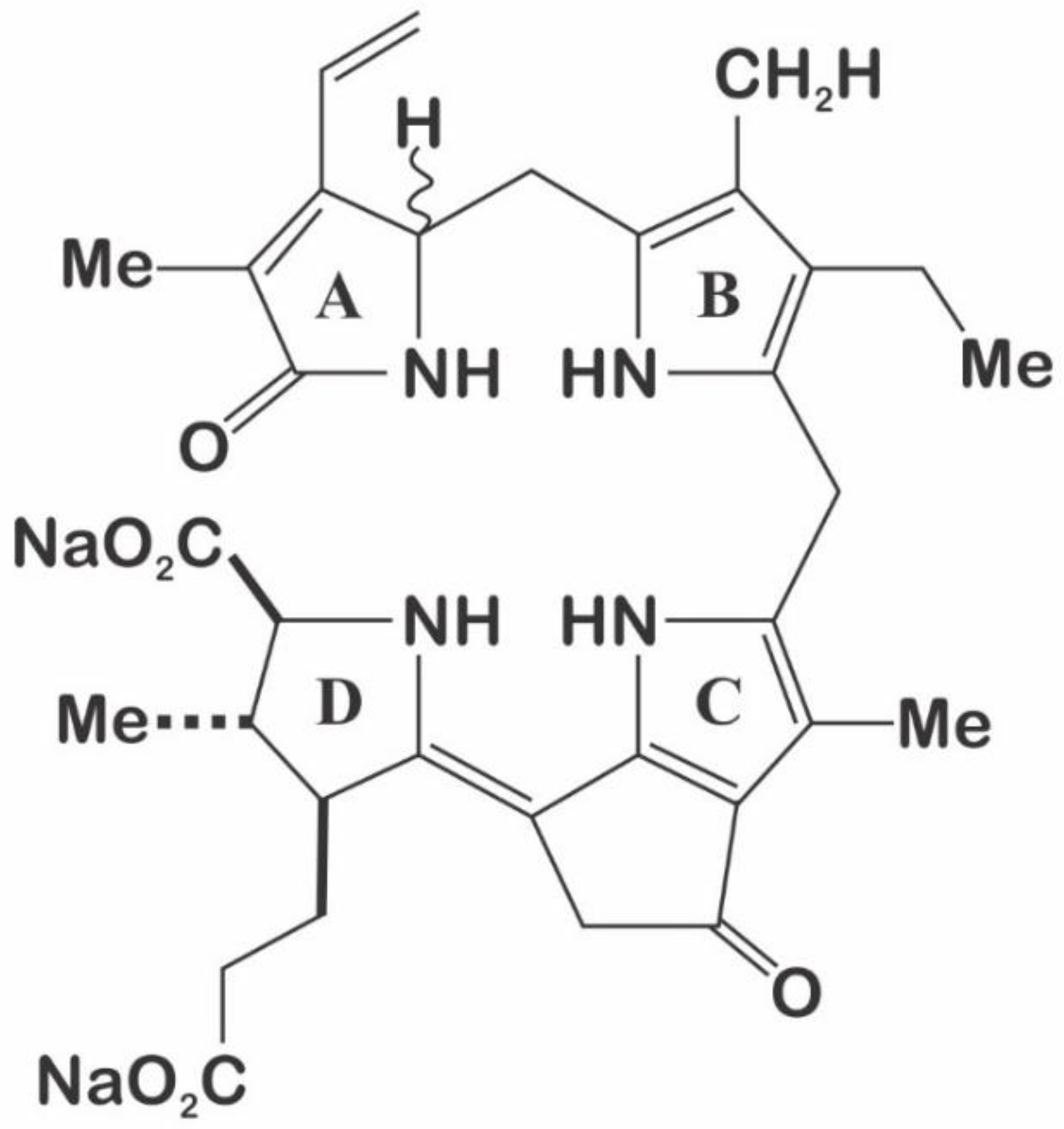
3.3. Luciferin
4. Glutathione S-Transferase (GST)
5. Function of Bioluminescence in Dinoflagellates
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| GSH | Glutathione |
| GST | Glutathione S-transferase |
| LBP | Luciferin-Binding Protein |
| LCF | Luciferase |
| TD-DFT | Time-Dependent Density Functional Theory |
| TICT | Twisted Intramolecular Charge Transfer |
References
- Widder, E. Marine bioluminescence. Biosci. Explain. 2001, 1, 1–9. [Google Scholar]
- Haddock, S.; Moline, M.; Case, J. Bioluminescence in the sea. Annu Rev. Mar. Sci. 2010, 2, 443–493. [Google Scholar] [CrossRef] [PubMed]
- Fuller, C.W.; Kreiss, P.; Seliger, H.H. Particulate Bioluininescence in Dinoflagellates: Dissociation and Partial Reconstitution. Science 1972, 177, 884–885. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Hastings, J.W. The structure and organization of the luciferase gene in the photosynthetic dinoflagellate Gonyaulax polyedra. Plant. Mol. Biol. 1998, 36, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Shimomura, O. Bioluminescence: Chemical Principles and Methods; Revised Edition; World Scientific Publishing Co. Pte. Ltd.: Singapore, 2012; ISBN 978-98-1436-608-3. [Google Scholar]
- Swift, E.; Taylor, W. Bioluminescence and chloroplast movement in the dinoflagellate Pyrocystis lunula. J. Phycol. 1967, 3, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Biggley, W.H.; Swift, E.; Buchanan, R.J.; Seliger, H.H. Stimulable and spontaneous bioluminescence in the marine dinoflagellates, Pyrodinium bahamense, Gonyaulax polyedra, and Pyrocystis lunula. J. Gen. Physiol. 1969, 54, 96–122. [Google Scholar] [CrossRef]
- Lecuyer, B.; Arrio, B.; Pincemin, J. Purification of Pyrocystis lunula luciferase. C. R. Acad. Sci. Hebd. Seances Acad. Sci. D 1974, 279, 1209–1212. [Google Scholar]
- Colepicolo, P.; Roenneberg, T.; Morse, D.; Taylor, W.; Hastings, J. Circadian regulation of bioluminescence in the dinoflagellate Pyrocyctis lunula. J. Phycol. 1993, 29, 173–179. [Google Scholar] [CrossRef]
- Heimann, K.; Klerks, P.; Hasenstein, K. Involvement of actin and microtubules in regulation of bioluminescence and translocation of chloroplasts in the dinoflagellate Pyrocystis lunula. Bot. Mar. 2009, 52, 170–177. [Google Scholar] [CrossRef]
- Stauber, J.; Binet, M.; Bao, V.; Boge, J.; Zhang, A.; Leung, K.; Adams, M. Comparison of the QwikLite algal bioluminescence test with marine algal growth rate inhibition bioassays. Environ. Toxicol. 2008, 23, 617–625. [Google Scholar] [CrossRef]
- Hildenbrand, Z.L.; Osorio, A.; Carlton, D.D.; Fontenot, B.E.; Walton, J.L.; Hunt, L.R.; Oka, H.; Hopkins, D.; Bjorndal, B.; Schug, K.A. Rapid analysis of eukaryotic bioluminescence to assess potential groundwater contamination events. J. Chem. 2015, 957608, 1–6. [Google Scholar] [CrossRef]
- Wu, C.; Kurinomaru, T. Development of the bioluminescent immunoassay for the detection of 5-hydroxymethylcytosine in dinoflagellate. Anal. Sci. 2019, 35, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Bachvaroff, T.R.; Gornik, S.G.; Concepcion, G.T.; Waller, R.F.; Mendez, G.S.; Lippmeier, J.C.; Delwiche, C.F. Dinoflagellate phylogeny revisited: Using ribosomal proteins to resolve deep branching dinoflagellate clades. Mol. Phylogenet. Evol. 2014, 70, 314–322. [Google Scholar] [CrossRef]
- Swift, E.; Remsen, C. The cell wall of Pyrocystis spp. (Dinococcales). J. Phycol. 1970, 6, 79–86. [Google Scholar] [CrossRef]
- Seo, K.; Fritz, L. Cell ultrastructural changes correlate with circadian rhythms in Pyrocystis lunula (Pyrrophyta). J. Phycol. 2000, 36, 351–358. [Google Scholar] [CrossRef]
- Steidinger, K.; Tangen, K. Dinoflagellates. In Identifying Marine Phytoplankton; Tomas, C., Ed.; Academic Press: New York, NY, USA, 1997; pp. 519–520. [Google Scholar]
- Elbrächter, M.; Drebes, G. Life cycles, phylogeny and taxonomy of Dissodinium and Pyrocystis (Dinophyta). Helgol. Mar. Res. 1978, 31, 347–366. [Google Scholar] [CrossRef]
- Seo, K.; Fritz, L. Evidence for sexual reproduction in the marine dinoflagellates, Pyrocystis noctiluca and Pyrocystis lunula (Dinophyta). J. Phycol. 2001, 37, 530–535. [Google Scholar] [CrossRef]
- Hastings, J.W. Circadian rhythms in dinoflagellates: What is the purpose of synthesis and destruction of proteins? Microorganisms 2013, 1, 26–32. [Google Scholar] [CrossRef]
- Shanks, A. An Identification Guide to the Larval Marine Invertebrates of the Pacific Northwest; Oregon State University Press: Corvalis, OR, USA, 2001; pp. 291–293. [Google Scholar]
- Yu, M.; Ohmiya, Y.; Naumov, P.; Liu, Y.J. Theoretical insight into the emission properties of the luciferin and oxyluciferin of Latia. Photochem. Photobiol. 2018, 94, 540–544. [Google Scholar] [CrossRef]
- Schmitter, R.; Njus, D.; Sulzman, F.; Gooch, V.; Hastings, J. Dinoflagellate bioluminescence: A comparative study of in vitro components. J. Cell. Physiol. 1976, 87, 123–134. [Google Scholar] [CrossRef]
- Johnson, C.; Inoue, S.; Flint, A.; Hastings, J. Compartmentalization of algal bioluminescence–autofluorescence of bioluminescent particles in the dinoflagellate Gonyaulax as studied with image-intensified video microscopy and flow cytometry. J. Cell Biol. 1985, 100, 1435–1446. [Google Scholar] [CrossRef]
- Knaust, R.; Urbig, T.; Li, L.; Taylor, W.; Hastings, J. The circadian rhythm of bioluminescence in Pyrocystis is not due to differences in the amount of luciferase: A comparative study of three bioluminescent marine dinoflagellates. J. Phycol. 1998, 34, 167–172. [Google Scholar] [CrossRef]
- Akimoto, H.; Wu, C.; Kinumi, T.; Ohmiya, Y. Biological rhythmicity in expressed proteins of the marine dinoflagellate Lingulodinium polyedrum demonstrated by chronological proteomics. Biochem. Biophys. Res. Commun. 2004, 315, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Machabee, S.; Wall, L.; Morse, D. Expression and genomic organization of a dinoflagellate gene family. Plant. Mol. Biol. 1994, 25, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Tanikawa, N.; Akimoto, H.; Ogoh, K.; Chun, W.; Ohmiya, Y. Expressed sequence tag analysis of the dinoflagellate Lingulodinium polyedrum during dark phase. Photochem. Photobiol. 2004, 80, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Fogel, M.; Hastings, J. A substrate-binding protein in the Gonyaulax bioluminescence reaction. Arch. Biochem. Biophys. 1971, 142, 310–321. [Google Scholar] [CrossRef]
- Morse, D.; Pappenheimer, A.; Hastings, J. Role of a luciferin-binding protein in the circadian bioluminescent reaction of Gonyaulax polyedra. J. Biol. Chem. 1989, 264, 11822–11826. [Google Scholar]
- Liu, L.; Hastings, J. Two different domains of the luciferase gene in the heterotrophic dinoflagellate Noctiluca scintillans occur as two separate genes in photosynthetic species. Proc. Natl. Acad. Sci. USA 2007, 104, 696–701. [Google Scholar] [CrossRef]
- Erdner, D.; Anderson, D. Global transcriptional profiling of the toxic dinoflagellate Alexandrium fundyense using massively parallel signature sequencing. BMC Genom. 2006, 7, 88. [Google Scholar] [CrossRef]
- Uribe, P.; Fuentes, D.; Valdés, J.; Shmaryahu, A.; Zúñiga, A.; Holmes, D.; Valenzuela, P. Preparation and analysis of an expressed sequence tag library from the toxic dinoflagellate Alexandrium catenella. Mar. Biotechnol. 2008, 10, 692–700. [Google Scholar] [CrossRef]
- Jaeckisch, N.; Yang, I.; Wohlrab, S.; Glöckner, G.; Kroymann, J.; Vogel, H.; Cembella, A.; John, U. Comparative genomic and transcriptomic characterization of the toxigenic marine dinoflagellate Alexandrium ostenfeldii. PLoS ONE 2011, 6, e28012. [Google Scholar] [CrossRef] [PubMed]
- Fajardo, C.; Amil-Ruiz, F.; Fuentes-Almagro, C.; De Donato, M.; Martínez-Rodriguez, G.; Escobar-Niño, A.; Carrasco, R.; Mancera, J.M.; Fernandez-Acero, F.J. An “omic” approach to Pyrocystis lunula: New insights related with this bioluminescent dinoflagellate. J. Proteom. 2019, 209, 103502. [Google Scholar] [CrossRef] [PubMed]
- Hackett, J.; Scheetz, T.; Yoon, H.; Soares, M.; Bonaldo, M.; Casavant, T.; Bhattacharya, D. Insights into a dinoflagellate genome through expressed sequence tag analysis. BMC Genom. 2005, 6, 80. [Google Scholar] [CrossRef] [PubMed]
- Valiadi, M.; Iglesias-Rodriguez, M. Diversity of the luciferin binding protein gene in bioluminescent dinoflagellates–Insights from a new gene in Noctiluca scintillans and sequences from Gonyaulacoid genera. J. Eukaryot. Microbiol. 2014, 61, 134–145. [Google Scholar] [CrossRef]
- Valiadi, M.; Iglesias-Rodriguez, M. Understanding bioluminescence in dinoflagellates: How far have we come? Microorganisms 2013, 1, 3–25. [Google Scholar] [CrossRef]
- Okamoto, O.; Liu, L.; Robertson, D.; Hastings, J. Members of a dinoflagellate luciferase gene family differ in synonymous substitution rates. Biochemistry 2001, 40, 15862–15868. [Google Scholar] [CrossRef]
- Nakamura, H.; Kishi, Y.; Shimomura, O.; Morse, D.; Hastings, J. Structure of dinoflagellate luciferin and its enzymic and nonenzymic air-oxidation products. J. Am. Chem. Soc. 1989, 111, 7607–7611. [Google Scholar] [CrossRef]
- Topalov, G.; Kishi, Y. Chlorophyll catabolism leading to the skeleton of dinoflagellate and krill luciferins: Hypothesis and model studies. Angew. Chem. Int. Ed. 2001, 40, 3892–3894. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Horiguchi, T. Culture of the heterotrophic dinoflagellate Protoperidinium crassipes (Dinophyceae) with noncellular food items. J. Phycol. 2008, 44, 1090–1092. [Google Scholar] [CrossRef]
- Wu, C.; Akimoto, H.; Ohmiya, Y. Tracer studies on dinoflagellate luciferin with [15N]-glycine and [15N]-L-glutamic acid in the dinoflagellate Pyrocystis lunula. Tetrahedron Lett. 2003, 44, 1263–1266. [Google Scholar] [CrossRef]
- Fresneau, C.; Arrio, B. Pyrocystis lunula bioluminescence: Physicochemical characterization of the luciferin precursor. Arch. Biochem. Biophys. 1988, 265, 22–27. [Google Scholar] [CrossRef]
- Edgar, R. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: Oxford, UK, 2000; ISBN 0-19-513584-9. [Google Scholar]
- Kelley, L.; Mezulis, S.; Yates, C.; Wass, M.; Sternberg, M. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Reynolds, C.; Islam, S.; Sternberg, M. EzMol: A web server wizard for the rapid visualization and image production of protein and nucleic acid structures. J. Mol. Biol. 2018, 430, 2244–2248. [Google Scholar] [CrossRef]
- Hastings, J.; Morin, J. Bioluminescence. In Neural and Integrative Animal Physiology; Prosser, C.L., Ed.; Wiley-Interscience: New York, NY, USA, 1991; pp. 131–170. [Google Scholar]
- Nicolas, M.; Nicolas, G.; Johnson, C.; Bassot, J.; Hastings, J. Characterization of the bioluminescent organelles in Gonyaulax polyedra (Dinoflagellates) after fast-freeze fixation and anti-luciferase immuno-gold staining. J. Cell Biol. 1987, 105, 723–735. [Google Scholar] [CrossRef]
- DeSa, R.; Hastings, J. The characterization of scintillons. Bioluminescent particles from the marine dinoflagellate Gonyaulax polyedra. J. Gen. Physiol. 1968, 51, 105–122. [Google Scholar] [CrossRef]
- Smith, S.; Morgan, D.; Musset, B.; Cherny, V.; Place, A.; Hastings, J.; Decoursey, T. Voltage gated proton channel in a dinoflagellate. Proc. Natl. Acad. Sci. USA 2011, 108, 18162–18167. [Google Scholar] [CrossRef]
- Rodriguez, J.; Haq, S.; Bachvaroff, T.; Nowak, K.; Nowak, S.; Morgan, D.; Cherny, V.; Sapp, M.; Bernstein, S.; Bolt, A.; et al. Identification of a vacuolar proton channel that triggers the bioluminescent flash in dinoflagellates. PLoS ONE 2017, 12, e0171594. [Google Scholar] [CrossRef]
- Seliger, H.; Biggley, W.; Swift, E. Absolute values of photon emission from the marine dinoflagellates Pyrodinium bahamense, Gonyaulax polyedra and Pyrocystis lunula. Photochem. Photobiol. 1969, 10, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Wilson, T.; Hastings, J. Bioluminescence. Ann. Rev. Cell Dev. Biol. 1998, 14, 197–230. [Google Scholar] [CrossRef] [PubMed]
- Hardeland, R. Circadian rhythms of bioluminescence in two species of Pyrocystis (Dinophyta). Measurements in cell populations and in single cells. J. Interdiscipl. Cycle Res. 1982, 13, 49–54. [Google Scholar] [CrossRef]
- Hardeland, R.; Nord, P. Visualization of free-running circadian rhythms in the dinoflagellate Pyrocystis noctiluca. Mar. Behav. Physiol. 1984, 11, 199–207. [Google Scholar] [CrossRef]
- Widder, E.; Case, J. Distribution of sub-cellular bioluminiscent sources in a dinoflagellate, Pyrocystis fusiformis. Biol Bull. 1982, 162, 423–448. [Google Scholar] [CrossRef]
- Eckert, R. Bioelectric control of bioluminescence in the dinoflagellate Noctiluca: Asynchronous flash initiation by a propagated triggering potential. Science 1965, 147, 1142–1145. [Google Scholar] [CrossRef]
- Widder, E.; Case, J. Bioluminescence excitation in a dinoflagellate. In Bioluminescence Current Perspectives; Nealson, K., Ed.; Burgess Pub. Co.: Minneapolis, MN, USA, 1981; pp. 125–132. [Google Scholar]
- Latz, M.; Bovard, M.; Groisman, A. Bioluminescent response of individual dinoflagellate cells to hydrodynamic stress measured with millisecond resolution in a microfluidic device. J. Exp. Biol. 2008, 211, 2865–2875. [Google Scholar] [CrossRef]
- Tesson, B.; Latz, M. Mechanosensitivity of a rapid bioluminescence reporter system assessed by atomic force microscopy. Biophys. J. 2015, 108, 1341–1351. [Google Scholar] [CrossRef]
- Morishita, H.; Ohashi, S.; Oku, T.; Nakajima, Y.; Kojima, S.; Ryufuku, M.; Ohmiya, Y. Cloning and characterization of an active fragment of luciferase from a luminescent marine alga, Pyrocystis lunula. Photochem. Photobiol. 2002, 75, 311–315. [Google Scholar] [CrossRef]
- Liu, L.; Wilson, T.; Hastings, J. Molecular evolution of dinoflagellate luciferases, enzymes with three catalytic domains in a single polypeptide. Proc. Natl. Acad. Sci. USA 2004, 101, 16555–16560. [Google Scholar] [CrossRef]
- Valiadi, M.; Iglesias-Rodriguez, D.; Amorim, A. Distribution and genetic diversity of the luciferase gene within marine dinoflagellates. J. Phycol. 2012, 48, 826–836. [Google Scholar] [CrossRef] [PubMed]
- Orr, R.; Murray, S.; Stuken, A.; Rhodes, L.; Jakobsen, K. When naked became armored: An eight-gene phylogeny reveals monophyletic origin of theca in dinoflagellates. PLoS ONE 2012, 7, e50004. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, L.; Hong, R.; Robertson, D.; Hastings, J. N-terminal intramolecularly conserved histidines of three domains in Gonylaulax luciferase are responsible for loss of activity in the alkaline region. Biochemistry 2001, 40, 1844–1849. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Hong, R.; Hastings, J. Three functional luciferase domains in a single polypeptide chain. Proc. Natl. Acad. Sci. USA 1997, 94, 8954–8958. [Google Scholar] [CrossRef]
- Schultz, L.; Liu, L.; Cegielski, M.; Hastings, J. Crystal structure of a pH-regulated luciferase catalyzing the bioluminescent oxidation of an open tetrapyrrole. Proc. Natl. Acad. Sci. USA 2005, 102, 1378–1383. [Google Scholar] [CrossRef]
- Wilson, T.; Hastings, J. Bioluminescence: Living Lights, Lights for Living; Harvard University Press: Cambridge, UK, 2013; ISBN 978-0674067165. [Google Scholar]
- Donnan, P.H.; Ngo, P.D.; Mansoorabadi, S.O. Constant pH Accelerated Molecular Dynamics Investigation of the pH Regulation Mechanism of Dinoflagellate Luciferase. Biochemistry 2017, 57, 295–299. [Google Scholar] [CrossRef]
- Jackson, A. Tandem gene arrays in Trypanosoma brucei: Comparative phylogenomic analysis of duplicate sequence variation. BMC Evol. Biol. 2007, 7, 54. [Google Scholar] [CrossRef]
- Lin, S. Genomic understanding of dinoflagellates. Res. Microbiol. 2011, 162, 551–569. [Google Scholar] [CrossRef]
- Lee, D.; Mittag, M.; Sczekan, S.; Morse, D.; Hastings, J. Molecular cloning and genomic organization of a gene for luciferin-binding protein from the dinoflagellate Gonyaulax polyedra. J. Biol. Chem. 1993, 268, 8842–8850. [Google Scholar]
- Nicolas, M.; Morse, D.; Bassot, J.; Hastings, J. Colocalization of luciferin binding protein and luciferase to the scintillons of Gonyaulax polyedra revealed by immunolabeling after fast-freeze fixation. Protoplasma 1991, 160, 159–166. [Google Scholar] [CrossRef]
- Bachvaroff, T.; Place, A. From stop to start: Tandem gene arrangement, copy number and trans-splicing sites in the dinoflagellate Amphidinium carterae. PLoS ONE 2008, 3, e2929. [Google Scholar] [CrossRef] [PubMed]
- Slamovits, C.; Keeling, P. Widespread recycling of processed cDNAs in dinoflagellates. Curr. Biol. 2008, 18, R550–R552. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Liu, Y. Theoretical study of dinoflagellate bioluminescence. Photochem. Photobiol. 2017, 93, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Dunlap, J.; Hastings, J. Biochemistry of dinoflagellate bioluminescence: Purification and characterization of dinoflagellate luciferin from Pyrocystis lunula. Biochemistry 1981, 20, 983–989. [Google Scholar] [CrossRef] [PubMed]
- Hamman, J.; Seliger, H. The mechanical triggering of bioluminescence in marine dinoflagellates: Chemical basis. J. Cell. Physiol. 1972, 80, 397–408. [Google Scholar] [CrossRef]
- Fresneau, C.; Hill, M.; Lescure, N.; Arrio, B.; Dupaix, A.; Volfin, P. Dinoflagellate luminescence: Purification of a NAD(P)H-dependent reductase and of its substrate. Arch. Biochem. Biophys. 1986, 251, 495–503. [Google Scholar] [CrossRef]
- Buchanan, B. Thioredoxin System and Glutaredoxin Systems; Raven Press: New York, NY, USA, 1985. [Google Scholar]
- Bennoun, P. Evidence for a respiratory chain in the chloroplast. Proc. Natl. Acad. Sci. USA 1982, 79, 4352–4356. [Google Scholar] [CrossRef]
- Peschek, G. The cytochrome oxidase-hydrogenase relationship in cyanobacteria. Naturwissensch 1982, 69, 599–600. [Google Scholar] [CrossRef]
- Omata, T.; Murata, N. Electron-transport reactions in cytoplasmic and thylakoid membranes prepared from the cyanobacteria (blue-green algae) Anacystis nidulans and Synechocystis PCC 6714. Biochim. Biophys. Acta 1985, 810, 354–361. [Google Scholar] [CrossRef]
- Doussiere, J.; Sainsard-Chanet, A.; Vignais, P. The respiratory chain of P. tetraurelia in wild-type and mutant Cl1. Biochim. Biophys. Acta 1979, 548, 236–252. [Google Scholar] [CrossRef]
- Ngo, P.; Mansoorabadi, S. Investigation of the Dinoflagellate Bioluminescence Mechanism: Chemically Initiated Electron Exchange Luminescence or Twisted Intramolecular Charge Transfer? ChemPhotoChem 2017, 1, 383–387. [Google Scholar] [CrossRef]
- Koo, J.; Schmidt, S.; Schuster, G. Bioluminescence of the firefly: Key steps in the formation of the electronically excited state for model systems. Proc. Natl. Acad. Sci. USA 1978, 75, 30–33. [Google Scholar] [CrossRef] [PubMed]
- Vacher, M.; Galvaán, I.; Ding, B.W.; Schramm, S.; Berraud-Pache, R.; Naumov, P.; Ferreé, N.; Liu, Y.J.; Navizet, I.; Roca-Sanjuaán, D.; et al. Chemi-and bioluminescence of cyclic peroxides. Chem. Rev. 2018, 118, 6927–6974. [Google Scholar] [CrossRef] [PubMed]
- Grabowski, Z.; Rotkiewicz, K.; Rettig, W. Structural changes accompanying intramolecular electron transfer: Focus on twisted intramolecular charge-transfer states and structures. Chem. Rev. 2003, 103, 3899–4032. [Google Scholar] [CrossRef]
- Janouskovec, J.; Gavelis, G.; Burki, F.; Dinh, D.; Bachvaroff, T.; Gornik, S.; Bright, K.; Imanian, B.; Strom, S.; Delwiche, C.; et al. Major transitions in dinoflagellate evolution unveiled by phylotranscriptomics. Proc. Natl. Acad. Sci. USA 2017, 114, E171–E180. [Google Scholar] [CrossRef]
- Gornik, S.; Febrimarsa; Cassin, A.M.; MacRae, J.; Ramaprasad, A.; Rchiad, Z.; McConville, M.; Bacic, A.; McFadden, G.; Pain, A.; et al. Endosymbiosis undone by stepwise elimination of the plastid in a parasitic dinoflagellate. Proc. Natl. Acad. Sci. USA 2015, 112, 5767–5772. [Google Scholar] [CrossRef]
- Martin, W.F.; Bryant, D.A.; Beatty, J.T. A physiological perspective on the origin and evolution of photosynthesis. FEMS Microbiol. Rev. 2018, 42, 205–231. [Google Scholar] [CrossRef]
- Fujita, Y.; Tsujimoto, R.; Aoki, R. Evolutionary Aspects and Regulation of Tetrapyrrole Biosynthesis in Cyanobacteria under Aerobic and Anaerobic Environments. Life 2015, 5, 1172–1203. [Google Scholar] [CrossRef]
- Okamoto, O.; Hastings, J. Genome-wide analysis of redox-regulated genes in a dinoflagellate. Gene 2003, 4, 73–81. [Google Scholar] [CrossRef]
- Allocati, N.; Federici, L.; Masulli, M.; Di Ilio, C. Glutathione transferases in bacteria. FEBS J. 2009, 276, 58–75. [Google Scholar] [CrossRef]
- Atkinson, H.; Babbitt, P. Glutathione transferases are structural and functional outliers in the thioredoxin fold. Biochemistry 2009, 48, 11108–11116. [Google Scholar] [CrossRef] [PubMed]
- Hartley, D.; Ruth, J.; Petersen, D. The hepatocellular metabolism of 4-hydroxynonenal by alcohol dehydrogenase, aldehyde dehydrogenase and glutathione S-transferase. Arch. Biochem. Biophys. 1995, 16, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, Y.; Hasegawa, A.; Taninaka, A.; Mizutani, M.; Sugimoto, Y. NADPH-dependent reductases involved in the detoxification of reactive carbonyls in plants. J. Biol. Chem. 2011, 286, 6999–7009. [Google Scholar] [CrossRef] [PubMed]
- Abrahams, M.; Townsend, L. Bioluminescence in dinoflagellates: A test of the burglar alarm hypothesis. Ecology 1993, 74, 258–260. [Google Scholar] [CrossRef]
- Buskey, E.; Mills, L.; Swift, E. The effects of dinoflagellate bioluminescence on the swimming behavior of a marine copepod. Limnol. Oceanogr. 1983, 28, 575–579. [Google Scholar] [CrossRef]
- Marcinko, C.; Painter, S.; Martin, A.; Allen, J. A review of the measurement and modelling of dinoflagellate bioluminescence. Prog. Oceanogr. 2013, 109, 117–129. [Google Scholar] [CrossRef]
- Widder, E. Bioluminescence in the ocean: Origins of biological, chemical, and ecological diversity. Science 2010, 328, 704–708. [Google Scholar] [CrossRef]
- Latz, M.; Case, J.; Gran, R. Excitation of bioluminescence by laminar fluid shear associated with simple Couette flow. Limnol. Oceanogr. 1994, 39, 1424–1439. [Google Scholar] [CrossRef]
- Maldonado, E.; Latz, M. Shear-stress dependence of dinoflagellate bioluminescence. Biol. Bull. 2007, 212, 242–249. [Google Scholar] [CrossRef]
- Buskey, E.; Mann, C.; Swift, E. Photophobic responses of calanoid copepods: Possible adaptive value. J. Plankton Res. 1987, 9, 857–870. [Google Scholar] [CrossRef]
- Burkenroad, M. A possible function of bioluminescence. J. Mar. Res. 1943, 5, 161–164. [Google Scholar]
- Prevett, A.; Lindström, J.; Xu, J.; Karlson, B.; Selander, E. Grazer-induced bioluminescence gives dinoflagellates a competitive edge. Curr. Biol. 2019, 29, R564–R565. [Google Scholar] [CrossRef] [PubMed]
- Lindström, J.; Grebner, W.; Rigby, K.; Selander, E. Effects of predator lipids on dinoflagellate defence mechanisms—increased bioluminescence capacity. Sci. Rep. 2017, 7, 13104. [Google Scholar] [CrossRef] [PubMed]
- Hanley, K.; Widder, E. Bioluminescence in dinoflagellates: Evidence that the adaptive value of bioluminescence in dinoflagellates is concentration dependent. Photochem. Photobiol. 2017, 93, 519–530. [Google Scholar] [CrossRef] [PubMed]


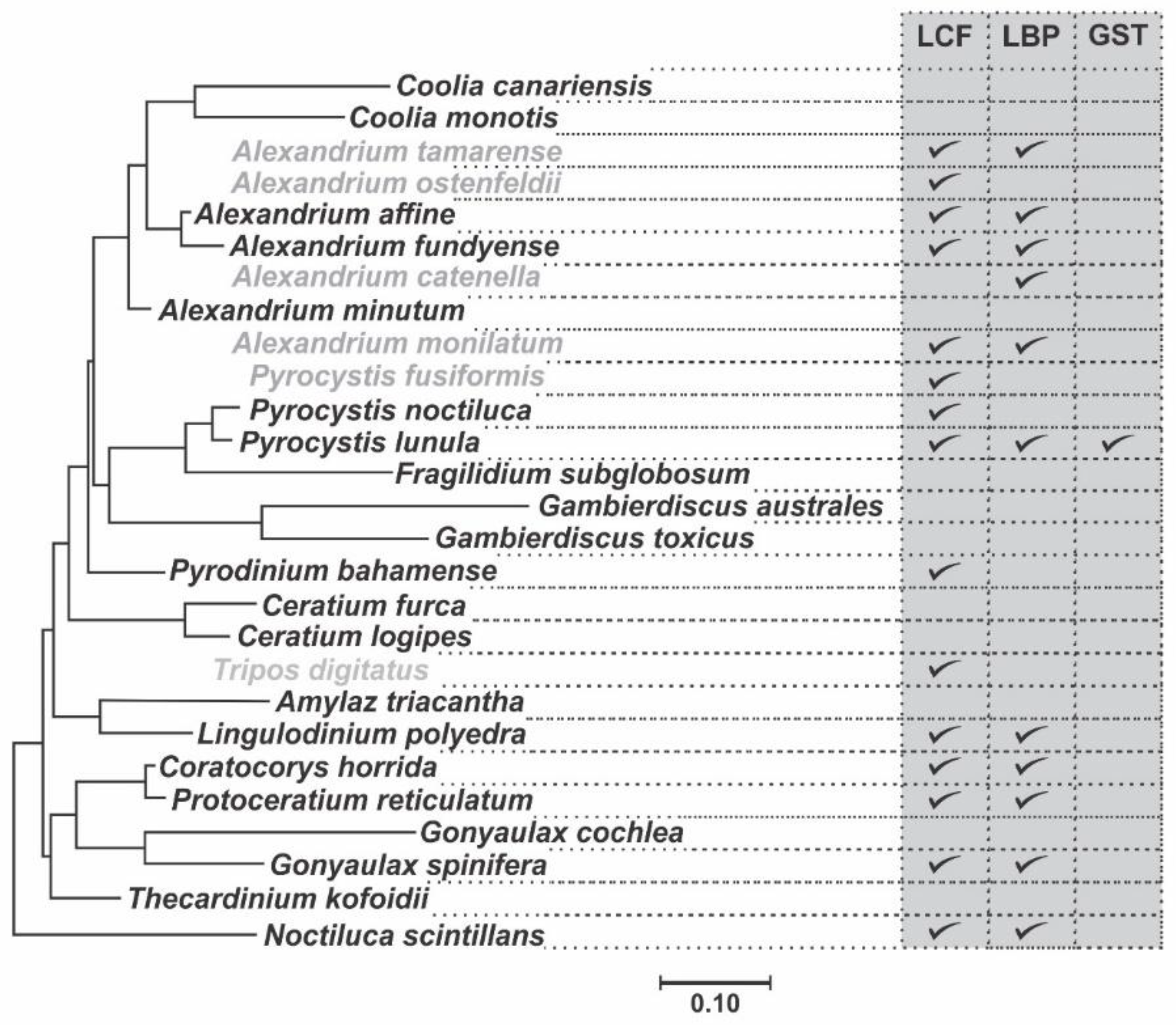
| Species | LCF | LBP | GST |
|---|---|---|---|
| Alexandrium affine | AAV35377 | AFN26992 | / |
| Alexandrium catenella | / | ABY78836 | / |
| Alexandrium fundyense | AEW67906 | AFN26994 | / |
| Alexandrium monilatum | AEW67931 | AFN26995 | / |
| Alexandrium ostenfeldii | AOG16037 | / | / |
| Alexandrium tamarense | AAV35378 | AFN27008 | / |
| Coratocorys horrida | AEW67919 | AFN27015 | / |
| Gonyaulax spinifera | ABO61069 | / | / |
| Lingulodinium polyedra | O77206 | AAA29165, AAA29166 | / |
| Noctiluca scintillans | AED02505 | AHB24369 | / |
| Protoceratium reticulatum | AAV35381 | AFN27016 | / |
| Pyrocystis fusiformis | AAV35379 | / | / |
| Pyrocystis lunula | AAL40676, AAL40677, AAL406778 | MN259726, MN259727 | AAN85429 |
| Pyrocystis noctiluca | AAV35380 | / | / |
| Pyrodinium bahamense | KX377172 | / | / |
| Tripos digitatus | AEW67915 | / | / |
| Substrates | Maximum Absorption (nm) | Maximum Fluorescence Excitation (nm) | Maximum Fluorescence Emission (nm) |
|---|---|---|---|
| P630 oxidized | 630-370-315-250 | 630 | 675 |
| P630 reduced | 390 | 390 | 480 |
| Luciferin | 390-250 | 390 | 480 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fajardo, C.; De Donato, M.; Rodulfo, H.; Martinez-Rodriguez, G.; Costas, B.; Mancera, J.M.; Fernandez-Acero, F.J. New Perspectives Related to the Bioluminescent System in Dinoflagellates: Pyrocystis lunula, a Case Study. Int. J. Mol. Sci. 2020, 21, 1784. https://doi.org/10.3390/ijms21051784
Fajardo C, De Donato M, Rodulfo H, Martinez-Rodriguez G, Costas B, Mancera JM, Fernandez-Acero FJ. New Perspectives Related to the Bioluminescent System in Dinoflagellates: Pyrocystis lunula, a Case Study. International Journal of Molecular Sciences. 2020; 21(5):1784. https://doi.org/10.3390/ijms21051784
Chicago/Turabian StyleFajardo, Carlos, Marcos De Donato, Hectorina Rodulfo, Gonzalo Martinez-Rodriguez, Benjamin Costas, Juan Miguel Mancera, and Francisco Javier Fernandez-Acero. 2020. "New Perspectives Related to the Bioluminescent System in Dinoflagellates: Pyrocystis lunula, a Case Study" International Journal of Molecular Sciences 21, no. 5: 1784. https://doi.org/10.3390/ijms21051784
APA StyleFajardo, C., De Donato, M., Rodulfo, H., Martinez-Rodriguez, G., Costas, B., Mancera, J. M., & Fernandez-Acero, F. J. (2020). New Perspectives Related to the Bioluminescent System in Dinoflagellates: Pyrocystis lunula, a Case Study. International Journal of Molecular Sciences, 21(5), 1784. https://doi.org/10.3390/ijms21051784










