The ArathEULS3 Lectin Ends up in Stress Granules and Can Follow an Unconventional Route for Secretion
Abstract
1. Introduction
2. Results
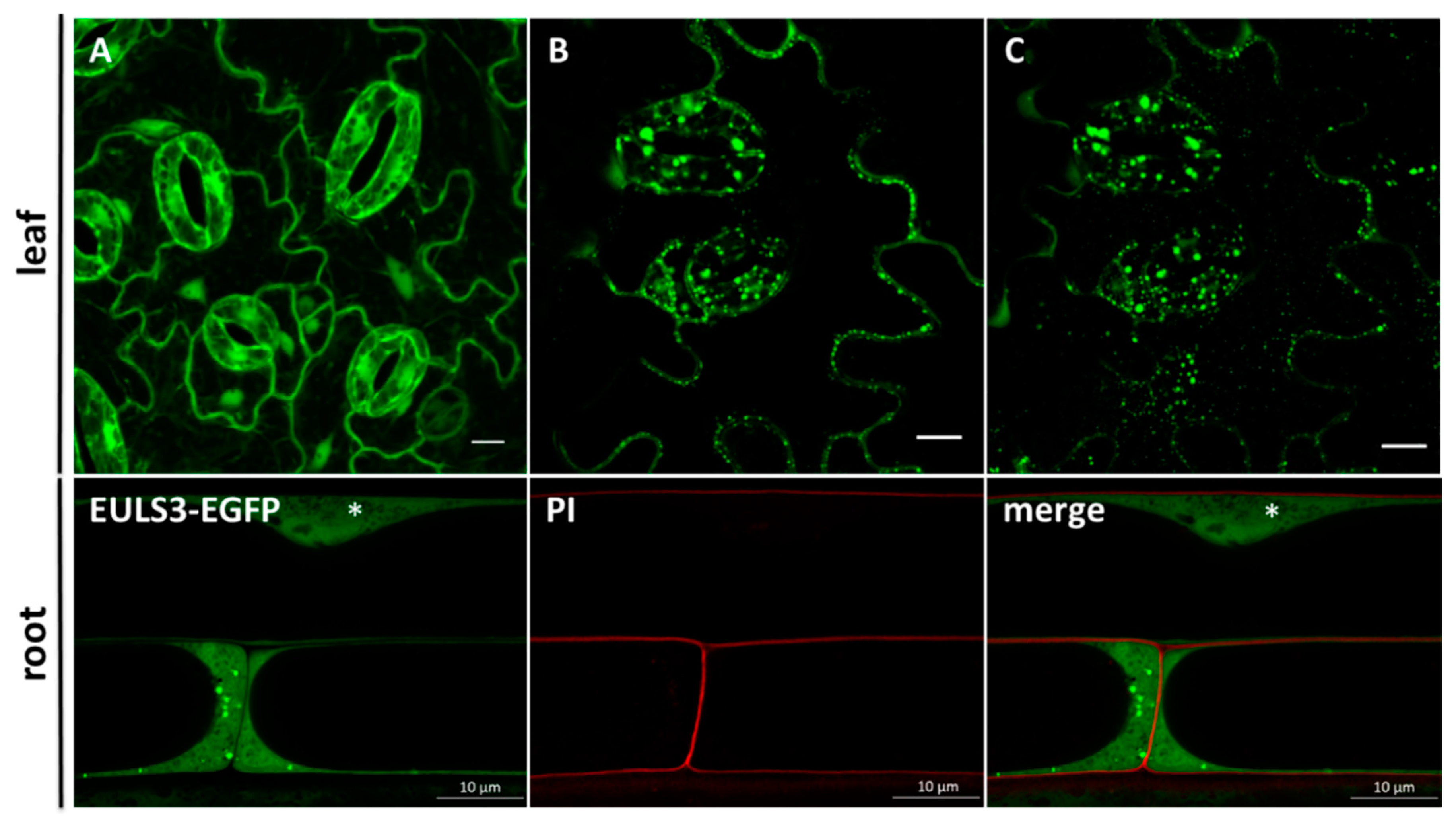
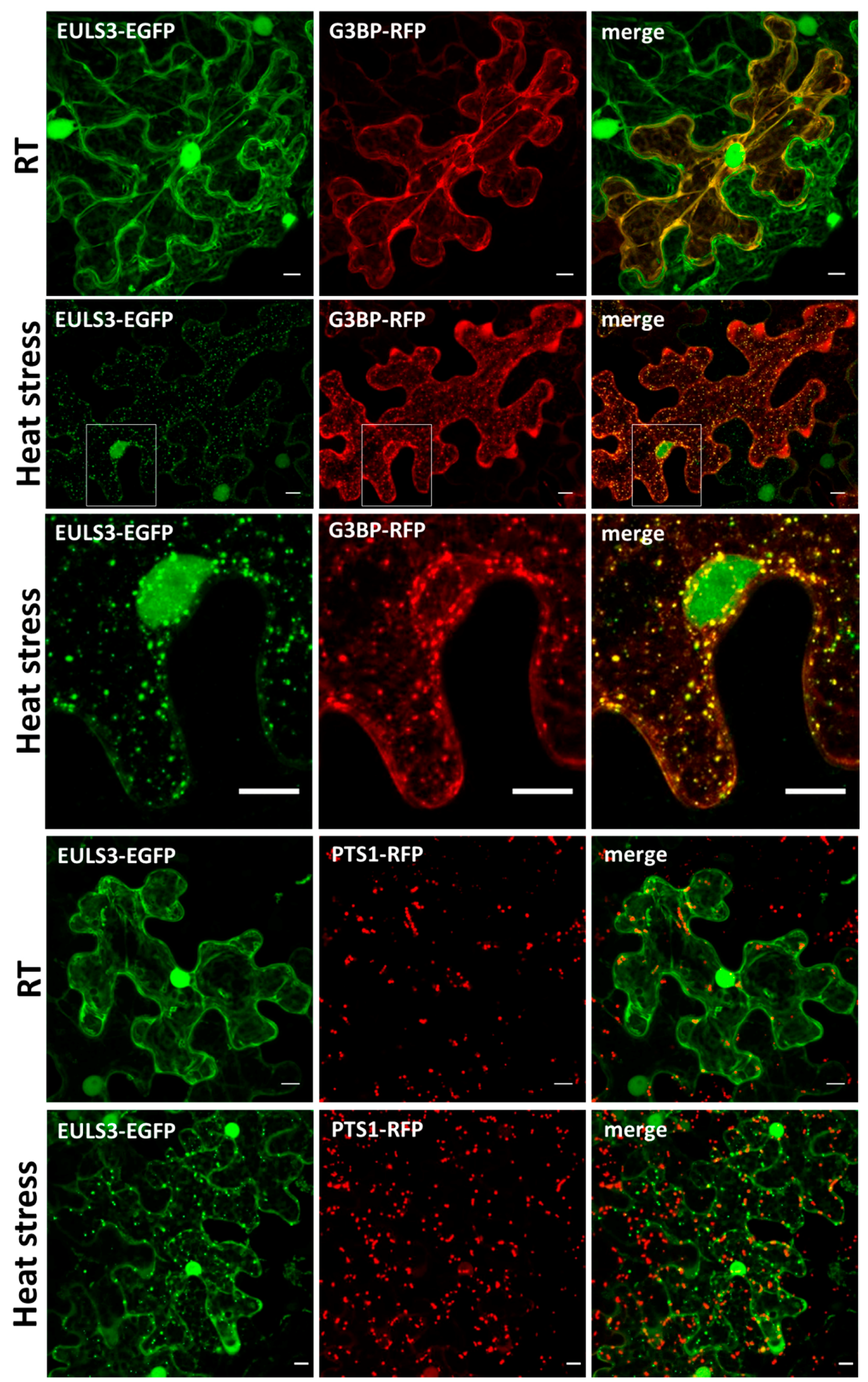
2.1. Nucleo-cytoplasmic ArathEULS3 Relocates to Stress Granules
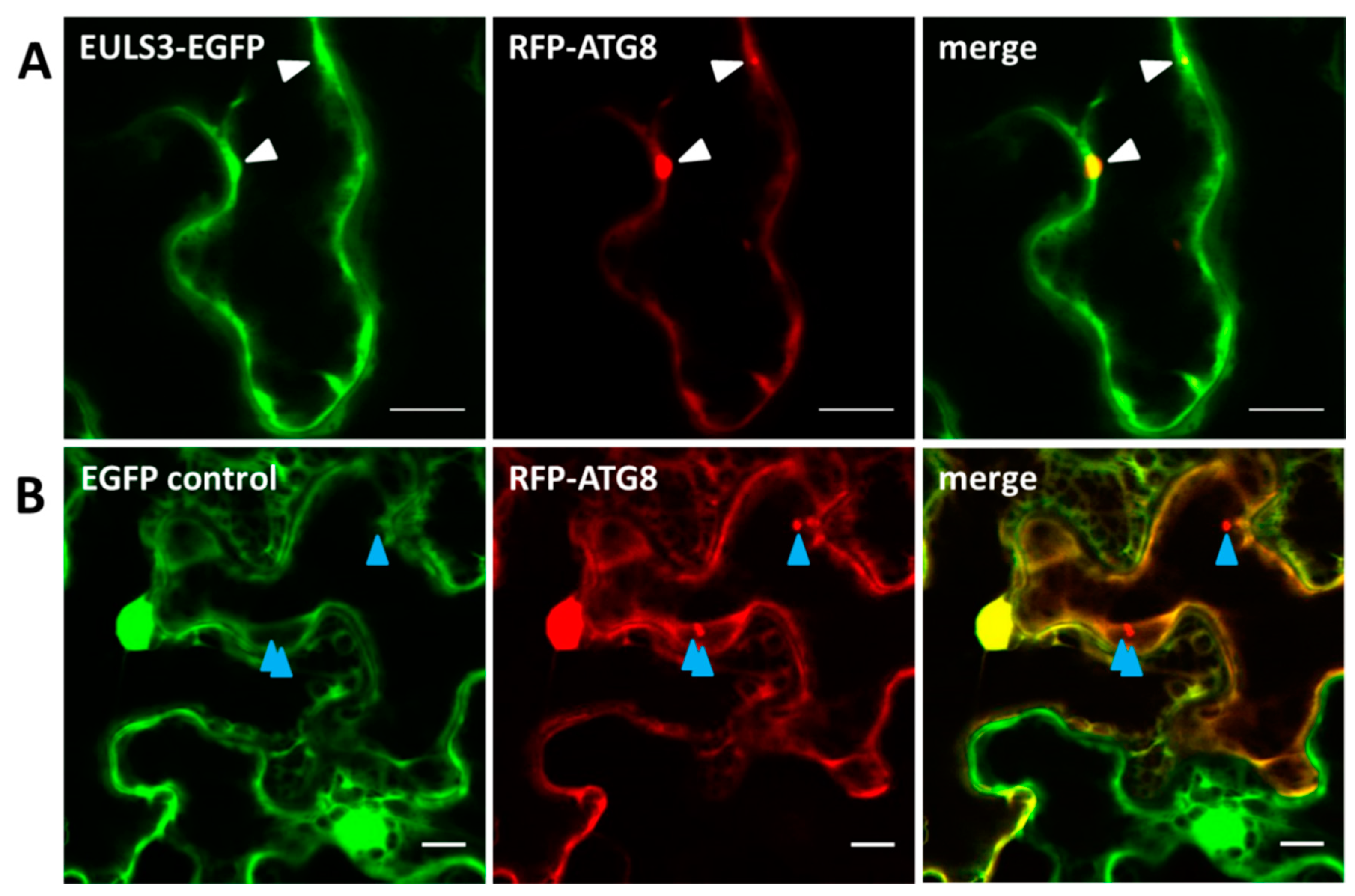
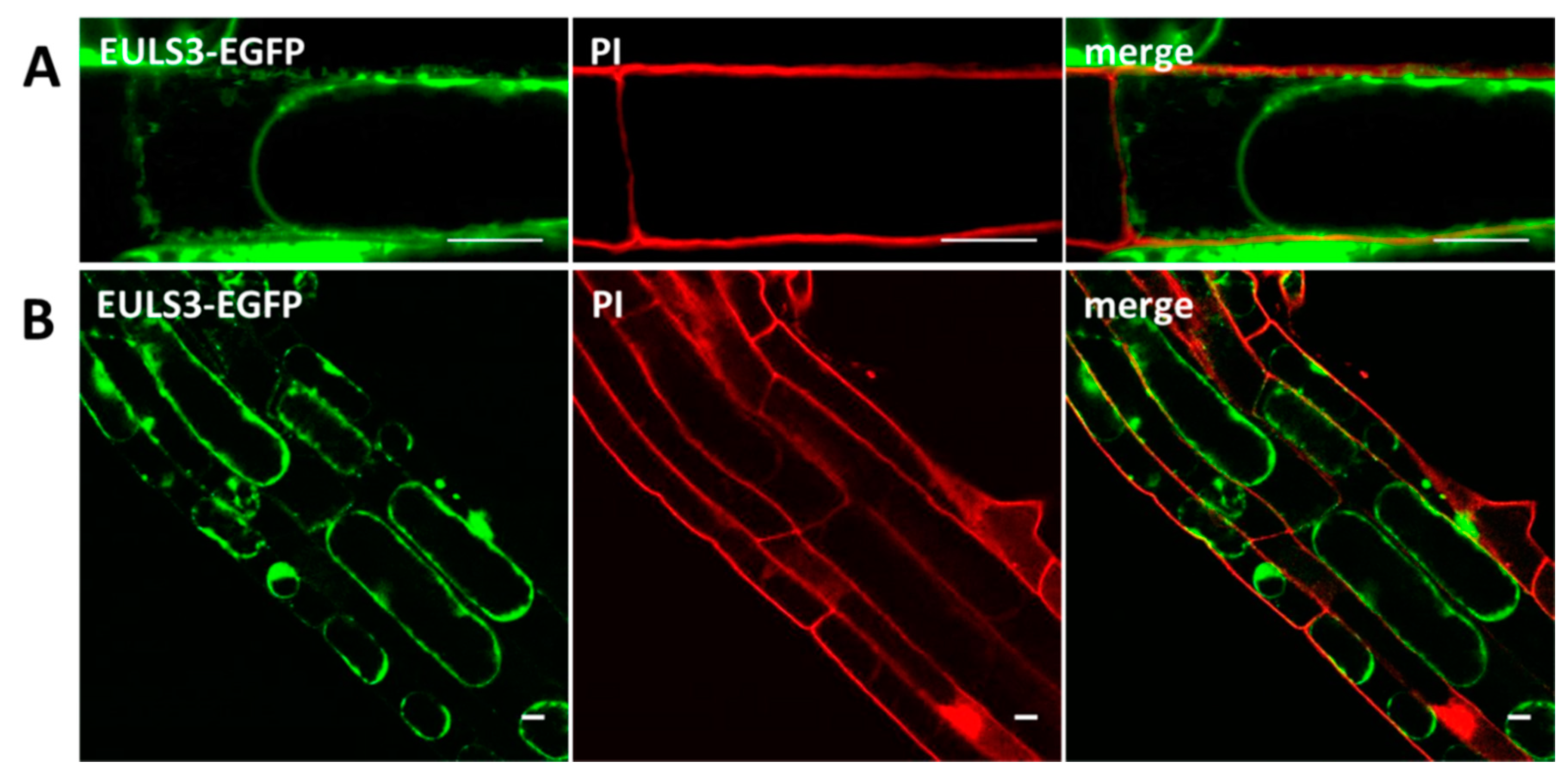
2.2. ArathEULS3 Is a Non-conventionally Secreted Protein
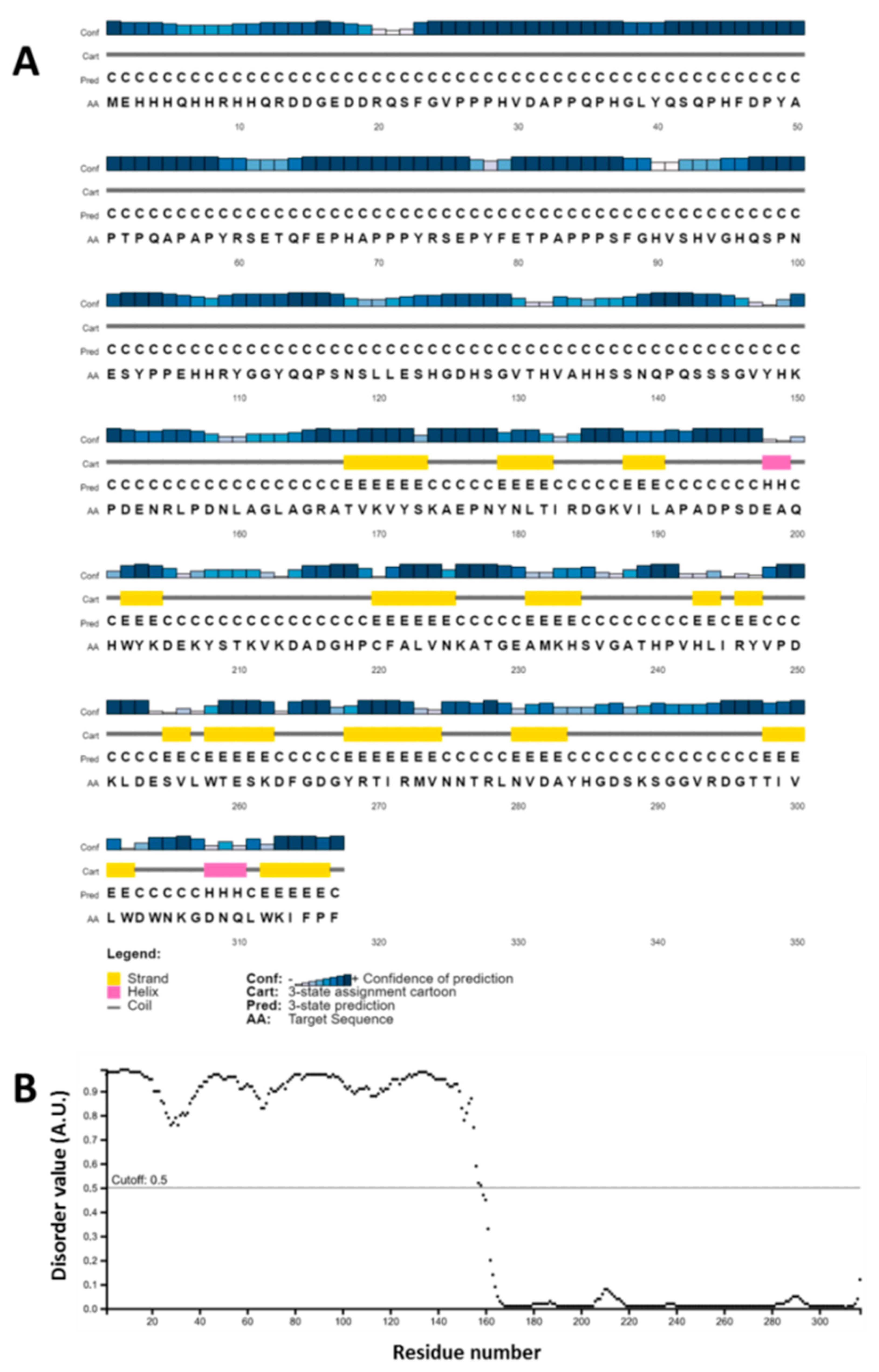
2.3. ArathEULS3 Sequence Analysis
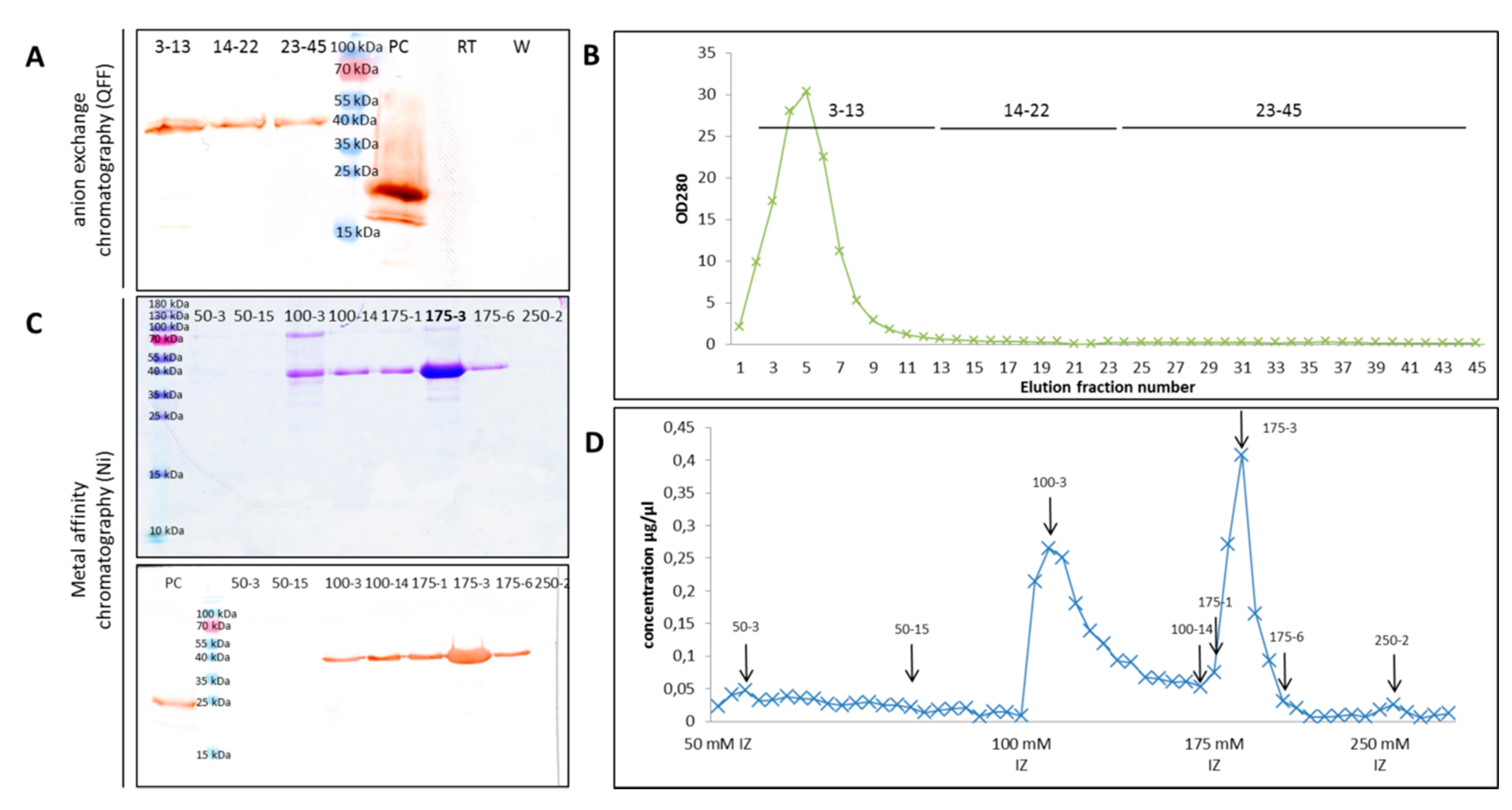
2.4. ArathEULS3 Protein Purification
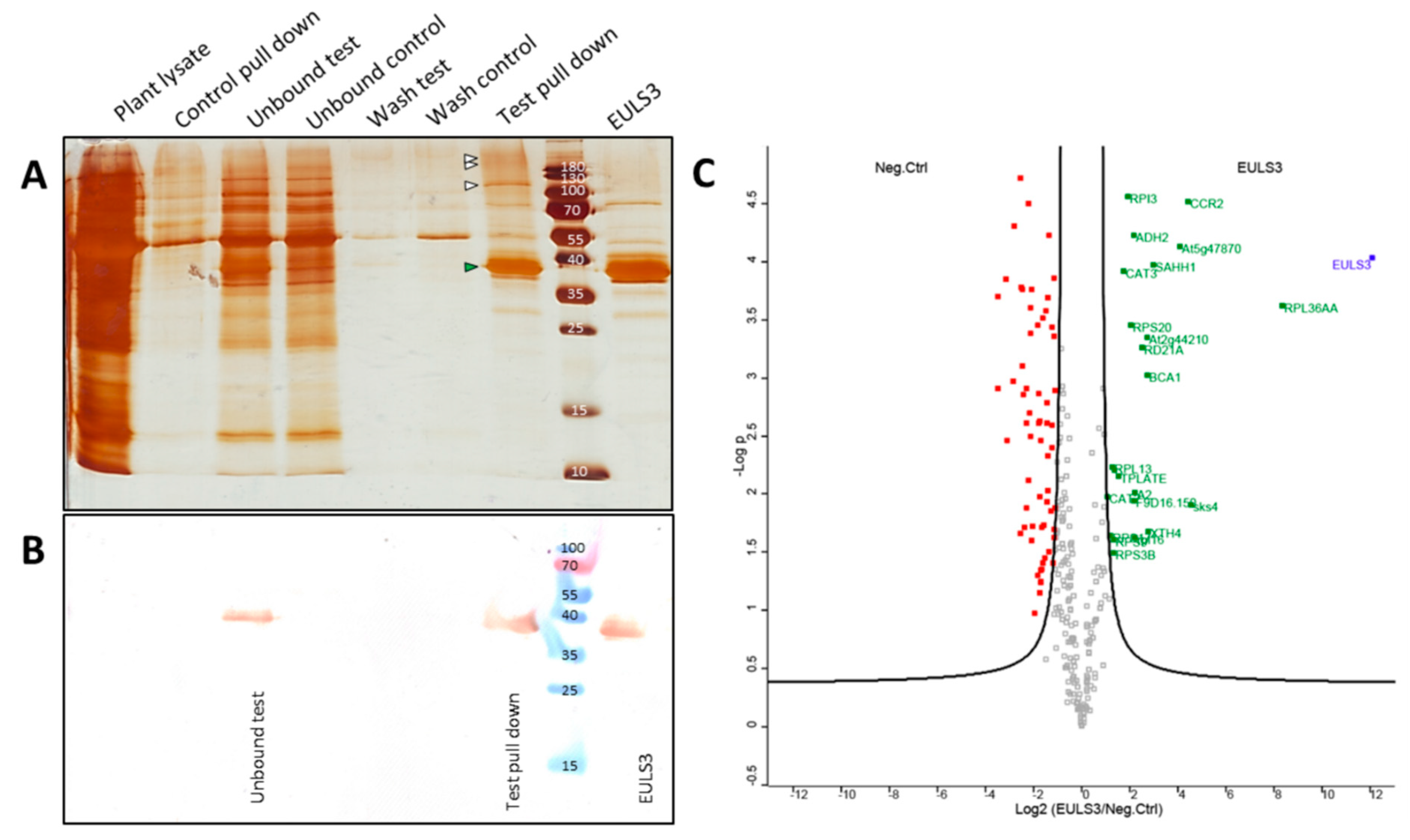
2.5. Pull-Down Analysis Reveals Several Novel Candidate Interactors for ArathEULS3
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Construction of Expression Vectors and Plant Transformation
4.3. Confocal Microscopy
4.4. Recombinant ArathEULS3 Protein Production and Purification
4.5. SDS-PAGE and Western Blot Analysis
4.6. Pull-down Analysis
4.7. LC−MS/MS Analysis
4.8. ArathEULS3 Sequence Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ABA | Abscisic Acid |
| EEA | Euonymus europeaus agglutinin |
| EGFP | enhanced Green Fluorescent Protein |
| EUL | Euonymus europaeus related lectins |
| EV | Extracellular Vesicle |
| EXPO | Exocyst-Positive Organelle |
| IDR | Intrinsically Disordered Region |
| LP | Leaderless Protein |
| MVB | Multivesicular Body |
| PB | Protein Body |
| PI | Propidium Iodide |
References
- Protter, D.S.W.; Parker, R. Principles and properties of stress granules. Trends Cell Biol. 2016, 26, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Beltran, E.; Moschou, P.N.; Smertenko, A.P.; Bozhkov, P.V. Tudor staphylococcal nuclease links formation of stress granules and processing bodies with mRNA catabolism in Arabidopsis. Plant Cell 2015, 27, 926–943. [Google Scholar] [CrossRef] [PubMed]
- Chantarachot, T.; Bailey-Serres, J. Polysomes, stress granules, and processing bodies: A dynamic triumvirate controlling cytoplasmic mRNA fate and function. Plant Physiol. 2018, 176, 254–269. [Google Scholar] [CrossRef] [PubMed]
- Nover, L.; Scharf, K.D.; Neumann, D. Formation of cytoplasmic heat shock granules in tomato cell cultures and leaves. Mol. Cell. Biol. 1983, 3, 1648–1655. [Google Scholar] [CrossRef] [PubMed]
- Collier, N.C.; Heuser, J.; Levy, M.A.; Schlesinger, M.J. Ultrastructural and biochemical analysis of the stress granule in chicken embryo fibroblasts. J. Cell Biol. 1988, 106, 1131–1139. [Google Scholar] [CrossRef]
- Hoyle, N.P.; Castelli, L.M.; Campbell, S.G.; Holmes, L.E.A.; Ashe, M.P. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J. Cell Biol. 2007, 179, 65–74. [Google Scholar] [CrossRef]
- Sorenson, R.; Bailey-Serres, J. Selective mRNA sequestration by OLIGOURIDYLATEBINDING PROTEIN 1 contributes to translational control during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 2373–2378. [Google Scholar] [CrossRef]
- Bhasin, H.; Hülskamp, M. ANGUSTIFOLIA, a plant homolog of CtBP/BARS localizes to stress granules and regulates their formation. Front. Plant Sci. 2017, 8, 1–17. [Google Scholar] [CrossRef]
- Marondedze, C.; Thomas, L.; Gehring, C.; Lilley, K.S. Changes in the Arabidopsis RNA-binding proteome reveal novel stress response mechanisms. BMC Plant Biol. 2019, 19, 1–11. [Google Scholar] [CrossRef]
- Kosmacz, M.; Gorka, M.; Schmidt, S.; Luzarowski, M.; Moreno, J.C.; Szlachetko, J.; Leniak, E.; Sokolowska, E.M.; Sofroni, K.; Schnittger, A.; et al. Protein and metabolite composition of Arabidopsis stress granules. New Phytol. 2019, 222, 1420–1433. [Google Scholar] [CrossRef]
- Abulfaraj, A.A.; Mariappan, K.; Bigeard, J.; Manickam, P.; Blilou, I.; Guo, X.; Al-Babili, S.; Pflieger, D.; Hirt, H.; Rayapuram, N. The Arabidopsis homolog of human G3BP1 is a key regulator of stomatal and apoplastic immunity. Life Sci. Alliance 2018, 1, 1–13. [Google Scholar] [CrossRef]
- Van Holle, S.; Van Damme, E.J.M. Signaling through plant lectins: Modulation of plant immunity and beyond. Biochem. Soc. Trans. 2018, 46, 217–233. [Google Scholar] [CrossRef]
- Van Damme, E.J.M.; Barre, A.; Rougé, P.; Peumans, W.J. Cytoplasmic/nuclear plant lectins: A new story. Trends Plant Sci. 2004, 9, 484–489. [Google Scholar] [CrossRef]
- Fouquaert, E.; Peumans, W.J.; Vandekerckhove, T.T.M.; Ongenaert, M.; Van Damme, E.J.M. Proteins with an Euonymus lectin-like domain are ubiquitous in Embryophyta. BMC Plant Biol. 2009, 9, 136. [Google Scholar] [CrossRef]
- Van Hove, J.; De Jaeger, G.; De Winne, N.; Guisez, Y.; Van Damme, E.J.M. The Arabidopsis lectin EULS3 is involved in stomatal closure. Plant Sci. 2015, 238, 312–322. [Google Scholar] [CrossRef] [PubMed]
- Van Hove, J.; Fouquaert, E.; Smith, D.F.; Proost, P.; Van Damme, E.J.M. Lectin activity of the nucleocytoplasmic EUL protein from Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2011, 414, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Krause, C.; Richter, S.; Knöll, C.; Jürgens, G. Plant secretome—From cellular process to biological activity. Biochim. Biophys. Acta Proteins Proteomics 2013, 1834, 2429–2441. [Google Scholar] [CrossRef] [PubMed]
- Halperin, W.; Jensen, W.A. Ultrastructural changes during growth and embryogenesis in carrot cell cultures. J. Ultrasructure Res. 1967, 18, 428–443. [Google Scholar] [CrossRef]
- Wang, J.; Ding, Y.; Wang, J.; Hillmer, S.; Miao, Y.; Lo, S.W.; Wang, X.; Robinson, D.G.; Jiang, L. EXPO, an exocyst-positive organelle distinct from multivesicular endosomes and autophagosomes, mediates cytosol to cell wall exocytosis in arabidopsis and tobacco cells. Plant Cell 2010, 22, 4009–4030. [Google Scholar] [CrossRef]
- Poulsen, C.P.; Dilokpimol, A.; Mouille, G.; Burow, M.; Geshi, N. Arabinogalactan glycosyltransferases target to a unique subcellular compartment that may function in unconventional secretion in plants. Traffic 2014, 15, 1219–1234. [Google Scholar] [CrossRef]
- Cui, Y.; Gao, J.; He, Y.; Jiang, L. Plant extracellular vesicles. Protoplasma 2019, 1, 10. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Wang, J.; Wang, J.; Stierhof, Y.D.; Robinson, D.G.; Jiang, L. Unconventional protein secretion. Trends Plant Sci. 2012, 17, 606–615. [Google Scholar] [CrossRef] [PubMed]
- Hatsugai, N.; Iwasaki, S.; Tamura, K.; Kondo, M.; Fuji, K.; Ogasawara, K.; Nishimura, M.; Hara-Nishimura, I. A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes Dev. 2009, 23, 2496–2506. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Qiao, L.; Wang, M.; He, B.; Lin, F.M.; Palmquist, J.; Huang, S.-D.; Jin, H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018, 360, 1126–1129. [Google Scholar] [CrossRef] [PubMed]
- An, Q.; Ehlers, K.; Kogel, K.H.; Van Bel, A.J.E.; Hückelhoven, R. Multivesicular compartments proliferate in susceptible and resistant MLA12-barley leaves in response to infection by the biotrophic powdery mildew fungus. New Phytol. 2006, 172, 563–576. [Google Scholar] [CrossRef]
- Wang, F.; Shang, Y.; Fan, B.; Yu, J.Q.; Chen, Z. Arabidopsis LIP5, a positive regulator of multivesicular body biogenesis, is a critical target of pathogen-responsive MAPK cascade in plant basal defense. PLoS Pathog. 2014, 10, 7. [Google Scholar] [CrossRef]
- Rutter, B.D.; Innes, R.W. Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol. 2017, 173, 728–741. [Google Scholar] [CrossRef]
- Movahed, N.; Cabanillas, D.G.; Wan, J.; Vali, H.; Laliberté, J.-F.; Zheng, H. Turnip mosaic virus components are released into the extracellular space by vesicles in infected leaves. Plant Physiol. 2019, 180, 1375–1388. [Google Scholar] [CrossRef]
- Regente, M.; Pinedo, M.; Clemente, H.S.; Balliau, T.; Jamet, E.; De La Canal, L. Plant extracellular vesicles are incorporated by a fungal pathogen and inhibit its growth. J. Exp. Bot. 2017, 68, 5485–5495. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, Z.; Mei, Q.; Chen, M. PSI: A Comprehensive and integrative approach for accurate plant subcellular localization prediction. PLoS ONE 2013, 8, 10. [Google Scholar] [CrossRef]
- Goldberg, T.; Hecht, M.; Hamp, T.; Karl, T.; Yachdav, G.; Ahmed, N.; Altermann, U.; Angerer, P.; Ansorge, S.; Balasz, K.; et al. LocTree3 prediction of localization. Nucleic Acids Res. 2014, 42, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Jamet, E.; Albenne, C.; Boudart, G.; Irshad, M.; Canut, H.; Pont-Lezica, R. Recent advances in plant cell wall proteomics. Proteomics 2008, 8, 893–908. [Google Scholar] [CrossRef] [PubMed]
- Oldfield, C.J.; Dunker, A.K. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem. 2014, 83, 553–584. [Google Scholar] [CrossRef] [PubMed]
- Van Der Lee, R.; Buljan, M.; Lang, B.; Weatheritt, R.J.; Daughdrill, G.W.; Dunker, A.K.; Fuxreiter, M.; Gough, J.; Gsponer, J.; Jones, D.T.; et al. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014, 114, 6589–6631. [Google Scholar] [CrossRef]
- Chen, J.W.; Romero, P.; Uversky, V.N.; Dunker, A.K. Conservation of intrinsic disorder in protein domains and families: II. Functions of conserved disorder. J. Proteome Res. 2006, 5, 888–898. [Google Scholar] [CrossRef]
- Koguchi, M.; Yamasaki, K.; Hirano, T.; Sato, M.H. Vascular plant one-zinc-finger protein 2 is localized both to the nucleus and stress granules under heat stress in Arabidopsis. Plant Signal. Behav. 2017, 12, 1–7. [Google Scholar] [CrossRef]
- Li, D.; Wang, X.; Yuan, D.; Zhang, L.; Jiang, X.; Tao, Z.; Li, Y.; Wang, J.; Li, X.; Yang, Y. Over-expression of ArathEULS3 confers ABA sensitivity and drought tolerance in Arabidopsis. Plant Cell. Tissue Organ Cult. 2014, 117, 431–442. [Google Scholar] [CrossRef]
- Reichel, M.; Liao, Y.; Rettel, M.; Ragan, C.; Evers, M.; Alleaume, A.M.; Horos, R.; Hentze, M.W.; Preiss, T.; Millar, A.A. In planta determination of the mRNA-binding proteome of Arabidopsis etiolated seedlings. Plant Cell 2016, 28, 2435–2452. [Google Scholar] [CrossRef]
- Masaki, S.; Yamada, T.; Hirasawa, T.; Todaka, D.; Kanekatsu, M. Proteomic analysis of RNA-binding proteins in dry seeds of rice after fractionation by ssDNA affinity column chromatography. Biotechnol. Lett. 2008, 30, 955–960. [Google Scholar] [CrossRef]
- Niklas, K.J.; Dunker, A.K.; Yruela, I. The evolutionary origins of cell type diversification and the role of intrinsically disordered proteins. J. Exp. Bot. 2018, 69, 1437–1446. [Google Scholar] [CrossRef]
- Tompa, P.; Kovacs, D. Intrinsically disordered chaperones in plants and animals. Biochem. Cell Biol. 2010, 88, 167–174. [Google Scholar] [CrossRef]
- Minezaki, Y.; Homma, K.; Kinjo, A.R.; Nishikawa, K. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J. Mol. Biol. 2006, 359, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Tsafou, K.; Tiwari, P.B.; Forman-Kay, J.D.; Metallo, S.J.; Toretsky, J.A. Targeting intrinsically disordered transcription factors: Changing the Paradigm. J. Mol. Biol. 2018, 430, 2321–2341. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Gee, H.Y.; Lee, M.G. Unconventional protein secretion—New insights into the pathogenesis and therapeutic targets of human diseases. J. Cell Sci. 2018, 131, jcs213686. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, L.; Gao, B.; Fan, H.; Jin, J.; Botella, M.A.; Jiang, L.; Lin, J. Golgi apparatus-localized synaptotagmin 2 is required for unconventional secretion in Arabidopsis. PLoS ONE 2011, 6, 11. [Google Scholar] [CrossRef]
- Cheng, F.Y.; Zamski, E.; Guo, W.W.; Pharr, D.M.; Williamson, J.D. Salicylic acid stimulates secretion of the normally symplastic enzyme mannitol dehydrogenase: A possible defense against mannitol-secreting fungal pathogens. Planta 2009, 230, 1093–1103. [Google Scholar] [CrossRef]
- Guarino, A.M.; Troiano, A.; Pizzo, E.; Bosso, A.; Vivo, M.; Pinto, G.; Amoresano, A.; Pollice, A.; La Mantia, G.; Calabrò, V. Oxidative stress causes enhanced secretion of YB-1 protein that restrains proliferation of receiving cells. Genes (Basel) 2018, 9, 513. [Google Scholar] [CrossRef]
- Keinath, N.F.; Kierszniowska, S.; Lorek, J.; Bourdais, G.; Kessler, S.A.; Shimosato-Asano, H.; Grossniklaus, U.; Schulze, W.X.; Robatzek, S.; Panstruga, R. PAMP (Pathogen-associated Molecular Pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J. Biol. Chem. 2010, 285, 39140–39149. [Google Scholar] [CrossRef]
- Gilbert, M.; Schulze, W.X. Global identification of protein complexes within the membrane proteome of Arabidopsis roots using a SEC-MS approach. J. Proteome Res. 2019, 18, 107–119. [Google Scholar] [CrossRef]
- Hervé, V.; Duruflé, H.; San Clemente, H.; Albenne, C.; Balliau, T.; Zivy, M.; Dunand, C.; Jamet, E. An enlarged cell wall proteome of Arabidopsis thaliana rosettes. Proteomics 2016, 16, 3183–3187. [Google Scholar] [CrossRef]
- Duruflé, H.; Clemente, H.S.; Balliau, T.; Zivy, M.; Dunand, C.; Jamet, E. Cell wall proteome analysis of Arabidopsis thaliana mature stems. Proteomics 2017, 17, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Duruflé, H.; Hervé, V.; Ranocha, P.; Balliau, T.; Zivy, M.; Chourré, J.; San Clemente, H.; Burlat, V.; Albenne, C.; Déjean, S.; et al. Cell wall modifications of two Arabidopsis thaliana ecotypes, Col and Sha, in response to sub-optimal growth conditions: An integrative study. Plant Sci. 2017, 263, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Seki, M.; Carninci, P.; Nishiyama, Y.; Hayashizaki, Y.; Shinozaki, K. High-efficiency cloning of Arabidopsis full-length cDNA by biotinylated CAP trapper. Plant J. 1998, 15, 707–720. [Google Scholar] [CrossRef] [PubMed]
- Zielinska, D.F.; Gnad, F.; Schropp, K.; Wiśniewski, J.R.; Mann, M. Mapping N-Glycosylation Sites across Seven Evolutionarily Distant Species Reveals a Divergent Substrate Proteome Despite a Common Core Machinery. Mol. Cell 2012, 46, 542–548. [Google Scholar] [CrossRef] [PubMed]
- Seki, M.; Mari, N.; AsakoIshida, K.; Satou, J.; Sakurai, M.; Nakajima, T.; Enju, M.; Akiyama, A.; Oono, K.; Muramatsu, Y.; et al. Functional annotation of a full-length Arabidopsis cDNA collection. Science 2002, 296, 141. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Wang, P.; Pleskot, R.; Zang, J.; Winkler, J.; Wang, J.; Yperman, K.; Zhang, T.; Wang, K.; Gong, J.; Guan, Y.; et al. Plant AtEH/Pan1 proteins drive autophagosome formation at ER-PM contact sites with actin and endocytic machinery. Nat. Commun. 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Krapp, S.; Greiner, E.; Amin, B.; Sonnewald, U.; Krenz, B. The stress granule component G3BP is a novel interaction partner for the nuclear shuttle proteins of the nanovirus pea necrotic yellow dwarf virus and geminivirus abutilon mosaic virus. Virus Res. 2017, 227, 6–14. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carrera, I.; Frise, E.; Verena, K.; Mark, L.; Tobias, P.; Stephan, P.; Curtis, R.; Stephan, S.; Benjamin, S.; et al. Fiji—An Open platform for biological image analysis. Nat. Methods 2012, 9, 676. [Google Scholar] [CrossRef]
- Jones, D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef]
- Jones, D.T.; Cozzetto, D. DISOPRED3: Precise disordered region predictions with annotated protein-binding activity. Bioinformatics 2015, 31, 857–863. [Google Scholar] [CrossRef] [PubMed]
- Cozzetto, D.; Minneci, F.; Currant, H.; Jones, D.T. FFPred 3: Feature-based function prediction for all Gene Ontology domains. Scientific Rep. 2016, 6, 31865. [Google Scholar] [CrossRef] [PubMed]
- Lobley, A.; Sadowski, M.I.; Jones, D.T. pGenTHREADER and pDomTHREADER: New methods for improved protein fold recognition and superfamily discrimination. Bioinformatics 2009, 25, 1761–1767. [Google Scholar] [CrossRef] [PubMed]
- Marchler-bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; Deweese-scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef]
- Buchan, D.W.A.; Jones, D.T. The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res. 2019, 47, W402–W407. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A.; Francisco, S. Protein structure modeling with MODELLER. In Functional Genomics; Humana Press: New York, NY, USA, 2017; pp. 39–54. [Google Scholar]
| Gene Locus | Full Name | MS Identifier | Score * |
|---|---|---|---|
| At2g39050 | Ricin B-like lectin EULS3 | EULS3 | 323.3 |
| At3g23390 | 60S ribosomal protein L36a | RPL36AA | 69.7 |
| At4g22010 | Pectin esterase-like protein SKU5 similar 4 | sks4 | 39.8 |
| At2g21660 | Glycine-rich RNA-binding protein 7 AtGRP7 | CCR2 | 37.5 |
| At5g47870 | DNA repair RAD52-like protein 2, chloroplastic | RAD52-2 | 26.3 |
| At4g13940 | Adenosyl homocysteinase | SAHH1 | 85.8 |
| At2g06850 | Xyloglucan endotransglucosylase /hydrolase | XTH4 | 26.8 |
| At3g01500 | Beta carbonic anhydrase 1 | BCA1 | 40.3 |
| At2g44210 | F4I1.2 uncharacterized protein | At2g44210 | 19.2 |
| At1g47128 | Cysteine proteinase | RD21A | 36.4 |
| AtCg00790 | 50S ribosomal protein L16 | rpl16 | 24.5 |
| At1g07920/30/40 | Elongation factor 1-alpha 1/2/3 | A1/A2/A3 | 107.2 |
| At4g23680 | At4g23680 | F9D16_150 | 12.4 |
| At5g43940 | S-(hydroxymethyl)glutathione dehydrogenase GSNORI | ADH2 | 33.1 |
| At3g15190 | 30S ribosomal protein S20 | RPS20 | 34.4 |
| At3G04790 | Probable ribose-5-phosphate isomerase 3 | RPI3 | 41.0 |
| At1G20620 | Catalase-3 | CAT3 | 170.6 |
| At3g01780 | Protein TPLATE | TPLATE | 25.4 |
| At2g20420 | Succinate--CoA ligase [ADP-forming] subunit beta | At2g20420 | 323.3 |
| At3g53870 | 40S ribosomal protein S3-2 | RPS3B | 66.0 |
| At1g74970 | 30S ribosomal protein S9 | RPS9 | 60.0 |
| At1g78630 | 50S ribosomal protein L13 | RPL13 | 107.9 |
| At1G79850 | 30S ribosomal protein S17 | RPS17 | 18.6 |
| At1g07660 | Histone H4 | At1g07660 | 77.8 |
| At4g35090 | Catalase-2 | CAT2 | 323.3 |
| AtCG00820 | 30S ribosomal protein S19 | rps19 | 69.7 |
| At1g11860 | Aminomethyltransferase, | GDCST | 39.8 |
| At1G07320 | 50S ribosomal protein L4 | RPL4 | 37.5 |
| At1G35680 | 50S ribosomal protein L21 | RPL21 | 26.3 |
| At3G09440 | Heat shock 70 kDa protein 3 | HSP70-3 | 85.8 |
| At5g54770 | Thiamine thiazole synthase | THI1 | 26.8 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dubiel, M.; De Coninck, T.; Osterne, V.J.S.; Verbeke, I.; Van Damme, D.; Smagghe, G.; Van Damme, E.J.M. The ArathEULS3 Lectin Ends up in Stress Granules and Can Follow an Unconventional Route for Secretion. Int. J. Mol. Sci. 2020, 21, 1659. https://doi.org/10.3390/ijms21051659
Dubiel M, De Coninck T, Osterne VJS, Verbeke I, Van Damme D, Smagghe G, Van Damme EJM. The ArathEULS3 Lectin Ends up in Stress Granules and Can Follow an Unconventional Route for Secretion. International Journal of Molecular Sciences. 2020; 21(5):1659. https://doi.org/10.3390/ijms21051659
Chicago/Turabian StyleDubiel, Malgorzata, Tibo De Coninck, Vinicius Jose Silva Osterne, Isabel Verbeke, Daniël Van Damme, Guy Smagghe, and Els J. M. Van Damme. 2020. "The ArathEULS3 Lectin Ends up in Stress Granules and Can Follow an Unconventional Route for Secretion" International Journal of Molecular Sciences 21, no. 5: 1659. https://doi.org/10.3390/ijms21051659
APA StyleDubiel, M., De Coninck, T., Osterne, V. J. S., Verbeke, I., Van Damme, D., Smagghe, G., & Van Damme, E. J. M. (2020). The ArathEULS3 Lectin Ends up in Stress Granules and Can Follow an Unconventional Route for Secretion. International Journal of Molecular Sciences, 21(5), 1659. https://doi.org/10.3390/ijms21051659








