MicroRNAs at the Crossroad of the Dichotomic Pathway Cell Death vs. Stemness in Neural Somatic and Cancer Stem Cells: Implications and Therapeutic Strategies
Abstract
1. Introduction
2. miRNAs Bridging Stemness and Cell Death in Neurogenesis
miRNA-Loaded Exosomes for Therapeutic Interventions in Neuropathologies
3. miRNAs Involved in the Regulation of Apoptosis in Human Neural/Neural Crest-Derived CSCs
3.1. Pro-Apoptotic miRNAs
3.2. Anti-Apoptotic miRNAs
3.3. Pro- and Anti-Apoptotic miRNAs and Their Therapeutic Implication in Tumors
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Aβ | amyloid β-peptide |
| ABC | ATP binding cassette transporter |
| ABCB1 | ATP binding cassette subfamily B member 1 |
| AD | Alzheimer’s disease |
| ADD3 | adducin 3 |
| ADSC | adipose-derived stem cell |
| Ago2 | eukaryotic translation initiation factor 2C 2 |
| AIS | acute ischemic stroke |
| AKT | serine-threonine protein kinase |
| ALS | amyotrophic lateral sclerosis |
| ApoE | apolipoprotein E |
| ASD | autism spectrum disorder |
| BAK1 | Bcl-2 antagonist/killer 1 |
| Bcl-2 | Bcl-2 apoptosis regulator |
| BCL2L12 | Bcl-2-like protein 12 |
| Bcl-w | Bcl-2-like protein 2 |
| Bcl-x | Bcl-2-like protein 1 |
| BH3 | Bcl-2 homology 3 domain |
| BIM | Bcl-2-interacting mediator of cell death |
| BLCAP | bladder cancer associated protein |
| BMI-1 | BMI1 proto-oncogene, polycomb ring finger |
| BMSC | bone marrow mesenchymal stem cell |
| BNIP3 | Bcl-2 interacting protein 3 |
| BRAF | B-Raf Proto-Oncogene, Serine/Threonine Kinase |
| BTG2 | nerve growth factor-inducible anti-proliferative |
| BTSC | brain tumor stem cell |
| CAB39 | calcium binding protein 39 |
| CASP2 | caspase-2 |
| CASP3 | caspase-3 |
| CASP7 | caspase-7 |
| CASP9 | caspase-9 |
| CCND1 | cyclin D1 |
| CD133 | prominin-1 |
| CDC25A | cell division cycle 25A |
| CDC42 | cell division cycle 42 |
| CDK6 | cyclin-dependent kinase 6 |
| CDKN1A/P21CIP1 | cyclin-dependent kinase inhibitor 1A |
| CDKN1B/P27KIP1 | cyclin-dependent kinase inhibitor 1B |
| CDKN1C/P57KIP2 | cyclin-dependent kinase inhibitor 1C |
| CDX1 | caudal-type homeobox 1 protein |
| c-FLIP | cellular FLICE-like inhibitory protein |
| CHOP | C/EBP homologous protein 10 |
| CIP2A | cellular inhibitor of PP2A |
| C-KIT/CD117 | V-Kit Hardy-Zuckerman 4 feline sarcoma viral oncogene |
| c-MET | MET proto-oncogene, receptor tyrosine kinase |
| c-MYC | myc proto-oncogene |
| CNS | central nervous system |
| CpG | cytosine-phosphate-guanine |
| CREB | cAMP response element-binding protein |
| CSC | cancer stem cell |
| CSF | cerebral spinal fluid |
| CTBP1 | C-terminal binding protein 1 |
| CXCL12 | C-X-C motif chemokine ligand 12 |
| CXCR4 | C-X-C motif chemokine receptor 4 |
| DAAM1 | disheveled-associated activator of morphogenesis 1 |
| DCA | dichloroacetate |
| DDIT3 | DNA damage-inducible transcript 3 |
| DFMO | difluoromethylornithine |
| Dll1 | Notch ligand Delta-like 1 |
| DMC | demethoxycurcumin |
| DOX | doxorubicin |
| DR5 | death receptor 5 |
| E2F | E2 factor family of DNA-binding transcription factor |
| E2F2 | E2F transcription factor 2 |
| ECOP | epidermal growth factor receptor-coamplified and overexpressed protein |
| EGFR | epidermal growth factor receptor |
| ELISA | enzyme-linked immunosorbent assay |
| EMT | epithelial-to-mesenchymal transition |
| ERK 1/2 | extracellular regulated kinase 1/2 |
| ESC | embryonic stem cell |
| ETS-1 | V-Ets avian erythroblastosis virus E26 oncogene homolog 1 |
| FASL | Fas cell surface death receptor ligand |
| FNDC3B | fibronectin type III domain containing 3B |
| FOXO1 | forkhead box O1 |
| FOXO3 | forkhead box O3 |
| GADD153 | growth arrest and DNA damage-inducible protein 153 |
| GAP43 | growth associated protein 43 |
| GBM | glioblastoma multiforme |
| GFAP | glial fibrillary acidic protein |
| GIC | glioma initiating cell |
| GLI1 | glioma-associated oncogene homolog 1 (zinc finger protein) |
| GLIPR | glioma pathogenesis-related protein |
| GPD1L | glycerol-3-phosphate dehydrogenase 1-like |
| GSC | glioma stem cell |
| GSEA | Gene Set Enrichment Analysis |
| HCELL/CD44 | hematopoietic cell E/L-selectin ligand |
| HES1 | class B basic helix-loop-helix protein 39 |
| HIF1α | hypoxia-inducible factor-1 alpha |
| HIF2α | hypoxia-inducible factor-2 alpha |
| hiPSC | human induced pluripotent stem cell |
| HMGA2 | high mobility group AT-hook 2 |
| HRAS | Harvey rat sarcoma viral oncogene homolog |
| HuR | Hu antigen R |
| IAP | inhibitor of apoptosis protein |
| IGF-1R | insulin-like growth factor-1 receptor |
| IGF2BP1 | insulin-like growth factor 2 MRNA binding protein 1 |
| IGF-2R | insulin-like growth factor 2 receptor |
| IκBα | NFκB inhibitor alpha |
| JAG1 | jagged canonical Notch ligand 1 |
| JAK | Janus kinase |
| JNK | c-Jun N-terminal kinase |
| KCNMA1 | potassium calcium-activated channel subfamily M alpha 1 |
| KLF4 | Krüppel-like factor 4 |
| KRAS | Kirsten rat sarcoma viral oncogene homolog |
| LGALS3 | galectin 3 |
| LIN28 | lin-28 homolog A |
| LIN28B | lin-28 homolog B |
| LNA | locked nucleic acid |
| lncRNA | long non-coding RNA |
| MAPK | mitogen-activated protein kinase |
| MB | medulloblastoma |
| MBNL1-3 | muscleblind-like splicing regulator 1 |
| MBP | myelin basic protein |
| MCAO | middle cerebral artery occlusion |
| MCL1 | myeloid cell leukemia sequence 1 (Bcl2-related) |
| MCSC | melanocyte stem cell |
| MEK1/2 | mitogen-activated protein kinase kinase 1/2 |
| mRNA | messenger RNA |
| MGMT | O6-methylguanine-DNA-methyltransferase |
| miRNA | microRNA |
| MIF | macrophage migration inhibitory factor |
| MITF | microphthalmia-associated transcription factor |
| MKI67 | marker of proliferation Ki-67 |
| MMP | matrix metalloproteinase |
| MMP-2 | matrix metalloproteinase-2 |
| MMP-9 | matrix metalloproteinase-9 |
| MMP-12 | matrix metalloproteinase-12 |
| MNT | MAX network transcriptional repressor |
| MS | multiple sclerosis |
| MSC | mesenchymal stem cell |
| MST1/2 | macrophage stimulating 1/2 |
| mTOR | mammalian target of rapamycin |
| mTORC2 | mammalian target of rapamycin complex 2 |
| MYCN | V-Myc avian myelocytomatosis viral oncogene neuroblastoma |
| NANOG | nanog homeobox |
| NB | neuroblastoma |
| NEAT1 | nuclear paraspeckle assembly transcript 1 |
| NEDD9 | neural precursor cell expressed, developmentally down-regulated 9 |
| NEFL | neurofilament light polypeptide |
| NF | neurofilament |
| NF-κB | nuclear factor kappa-light-chain-enhancer of activated B cells |
| NKIRAS2 | NF-κB inhibitor interacting RAS-like 2 |
| NOTCH1/2 | Notch receptor 1/2 |
| NPC | neural progenitor cell |
| NSC | neural stem cell |
| NTSR1 | neurotensin receptor 1 |
| OCT4 | octamer-binding transcription factor 4 |
| OPC | oligodendrocyte precursor cell |
| p38MAPK | p38 mitogen-activated protein kinase |
| PAX6 | paired box 6 |
| PD | Parkinson’s disease |
| PDCD4 | programmed cell death 4 |
| PDGFR | platelet-derived growth factor receptor |
| PI3K | phosphoinositide-3-kinase C |
| PIK3R3 | phosphatidylinositol-3-kinase regulatory subunit 3 |
| PIN1 | protein interacting with never in mitosis A1 |
| PLD2 | phospholipase D2 |
| PRPS1 | phosphoribosyl pyrophosphate synthetase 1 |
| PTCH1 | Patched 1 |
| PTEN | phosphatase and tensin homolog |
| PUMA | p53-upregulated modulator of apoptosis |
| QKI-6 | Quaking gene isoform 6 |
| RELA | RELA proto-oncogene, NF-KB p65 subunit |
| RB | retinoblastoma protein |
| RICTOR | rapamycin-insensitive companion of mTOR |
| rmTBI | repetitive mild traumatic brain injury |
| ROCK1 | rho associated coiled-coil containing protein kinase 1 |
| ROD1 | regulator of differentiation 1 |
| RSRC1 | arginine and serine rich coiled-coil 1 |
| RTVP-1 | related to testis-specific, vespid, and pathogenesis proteins 1 |
| SART3 | spliceosome associated factor 3, U4/U6 recycling protein |
| SAV1 | protein salvador homolog 1 |
| SCI | spinal cord injury |
| Shh | sonic hedgehog |
| shRNA | short hairpin RNA |
| SIX1 | sine oculis homeobox 1 |
| SMAD | mothers against decapentaplegic homolog |
| SMAD3 | SMAD family member 3 |
| SMO | smoothened, frizzled class receptor |
| SOCS3 | suppressor of cytokine signaling 3 |
| SODDs | silencer of death domains |
| SOX2 | sex determining region Y-box 2 |
| SOX9 | SRY-box transcription factor 9 |
| SSEA-1/CD15 | stage specific embryonic antigen 1 |
| ST7L | suppression of tumorigenicity 7-like |
| STAT3 | signal transducer and activator of transcription 3 |
| TBI | traumatic brain injury |
| TCF-4 | transcription factor 4 |
| TCGA | The Cancer Genome Atlas |
| TGF-β | transforming growth factor-β |
| TIMP3 | tissue inhibitor of metalloproteinases 3 |
| TLR4 | Toll-like receptor 4 |
| TMZ | temozolomide |
| TNFAIP3 | tumor necrosis factor alpha-induced protein 3 |
| TNFα | tumor necrosis factor alpha |
| TP53 | tumor protein p53 |
| TP53INP2 | TP53-inducible nuclear protein 2 |
| TRAIL | tumor necrosis factor apoptosis inducing ligand |
| TUNEL | terminal deoxynucleotidyl transferase (TdT) dUTP nick- end labeling |
| TUSC2 | tumor suppressor candidate 2 |
| Wnt | wingless-related integration site |
| WT1 | Wilms tumor protein 1 |
| YB-1 | human Y-box binding protein 1 |
References
- Im, H.I.; Kenny, P.J. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012, 35, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Diana, A.; Gaido, G.; Murtas, D. MicroRNA Signature in human normal and tumoral neural stem cells. Int. J. Mol. Sci. 2019, 20, 4123. [Google Scholar] [CrossRef] [PubMed]
- Banelli, B.; Forlani, A.; Allemanni, G.; Morabito, A.; Pistillo, M.P.; Romani, M. MicroRNA in glioblastoma: An overview. Int. J. Genom. 2017, 2017, 7639084. [Google Scholar] [CrossRef]
- Jin, Y.; Wang, J.; Zhang, M.; Zhang, S.; Lei, C.; Chen, H.; Guo, W.; Lan, X. Role of bta-miR-204 in the regulation of adipocyte proliferation, differentiation, and apoptosis. J. Cell Physiol. 2019, 234, 11037–11046. [Google Scholar] [CrossRef]
- Su, Z.; Yang, Z.; Xu, Y.; Chen, Y.; Yu, Q. MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget 2015, 6, 8474–8490. [Google Scholar] [CrossRef]
- Su, Y.; Wu, H.; Pavlosky, A.; Zou, L.L.; Deng, X.; Zhang, Z.X.; Jevnikar, A.M. Regulatory non-coding RNA: New instruments in the orchestration of cell death. Cell Death Dis. 2016, 7, e2333. [Google Scholar] [CrossRef]
- Pollock, A.; Bian, S.; Zhang, C.; Chen, Z.; Sun, T. Growth of the developing cerebral cortex is controlled by microRNA-7 through the p53 pathway. Cell Rep. 2014, 7, 1184–1196. [Google Scholar] [CrossRef]
- de Chevigny, A.; Coré, N.; Follert, P.; Gaudin, M.; Barbry, P.; Béclin, C.; Cremer, H. miR-7a regulation of Pax6 controls spatial origin of forebrain dopaminergic neurons. Nat. Neurosci. 2012, 15, 1120–1126. [Google Scholar] [CrossRef]
- Zhao, C.; Sun, G.; Li, S.; Shi, Y.A. Feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 2009, 16, 365–371. [Google Scholar] [CrossRef]
- Shibata, M.; Nakao, H.; Kiyonari, H.; Abe, T.; Aizawa, S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J. Neurosci. 2011, 31, 3407–3422. [Google Scholar] [CrossRef] [PubMed]
- Shibata, M.; Kurokawa, D.; Nakao, H.; Ohmura, T.; Aizawa, S. MicroRNA-9 modulates Cajal-Retzius cell differentiation by suppressing Foxg1 expression in mouse medial pallium. J. Neurosci. 2008, 28, 10415–10421. [Google Scholar] [CrossRef] [PubMed]
- Dajas-Bailador, F.; Bonev, B.; Garcez, P.; Stanley, P.; Guillemot, F.; Papalopulu, N. microRNA-9 regulates axon extension and branching by targeting Map1b in mouse cortical neurons. Nat. Neurosci. 2012, 15, 697–699. [Google Scholar] [CrossRef] [PubMed]
- Clovis, Y.M.; Enard, W.; Marinaro, F.; Huttner, W.B.; De Pietri Tonelli, D. Convergent repression of Foxp2 3′UTR by miR-9 and miR-132 in embryonic mouse neocortex: Implications for radial migration of neurons. Development 2012, 139, 3332–3342. [Google Scholar] [CrossRef] [PubMed]
- Shu, P.; Wu, C.; Ruan, X.; Liu, W.; Hou, L.; Fu, H.; Wang, M.; Liu, C.; Zeng, Y.; Chen, P.; et al. Opposing gradients of microRNA expression temporally pattern layer formation in the developing neocortex. Dev. Cell 2019, 49, 764–785. [Google Scholar] [CrossRef] [PubMed]
- Makeyev, E.V.; Zhang, J.; Carrasco, M.A.; Maniatis, T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 2007, 27, 435–448. [Google Scholar] [CrossRef] [PubMed]
- Volvert, M.L.; Prévot, P.P.; Close, P.; Laguesse, S.; Pirotte, S.; Hemphill, J.; Rogister, F.; Kruzy, N.; Sacheli, R.; Moonen, G.; et al. MicroRNA targeting of CoREST controls polarization of migrating cortical neurons. Cell Rep. 2014, 7, 1168–1183. [Google Scholar] [CrossRef]
- Cheng, L.C.; Pastrana, E.; Tavazoie, M.; Doetsch, F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 2009, 12, 399–408. [Google Scholar] [CrossRef]
- Szulwach, K.E.; Li, X.; Smrt, R.D.; Li, Y.; Luo, Y.; Lin, L.; Santistevan, N.J.; Li, W.; Zhao, X.; Jin, P. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J. Cell. Biol. 2010, 189, 127–141. [Google Scholar] [CrossRef]
- Sun, G.; Ye, P.; Murai, K.; Lang, M.F.; Li, S.; Zhang, H.; Li, W.; Fu, C.; Yin, J.; Wang, A.; et al. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat. Commun. 2011, 2, 529. [Google Scholar] [CrossRef]
- Chua, C.; Tang, B.L. miR-34a in Neurophysiology and Neuropathology. J. Mol. Neurosci. 2019, 67, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Aranha, M.M.; Santos, D.M.; Xavier, J.M.; Low, W.C.; Steer, C.J.; Solá, S.; Rodrigues, C.M. Apoptosis-associated microRNAs are modulated in mouse, rat and human neural differentiation. BMC Genom. 2010, 11, 514. [Google Scholar] [CrossRef] [PubMed]
- Kole, A.J.; Swahari, V.; Hammond, S.M.; Deshmukh, M. miR-29b is activated during neuronal maturation and targets BH3-only genes to restrict apoptosis. Genes Dev. 2011, 25, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Xiong, L.; Huang, X.; Zhao, T.; Wu, L.Y.; Liu, Z.H.; Ding, X.; Liu, S.; Wu, Y.; Zhao, Y.; et al. miR-210 suppresses BNIP3 to protect against the apoptosis of neural progenitor cells. Stem Cell Res. 2013, 11, 657–667. [Google Scholar] [CrossRef]
- Konovalova, J.; Gerasymchuk, D.; Parkkinen, I.; Chmielarz, P.; Domanskyi, A. Interplay between microRNAs and oxidative stress in neurodegenerative diseases. Int. J. Mol. Sci. 2019, 20, 6055. [Google Scholar] [CrossRef]
- Wei, W.; Wang, Z.Y.; Ma, L.N.; Zhang, T.T.; Cao, Y.; Li, H. MicroRNAs in Alzheimer’s disease: Function and potential applications as diagnostic biomarkers. Front. Mol. Neurosci. 2020, 13, 160. [Google Scholar] [CrossRef]
- Kou, X.; Chen, D.; Chen, N. The Regulation of microRNAs in Alzheimer’s Disease. Front. Neurol. 2020, 11, 288. [Google Scholar] [CrossRef]
- Praticò, D. The functional role of microRNAs in the pathogenesis of tauopathy. Cells 2020, 9, 2262. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, L. MicroRNAs in amyotrophic lateral sclerosis: From pathogenetic involvement to diagnostic biomarker and therapeutic agent development. Neurol. Sci. 2020, 1–9. [Google Scholar] [CrossRef]
- Sadlon, A.; Takousis, P.; Alexopoulos, P.; Evangelou, E.; Prokopenko, I.; Perneczky, R. miRNAs identify shared pathways in Alzheimer’s and Parkinson’s diseases. Trends Mol. Med. 2019, 25, 662–672. [Google Scholar] [CrossRef]
- Doxakis, E. Cell-free microRNAs in Parkinson’s disease: Potential biomarkers that provide new insights into disease pathogenesis. Ageing Res. Rev. 2020, 58, 101023. [Google Scholar] [CrossRef] [PubMed]
- Ravnik-Glavač, M.; Glavač, D. Circulating RNAs as potential biomarkers in amyotrophic lateral sclerosis. Int. J. Mol. Sci. 2020, 21, 1714. [Google Scholar] [CrossRef] [PubMed]
- Martinez, B.; Peplow, P.V. MicroRNAs in blood and cerebrospinal fluid as diagnostic biomarkers of multiple sclerosis and to monitor disease progression. Neural Regen. Res. 2020, 15, 606–619. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Taheri, M. A comprehensive review of non-coding RNAs functions in multiple sclerosis. Eur. J. Pharmacol. 2020, 879, 173127. [Google Scholar] [CrossRef]
- Atif, H.; Hicks, S.D. A review of MicroRNA biomarkers in traumatic brain injury. J. Exp. Neurosci. 2019, 13, 1179069519832286. [Google Scholar] [CrossRef]
- Guedes, V.A.; Devoto, C.; Leete, J.; Sass, D.; Acott, J.D.; Mithani, S.; Gill, J.M. Extracellular vesicle proteins and microRNAs as biomarkers for traumatic brain injury. Front. Neurol. 2020, 11, 663. [Google Scholar] [CrossRef]
- Salloum-Asfar, S.; Satheesh, N.J.; Abdulla, S.A. Circulating miRNAs, Small but Promising Biomarkers for Autism Spectrum Disorder. Front. Mol. Neurosci. 2019, 12, 253. [Google Scholar] [CrossRef]
- Wu, X.; Li, W.; Zheng, Y. Recent progress on relevant microRNAs in autism spectrum disorders. Int. J. Mol. Sci. 2020, 21, 5904. [Google Scholar] [CrossRef]
- Fauré, J.; Lachenal, G.; Court, M.; Hirrlinger, J.; Chatellard-Causse, C.; Blot, B.; Grange, J.; Schoehn, G.; Goldberg, Y.; Boyer, V.; et al. Exosomes are released by cultured cortical neurones. Mol. Cell. Neurosci. 2006, 31, 642–648. [Google Scholar] [CrossRef]
- Lachenal, G.; Pernet-Gallay, K.; Chivet, M.; Hemming, F.J.; Belly, A.; Bodon, G.; Blot, B.; Haase, G.; Goldberg, Y.; Sadoul, R. Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. Mol. Cell. Neurosci. 2011, 46, 409–418. [Google Scholar] [CrossRef]
- Frühbeis, C.; Fröhlich, D.; Kuo, W.P.; Amphornrat, J.; Thilemann, S.; Saab, A.S.; Kirchhoff, F.; Möbius, W.; Goebbels, S.; Nave, K.A.; et al. Neurotransmitter-triggered transfer of exosomes mediates oligodendrocyte-neuron communication. PLoS Biol. 2013, 11, e1001604. [Google Scholar] [CrossRef] [PubMed]
- Chivet, M.; Javalet, C.; Laulagnier, K.; Blot, B.; Hemming, F.J.; Sadoul, R. Exosomes secreted by cortical neurons upon glutamatergic synapse activation specifically interact with neurons. J. Extracell. Vesicles 2014, 3, 24722. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Mesci, P.; Carromeu, C.; McClatchy, D.R.; Schiapparelli, L.; Yates, J.R., 3rd; Muotri, A.R.; Cline, H.T. Exosomes regulate neurogenesis and circuit assembly. Proc. Natl. Acad. Sci. USA 2019, 116, 16086–16094. [Google Scholar] [CrossRef]
- Andjus, P.; Kosanović, M.; Milićević, K.; Gautam, M.; Vainio, S.J.; Jagečić, D.; Kozlova, E.N.; Pivoriūnas, A.; Chachques, J.C.; Sakaj, M.; et al. Extracellular vesicles as innovative tool for diagnosis, regeneration and protection against neurological damage. Int. J. Mol. Sci. 2020, 21, 6859. [Google Scholar] [CrossRef]
- Jahangard, Y.; Monfared, H.; Moradi, A.; Zare, M.; Mirnajafi-Zadeh, J.; Mowla, S.J. Therapeutic effects of transplanted exosomes containing miR-29b to a rat model of Alzheimer’s disease. Front. Neurosci. 2020, 14, 564. [Google Scholar] [CrossRef]
- Marzano, M.; Bejoy, J.; Cheerathodi, M.R.; Sun, L.; York, S.B.; Zhao, J.; Kanekiyo, T.; Bu, G.; Meckes, D.G., Jr.; Li, Y. Differential effects of extracellular vesicles of lineage-specific human pluripotent stem cells on the cellular behaviors of isogenic cortical spheroids. Cells 2019, 8, 993. [Google Scholar] [CrossRef]
- Monni, E.; Congiu, T.; Massa, D.; Nat, R.; Diana, A. Human neurospheres: From stained sections to three-dimensional assembly. Transl. Neurosci. 2011, 2, 43–48. [Google Scholar] [CrossRef]
- Massa, D.; Pillai, R.; Monni, E.; Kokaia, Z.; Diana, A. Expression analysis of pluripotency-associated genes in human fetal cortical and striatal neural stem cells during differentiation. Transl. Neurosci. 2012, 3, 242–248. [Google Scholar] [CrossRef]
- Pusic, K.M.; Pusic, A.D.; Kraig, R.P. Environmental enrichment stimulates immune cell secretion of exosomes that promote CNS myelination and may regulate inflammation. Cell. Mol. Neurobiol. 2016, 36, 313–325. [Google Scholar] [CrossRef]
- Geng, W.; Tang, H.; Luo, S.; Lv, Y.; Liang, D.; Kang, X.; Hong, W. Exosomes from miRNA-126-modified ADSCs promotes functional recovery after stroke in rats by improving neurogenesis and suppressing microglia activation. Am. J. Transl. Res. 2019, 11, 780–792. [Google Scholar]
- Xin, H.; Wang, F.; Li, Y.; Lu, Q.E.; Cheung, W.L.; Zhang, Y.; Zhang, Z.G.; Chopp, M. Secondary release of exosomes from astrocytes contributes to the increase in neural plasticity and improvement of functional recovery after stroke in rats treated with exosomes harvested from MicroRNA 133b-overexpressing multipotent mesenchymal stromal cells. Cell Transplant. 2017, 26, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Katakowski, M.; Wang, F.; Qian, J.Y.; Liu, X.S.; Ali, M.M.; Buller, B.; Zhang, Z.G.; Chopp, M. MicroRNA cluster miR-17-92 cluster in exosomes enhance neuroplasticity and functional recovery after stroke in rats. Stroke 2017, 48, 747–753, Erratum in Stroke 2017, 48, e137. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Wang, F.; Cao, J.; Wang, C. Exosomes derived from microRNA-146a-5p-enriched bone marrow mesenchymal stem cells alleviate intracerebral hemorrhage by inhibiting neuronal apoptosis and microglial m1 polarization. Drug Des. Dev. Ther. 2020, 14, 3143–3158. [Google Scholar] [CrossRef] [PubMed]
- Ling, X.; Zhang, G.; Xia, Y.; Zhu, Q.; Zhang, J.; Li, Q.; Niu, X.; Hu, G.; Yang, Y.; Wang, Y.; et al. Exosomes from human urine-derived stem cells enhanced neurogenesis via miR-26a/HDAC6 axis after ischaemic stroke. J. Cell. Mol. Med. 2020, 24, 640–654. [Google Scholar] [CrossRef]
- Huang, S.; Ge, X.; Yu, J.; Han, Z.; Yin, Z.; Li, Y.; Chen, F.; Wang, H.; Zhang, J.; Lei, P. Increased miR-124-3p in microglial exosomes following traumatic brain injury inhibits neuronal inflammation and contributes to neurite outgrowth via their transfer into neurons. FASEB J. 2018, 32, 512–528, Erratum in FASEB J. 2018, 32, 2315. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ye, Y.; Kong, C.; Su, X.; Zhang, X.; Bai, W.; He, X. MiR-124 enriched exosomes promoted the M2 polarization of microglia and enhanced hippocampus neurogenesis after traumatic brain injury by inhibiting TLR4 pathway. Neurochem. Res. 2019, 44, 811–828. [Google Scholar] [CrossRef]
- Li, D.; Huang, S.; Yin, Z.; Zhu, J.; Ge, X.; Han, Z.; Tan, J.; Zhang, S.; Zhao, J.; Chen, F.; et al. Increases in miR-124-3p in microglial exosomes confer neuroprotective effects by targeting fip200-mediated neuronal autophagy following traumatic brain injury. Neurochem. Res. 2019, 44, 1903–1923. [Google Scholar] [CrossRef]
- Ge, X.; Guo, M.; Hu, T.; Li, W.; Huang, S.; Yin, Z.; Li, Y.; Chen, F.; Zhu, L.; Kang, C.; et al. Increased microglial exosomal miR-124-3p alleviates neurodegeneration and improves cognitive outcome after rmTBI. Mol. Ther. 2020, 28, 503–522. [Google Scholar] [CrossRef]
- Kang, J.; Li, Z.; Zhi, Z.; Wang, S.; Xu, G. MiR-21 derived from the exosomes of MSCs regulates the death and differentiation of neurons in patients with spinal cord injury. Gene Ther. 2019, 26, 491–503. [Google Scholar] [CrossRef]
- Xu, G.; Ao, R.; Zhi, Z.; Jia, J.; Yu, B. miR-21 and miR-19b delivered by hMSC-derived EVs regulate the apoptosis and differentiation of neurons in patients with spinal cord injury. J. Cell. Physiol. 2019, 234, 10205–10217. [Google Scholar] [CrossRef]
- Zhou, X.; Chu, X.; Yuan, H.; Qiu, J.; Zhao, C.; Xin, D.; Li, T.; Ma, W.; Wang, H.; Wang, Z.; et al. Mesenchymal stem cell derived EVs mediate neuroprotection after spinal cord injury in rats via the microRNA-21-5p/FasL gene axis. Biomed. Pharmacother. 2019, 115, 108818. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, P.; Yao, X.; Li, H.; Shen, H.; Li, X.; Wu, J.; Lu, X. Exosomes derived from miR-133b-modified mesenchymal stem cells promote recovery after spinal cord injury. Front. Neurosci. 2018, 12, 845. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.W.; Zhou, J.G.; Xiong, Z.K.; Zhu, F.Z.; Guo, X.D. Effect of exosomes derived from MiR-133b-modified ADSCs on the recovery of neurological function after SCI. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 52–60. [Google Scholar] [CrossRef]
- Yu, T.; Zhao, C.; Hou, S.; Zhou, W.; Wang, B.; Chen, Y. Exosomes secreted from miRNA-29b-modified mesenchymal stem cells repaired spinal cord injury in rats. Braz. J. Med. Biol. Res. 2019, 52, e8735. [Google Scholar] [CrossRef]
- Li, C.; Li, X.; Zhao, B.; Wang, C. Exosomes derived from miR-544-modified mesenchymal stem cells promote recovery after spinal cord injury. Arch. Physiol. Biochem. 2020, 126, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Eiriz, M.F.; Malva, J.O.; Agasse, F.; Bernardino, L. Histamine in the neural and cancer stem cell niches. In Stem Cells and Cancer Stem Cells; Hayat, M., Ed.; Springer: Dordrecht, The Netherlands, 2014; Volume 12, pp. 3–17. [Google Scholar] [CrossRef]
- Snyder, E.Y.; Stephen Yip, S.; Pernia, C.; Lopez, C.A.; Liu, Y.; Sajti, E. Stem Cell Biology. In Fetal and Neonatal Physiology, 3rd ed.; Polin, R.A., Fox, W.W., Abman, S.H., Rowitch, D.H., Benitz, W.E., Eds.; Elsevier: Philadelphia, PA, USA, 2017; Volume 1, Chapter 6; pp. 54–75. [Google Scholar]
- Wigle, J.Y.; Eisenstat, D.E. Common signaling pathways used during development. In The Developing Human, Clinically Oriented Embryology, 11th ed.; Moore, K., Persaud, T.V.N., Torchia, M., Eds.; Elsevier: Philadelphia, PA, USA, 2020; Chapter 21; pp. 463–495. [Google Scholar]
- Dirks, P.B. Brain tumor stem cells: The cancer stem cell hypothesis writ large. Mol. Oncol. 2010, 4, 420–430. [Google Scholar] [CrossRef]
- Bayin, N.S.; Modrek, A.S.; Placantonakis, D.G. Brain tumor stem cells. In Molecular Pathology of Nervous System Tumors. Biological Stratification and Targeted Therapies; Karajannis, M.A., Zagzag, D., Eds.; Springer: New York, NY, USA, 2015; Chapter 2; pp. 23–34. [Google Scholar] [CrossRef]
- Aghajani, M.; Mansoori, B.; Mohammadi, A.; Asadzadeh, Z.; Baradaran, B. New emerging roles of CD133 in cancer stem cell: Signaling pathway and miRNA regulation. J. Cell. Physiol. 2019, 234, 21642–21661. [Google Scholar] [CrossRef]
- Piras, F.; Perra, M.T.; Murtas, D.; Minerba, L.; Floris, C.; Maxia, C.; Demurtas, P.; Ugalde, J.; Ribatti, D.; Sirigu, P. Combinations of apoptosis and cell-cycle control biomarkers predict the outcome of human melanoma. Oncol. Rep. 2008, 20, 271–277. [Google Scholar] [CrossRef][Green Version]
- Pessoa, I.A.; da Silva, F.P.E.; Anselmo, N.P.; de Oliveira, E.H.C. Alterations in TP53 gene–Implications in tumorigenesis. Process and prognosis in central nervous system cancer. In Tumors of the Central Nervous System-Primary and Secondary; Morgan, L.R., Ed.; IntechOpen: London, UK, 2014; Chapter 6; pp. 127–174. [Google Scholar] [CrossRef]
- Speciale, A.; Monti, P.; Fronza, G.; Izzotti, A.; Menichini, P. MicroRNA-mutant P53 crosstalk in chemoresistance: A hint to monitor therapy outcome. MicroRNA 2020. Advance online publication. [Google Scholar] [CrossRef]
- An, X.; Sarmiento, C.; Tan, T.; Zhu, H. Regulation of multidrug resistance by microRNAs in anti-cancer therapy. Acta Pharm. Sin. B 2017, 7, 38–51. [Google Scholar] [CrossRef]
- Safa, A.R. Resistance to cell death and its modulation in cancer stem cells. Crit. Rev. Oncog. 2016, 21, 203–219. [Google Scholar] [CrossRef] [PubMed]
- Ukrainskaya, V.M.; Stepanov, A.V.; Glagoleva, I.S.; Knorre, V.D.; Belogurov, A.A.J.; Gabibov, A.G. Death receptors: New opportunities in cancer therapy. Acta Nat. 2017, 9, 55–63. [Google Scholar] [CrossRef]
- Brower, J.V.; Clark, P.A.; Lyon, W.; Kuo, J.S. MicroRNAs in cancer: Glioblastoma and glioblastoma cancer stem cells. Neurochem. Int. 2014, 77, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Gorain, M.; Kundu, G.; Kundu, G.C. Therapeutic implications of cellular and molecular biology of cancer stem cells in melanoma. Mol. Cancer 2017, 16, 7. [Google Scholar] [CrossRef] [PubMed]
- Aravindan, N.; Jain, D.; Somasundaram, D.B.; Herman, T.S.; Aravindan, S. Cancer stem cells in neuroblastoma therapy resistance. Cancer Drug Resist. 2019, 2, 948–967. [Google Scholar] [CrossRef] [PubMed]
- Islam, F.; Gopalan, V.; Lam, A.K.Y. Cancer stem cells: Role in tumor progression and treatment resistance. In Oncogenomics. From Basic Research to Precision Medicine; Dammacco, F., Silvestris, F., Eds.; Elsevier: Philadelphia, PA, USA, 2019; Chapter 6; pp. 77–87. [Google Scholar] [CrossRef]
- Mens, M.M.J.; Ghanbari, M. Cell cycle regulation of stem cells by microRNAs. Stem Cell Rev. Rep. 2018, 14, 309–322. [Google Scholar] [CrossRef]
- Bhere, D.; Tamura, K.; Wakimoto, H.; Choi, S.H.; Purow, B.; Debatisse, J.; Shah, K. microRNA-7 upregulates death receptor 5 and primes resistant brain tumors to caspase-mediated apoptosis. Neuro Oncol. 2018, 20, 215–224. [Google Scholar] [CrossRef]
- Geng, J.; Luo, H.; Pu, Y.; Zhou, Z.; Wu, X.; Xu, W.; Yang, Z. Methylation mediated silencing of miR-23b expression and its role in glioma stem cells. Neurosci. Lett. 2012, 528, 185–189. [Google Scholar] [CrossRef]
- Reuland, S.N.; Smith, S.M.; Bemis, L.T.; Goldstein, N.B.; Almeida, A.R.; Partyka, K.A.; Marquez, V.E.; Zhang, Q.; Norris, D.A.; Shellman, Y.G. MicroRNA-26a is strongly downregulated in melanoma and induces cell death through repression of silencer of death domains (SODD). J. Investig. Dermatol. 2013, 133, 1286–1293. [Google Scholar] [CrossRef]
- Qian, H.; Yang, C.; Yang, Y. MicroRNA-26a inhibits the growth and invasiveness of malignant melanoma and directly targets on MITF gene. Cell Death Discov. 2017, 3, 17028. [Google Scholar] [CrossRef]
- Valdés-Rives, S.A.; Casique-Aguirre, D.; Germán-Castelán, L.; Velasco-Velázquez, M.A.; González-Arenas, A. Apoptotic signaling pathways in glioblastoma and therapeutic implications. Biomed. Res. Int. 2017, 2017, 7403747. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, N.; Yan, Z.; Li, C.; Zhao, Z. MiR-29a-mediated CD133 expression contributes to cisplatin resistance in CD133+ glioblastoma stem cells. J. Mol. Neurosci. 2018, 66, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Rokavec, M.; Li, H.; Jiang, L.; Hermeking, H. The p53/miR-34 axis in development and disease. J. Mol. Cell Biol. 2014, 6, 214–230. [Google Scholar] [CrossRef] [PubMed]
- de Antonellis, P.; Medaglia, C.; Cusanelli, E.; Andolfo, I.; Liguori, L.; De Vita, G.; Carotenuto, M.; Bello, A.; Formiggini, F.; Galeone, A.; et al. MiR-34a targeting of Notch ligand delta-like 1 impairs CD15+/CD133+ tumor-propagating cells and supports neural differentiation in medulloblastoma. PLoS ONE 2011, 6, e24584. [Google Scholar] [CrossRef] [PubMed]
- Guessous, F.; Zhang, Y.; Kofman, A.; Catania, A.; Li, Y.; Schiff, D.; Purow, B.; Abounader, R. MicroRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle 2010, 9, 1031–1036. [Google Scholar] [CrossRef]
- Sun, L.; Wu, Z.; Shao, Y.; Pu, Y.; Miu, W.; Yao, J.; Wu, Y.; Yang, Z. MicroRNA-34a suppresses cell proliferation and induces apoptosis in U87 glioma stem cells. Technol. Cancer Res. Treat. 2012, 11, 483–490. [Google Scholar] [CrossRef]
- Si, W.; Shen, J.; Zheng, H.; Fan, W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin. Epigenetics 2019, 11, 25. [Google Scholar] [CrossRef]
- Prokopi, M.; Kousparou, C.A.; Epenetos, A.A. The Secret Role of microRNAs in cancer stem cell development and potential therapy: A notch-pathway approach. Front. Oncol. 2015, 4, 389. [Google Scholar] [CrossRef]
- Lodygin, D.; Tarasov, V.; Epanchintsev, A.; Berking, C.; Knyazeva, T.; Körner, H.; Knyazev, P.; Diebold, J.; Hermeking, H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle (Georget. Tex.) 2008, 7, 2591–2600. [Google Scholar] [CrossRef]
- Chen, L.; Chen, X.R.; Chen, F.F.; Liu, Y.; Li, P.; Zhang, R.; Yan, K.; Yi, Y.J.; Xu, Z.M.; Jiang, X.D. MicroRNA-107 inhibits U87 glioma stem cells growth and invasion. Cell. Mol. Neurobiol. 2013, 33, 651–657. [Google Scholar] [CrossRef]
- Silber, J.; Lim, D.A.; Petritsch, C.; Persson, A.I.; Maunakea, A.K.; Yu, M.; Vandenberg, S.R.; Ginzinger, D.G.; James, C.D.; Costello, J.F.; et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008, 6, 14. [Google Scholar] [CrossRef] [PubMed]
- Meza-Sosa, K.F.; Pedraza-Alva, G.; Pérez-Martínez, L. microRNAs: Key triggers of neuronal cell fate. Front. Cell. Neurosci. 2014, 8, 175. [Google Scholar] [CrossRef] [PubMed]
- Bier, A.; Giladi, N.; Kronfeld, N.; Lee, H.K.; Cazacu, S.; Finniss, S.; Xiang, C.; Poisson, L.; de Carvalho, A.C.; Slavin, S.; et al. MicroRNA-137 is downregulated in glioblastoma and inhibits the stemness of glioma stem cells by targeting RTVP-1. Oncotarget 2013, 4, 665–676. [Google Scholar] [CrossRef]
- Deng, Y.; Deng, H.; Bi, F.; Liu, J.; Bemis, L.T.; Norris, D.; Wang, X.J.; Zhang, Q. MicroRNA-137 targets carboxyl-terminal binding protein 1 in melanoma cell lines. Int. J. Biol. Sci. 2011, 7, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Wang, W.W.; Chen, W.; Lu, W.Y.; Shang, A.Q. Mechanism of miR-137 regulating migration and invasion of melanoma cells by targeting PIK3R3 gene. J. Cell. Biochem. 2018. [Google Scholar] [CrossRef] [PubMed]
- Varrone, F.; Caputo, E. The miRNAs Role in Melanoma and in Its Resistance to Therapy. Int. J. Mol. Sci. 2020, 21, 878. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, S.; Ma, P.; Jing, Y.; Peng, H.; Gao, W.Q.; Zhuang, G. Lin28B promotes melanoma growth by mediating a microRNA regulatory circuit. Carcinogenesis 2015, 36, 937–945. [Google Scholar] [CrossRef]
- Afrang, N.; Imani, M.; Honardoost, M. Melanoma and Associated MicroRNAs. J. Skin Stem Cell 2016, 3, e63854. [Google Scholar] [CrossRef]
- Garg, N.; Vijayakumar, T.; Bakhshinyan, D.; Venugopal, C.; Singh, S.K. MicroRNA regulation of brain tumour initiating cells in central nervous system tumours. Stem Cells Int. 2015, 2015, 141793. [Google Scholar] [CrossRef]
- Shi, L.; Zhang, J.; Pan, T.; Zhou, J.; Gong, W.; Liu, N.; Fu, Z.; You, Y. MiR-125b is critical for the suppression of human U251 glioma stem cell proliferation. Brain Res. 2010, 1312, 120–126. [Google Scholar] [CrossRef]
- Zhao, B.; Bian, E.B.; Li, J.; Li, J. New advances of microRNAs in glioma stem cells, with special emphasis on aberrant methylation of microRNAs. J. Cell. Physiol. 2014, 229, 1141–1147. [Google Scholar] [CrossRef] [PubMed]
- Macharia, L.W.; Wanjiru, C.M.; Mureithi, M.W.; Pereira, C.M.; Ferrer, V.P.; Moura-Neto, V. MicroRNAs, hypoxia and the stem-like state as contributors to cancer aggressiveness. Front. Genet. 2019, 10, 125. [Google Scholar] [CrossRef] [PubMed]
- Haemmig, S.; Baumgartner, U.; Glück, A.; Zbinden, S.; Tschan, M.P.; Kappeler, A.; Mariani, L.; Vajtai, I.; Vassella, E. miR-125b controls apoptosis and temozolomide resistance by targeting TNFAIP3 and NKIRAS2 in glioblastomas. Cell Death Dis. 2014, 5, e1279. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Fu, X.; Wan, Y.; Wang, Z.; Jiang, D.; Shi, L. miR-125b inhibitor enhance the chemosensitivity of glioblastoma stem cells to temozolomide by targeting Bak1. Tumour Biol. 2014, 35, 6293–6302. [Google Scholar] [CrossRef]
- Wang, Y.; Zeng, G.; Jiang, Y. The emerging roles of miR-125b in cancers. Cancer Manag. Res. 2020, 12, 1079–1088. [Google Scholar] [CrossRef]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluids-the mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef]
- Le, M.T.; Teh, C.; Shyh-Chang, N.; Xie, H.; Zhou, B.; Korzh, V.; Lodish, H.F.; Lim, B. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009, 23, 862–876. [Google Scholar] [CrossRef]
- Rambow, F.; Bechadergue, A.; Luciani, F.; Gros, G.; Domingues, M.; Bonaventure, J.; Meurice, G.; Marine, J.C.; Larue, L. Regulation of Melanoma Progression through the TCF4/miR-125b/NEDD9 Cascade. J. Investig. Dermatol. 2016, 136, 1229–1237. [Google Scholar] [CrossRef]
- Wozniak, M. microRNA in the control of stem-like phenotype of cancer cells. Cent. Eur. J. Biol. 2013, 8, 931–942. [Google Scholar] [CrossRef]
- Godlewski, J.; Nowicki, M.O.; Bronisz, A.; Williams, S.; Otsuki, A.; Nuovo, G.; Raychaudhury, A.; Newton, H.B.; Chiocca, E.A.; Lawler, S. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008, 68, 9125–9130. [Google Scholar] [CrossRef]
- Fang, Y.; Zhang, L.; Li, Z.; Li, Y.; Huang, C.; Lu, X. MicroRNAs in DNA damage response, carcinogenesis, and chemoresistance. In International Review of Cell and Molecular Biology. MiRNAs in Differentiation and Development, 1st ed.; Galluzzi, L., Vitale, I., Eds.; Academic Press (Elsevier): Cambridge, MA, USA, 2017; Volume 333, pp. 1–49. [Google Scholar] [CrossRef]
- Gao, X.; Jin, W. The emerging role of tumor-suppressive microRNA-218 in targeting glioblastoma stemness. Cancer Lett. 2014, 353, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.M.; Sun, C.Y.; Yu, S.Z.; Wang, Q.; An, T.L.; Li, Y.Y.; Kong, Y.L.; Wen, Y.J. Relationship between miR-218 and CDK6 expression and their biological impact on glioma cell proliferation and apoptosis. Zhonghua Bing Li Xue Za Zhi 2011, 40, 454–459. [Google Scholar] [PubMed]
- Xia, H.; Yan, Y.; Hu, M.; Wang, Y.; Wang, Y.; Dai, Y.; Chen, J.; Di, G.; Chen, X.; Jiang, X. MiR-218 sensitizes glioma cells to apoptosis and inhibits tumorigenicity by regulating ECOP-mediated suppression of NF-κB activity. Neuro Oncol. 2013, 15, 413–422. [Google Scholar] [CrossRef]
- Xu, H.; Hu, Y.; Qiu, W. Potential mechanisms of microRNA-129-5p in inhibiting cell processes including viability, proliferation, migration and invasiveness of glioblastoma cells U87 through targeting FNDC3B. Biomed. Pharmacother. 2017, 87, 405–411. [Google Scholar] [CrossRef]
- Niu, C.S.; Yang, Y.; Cheng, C.D. MiR-134 regulates the proliferation and invasion of glioblastoma cells by reducing Nanog expression. Int. J. Oncol. 2013, 42, 1533–1540. [Google Scholar] [CrossRef]
- Qiu, H.; Chen, F.; Chen, M. MicroRNA-138 negatively regulates the hypoxia-inducible factor 1α to suppress melanoma growth and metastasis. Biol. Open 2019, 8, bio042937. [Google Scholar] [CrossRef]
- Gao, X.; Zhu, X.; Sun, Y.; Liu, J. MicroRNA‑141 inhibits the self‑renewal of glioblastoma stem cells via Jagged1. Mol. Med. Rep. 2017, 16, 167–173. [Google Scholar] [CrossRef]
- Du, Y.; Li, J.; Xu, T.; Zhou, D.D.; Zhang, L.; Wang, X. MicroRNA-145 induces apoptosis of glioma cells by targeting BNIP3 and Notch signaling. Oncotarget 2017, 8, 61510–61527. [Google Scholar] [CrossRef]
- Qian, C.; Wang, B.; Zou, Y.; Zhang, Y.; Hu, X.; Sun, W.; Xiao, H.; Liu, H.; Shi, L. MicroRNA 145 enhances chemosensitivity of glioblastoma stem cells to demethoxycurcumin. Cancer Manag. Res. 2019, 11, 6829–6840. [Google Scholar] [CrossRef]
- Rani, S.B.; Rathod, S.S.; Karthik, S.; Kaur, N.; Muzumdar, D.; Shiras, A.S. MiR-145 functions as a tumor-suppressive RNA by targeting Sox9 and adducin 3 in human glioma cells. Neuro Oncol. 2013, 15, 1302–1316. [Google Scholar] [CrossRef]
- Yang, W.; Yu, H.; Shen, Y.; Liu, Y.; Yang, Z.; Sun, T. MiR-146b-5p overexpression attenuates stemness and radioresistance of glioma stem cells by targeting HuR/lincRNA-p21/β-catenin pathway. Oncotarget 2016, 7, 41505–41526. [Google Scholar] [CrossRef] [PubMed]
- Mao, F.; Zhang, J.; Cheng, X.; Xu, Q. miR-149 inhibits cell proliferation and enhances chemosensitivity by targeting CDC42 and BCL2 in neuroblastoma. Cancer Cell Int. 2019, 19, 357. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Deng, Y.; Liu, Y.; Chen, X.; Yang, G.; Mu, Y.; Zhang, D.; Kang, J.; Wu, Z. MicroRNA-153 is tumor suppressive in glioblastoma stem cells. Mol. Biol. Rep. 2013, 40, 2789–2798. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Yao, Y.; Wang, P.; Liu, Y.; Zhao, L.; Li, Z.; Li, Z.; Xue, Y. MiR-152 functions as a tumor suppressor in glioblastoma stem cells by targeting Krüppel-like factor 4. Cancer Lett. 2014, 355, 85–95. [Google Scholar] [CrossRef]
- Sasayama, T.; Tanaka, K.; Kohmura, E. The roles of microRNAs in glioblastoma biology and biomarker. In Neurooncology-Newer Developments; Agrawal, A., Ed.; IntechOpen: London, UK, 2016; Chapter 2; pp. 27–66. [Google Scholar] [CrossRef]
- Huang, S.X.; Zhao, Z.Y.; Weng, G.H.; He, X.Y.; Wu, C.J.; Fu, C.Y.; Sui, Z.Y.; Ma, Y.S.; Liu, T. Upregulation of miR-181a suppresses the formation of glioblastoma stem cells by targeting the Notch2 oncogene and correlates with good prognosis in patients with glioblastoma multiforme. Biochem. Biophys. Res. Commun. 2017, 486, 1129–1136. [Google Scholar] [CrossRef]
- González-Gómez, P.; Sánchez, P.; Mira, H. MicroRNAs as regulators of neural stem cell-related pathways in glioblastoma multiforme. Mol. Neurobiol. 2011, 44, 235–249. [Google Scholar] [CrossRef]
- Kouri, F.M.; Hurley, L.A.; Daniel, W.L.; Day, E.S.; Hua, Y.; Hao, L.; Peng, C.Y.; Merkel, T.J.; Queisser, M.A.; Ritner, C.; et al. miR-182 integrates apoptosis, growth, and differentiation programs in glioblastoma. Genes Dev. 2015, 29, 732–745. [Google Scholar] [CrossRef]
- Matarredona, E.R.; Pastor, A.M. Neural stem cells of the subventricular zone as the origin of human glioblastoma stem cells. Therapeutic implications. Front. Oncol. 2019, 9, 779. [Google Scholar] [CrossRef]
- Zhao, C.; Ma, Z.G.; Mou, S.L.; Yang, Y.X.; Zhang, Y.H.; Yao, W.C. Targeting effect of microRNA on CD133 and its impact analysis on proliferation and invasion of glioma cells. Genet. Mol. Res. 2017, 16. [Google Scholar] [CrossRef]
- Deng, Y.; Zhu, G.; Luo, H.; Zhao, S. MicroRNA-203 as a stemness inhibitor of glioblastoma stem cells. Mol. Cells 2016, 39, 619–624. [Google Scholar] [CrossRef]
- Asuthkar, S.; Velpula, K.K.; Chetty, C.; Gorantla, B.; Rao, J.S. Epigenetic regulation of miRNA-211 by MMP-9 governs glioma cell apoptosis, chemosensitivity and radiosensitivity. Oncotarget 2012, 3, 1439–1454. [Google Scholar] [CrossRef]
- Aftab, M.N.; Dinger, M.E.; Perera, R.J. The role of microRNAs and long non-coding RNAs in the pathology, diagnosis, and management of melanoma. Arch. Biochem. Biophys. 2014, 563, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Mazar, J.; DeYoung, K.; Khaitan, D.; Meister, E.; Almodovar, A.; Goydos, J.; Ray, A.; Perera, R.J. The regulation of miRNA-211 expression and its role in melanoma cell invasiveness. PLoS ONE 2010, 5, e13779. [Google Scholar] [CrossRef] [PubMed]
- Levy, C.; Khaled, M.; Iliopoulos, D.; Janas, M.M.; Schubert, S.; Pinner, S.; Chen, P.H.; Li, S.; Fletcher, A.L.; Yokoyama, S.; et al. Intronic miR-211 assumes the tumor suppressive function of its host gene in melanoma. Mol. Cell. 2010, 40, 841–849. [Google Scholar] [CrossRef] [PubMed]
- Margue, C.; Philippidou, D.; Reinsbach, S.E.; Schmitt, M.; Behrmann, I.; Kreis, S. New target genes of MITF-induced microRNA-211 contribute to melanoma cell invasion. PLoS ONE 2013, 8, e73473. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Du, Y.; Chen, X.; Li, P.; Wang, Y.; Zang, W.; Zhao, L.; Li, Z.; Zhao, G. Expression patterns of microRNA-218 and its potential functions by targeting CIP2A and BMI1 genes in melanoma. Tumor. Biol. 2014, 35, 8007–8015. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Menggen, Q.; Wuren, Q.; Shi, Q.; Pi, X. MiR-219-5p inhibits the growth and metastasis of malignant melanoma by targeting BCL-2. BioMed Res. Int. 2017, 2017, 9032502. [Google Scholar] [CrossRef]
- Fareh, M.; Turchi, L.; Virolle, V.; Debruyne, D.; Almairac, F.; de-la-Forest Divonne, S.; Paquis, P.; Preynat-Seauve, O.; Krause, K.H.; Chneiweiss, H.; et al. The miR 302-367 cluster drastically affects self-renewal and infiltration properties of glioma-initiating cells through CXCR4 repression and consequent disruption of the SHH-GLI-NANOG network. Cell Death Differ. 2012, 19, 232–244. [Google Scholar] [CrossRef]
- Kefas, B.; Comeau, L.; Floyd, D.H.; Seleverstov, O.; Godlewski, J.; Schmittgen, T.; Jiang, J.; diPierro, C.G.; Li, Y.; Chiocca, E.A.; et al. The neuronal microRNA miR-326 acts in a feedback loop with notch and has therapeutic potential against brain tumors. J. Neurosci. 2009, 29, 15161–15168. [Google Scholar] [CrossRef]
- Du, W.; Liu, X.; Chen, L.; Dou, Z.; Lei, X.; Chang, L.; Cai, J.; Cui, Y.; Yang, D.; Sun, Y.; et al. Targeting the SMO oncogene by miR-326 inhibits glioma biological behaviors and stemness. Neuro Oncol. 2015, 17, 243–253. [Google Scholar] [CrossRef]
- Miele, E.; Po, A.; Begalli, F.; Antonucci, L.; Mastronuzzi, A.; Marras, C.E.; Carai, A.; Cucchi, D.; Abballe, L.; Besharat, Z.M.; et al. β-arrestin1-mediated acetylation of Gli1 regulates Hedgehog/Gli signaling and modulates self-renewal of SHH medulloblastoma cancer stem cells. BMC Cancer 2017, 17, 488. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Shi, P.; Zhao, G.; Xu, J.; Peng, W.; Zhang, J.; Zhang, G.; Wang, X.; Dong, Z.; Chen, F.; et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target. Ther. 2020, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Qiu, S.; Ge, R.; He, L.; Li, M.; Li, Y.; Peng, Y. miR-340 suppresses glioblastoma multiforme. Oncotarget 2015, 6, 9257–9270. [Google Scholar] [CrossRef] [PubMed]
- Nan, Y.; Han, L.; Zhang, A.; Wang, G.; Jia, Z.; Yang, Y.; Yue, X.; Pu, P.; Zhong, Y.; Kang, C. MiRNA-451 plays a role as tumor suppressor in human glioma cells. Brain Res. 2010, 1359, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Gal, H.; Pandi, G.; Kanner, A.A.; Ram, Z.; Lithwick-Yanai, G.; Amariglio, N.; Rechavi, G.; Givol, D. MIR-451 and Imatinib mesylate inhibit tumor growth of Glioblastoma stem cells. Biochem. Biophys. Res. Commun. 2008, 376, 86–90. [Google Scholar] [CrossRef]
- Tian, Y.; Nan, Y.; Han, L.; Zhang, A.; Wang, G.; Jia, Z.; Hao, J.; Pu, P.; Zhong, Y.; Kang, C. MicroRNA miR-451 downregulates the PI3K/AKT pathway through CAB39 in human glioma. Int. J. Oncol 2012, 40, 1105–1112. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, X.; Lian, H.; Liu, J.; Zhou, B.; Han, S.; Peng, B.; Yin, J.; Liu, W.; He, X. MicroRNA-503 acts as a tumor suppressor in glioblastoma for multiple antitumor effects by targeting IGF-1R. Oncol. Rep. 2014, 31, 1445–1452. [Google Scholar] [CrossRef]
- Wang, Z.; Xue, Y.; Wang, P.; Zhu, J.; Ma, J. MiR-608 inhibits the migration and invasion of glioma stem cells by targeting macrophage migration inhibitory factor. Oncol. Rep. 2016, 35, 2733–2742. [Google Scholar] [CrossRef]
- Fang, W.; Fan, Y.; Fa, Z.; Xu, J.; Yu, H.; Li, P.; Gu, J. microRNA-625 inhibits tumorigenicity by suppressing proliferation, migration and invasion in malignant melanoma. Oncotarget 2017, 8, 13253–13263. [Google Scholar] [CrossRef]
- Wang, R.J.; Li, J.W.; Bao, B.H.; Wu, H.C.; Du, Z.H.; Su, J.L.; Zhang, M.H.; Liang, H.Q. MicroRNA-873 (miRNA-873) inhibits glioblastoma tumorigenesis and metastasis by suppressing the expression of IGF2BP1. J. Biol. Chem. 2015, 290, 8938–8948. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, X.; Greshock, J.; Shen, L.; Yang, X.; Shao, Z.; Liang, S.; Tanyi, J.L.; Sood, A.K.; Zhang, L. Genomic DNA copy-number alterations of the let-7 family in human cancers. PLoS ONE 2012, 7, e44399. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Zheng, J.; Liu, X.; Ma, J.; Liu, Y.; Xue, Y. Knockdown of NEAT1 restrained the malignant progression of glioma stem cells by activating microRNA let-7e. Oncotarget 2016, 7, 62208–62223. [Google Scholar] [CrossRef] [PubMed]
- Po, A.; Abballe, L.; Sabato, C.; Gianno, F.; Chiacchiarini, M.; Catanzaro, G.; De Smaele, E.; Giangaspero, F.; Ferretti, E.; Miele, E.; et al. Sonic Hedgehog medulloblastoma cancer stem cells mirnome and transcriptome highlight novel functional networks. Int. J. Mol. Sci. 2018, 19, 2326. [Google Scholar] [CrossRef] [PubMed]
- Chirshev, E.; Oberg, K.C.; Ioffe, Y.J.; Unternaehrer, J.J. Let-7 as biomarker, prognostic indicator, and therapy for precision medicine in cancer. Clin. Transl. Med. 2019, 8, 24. [Google Scholar] [CrossRef]
- Bahmad, H.F.; Chamaa, F.; Assi, S.; Chalhoub, R.M.; Abou-Antoun, T.; Abou-Kheir, W. Cancer stem cells in neuroblastoma: Expanding the therapeutic frontier. Front. Mol. Neurosci. 2019, 12, 131. [Google Scholar] [CrossRef]
- Lozier, A.M.; Rich, M.E.; Grawe, A.P.; Peck, A.S.; Zhao, P.; Chang, A.T.; Bond, J.P.; Sholler, G.S. Targeting ornithine decarboxylase reverses the LIN28/Let-7 axis and inhibits glycolytic metabolism in neuroblastoma. Oncotarget 2015, 6, 196–206. [Google Scholar] [CrossRef]
- Coolen, M.; Katz, S.; Bally-Cuif, L. miR-9: A versatile regulator of neurogenesis. Front. Cell. Neurosci. 2013, 7, 220. [Google Scholar] [CrossRef]
- Ali Hosseini Rad, S.M.; Bavarsad, M.S.; Arefian, E.; Jaseb, K.; Shahjahani, M.; Saki, N. The role of micrornas in stemness of cancer stem cells. Oncol. Rev. 2013, 7, e8. [Google Scholar] [CrossRef][Green Version]
- Munoz, J.L.; Rodriguez-Cruz, V.; Rameshwar, P. High expression of miR-9 in CD133+ glioblastoma cells in chemoresistance to temozolomide. J. Cancer Stem Cell Res. 2015, 3, e1003. [Google Scholar] [CrossRef]
- Jeon, H.M.; Sohn, Y.W.; Oh, S.Y.; Kim, S.H.; Beck, S.; Kim, S.; Kim, H. ID4 imparts chemoresistance and cancer stemness to glioma cells by derepressing miR-9*-mediated suppression of SOX2. Cancer Res. 2011, 71, 3410–3421. [Google Scholar] [CrossRef]
- Guessous, F.; Alvarado-Velez, M.; Marcinkiewicz, L.; Zhang, Y.; Kim, J.; Heister, S.; Kefas, B.; Godlewski, J.; Schiff, D.; Purow, B.; et al. Oncogenic effects of miR-10b in glioblastoma stem cells. J. Neurooncol. 2013, 112, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Teplyuk, N.M.; Uhlmann, E.J.; Gabriely, G.; Volfovsky, N.; Wang, Y.; Teng, J.; Karmali, P.; Marcusson, E.; Peter, M.; Mohan, A.; et al. Therapeutic potential of targeting microRNA-10b in established intracranial glioblastoma: First steps toward the clinic. EMBO Mol. Med. 2016, 8, 268–287. [Google Scholar] [CrossRef] [PubMed]
- Gabriely, G.; Wurdinger, T.; Kesari, S.; Esau, C.C.; Burchard, J.; Linsley, P.S.; Krichevsky, A.M. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol. Cell. Biol. 2008, 28, 5369–5380. [Google Scholar] [CrossRef]
- Gaur, A.B.; Holbeck, S.L.; Colburn, N.H.; Israel, M.A. Downregulation of Pdcd4 by mir-21 facilitates glioblastoma proliferation in vivo. Neuro Oncol. 2011, 13, 580–590. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wang, J.J.; Tang, H.M.; To, S.S. Targeting strategies on miRNA-21 and PDCD4 for glioblastoma. Arch. Biochem. Biophys. 2015, 580, 64–74. [Google Scholar] [CrossRef]
- Sekar, D.; Krishnan, R.; Panagal, M.; Sivakumar, P.; Gopinath, V.; Basam, V. Deciphering the role of microRNA 21 in cancer stem cells (CSCs). Genes Dis. 2016, 3, 277–281. [Google Scholar] [CrossRef]
- Gaziel-Sovran, A.; Segura, M.F.; Di Micco, R.; Collins, M.K.; Hanniford, D.; Vega-Saenz de Miera, E.; Rakus, J.F.; Dankert, J.F.; Shang, S.; Kerbel, R.S.; et al. miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell 2011, 20, 104–118. [Google Scholar] [CrossRef]
- Jiang, L.; Lv, X.; Li, J.; Li, J.; Li, X.; Li, W.; Li, Y. The status of microRNA-21 expression and its clinical significance in human cutaneous malignant melanoma. Acta Histochem. 2012, 114, 582–588. [Google Scholar] [CrossRef]
- Martin del Campo, S.E.; Latchana, N.; Levine, K.M.; Grignol, V.P.; Fairchild, E.T.; Jaime-Ramirez, A.C.; Dao, T.V.; Karpa, V.I.; Carson, M.; Ganju, A. MiR-21 enhances melanoma invasiveness via inhibition of tissue inhibitor of metalloproteinases 3 expression: In vivo effects of MiR-21 inhibitor. PLoS ONE 2015, 10, e0115919. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, A.; Li, Y.; Zhang, K.; Han, L.; Du, W.; Yan, W.; Li, R.; Wang, Y.; Wang, K.; et al. MiR-24 regulates the proliferation and invasion of glioma by ST7L via β-catenin/Tcf-4 signaling. Cancer Lett. 2013, 329, 174–180. [Google Scholar] [CrossRef]
- Chen, L.; Li, H.; Han, L.; Zhang, K.; Wang, G.; Wang, Y.; Liu, Y.; Zheng, Y.; Jiang, T.; Pu, P.; et al. Expression and function of miR-27b in human glioma. Oncol. Rep. 2011, 26, 1617–1621. [Google Scholar] [CrossRef] [PubMed]
- Che, S.; Sun, T.; Wang, J.; Jiao, Y.; Wang, C.; Meng, Q.; Qi, W.; Yan, Z. miR-30 overexpression promotes glioma stem cells by regulating Jak/STAT3 signaling pathway. Tumour Biol. 2015, 36, 6805–6811. [Google Scholar] [CrossRef] [PubMed]
- Li, X.T.; Wang, H.Z.; Wu, Z.W.; Yang, T.Q.; Zhao, Z.H.; Chen, G.L.; Xie, X.S.; Li, B.; Wei, Y.X.; Huang, Y.L.; et al. miR-494-3p Regulates cellular proliferation, invasion, migration, and apoptosis by PTEN/AKT signaling in human glioblastoma cells. Cell. Mol. Neurobiol. 2015, 35, 679–687. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.F.; Liao, F.; Wu, H.; Dai, J. Glioma stem cells-derived exosomal miR-26a promotes angiogenesis of microvessel endothelial cells in glioma. J. Exp. Clin. Cancer Res. 2019, 38, 201. [Google Scholar] [CrossRef]
- Wu, Z.B.; Cai, L.; Lin, S.J.; Lu, J.L.; Yao, Y.; Zhou, L.F. The miR-92b functions as a potential oncogene by targeting on Smad3 in glioblastomas. Brain Res. 2013, 1529, 16–25. [Google Scholar] [CrossRef]
- Sampath, P. miR-138: A prosurvival oncomiR for glioma stem cells and its therapeutic implications. Future Neurol. 2013, 8, 119–121. [Google Scholar] [CrossRef]
- Shen, X.; Li, J.; Liao, W.; Wang, J.; Chen, H.; Yao, Y.; Liu, H.; Ding, K. microRNA-149 targets caspase-2 in glioma progression. Oncotarget 2016, 7, 26388–26399. [Google Scholar] [CrossRef]
- Shirjang, S.; Mansoori, B.; Asghari, S.; Duijf, P.; Mohammadi, A.; Gjerstorff, M.; Baradaran, B. MicroRNAs in cancer cell death pathways: Apoptosis and necroptosis. Free Radic. Biol. Med. 2019, 139, 1–15. [Google Scholar] [CrossRef]
- Wu, D.; Wang, C. miR-155 regulates the proliferation of glioma cells through PI3K/AKT signaling. Front. Neurol. 2020, 11, 297. [Google Scholar] [CrossRef]
- Chen, G.; Chen, Z.; Zhao, H. MicroRNA-155-3p promotes glioma progression and temozolomide resistance by targeting Six1. J. Cell. Mol. Med. 2020, 24, 5363–5374. [Google Scholar] [CrossRef]
- Segura, M.F.; Hanniford, D.; Menendez, S.; Reavie, L.; Zou, X.; Alvarez-Diaz, S.; Zakrzewski, J.; Blochin, E.; Rose, A.; Bogunovic, D.; et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc. Natl. Acad. Sci. USA 2009, 106, 1814–1819. [Google Scholar] [CrossRef]
- Liu, S.; Howell, P.M.; Riker, A.I. Up-regulation of miR-182 expression after epigenetic modulation of human melanoma cells. Ann. Surg. Oncol. 2013, 20, 1745–1752. [Google Scholar] [CrossRef]
- Gajos-Michniewicz, A.; Czyz, M. Role of miRNAs in Melanoma Metastasis. Cancers 2019, 11, 326. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Han, D.; Chen, X.; Zhang, D.; Wang, L.; Shi, C.; Zhang, W.; Li, C.; Chen, X.; Liu, H.; et al. MiR-196a exerts its oncogenic effect in glioblastoma multiforme by inhibition of IκBα both in vitro and in vivo. Neuro Oncol. 2014, 16, 652–661. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, Y.; Zheng, J.; Liu, X.; Chen, J.; Liu, L.; Wang, P.; Xue, Y. GAS5 suppresses malignancy of human glioma stem cells via a miR-196a-5p/FOXO1 feedback loop. Biochim. Biophys. Acta Mol. Cell. Res. 2017, 1864, 1605–1617. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Martínez, M.; Benito-Jardón, L.; Alonso, L.; Koetz-Ploch, L.; Hernando, E.; Teixidó, J. miR-204-5p and miR-211-5p contribute to BRAF inhibitor resistance in melanoma. Cancer Res. 2018, 78, 1017–1030. [Google Scholar] [CrossRef] [PubMed]
- Motti, M.L.; Minopoli, M.; Di Carluccio, G.; Ascierto, P.A.; Carriero, M.V. MicroRNAs as key players in melanoma cell resistance to MAPK and immune checkpoint inhibitors. Int. J. Mol. Sci. 2020, 21, 4544. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Wei, J.; Guo, T.; Shen, Y.; Liu, F. Knockdown of miR-210 decreases hypoxic glioma stem cells stemness and radioresistance. Exp. Cell Res. 2014, 326, 22–35. [Google Scholar] [CrossRef]
- Bavelloni, A.; Ramazzotti, G.; Poli, A.; Piazzi, M.; Focaccia, E.; Blalock, W.; Faenza, I. MiRNA-210: A current overview. Anticancer Res. 2017, 37, 6511–6521. [Google Scholar] [CrossRef]
- Zhang, S.; Lai, N.; Liao, K.; Sun, J.; Lin, Y. MicroRNA-210 regulates cell proliferation and apoptosis by targeting regulator of differentiation 1 in glioblastoma cells. Folia Neuropathol. 2015, 53, 236–244. [Google Scholar] [CrossRef]
- Medina, R.; Zaidi, S.K.; Liu, C.G.; Stein, J.L.; van Wijnen, A.J.; Croce, C.M.; Stein, G.S. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res. 2008, 68, 2773–2780. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Z.; Zhang, J.X.; Zhang, A.L.; Shi, Z.D.; Han, L.; Jia, Z.F.; Yang, W.D.; Wang, G.X.; Jiang, T.; You, Y.P.; et al. MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol. Cancer 2010, 9, 229. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, J.; Han, L.; Zhang, A.; Zhang, C.; Zheng, Y.; Jiang, T.; Pu, P.; Jiang, C.; Kang, C. Downregulation of miR-221/222 sensitizes glioma cells to temozolomide by regulating apoptosis independently of p53 status. Oncol. Rep. 2012, 27, 854–860. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Kang, C.; You, Y.; Pu, P.; Yang, W.; Zhao, P.; Wang, G.; Zhang, A.; Jia, Z.; Han, L.; et al. Co-suppression of miR-221/222 cluster suppresses human glioma cell growth by targeting p27kip1 in vitro and in vivo. Int. J. Oncol. 2009, 34, 1653–1660. [Google Scholar] [CrossRef] [PubMed]
- Errico, M.C. Melanoma Stem-Like Cells and Melanoma Cell liNes: Main Molecular Pathways and Possible Microrna Involvement. Ph.D. Thesis, University of Catania, Catania, Italy, 2010. [Google Scholar]
- Wang, H.; Boussouar, A.; Mazelin, L.; Tauszig-Delamasure, S.; Sun, Y.; Goldschneider, D.; Paradisi, A.; Mehlen, P. The proto-oncogene c-Kit inhibits tumor growth by behaving as a dependence receptor. Mol. Cell 2018, 72, 413–425. [Google Scholar] [CrossRef]
- Gajos-Michniewicz, A.; Duechler, M.; Czyz, M. MiRNA in melanoma-derived exosomes. Cancer Lett. 2014, 347, 29–37. [Google Scholar] [CrossRef]
- Felicetti, F.; Errico, M.C.; Bottero, L.; Segnalini, P.; Stoppacciaro, A.; Biffoni, M.; Felli, N.; Mattia, G.; Petrini, M.; Colombo, M.P.; et al. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 2008, 68, 2745–2754. [Google Scholar] [CrossRef]
- Mirzaei, H.; Gholamin, S.; Shahidsales, S.; Sahebkar, A.; Jaafari, M.R.; Mirzaei, H.R.; Hassanian, S.M.; Avan, A. MicroRNAs as potential diagnostic and prognostic biomarkers in melanoma. Eur. J. Cancer 2016, 53, 25–32. [Google Scholar] [CrossRef]
- Floyd, D.H.; Zhang, Y.; Dey, B.K.; Kefas, B.; Breit, H.; Marks, K.; Dutta, A.; Herold-Mende, C.; Synowitz, M.; Glass, R.; et al. Novel anti-apoptotic microRNAs 582-5p and 363 promote human glioblastoma stem cell survival via direct inhibition of caspase 3, caspase 9, and Bim. PLoS ONE 2014, 9, e96239. [Google Scholar] [CrossRef]
- Shu, M.; Zheng, X.; Wu, S.; Lu, H.; Leng, T.; Zhu, W.; Zhou, Y.; Ou, Y.; Lin, X.; Lin, Y.; et al. Targeting oncogenic miR-335 inhibits growth and invasion of malignant astrocytoma cells. Mol. Cancer 2011, 10, 59. [Google Scholar] [CrossRef]
- Cheng, Q.; Cao, H.; Chen, Z.; Ma, Z.; Wan, X.; Peng, R.; Jiang, B. PAX6, a novel target of miR-335, inhibits cell proliferation and invasion in glioma cells. Mol. Med. Rep. 2014, 10, 399–404. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shea, A.; Harish, V.; Afzal, Z.; Chijioke, J.; Kedir, H.; Dusmatova, S.; Roy, A.; Ramalinga, M.; Harris, B.; Blancato, J.; et al. MicroRNAs in glioblastoma multiforme pathogenesis and therapeutics. Cancer Med. 2016, 5, 1917–1946. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kong, D.; Ahmad, A.; Bao, B.; Sarkar, F.H. miRNAs in cancer stem cells. In MicroRNA in Regenerative Medicine; Sen, C.K., Ed.; Academic Press (Elsevier): London, UK, 2015; Chapter 5; pp. 137–161. [Google Scholar]
- Swarbrick, A.; Woods, S.L.; Shaw, A.; Balakrishnan, A.; Phua, Y.; Nguyen, A.; Chanthery, Y.; Lim, L.; Ashton, L.J.; Judson, R.L.; et al. miR-380-5p represses p53 to control cellular survival and is associated with poor outcome in MYCN-amplified neuroblastoma. Nat. Med. 2010, 16, 1134–1140. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A.; Schmitz, U.; Raatz, Y.; Schönherr, M.; Kottek, T.; Schauer, M.; Franz, S.; Saalbach, A.; Anderegg, U.; Wolkenhauer, O.; et al. miR-638 promotes melanoma metastasis and protects melanoma cells from apoptosis and autophagy. Oncotarget 2015, 6, 2966–2980. [Google Scholar] [CrossRef]
- Zhu, G.; Wang, Y.; Mijiti, M.; Wang, Z.; Wu, P.F.; Jiafu, D. Upregulation of miR-130b enhances stem cell-like phenotype in glioblastoma by inactivating the Hippo signaling pathway. Biochem. Biophys. Res. Commun. 2015, 465, 194–199. [Google Scholar] [CrossRef]
- Chang, T.C.; Wentzel, E.A.; Kent, O.A.; Ramachandran, K.; Mullendore, M.; Lee, K.H.; Feldmann, G.; Yamakuchi, M.; Ferlito, M.; Lowenstein, C.J.; et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007, 26, 745–752. [Google Scholar] [CrossRef]
- Bommer, G.T.; Gerin, I.; Feng, Y.; Kaczorowski, A.J.; Kuick, R.; Love, R.E.; Zhai, Y.; Giordano, T.J.; Qin, Z.S.; Moore, B.B.; et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr. Biol. 2007, 17, 1298–1307. [Google Scholar] [CrossRef]
- Okada, N.; Lin, C.P.; Ribeiro, M.C.; Biton, A.; Lai, G.; He, X.; Bu, P.; Vogel, H.; Jablons, D.M.; Keller, A.C.; et al. A positive feedback between p53 and miR-34miRNAs mediates tumor suppression. Genes Dev. 2014, 28, 438–450. [Google Scholar] [CrossRef]
- Rathod, S.S.; Rani, S.B.; Khan, M.; Muzumdar, D.; Shiras, A. Tumor suppressive miRNA-34a suppresses cell proliferation and tumor growth of glioma stem cells by targeting Akt and Wnt signaling pathways. FEBS Open Bio 2014, 4, 485–495. [Google Scholar] [CrossRef]
- Kawamura, Y.; Takouda, J.; Yoshimoto, K.; Nakashima, K. New aspects of glioblastoma multiforme revealed by similarities between neural and glioblastoma stem cells. Cell. Biol. Toxicol. 2018, 34, 425–440. [Google Scholar] [CrossRef]
- Yao, M.; Li, S.; Wu, X.; Diao, S.; Zhang, G.; He, H.; Bian, L.; Lu, Y. Cellular origin of glioblastoma and its implication in precision therapy. Cell. Mol. Immunol. 2018, 15, 737–739. [Google Scholar] [CrossRef] [PubMed]
- Gimple, R.C.; Bhargava, S.; Dixit, D.; Rich, J.N. Glioblastoma stem cells: Lessons from the tumor hierarchy in a lethal cancer. Genes Dev. 2019, 33, 591–609. [Google Scholar] [CrossRef] [PubMed]
- Genovesi, L.A.; Carter, K.W.; Gottardo, N.G.; Giles, K.M.; Dallas, P.B. Integrated analysis of miRNA and mRNA expression in childhood medulloblastoma compared with neural stem cells. PLoS ONE 2011, 6, e23935. [Google Scholar] [CrossRef] [PubMed]
- Andolfo, I.; Liguori, L.; De Antonellis, P.; Cusanelli, E.; Marinaro, F.; Pistollato, F.; Garzia, L.; De Vita, G.; Petrosino, G.; Accordi, B.; et al. The micro-RNA 199b-5p regulatory circuit involves Hes1, CD15, and epigenetic modifications in medulloblastoma. Neuro Oncol. 2012, 14, 596–612. [Google Scholar] [CrossRef] [PubMed]
- Azzarelli, R.; Simons, B.D.; Philpott, A. The developmental origin of brain tumours: A cellular and molecular framework. Development 2018, 145, dev162693. [Google Scholar] [CrossRef]
- Agnihotri, S.; Munoz, D.; Zadeh, G.; Guha, A. Brain tumor-initiating cells and cells of origin in glioblastoma. Transl. Neurosci. 2011, 2, 331–338. [Google Scholar] [CrossRef][Green Version]
- Sun, X.; Jiao, X.; Pestell, T.G.; Fan, C.; Qin, S.; Mirabelli, E.; Ren, H.; Pestell, R.G. MicroRNAs and cancer stem cells: The sword and the shield. Oncogene 2014, 33, 4967–4977. [Google Scholar] [CrossRef]
- Kong, D.S. Cancer stem cells in brain tumors and their lineage hierarchy. Int. J. Stem Cells 2012, 5, 12–15. [Google Scholar] [CrossRef]
- Ahmed, A.U.; Auffinger, B.; Lesniak, M.S. Understanding glioma stem cells: Rationale, clinical relevance and therapeutic strategies. Expert Rev. Neurother. 2013, 13, 545–555. [Google Scholar] [CrossRef]
- Lathia, J.D.; Mack, S.C.; Mulkearns-Hubert, E.E.; Valentim, C.L.; Rich, J.N. Cancer stem cells in glioblastoma. Genes Dev. 2015, 29, 1203–1217. [Google Scholar] [CrossRef]
- Godlewski, J.; Ferrer-Luna, R.; Rooj, A.K.; Mineo, M.; Ricklefs, F.; Takeda, Y.S.; Nowicki, M.O.; Salińska, E.; Nakano, I.; Lee, H.; et al. MicroRNA signatures and molecular subtypes of glioblastoma: The role of extracellular transfer. Stem Cell Rep. 2017, 8, 1497–1505. [Google Scholar] [CrossRef]
- Nakagawara, A.; Li, Y.; Izumi, H.; Muramori, K.; Inada, H.; Nishi, M. Neuroblastoma. Jpn. J. Clin. Oncol. 2018, 48, 214–241. [Google Scholar] [CrossRef] [PubMed]
- Buhagiar, A.; Ayers, D. Chemoresistance, cancer stem cells, and miRNA influences: The case for neuroblastoma. Anal. Cell. Pathol. (Amst) 2015, 2015, 150634. [Google Scholar] [CrossRef] [PubMed]
- Veschi, V.; Verona, F.; Thiele, C.J. Cancer stem cells and neuroblastoma: Characteristics and therapeutic targeting options. Front. Endocrinol. (Lausanne) 2019, 10, 782. [Google Scholar] [CrossRef] [PubMed]
- Zage, P.E.; Whittle, S.B.; Shohet, J.M. CD114: A new member of the neural crest-derived cancer stem cell marker family. J. Cell. Biochem. 2017, 118, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Moon, H.; Donahue, L.R.; Choi, E.; Scumpia, P.O.; Lowry, W.E.; Grenier, J.K.; Zhu, J.; White, A.C. Melanocyte Stem Cell Activation and Translocation Initiate Cutaneous Melanoma in Response to UV Exposure. Cell Stem Cell 2017, 21, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Murtas, D.; Pilloni, L.; Diana, A.; Casula, L.; Tomei, S.; Piras, F.; Ferreli, C.; Maxia, C.; Perra, M.T. Tyrosinase and nestin immunohistochemical expression in melanocytic nevi as a histopathologic pattern to trace melanocyte differentiation and nevogenesis. Histochem. Cell. Biol. 2019, 151, 175–185. [Google Scholar] [CrossRef]
- Fang, D.; Nguyen, T.K.; Leishear, K.; Finko, R.; Kulp, A.N.; Hotz, S.; Van Belle, P.A.; Xu, X.; Elder, D.E.; Herlyn, M. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005, 65, 9328–9337. [Google Scholar] [CrossRef]
- Piras, F.; Perra, M.T.; Murtas, D.; Minerba, L.; Floris, C.; Maxia, C.; Demurtas, P.; Ugalde, J.; Ribatti, D.; Sirigu, P. The stem cell marker nestin predicts poor prognosis in human melanoma. Oncol. Rep. 2010, 23, 17–24. [Google Scholar] [CrossRef]
- Ramgolam, K.; Lauriol, J.; Lalou, C.; Lauden, L.; Michel, L.; de la Grange, P.; Khatib, A.M.; Aoudjit, F.; Charron, D.; Alcaide-Loridan, C.; et al. Melanoma spheroids grown under neural crest cell conditions are highly plastic migratory/invasive tumor cells endowed with immunomodulator function. PLoS ONE 2011, 6, e18784. [Google Scholar] [CrossRef]
- Lin, J.; Zhang, D.; Fan, Y.; Chao, Y.; Chang, J.; Li, N.; Han, L.; Han, C. Regulation of Cancer Stem Cell Self-Renewal by HOXB9 Antagonizes Endoplasmic Reticulum Stress-Induced Melanoma Cell Apoptosis via the miR-765-FOXA2 Axis. J. Investig. Dermatol. 2018, 138, 1609–1619. [Google Scholar] [CrossRef] [PubMed]
- Marzagalli, M.; Moretti, R.M.; Messi, E.; Marelli, M.M.; Fontana, F.; Anastasia, A.; Bani, M.R.; Beretta, G.; Limonta, P. Targeting melanoma stem cells with the Vitamin E derivative δ-tocotrienol. Sci. Rep. 2018, 8, 587. [Google Scholar] [CrossRef] [PubMed]
- Ross, R.A.; Spengler, B.A.; Domènech, C.; Porubcin, M.; Rettig, W.J.; Biedler, J.L. Human neuroblastoma I-type cells are malignant neural crest stem cells. Cell Growth Differ. 1995, 6, 449–456. [Google Scholar] [PubMed]
- Ross, R.A. Cellular Heterogeneity. In Neuroblastoma; Cheung, N.K.V., Cohn, S.L., Eds.; Springer: Berlin/Heidelberg, Germany, 2005; Chapter 6; pp. 55–61. [Google Scholar] [CrossRef]
- Masserot, C.; Liu, Q.; Nguyen, E.; Gattolliat, C.H.; Valteau-Couanet, D.; Bénard, J.; Huber, C.; Ségal-Bendirdjian, E. WT1 expression is inversely correlated with MYCN amplification or expression and associated with poor survival in non-MYCN-amplified neuroblastoma. Mol. Oncol. 2016, 10, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Sztiller-Sikorska, M.; Koprowska, K.; Jakubowska, J.; Zalesna, I.; Stasiak, M.; Duechler, M.; Czyz, M.E. Sphere formation and self-renewal capacity of melanoma cells is affected by the microenvironment. Melanoma Res. 2012, 22, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Sahranavardfard, P.; Firouzi, J.; Azimi, M.; Khosravani, P.; Heydari, R.; Emami Razavi, A.; Dorraj, M.; Keighobadi, F.; Ebrahimi, M. MicroRNA-203 reinforces stemness properties in melanoma and augments tumorigenesis in vivo. J. Cell. Physiol. 2019, 234, 20193–20205. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wang, J.; Zhao, H.; Wang, J.; Tony To, S.S. The role of Myc and let-7a in glioblastoma, glucose metabolism and response to therapy. Arch. Biochem. Biophys. 2015, 580, 84–92. [Google Scholar] [CrossRef]
- Molenaar, J.J.; Domingo-Fernández, R.; Ebus, M.E.; Lindner, S.; Koster, J.; Drabek, K.; Mestdagh, P.; van Sluis, P.; Valentijn, L.J.; van Nes, J.; et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat. Genet. 2012, 44, 1199–1206. [Google Scholar] [CrossRef]
- Dong, Z.; Lei, Q.; Yang, R.; Zhu, S.; Ke, X.X.; Yang, L.; Cui, H.; Yi, L. Inhibition of neurotensin receptor 1 induces intrinsic apoptosis via let-7a-3p/Bcl-w axis in glioblastoma. Br. J. Cancer 2017, 116, 1572–1584. [Google Scholar] [CrossRef]
- Inomistova, M.; Khranovska, N.; Skachkova, O. Role of genetic and epigenetic alterations in pathogenesis of neuroblastoma. In Neuroblastoma, Molecular Mechanisms and Therapeutic Interventions; Ray, S.K., Ed.; Academic Press (Elsevier): London, UK, 2019; Chapter 2; pp. 23–41. [Google Scholar]
- Yang, L.; Yan, Z.; Wang, Y.; Ma, W.; Li, C. Down-expression of miR-154 suppresses tumourigenesis in CD133 (+) glioblastoma stem cells. Cell Biochem. Funct. 2016, 34, 404–413. [Google Scholar] [CrossRef]
- Gillies, J.K.; Lorimer, I.A. Regulation of p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle 2007, 6, 2005–2009. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, J.; Xu, G.; Wang, W.; Liu, C.; Yang, H.; Yu, Z.; Lei, Q.; Xiao, L.; Xiong, J.; et al. Targeting miR-381-NEFL axis sensitizes glioblastoma cells to temozolomide by regulating stemness factors and multidrug resistance factors. Oncotarget 2015, 6, 3147–3164. [Google Scholar] [CrossRef] [PubMed]
- Streicher, K.L.; Zhu, W.; Lehmann, K.P.; Georgantas, R.W.; Morehouse, C.A.; Brohawn, P.; Carrasco, R.A.; Xiao, Z.; Tice, D.A.; Higgs, B.W.; et al. A novel oncogenic role for the miRNA-506-514 cluster in initiating melanocyte transformation and promoting melanoma growth. Oncogene 2012, 31, 1558–1570. [Google Scholar] [CrossRef] [PubMed]
- Heddleston, J.M.; Li, Z.; McLendon, R.E.; Hjelmeland, A.B.; Rich, J.N. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle 2009, 8, 3274–3284. [Google Scholar] [CrossRef]
- Méndez, O.; Zavadil, J.; Esencay, M.; Lukyanov, Y.; Santovasi, D.; Wang, S.C.; Newcomb, E.W.; Zagzag, D. Knock down of HIF-1alpha in glioma cells reduces migration in vitro and invasion in vivo and impairs their ability to form tumor spheres. Mol. Cancer 2010, 9, 133. [Google Scholar] [CrossRef]
- Heddleston, J.M.; Wu, Q.; Rivera, M.; Minhas, S.; Lathia, J.D.; Sloan, A.E.; Iliopoulos, O.; Hjelmeland, A.B.; Rich, J.N. Hypoxia-induced mixed-lineage leukemia 1 regulates glioma stem cell tumorigenic potential. Cell Death Differ. 2012, 19, 428–439. [Google Scholar] [CrossRef]
- Qiang, L.; Wu, T.; Zhang, H.W.; Lu, N.; Hu, R.; Wang, Y.J.; Zhao, L.; Chen, F.H.; Wang, X.T.; You, Q.D.; et al. HIF-1α is critical for hypoxia-mediated maintenance of glioblastoma stem cells by activating Notch signaling pathway. Cell Death Differ. 2012, 19, 284–294. [Google Scholar] [CrossRef]
- Auffinger, B.; Spencer, D.; Pytel, P.; Ahmed, A.U.; Lesniak, M.S. The role of glioma stem cells in chemotherapy resistance and glioblastoma multiforme recurrence. Expert Rev. Neurother. 2015, 15, 741–752. [Google Scholar] [CrossRef]
- Casciati, A.; Tanori, M.; Manczak, R.; Saada, S.; Tanno, B.; Giardullo, P.; Porcù, E.; Rampazzo, E.; Persano, L.; Viola, G.; et al. Human medulloblastoma cell lines: Investigating on cancer stem cell-like phenotype. Cancers 2020, 12, 226. [Google Scholar] [CrossRef]
- Qian, X.; Ren, Y.; Shi, Z.; Long, L.; Pu, P.; Sheng, J.; Yuan, X.; Kang, C. Sequence-dependent synergistic inhibition of human glioma cell lines by combined temozolomide and miR-21 inhibitor gene therapy. Mol. Pharm. 2012, 9, 2636–2645. [Google Scholar] [CrossRef]
- Gambari, R.; Brognara, E.; Spandidos, D.A.; Fabbri, E. Targeting oncomiRNAs and mimicking tumor suppressor miRNAs: New trends in the development of miRNA therapeutic strategies in oncology (Review). Int. J. Oncol. 2016, 49, 5–32. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, M.; Sekar, T.V.; Ananta, J.S.; Devulapally, R.; Afjei, R.; Babikir, H.A.; Paulmurugan, R.; Massoud, T.F. Targeted nanoparticle delivery of therapeutic antisense microRNAs presensitizes glioblastoma cells to lower effective doses of temozolomide in vitro and in a mouse model. Oncotarget 2018, 9, 21478–21494. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Costa, P.M.; Cardoso, A.L.; Custódia, C.; Cunha, P.; Pereira de Almeida, L.; Pedroso de Lima, M.C. MiRNA-21 silencing mediated by tumor-targeted nanoparticles combined with sunitinib: A new multimodal gene therapy approach for glioblastoma. J. Control. Release 2015, 207, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Rodova, M.; Nanta, R.; Meeker, D.; Van Veldhuizen, P.J.; Srivastava, R.K.; Shankar, S. NPV-LDE-225 (Erismodegib) inhibits epithelial mesenchymal transition and self-renewal of glioblastoma initiating cells by regulating miR-21, miR-128, and miR-200. Neuro Oncol. 2013, 15, 691–706. [Google Scholar] [CrossRef]
- Anthiya, S.; Griveau, A.; Loussouarn, C.; Baril, P.; Garnett, M.; Issartel, J.P.; Garcion, E. MicroRNA-based drugs for brain tumors. Trends Cancer 2018, 4, 222–238. [Google Scholar] [CrossRef]
- Shi, L.; Fei, X.; Wang, Z. PI3K inhibitor combined with miR-125b inhibitor sensitize TMZ-induced anti-glioma stem cancer effects through inactivation of Wnt/β-catenin signaling pathway. In Vitro Cell. Dev. Biol. Anim. 2015, 51, 1047–1055. [Google Scholar] [CrossRef]
- Jiang, W.; Finniss, S.; Cazacu, S.; Xiang, C.; Brodie, Z.; Mikkelsen, T.; Poisson, L.; Shackelford, D.B.; Brodie, C. Repurposing phenformin for the targeting of glioma stem cells and the treatment of glioblastoma. Oncotarget 2016, 7, 56456–56470. [Google Scholar] [CrossRef]
- Upraity, S.; Kazi, S.; Padul, V.; Shirsat, N.V. MiR-224 expression increases radiation sensitivity of glioblastoma cells. Biochem. Biophys. Res. Commun. 2014, 448, 225–230. [Google Scholar] [CrossRef]
- Bahreyni-Toossi, M.T.; Dolat, E.; Khanbabaei, H.; Zafari, N.; Azimian, H. microRNAs: Potential glioblastoma radiosensitizer by targeting radiation-related molecular pathways. Mutat. Res. 2019, 816, 111679. [Google Scholar] [CrossRef]
- Wu, H.; Liu, Q.; Cai, T.; Chen, Y.D.; Wang, Z.F. Induction of microRNA-146a is involved in curcumin-mediated enhancement of temozolomide cytotoxicity against human glioblastoma. Mol. Med. Rep. 2015, 12, 5461–5466. [Google Scholar] [CrossRef]
- Yin, S.; Du, W.; Wang, F.; Han, B.; Cui, Y.; Yang, D.; Chen, H.; Liu, D.; Liu, X.; Zhai, X.; et al. MicroRNA-326 sensitizes human glioblastoma cells to curcumin via the SHH/GLI1 signaling pathway. Cancer Biol. Ther. 2018, 19, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Xu, C.; Zhang, X.; Yu, A.; Shu, L. Shrimp miR-965 induced the human melanoma stem-like cell apoptosis and inhibited their stemness by disrupting the MCL-1-ER stress-XBP1 feedback loop in a cross-species manner. Stem Cell Res. Ther. 2020, 11, 248. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Wei, J.; Zhang, S.; Zhang, X. Shrimp miR-S8 suppresses the stemness of human melanoma stem-like cells by targeting the transcription factor YB-1. Cancer Res. 2017, 77, 5543–5553. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, X. Shrimp miRNA suppresses the stemness of human cancer stem cells via the PIN1 pathway. FASEB J. 2019, 33, 10767–10779. [Google Scholar] [CrossRef] [PubMed]
- Garg, M. Emerging role of microRNAs in cancer stem cells: Implications in cancer therapy. World J. Stem Cells 2015, 7, 1078–1089. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Tu, Y. Systematic review of microRNAs and its therapeutic potential in glioma. Cancer Transl. Med. 2015, 1, 50–66. [Google Scholar] [CrossRef]
- Halle, B.; Thomassen, M.; Venkatesan, R.; Kaimal, V.; Marcusson, E.G.; Munthe, S.; Sørensen, M.D.; Aaberg-Jessen, C.; Jensen, S.S.; Meyer, M.; et al. Shift of microRNA profile upon orthotopic xenografting of glioblastoma spheroid cultures. J. Neurooncol. 2016, 128, 395–404. [Google Scholar] [CrossRef]
- Rodrigues, A.R.; Neto, F.S.L.; Lourenço, L.G.; Trevisan, F.A.; Cirino, M.L.A.; Nery, B.; Peria, F.M.; Pereira-da-Silva, G.; Tazima, M.F.G.S.; Tirapelli, L.F.; et al. Expression of oncogenic microRNA-21 in neurospheres and attached cells of a glioblastoma cell line increased after treatment with temozolomide and ionizing radiation. Genet. Mol. Res. 2019, 18, gmr18095. [Google Scholar] [CrossRef]
- Yin, P.T.; Shah, B.P.; Lee, K.B. Combined magnetic nanoparticle-based microRNA and hyperthermia therapy to enhance apoptosis in brain cancer cells. Small 2014, 10, 4106–4112. [Google Scholar] [CrossRef]
- Klinger, N.V.; Mittal, S. Therapeutic potential of curcumin for the treatment of brain tumors. Oxid. Med. Cell. Longev. 2016, 2016, 9324085. [Google Scholar] [CrossRef]
- Bertoli, G.; Cava, C.; Castiglioni, I. MicroRNAs: New biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics 2015, 5, 1122–1143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, J.; Hao, J.; Shi, Z.; Wang, Y.; Han, L.; Yu, S.; You, Y.; Jiang, T.; Wang, J.; et al. High level of miR-221/222 confers increased cell invasion and poor prognosis in glioma. J. Transl. Med. 2012, 10, 119. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Sun, X.; Wang, W.; Jin, X.; Bo, X.; Li, Z.; Bian, A.; Jiu, J.; Wang, X.; Liu, D.; et al. Tumor microRNA-335 expression is associated with poor prognosis in human glioma. Med. Oncol. 2012, 29, 3472–3477. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Lian, Y.J.; Ma, Y.Q.; Wu, C.J.; Zheng, Y.K.; Xie, N.C. LncRNA SNHG1 promotes α-synuclein aggregation and toxicity by targeting miR-15b-5p to activate SIAH1 in human neuroblastoma SH-SY5Y cells. Neurotoxicology 2018, 68, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Aravindan, N.; Subramanian, K.; Somasundaram, D.B.; Herman, T.S.; Aravindan, S. MicroRNAs in neuroblastoma tumorigenesis, therapy resistance, and disease evolution. Cancer Drug Resist. 2019, 2, 1086–1105. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Doss, C.; Lee, S.S. Therapeutic miRNA and siRNA: Moving from bench to clinic as next generation medicine. Mol. Ther. Nucleic Acids 2017, 8, 132–143. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Hanna, J.; Hossain, G.S.; Kocerha, J. The Potential for microRNA Therapeutics and Clinical Research. Front. Genet. 2019, 10, 478. [Google Scholar] [CrossRef]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Lee, S.S. Therapeutic advances of miRNAs: A preclinical and clinical update. J. Adv. Res. 2020, in press. [Google Scholar] [CrossRef]
- Hong, D.S.; Kang, Y.K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.L.; Kim, T.Y.; et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 2020, 122. [Google Scholar] [CrossRef]
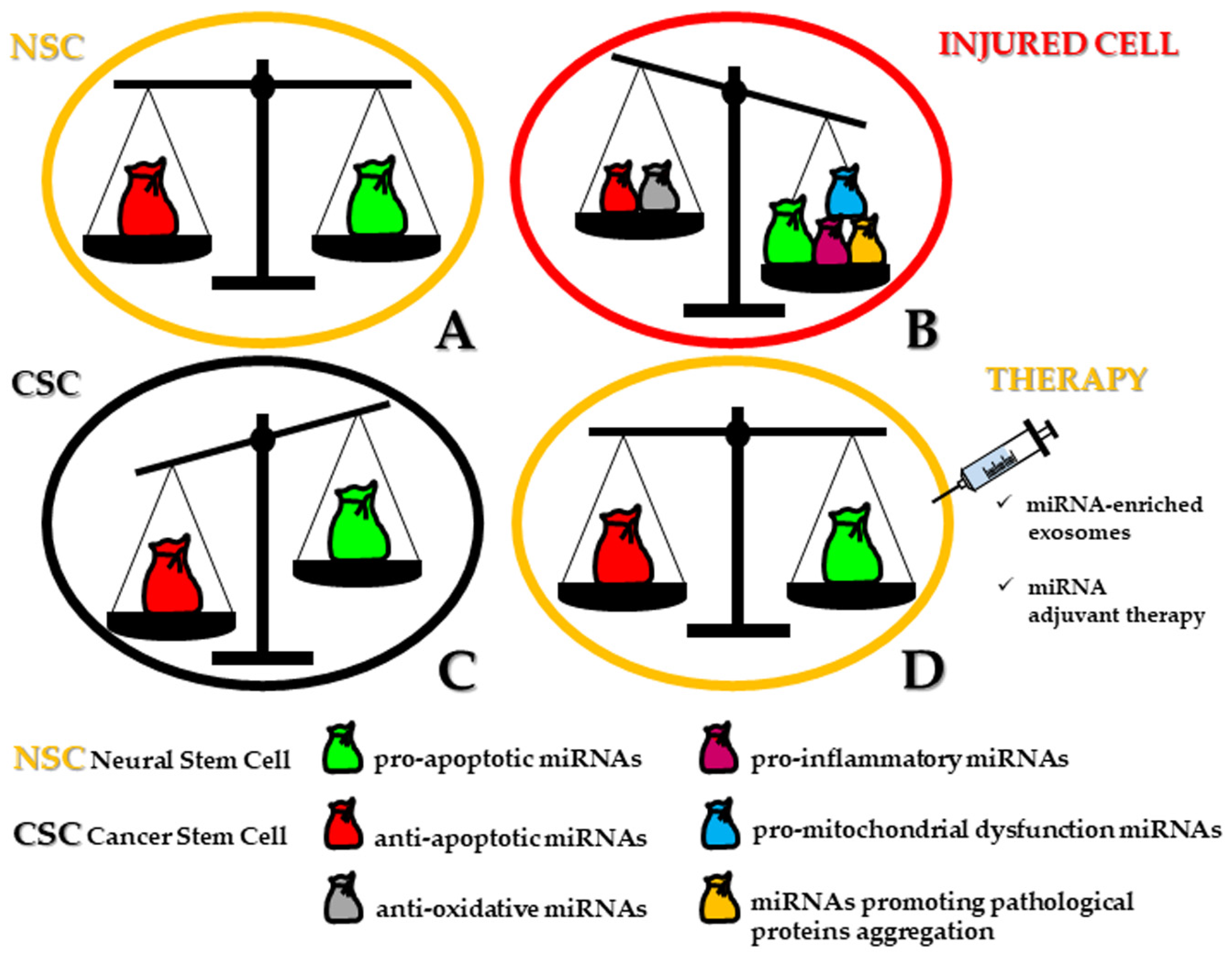
| miRNA | Target mRNA/Pathway | miRNA Effect on Apoptosis | miRNA Expression in CSCs | Cancer Type | Ref. |
|---|---|---|---|---|---|
| miR-7 | EGFR, AKT, DR5 | ↑ | down | glioma | [83] |
| miR-23b | HMGA2 | ↑ | down | glioma | [84] |
| miR-26a | SODDs | ↑ | down | melanoma | [85,86] |
| miR-29a | QKI-6 | ↑ | down | glioma | [87,88] |
| miR-34a | BCL2, TP53, NOTCH1/NOTCH2, Dll1, RICTOR, AKT, Wnt/β-catenin | ↑ | down | glioma, MB | [89,90,91,92] |
| TP53, NOTCH | ↑ | down | melanoma | [79,93,94,95] | |
| miR-107 | NOTCH2, MMP-12 | ↑ | down | glioma | [96] |
| miR-124 | CDK6, RB | ↑ | down | glioma | [4,97] |
| miR-137 | CDK6, RB, GLIPR | ↑ | down | glioma | [4,97,98,99] |
| CTBP1, PIK3R3 | ↑ | down | melanoma | [100,101,102] | |
| miR-125a-5p | LIN28B | ↑ | down | melanoma | [103,104] |
| miR-125b | CDK6, CDC25A | ↑ | down | glioma | [105,106,107,108] |
| TNFAIP3, NKIRAS2, TP53, p38MAPK, BAK1 | ↓ | up | glioma | [109,110,111] | |
| TP53 | ↓ | up | NB | [112,113] | |
| NEDD9 | ↓ | up | melanoma | [114] | |
| miR-128 | EGFR/PDGFR/AKT, BMI-1 | ↑ | down | glioma | [105,115,116,117] |
| miR-218 | CDK6, MKI67, ECOP | ↑ | down | glioma | [108,118,119,120] |
| miR-129-5p | FNDC3B | ↑ | down | glioma | [121] |
| miR-134 | NANOG | ↑ | down | glioma | [122] |
| miR-138 | HIF1α | ↑ | down | melanoma | [123] |
| miR-141 | JAG1 | ↑ | down | glioma | [124] |
| miR-145 | BNIP3, NOTCH, SOX9, ADD3 | ↑ | down | glioma | [125,126,127] |
| miR-146b-5p | HuR | ↑ | down | glioma | [128] |
| miR-149 | BCL2, CDC42 | ↑ | down | NB | [129] |
| miR-152/153 | KLF4, LGALS3 | ↑ | down | glioma | [105,130,131,132] |
| miR-181 | NOTCH2 | ↑ | down | glioma | [133,134] |
| miR-182 | BCL2L12, c-MET, HIF2α | ↑ | down | glioma | [135] |
| miR-199b-5p | HES1 | ↑ | down | MB | [136,117] |
| miR-200b | CD133 | ↑ | down | glioma | [137] |
| miR-203 | PLD2 | ↑ | down | glioma | [138] |
| miR-211 | MMP-9, BCL2 | ↑ | down | glioma | [139] |
| KCNMA1 | ↑ | down | melanoma | [140,141,142,143] | |
| miR-218 | CIP2A, BMI-1 | ↑ | down | melanoma | [102,144] |
| miR-219-5p | BCL2 | ↑ | down | melanoma | [145] |
| miR 302-367 | CXCR4, CXCL12 | ↑ | down | glioma | [146] |
| miR-326 | NOTCH, SMO, GLI1 | ↑ | down | glioma, MB | [108,117,147,148,149,150] |
| miR-340 | ROCK1 | ↑ | down | glioma | [151] |
| miR-451 | AKT1, CCND1, MMP-2, MMP-9, BCL2, CAB39 | ↑ | down | glioma | [152,153,154] |
| miR-503 | IGF-1R/PI3K/AKT | ↑ | down | glioma | [155] |
| miR-608 | MIF | ↑ | down | glioma | [3,156] |
| miR-625 | SOX2 | ↑ | down | melanoma | [104,157] |
| miR-873 | IGF2BP1 | ↑ | down | glioma | [158] |
| let-7 | KRAS, PI3K/AKT, MAPK/ERK, LIN28/let-7/c-MYC, NEAT1 | ↑ | down | glioma, MB | [159,160,161,162] |
| MYCN | ↑ | down | NB | [163,164] | |
| miR-9 | PTCH1 | ↓ | up | glioma | [71,165,166,167] |
| miR-9* | SOX2 | ↑ | down | glioma | [168,165] |
| miR-10b | MBNL1-3, SART3, RSRC1 | ↓ | up | glioma | [3,108,169,170] |
| miR-21 | FASL, TP53, TGF-β, PTEN, PDCD4, TIMP3 | ↓ | up | glioma | [3,87,134,171,172,173,174] |
| TIMP3 | ↓ | up | melanoma | [140,175,176,177] | |
| miR-24 | ST7L, β-catenin/TCF-4 | ↓ | up | glioma | [178] |
| miR-27b | β-catenin/TCF-4 | ↓ | up | glioma | [179] |
| miR-30 | SOCS3, JAK/STAT3 | ↓ | up | glioma | [150,180] |
| miR-17-92 | PTEN, PI3K/AKT | ↓ | up | glioma | [3,132,134,181,182] |
| miR-92b | SMAD3, TGF-β/P21 | ↓ | up | glioma | [183] |
| miR-138 | BLCAP, CASP3, BTG2, TUSC2 | ↓ | up | glioma | [126,184] |
| miR-149 | CASP2 | ↓ | up | glioma | [185,186] |
| miR-155 | PTEN, CASP3, CASP9, PI3K/AKT, CDX1, SIX1 | ↓ | up | glioma | [3,187,188] |
| miR-182 | FOXO3, MITF | ↓ | up | melanoma | [140,189,190,191] |
| miR-196a | IκBα | ↓ | up | glioma | [192] |
| miR-196a-5p | FOXO1 | ↓ | up | glioma | [193] |
| miR-211–5p | CHOP, IGF-2R | ↓ | up | melanoma | [194,195] |
| miR-210 | MNT,HIF1α/miR-210/GPD1L, ROD1 | ↓ | up | glioma | [186,196,197,198] |
| miR-221/222 | P27, P57, PUMA | ↓ | up | glioma | [199,200,201,202] |
| C-KIT, ETS-1, P27 | ↓ | up | melanoma | [140,203,204,205,206,207] | |
| miR-330, -363, -582-5p | CASP3, CASP9, BIM | ↓ | up | glioma | [87,208] |
| miR-335 | DAAM1, PAX6 | ↓ | up | glioma | [209,210,211] |
| miR-380-5p | TP53, HRAS | ↓ | up | NB | [212,213] |
| miR-638 | TP53INP2 | ↓ | up | melanoma | [214] |
| miR-130b | MST1/2, SAV1, Hippo pathway | ↓ | up | glioma | [215] |
| Therapeutic Agent | Combined Treatment | Cancer Type | Ref. |
|---|---|---|---|
| antagomiR | |||
| anti-miR-381 | TMZ | GBM | [254] |
| anti-miR-10b + anti-miR-21 | TMZ | glioma, GBM | [262,263,264] |
| anti-miR-21 | Sunitinib | GBM | [265] |
| anti-miR-21 | Erismodegib | glioma | [266,267] |
| anti-miR-125b | LY294002 + TMZ | GBM | [268] |
| miRNA mimics | |||
| miR-145 | DMC | GBM | [126] |
| miR-146b-5p | Radiotherapy | glioma | [128] |
| miR-181a | Radiotherapy | GBM | [134] |
| miR-211 | Radiotherapy + TMZ | GBM | [139] |
| let-7 | Phenformin + TMZ/DCA | glioma | [269] |
| miR-224 | Radiotherapy | GBM | [270] |
| miR-153 | Radiotherapy | GBM | [271] |
| miR-146a | Curcumin + TMZ | GBM | [272] |
| miR-326 | Curcumin | GBM | [273] |
| Cross-phylum derived miRNAs | |||
| Shrimp-derived miR-S8, -35-3p, -965 | melanoma | [274,275,276] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diana, A.; Gaido, G.; Maxia, C.; Murtas, D. MicroRNAs at the Crossroad of the Dichotomic Pathway Cell Death vs. Stemness in Neural Somatic and Cancer Stem Cells: Implications and Therapeutic Strategies. Int. J. Mol. Sci. 2020, 21, 9630. https://doi.org/10.3390/ijms21249630
Diana A, Gaido G, Maxia C, Murtas D. MicroRNAs at the Crossroad of the Dichotomic Pathway Cell Death vs. Stemness in Neural Somatic and Cancer Stem Cells: Implications and Therapeutic Strategies. International Journal of Molecular Sciences. 2020; 21(24):9630. https://doi.org/10.3390/ijms21249630
Chicago/Turabian StyleDiana, Andrea, Giuseppe Gaido, Cristina Maxia, and Daniela Murtas. 2020. "MicroRNAs at the Crossroad of the Dichotomic Pathway Cell Death vs. Stemness in Neural Somatic and Cancer Stem Cells: Implications and Therapeutic Strategies" International Journal of Molecular Sciences 21, no. 24: 9630. https://doi.org/10.3390/ijms21249630
APA StyleDiana, A., Gaido, G., Maxia, C., & Murtas, D. (2020). MicroRNAs at the Crossroad of the Dichotomic Pathway Cell Death vs. Stemness in Neural Somatic and Cancer Stem Cells: Implications and Therapeutic Strategies. International Journal of Molecular Sciences, 21(24), 9630. https://doi.org/10.3390/ijms21249630






