iPSC-Derived Microglia for Modeling Human-Specific DAMP and PAMP Responses in the Context of Alzheimer’s Disease
Abstract
1. Introduction
2. Results
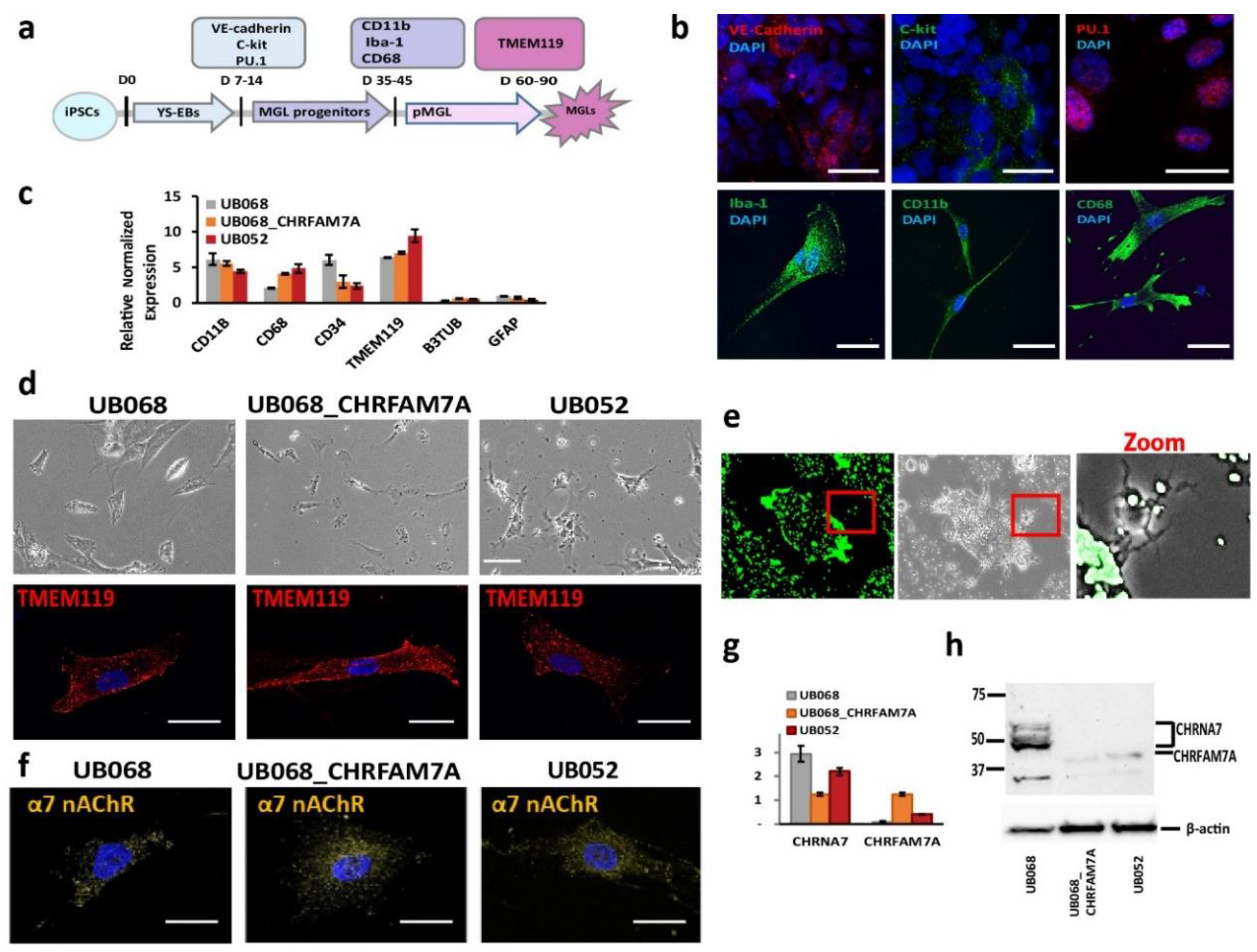
2.1. CHRNA7 and CHRFAM7A Expression in Functional Microglia Like Cells
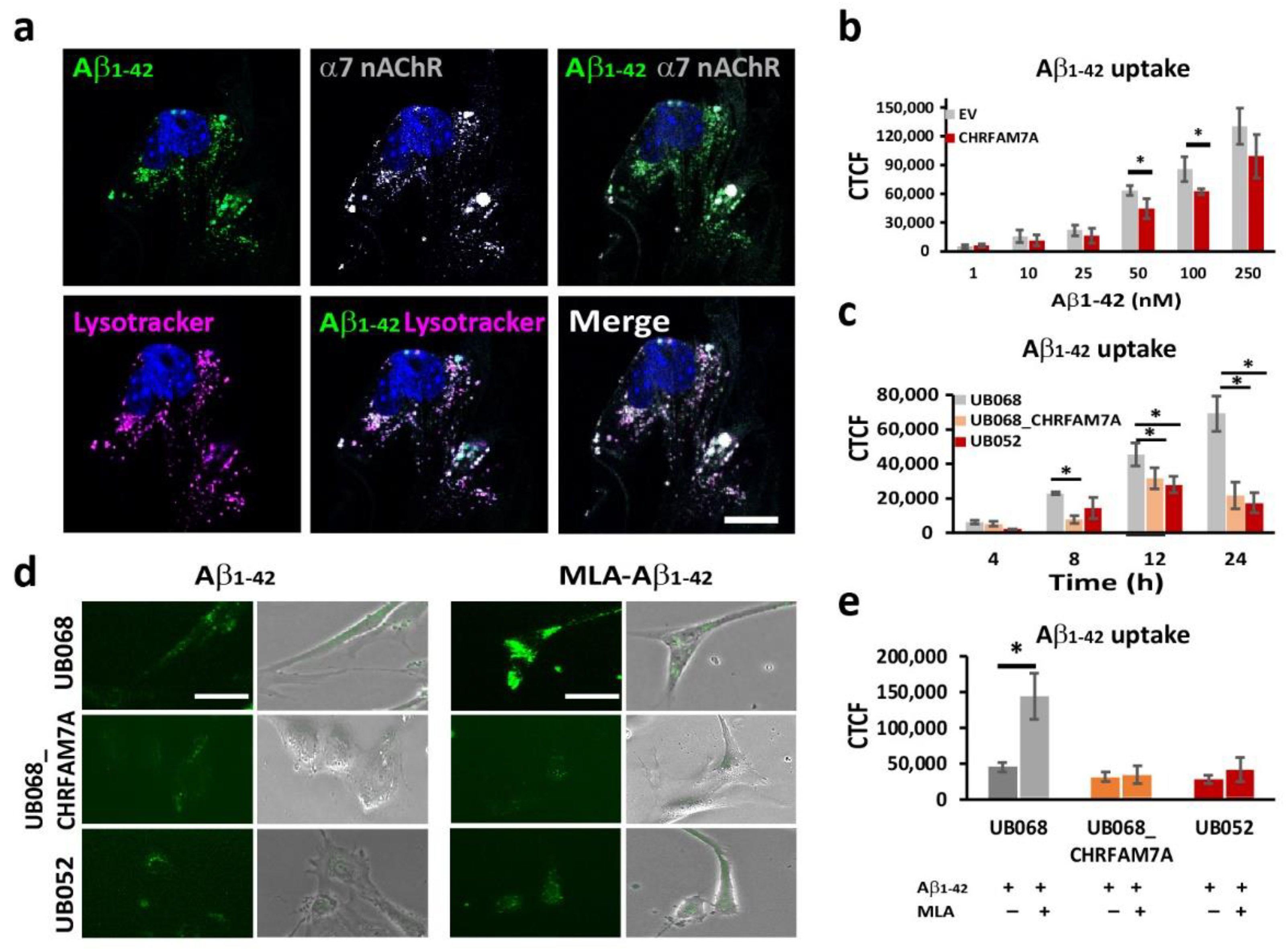
2.2. CHRFAM7A Mitigated the α7-nAChR-Dependent Aβ1–42 Uptake
2.3. Immune Surveillance to DAMP: CHRFAM7A Facilitated Microglia Activation
2.4. CHRFAM7A Heightened the Immune Responsiveness to PAMP
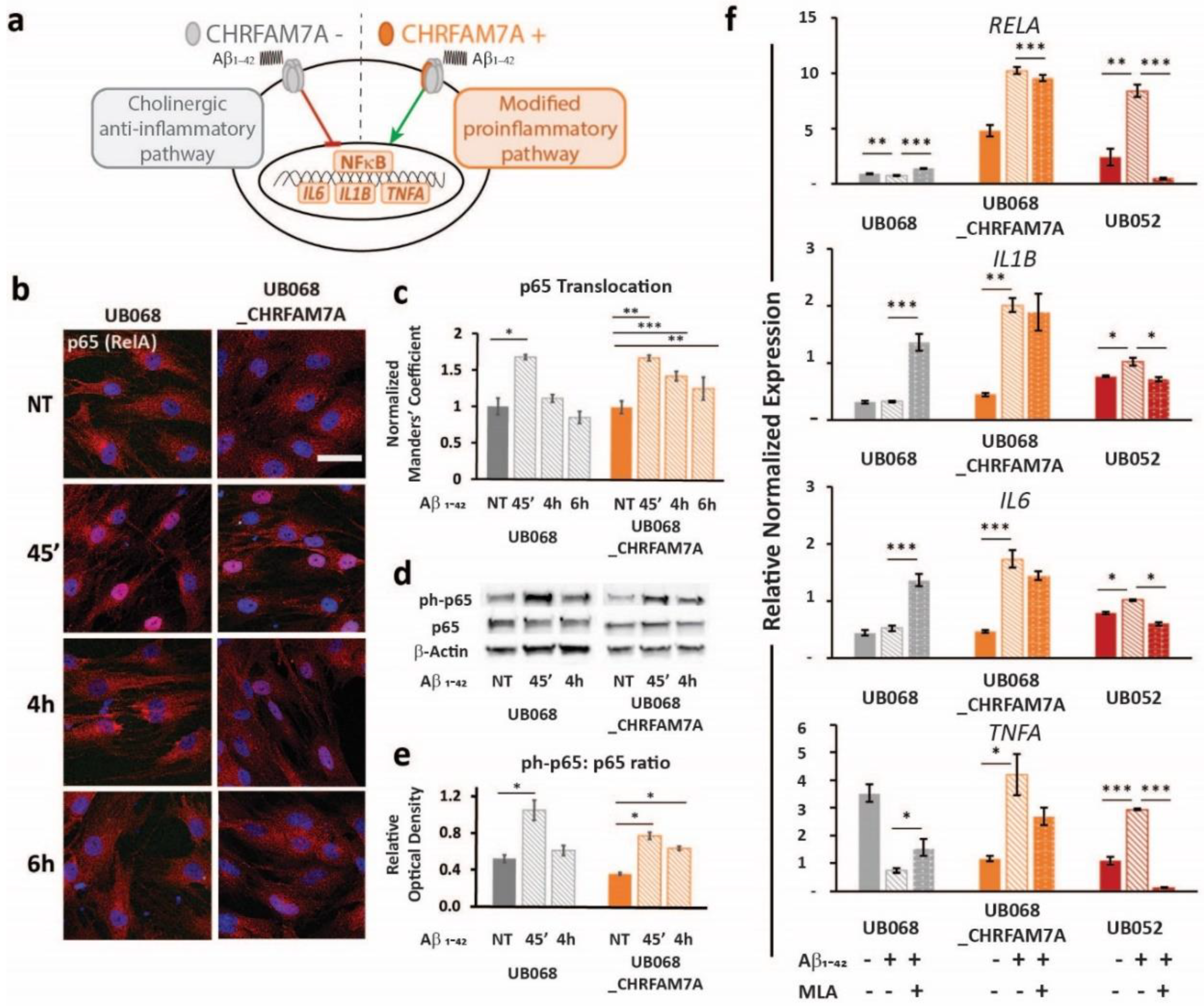
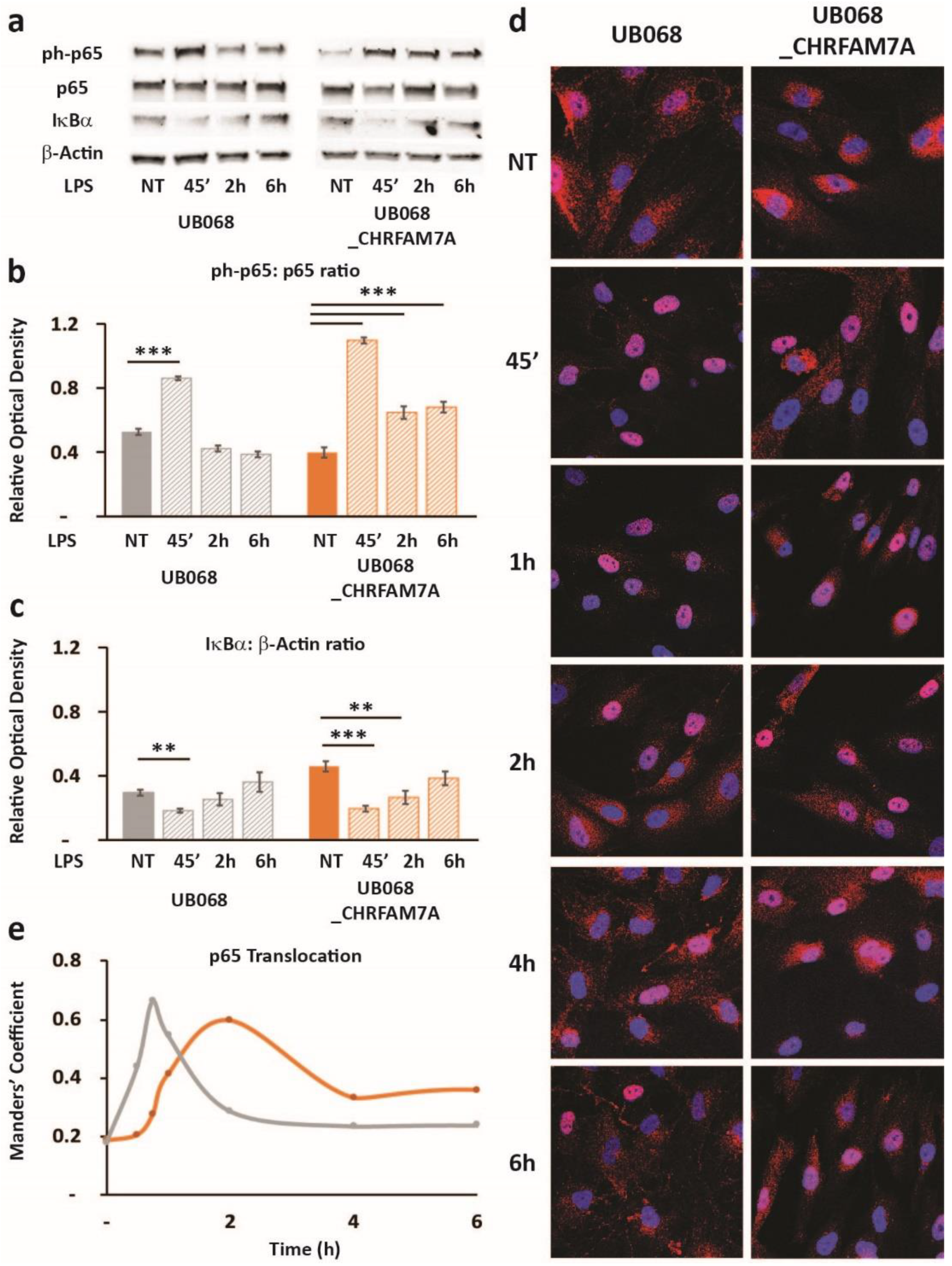
2.5. CHRFAM7A Affected the NF-κB Translocation Dynamics, Resulting in Innate Immune Activation
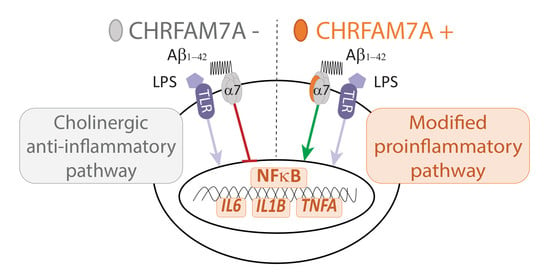
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Microglia Differentiation
4.2. Transfection
4.3. Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
4.4. Immunocytochemistry
4.5. Total Cell Lysate Preparation and Immunoblotting
4.6. Phagocytosis Assay
4.7. Cytokine Profiler
4.8. Amyloid Beta Uptake and Cell Counts
4.9. Treatment with LPS and α7 nAChR Agonist/Antagonist
4.10. ELISA
4.11. NF-κB Binding Assay
4.12. NF-κB Translocation and Quantification
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| α7 nAChR | Alpha7 nicotinic acetylcholine receptor |
| AD | Alzheimer’s disease |
| AB | Amyloid beta |
| DAM | Disease-associated microglia |
| DAMPs | Damage-associated molecular patterns |
| PAMPs | Pathogen-associated molecular patterns |
| MLA | Methyllycaconitine |
| LPS | Lipopolysaccharides |
References
- Zhang, Z.G.; Li, Y.; Ng, C.T.; Song, Y.Q. Inflammation in Alzheimer’s Disease and Molecular Genetics: Recent Update. Arch Immunol. Ther. Exp. (Warsz) 2015, 63, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, T.W.; Soreq, H.; Poirier, J.; Spreng, R.N. Longitudinal Basal Forebrain Degeneration Interacts with TREM2/C3 Biomarkers of Inflammation in Presymptomatic Alzheimer’s Disease. J. Neurosci. 2020, 40, 1931–1942. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yu, M.; Ochani, M.; Amella, C.A.; Tanovic, M.; Susarla, S.; Li, J.H.; Wang, H.; Yang, H.; Ulloa, L.; et al. Nicotinic acetylcholine receptor alpha7 subunit is an essential regulator of inflammation. Nature 2003, 421, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Shytle, R.D.; Mori, T.; Townsend, K.; Vendrame, M.; Sun, N.; Zeng, J.; Ehrhart, J.; Silver, A.A.; Sanberg, P.R.; Tan, J. Cholinergic modulation of microglial activation by alpha 7 nicotinic receptors. J. Neurochem. 2004, 89, 337–343. [Google Scholar] [CrossRef]
- De Simone, R.; Ajmone-Cat, M.A.; Carnevale, D.; Minghetti, L. Activation of alpha7 nicotinic acetylcholine receptor by nicotine selectively up-regulates cyclooxygenase-2 and prostaglandin E2 in rat microglial cultures. J. Neuroinflamm. 2005, 2, 4. [Google Scholar] [CrossRef]
- Kalkman, H.O.; Feuerbach, D. Modulatory effects of alpha7 nAChRs on the immune system and its relevance for CNS disorders. Cell Mol. Life Sci. 2016, 73, 2511–2530. [Google Scholar] [CrossRef]
- Pohanka, M. Alpha7 nicotinic acetylcholine receptor is a target in pharmacology and toxicology. Int. J. Mol. Sci. 2012, 13, 2219–2238. [Google Scholar] [CrossRef]
- Wang, H.Y.; Lee, D.H.; Davis, C.B.; Shank, R.P. Amyloid peptide Abeta(1-42) binds selectively and with picomolar affinity to alpha7 nicotinic acetylcholine receptors. J. Neurochem. 2000, 75, 1155–1161. [Google Scholar] [CrossRef]
- Lewis, A.S.; van Schalkwyk, G.I.; Bloch, M.H. Alpha-7 nicotinic agonists for cognitive deficits in neuropsychiatric disorders: A translational meta-analysis of rodent and human studies. Prog. Neuropsychopharmacol. Biol. Psychiatry 2017, 75, 45–53. [Google Scholar] [CrossRef]
- Zhou, Y.; Song, W.M.; Andhey, P.S.; Swain, A.; Levy, T.; Miller, K.R.; Poliani, P.L.; Cominelli, M.; Grover, S.; Gilfillan, S.; et al. Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 2020, 26, 131–142. [Google Scholar] [CrossRef]
- Fortna, A.; Kim, Y.; MacLaren, E.; Marshall, K.; Hahn, G.; Meltesen, L.; Brenton, M.; Hink, R.; Burgers, S.; Hernandez-Boussard, T.; et al. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2004, 2, E207. [Google Scholar] [CrossRef] [PubMed]
- Sudmant, P.H.; Kitzman, J.O.; Antonacci, F.; Alkan, C.; Malig, M.; Tsalenko, A.; Sampas, N.; Bruhn, L.; Shendure, J.; Genomes, P.; et al. Diversity of human copy number variation and multicopy genes. Science 2010, 330, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Charrier, C.; Joshi, K.; Coutinho-Budd, J.; Kim, J.E.; Lambert, N.; de Marchena, J.; Jin, W.L.; Vanderhaeghen, P.; Ghosh, A.; Sassa, T.; et al. Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell 2012, 149, 923–935. [Google Scholar] [CrossRef] [PubMed]
- Florio, M.; Albert, M.; Taverna, E.; Namba, T.; Brandl, H.; Lewitus, E.; Haffner, C.; Sykes, A.; Wong, F.K.; Peters, J.; et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 2015, 347, 1465–1470. [Google Scholar] [CrossRef]
- Dennis, M.Y.; Eichler, E.E. Human adaptation and evolution by segmental duplication. Curr. Opin. Genet. Dev. 2016, 41, 44–52. [Google Scholar] [CrossRef]
- Bitar, M.; Kuiper, S.; O’Brien, E.A.; Barry, G. Genes with human-specific features are primarily involved with brain, immune and metabolic evolution. BMC Bioinform. 2019, 20 (Suppl 9), 406. [Google Scholar] [CrossRef]
- Sinkus, M.L.; Graw, S.; Freedman, R.; Ross, R.G.; Lester, H.A.; Leonard, S. The human CHRNA7 and CHRFAM7A genes: A review of the genetics, regulation, and function. Neuropharmacology 2015, 96, 274–288. [Google Scholar] [CrossRef]
- Costantini, T.W.; Chan, T.W.; Cohen, O.; Langness, S.; Treadwell, S.; Williams, E.; Eliceiri, B.P.; Baird, A. Uniquely human CHRFAM7A gene increases the hematopoietic stem cell reservoir in mice and amplifies their inflammatory response. Proc. Natl. Acad. Sci. USA 2019, 116, 7932–7940. [Google Scholar] [CrossRef]
- Jiang, Y.; Yuan, H.; Huang, L.; Hou, X.; Zhou, R.; Dang, X. Global proteomic profiling of the uniquely human CHRFAM7A gene in transgenic mouse brain. Gene 2019, 714, 143996. [Google Scholar] [CrossRef]
- Ihnatovych, I.; Nayak, T.K.; Ouf, A.; Sule, N.; Birkaya, B.; Chaves, L.; Auerbach, A.; Szigeti, K. iPSC model of CHRFAM7A effect on alpha7 nicotinic acetylcholine receptor function in the human context. Transl. Psychiatry 2019, 9, 59. [Google Scholar] [CrossRef]
- Szigeti, K.; Ihnatovych, I.; Birkaya, B.; Chen, Z.; Ouf, A.; Indurthi, D.C.; Bard, J.E.; Kann, J.; Adams, A.; Chaves, L.; et al. CHRFAM7A: A human specific fusion gene, accounts for the translational gap for cholinergic strategies in Alzheimer’s disease. EBioMedicine 2020, 59, 102892. [Google Scholar] [CrossRef] [PubMed]
- Kabbani, N.; Nichols, R.A. Beyond the Channel: Metabotropic Signaling by Nicotinic Receptors. Trends Pharmacol. Sci. 2018, 39, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Rosas-Ballina, M.; Tracey, K.J. Cholinergic control of inflammation. J. Intern. Med. 2009, 265, 663–679. [Google Scholar] [CrossRef] [PubMed]
- Venkataraman, L.; Fair, S.R.; McElroy, C.A.; Hester, M.E.; Fu, H. Modeling neurodegenerative diseases with cerebral organoids and other three-dimensional culture systems: Focus on Alzheimer’s disease. Stem. Cell Rev. Rep. 2020. [Google Scholar] [CrossRef]
- Garcia-Leon, J.A.; Caceres-Palomo, L.; Sanchez-Mejias, E.; Mejias-Ortega, M.; Nunez-Diaz, C.; Fernandez-Valenzuela, J.J.; Sanchez-Varo, R.; Davila, J.C.; Vitorica, J.; Gutierrez, A. Human Pluripotent Stem Cell-Derived Neural Cells as a Relevant Platform for Drug Screening in Alzheimer’s Disease. Int. J. Mol. Sci. 2020, 21, 6867. [Google Scholar] [CrossRef]
- Lee, C.T.; Bendriem, R.M.; Wu, W.W.; Shen, R.F. 3D brain Organoids derived from pluripotent stem cells: Promising experimental models for brain development and neurodegenerative disorders. J. Biomed. Sci. 2017, 24, 59. [Google Scholar] [CrossRef]
- Cenini, G.; Hebisch, M.; Iefremova, V.; Flitsch, L.J.; Breitkreuz, Y.; Tanzi, R.E.; Kim, D.Y.; Peitz, M.; Brustle, O. Dissecting Alzheimer’s disease pathogenesis in human 2D and 3D models. Mol. Cell Neurosci. 2020, 103568. [Google Scholar] [CrossRef]
- Muffat, J.; Li, Y.; Yuan, B.; Mitalipova, M.; Omer, A.; Corcoran, S.; Bakiasi, G.; Tsai, L.H.; Aubourg, P.; Ransohoff, R.M.; et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat. Med. 2016, 22, 1358–1367. [Google Scholar] [CrossRef]
- Kierdorf, K.; Prinz, M. Factors regulating microglia activation. Front. Cell Neurosci. 2013, 7, 44. [Google Scholar] [CrossRef]
- Smith, A.M.; Gibbons, H.M.; Oldfield, R.L.; Bergin, P.M.; Mee, E.W.; Faull, R.L.; Dragunow, M. The transcription factor PU.1 is critical for viability and function of human brain microglia. Glia 2013, 61, 929–942. [Google Scholar] [CrossRef]
- Lane, K.; Van Valen, D.; DeFelice, M.M.; Macklin, D.N.; Kudo, T.; Jaimovich, A.; Carr, A.; Meyer, T.; Pe’er, D.; Boutet, S.C.; et al. Measuring Signaling and RNA-Seq in the Same Cell Links Gene Expression to Dynamic Patterns of NF-kappaB Activation. Cell Syst. 2017, 4, 458–469.e5. [Google Scholar] [CrossRef] [PubMed]
- Costantini, T.W.; Dang, X.; Coimbra, R.; Eliceiri, B.P.; Baird, A. CHRFAM7A, a human-specific and partially duplicated alpha7-nicotinic acetylcholine receptor gene with the potential to specify a human-specific inflammatory response to injury. J. Leukoc. Biol. 2015, 97, 247–257. [Google Scholar] [CrossRef]
- Maldifassi, M.C.; Martin-Sanchez, C.; Atienza, G.; Cedillo, J.L.; Arnalich, F.; Bordas, A.; Zafra, F.; Gimenez, C.; Extremera, M.; Renart, J.; et al. Interaction of the alpha7-nicotinic subunit with its human-specific duplicated dupalpha7 isoform in mammalian cells: Relevance in human inflammatory responses. J. Biol. Chem. 2018, 293, 13874–13888. [Google Scholar] [CrossRef] [PubMed]
- Ries, M.; Sastre, M. Mechanisms of Abeta Clearance and Degradation by Glial Cells. Front. Aging Neurosci. 2016, 8, 160. [Google Scholar] [CrossRef]
- Jefferson, W.K.; Shane, M.B.; Andrew, S.M.; Amanda, M.L.; Arnold, M.S.; Bruce, T.L. Inflammation as a central mechanism in Alzheimer’s disease. Alzheimers Dement. (N. Y.) 2018, 4, 575–590. [Google Scholar]
- Benfante, R.; Antonini, R.A.; De Pizzol, M.; Gotti, C.; Clementi, F.; Locati, M.; Fornasari, D. Expression of the alpha7 nAChR subunit duplicate form (CHRFAM7A) is down-regulated in the monocytic cell line THP-1 on treatment with LPS. J. Neuroimmunol. 2011, 230, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Cirrito, J.R.; May, P.C.; O’Dell, M.A.; Taylor, J.W.; Parsadanian, M.; Cramer, J.W.; Audia, J.E.; Nissen, J.S.; Bales, K.R.; Paul, S.M.; et al. In vivo assessment of brain interstitial fluid with microdialysis reveals plaque-associated changes in amyloid-beta metabolism and half-life. J. Neurosci. 2003, 23, 8844–8853. [Google Scholar] [CrossRef]
- de Lucas-Cerrillo, A.M.; Maldifassi, M.C.; Arnalich, F.; Renart, J.; Atienza, G.; Serantes, R.; Cruces, J.; Sanchez-Pacheco, A.; Andres-Mateos, E.; Montiel, C. Function of partially duplicated human alpha77 nicotinic receptor subunit CHRFAM7A gene: Potential implications for the cholinergic anti-inflammatory response. J. Biol. Chem. 2011, 286, 594–606. [Google Scholar] [CrossRef]
- Bencherif, M.; Lippiello, P.M.; Lucas, R.; Marrero, M.B. Alpha7 nicotinic receptors as novel therapeutic targets for inflammation-based diseases. Cell Mol. Life Sci. 2011, 68, 931–949. [Google Scholar] [CrossRef]
- Walana, W.; Wang, J.J.; Yabasin, I.B.; Ntim, M.; Kampo, S.; Al-Azab, M.; Elkhider, A.; Dogkotenge Kuugbee, E.; Cheng, J.W.; Gordon, J.R.; et al. IL-8 analogue CXCL8 (3-72) K11R/G31P, modulates LPS-induced inflammation via AKT1-NF-kbeta and ERK1/2-AP-1 pathways in THP-1 monocytes. Hum. Immunol. 2018, 79, 809–816. [Google Scholar] [CrossRef]
- Falvo, J.V.; Tsytsykova, A.V.; Goldfeld, A.E. Transcriptional Control of the TNF Gene. Curr. Dir. Autoimmun. 2010, 11, 27–60. [Google Scholar] [PubMed]
- Laffer, B.; Bauer, D.; Wasmuth, S.; Busch, M.; Jalilvand, T.V.; Thanos, S.; Zu Horste, G.M.; Loser, K.; Langmann, T.; Heiligenhaus, A.; et al. Loss of IL-10 Promotes Differentiation of Microglia to a M1 Phenotype. Front. Cell Neurosci. 2019, 13. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Saredy, J.; Zhang, R.; Shao, Y.; Sun, Y.; Yang, W.Y.; Wang, J.; Liu, L.; Drummer, C., IV; Johnson, C.; et al. Approaching Inflammation Paradoxes-Proinflammatory Cytokine Blockages Induce Inflammatory Regulators. Front. Immunol. 2020, 11, 554301. [Google Scholar] [CrossRef] [PubMed]
- Lobo-Silva, D.; Carriche, G.M.; Castro, A.G.; Roque, S.; Saraiva, M. Balancing the immune response in the brain: IL-10 and its regulation. J. Neuroinflamm. 2016, 13. [Google Scholar] [CrossRef]
- Zambrano, S.; Bianchi, M.E.; Agresti, A. High-throughput analysis of NF-kappaB dynamics in single cells reveals basal nuclear localization of NF-kappaB and spontaneous activation of oscillations. PLoS ONE 2014, 9, e90104. [Google Scholar] [CrossRef]
- Maity, A.; Wollman, R. Information transmission from NFkB signaling dynamics to gene expression. PLoS Comput. Biol. 2020, 16, e1008011. [Google Scholar] [CrossRef]
- Dorrington, M.G.; Fraser, I.D.C. NF-kappaB Signaling in Macrophages: Dynamics, Crosstalk, and Signal Integration. Front. Immunol. 2019, 10, 705. [Google Scholar] [CrossRef]
- Zambrano, S.; De Toma, I.; Piffer, A.; Bianchi, M.E.; Agresti, A. NF-kappaB oscillations translate into functionally related patterns of gene expression. Elife 2016, 5, e09100. [Google Scholar] [CrossRef]
- Lin, Y.T.; Seo, J.; Gao, F.; Feldman, H.M.; Wen, H.L.; Penney, J.; Cam, H.P.; Gjoneska, E.; Raja, W.K.; Cheng, J.; et al. APOE4 Causes Widespread Molecular and Cellular Alterations Associated with Alzheimer’s Disease Phenotypes in Human iPSC-Derived Brain Cell Types. Neuron 2018, 98, 1141–1154.e7. [Google Scholar] [CrossRef]
- Wang, Y.; Xiao, C.; Indersmitten, T.; Freedman, R.; Leonard, S.; Lester, H.A. The duplicated alpha7 subunits assemble and form functional nicotinic receptors with the full-length alpha7. J. Biol. Chem. 2014, 289, 26451–26463. [Google Scholar] [CrossRef]
- Warren, L.; Manos, P.D.; Ahfeldt, T.; Loh, Y.H.; Li, H.; Lau, F.; Ebina, W.; Mandal, P.K.; Smith, Z.D.; Meissner, A.; et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 2010, 7, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Crick, S.L.; Bu, G.; Frieden, C.; Pappu, R.V.; Lee, J.M. Amyloid seeds formed by cellular uptake, concentration, and aggregation of the amyloid-beta peptide. Proc. Natl. Acad. Sci. USA 2009, 106, 20324–20329. [Google Scholar] [CrossRef] [PubMed]
- Trask, O.J., Jr. Nuclear Factor Kappa B (NF-kappaB) Translocation Assay Development and Validation for High Content Screening. In Assay Guidance Manual; Markossian, S., Sittampalam, G.S., Grossman, A., Brimacombe, K., Arkin, M., Auld, D., Austin, C.P., Baell, J., Caaveiro, J.M.M., Chung, T.D.Y., et al., Eds.; Bethesda: Rockville, MD, USA, 2004. [Google Scholar]
- Bagaev, A.V.; Garaeva, A.Y.; Lebedeva, E.S.; Pichugin, A.V.; Ataullakhanov, R.I.; Ataullakhanov, F.I. Elevated pre-activation basal level of nuclear NF-kappaB in native macrophages accelerates LPS-induced translocation of cytosolic NF-kappaB into the cell nucleus. Sci. Rep. 2019, 9, 4563. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ihnatovych, I.; Birkaya, B.; Notari, E.; Szigeti, K. iPSC-Derived Microglia for Modeling Human-Specific DAMP and PAMP Responses in the Context of Alzheimer’s Disease. Int. J. Mol. Sci. 2020, 21, 9668. https://doi.org/10.3390/ijms21249668
Ihnatovych I, Birkaya B, Notari E, Szigeti K. iPSC-Derived Microglia for Modeling Human-Specific DAMP and PAMP Responses in the Context of Alzheimer’s Disease. International Journal of Molecular Sciences. 2020; 21(24):9668. https://doi.org/10.3390/ijms21249668
Chicago/Turabian StyleIhnatovych, Ivanna, Barbara Birkaya, Emily Notari, and Kinga Szigeti. 2020. "iPSC-Derived Microglia for Modeling Human-Specific DAMP and PAMP Responses in the Context of Alzheimer’s Disease" International Journal of Molecular Sciences 21, no. 24: 9668. https://doi.org/10.3390/ijms21249668
APA StyleIhnatovych, I., Birkaya, B., Notari, E., & Szigeti, K. (2020). iPSC-Derived Microglia for Modeling Human-Specific DAMP and PAMP Responses in the Context of Alzheimer’s Disease. International Journal of Molecular Sciences, 21(24), 9668. https://doi.org/10.3390/ijms21249668