The Role of Epigenomics in Osteoporosis and Osteoporotic Vertebral Fracture
Abstract
1. Introduction
2. Search Strategy
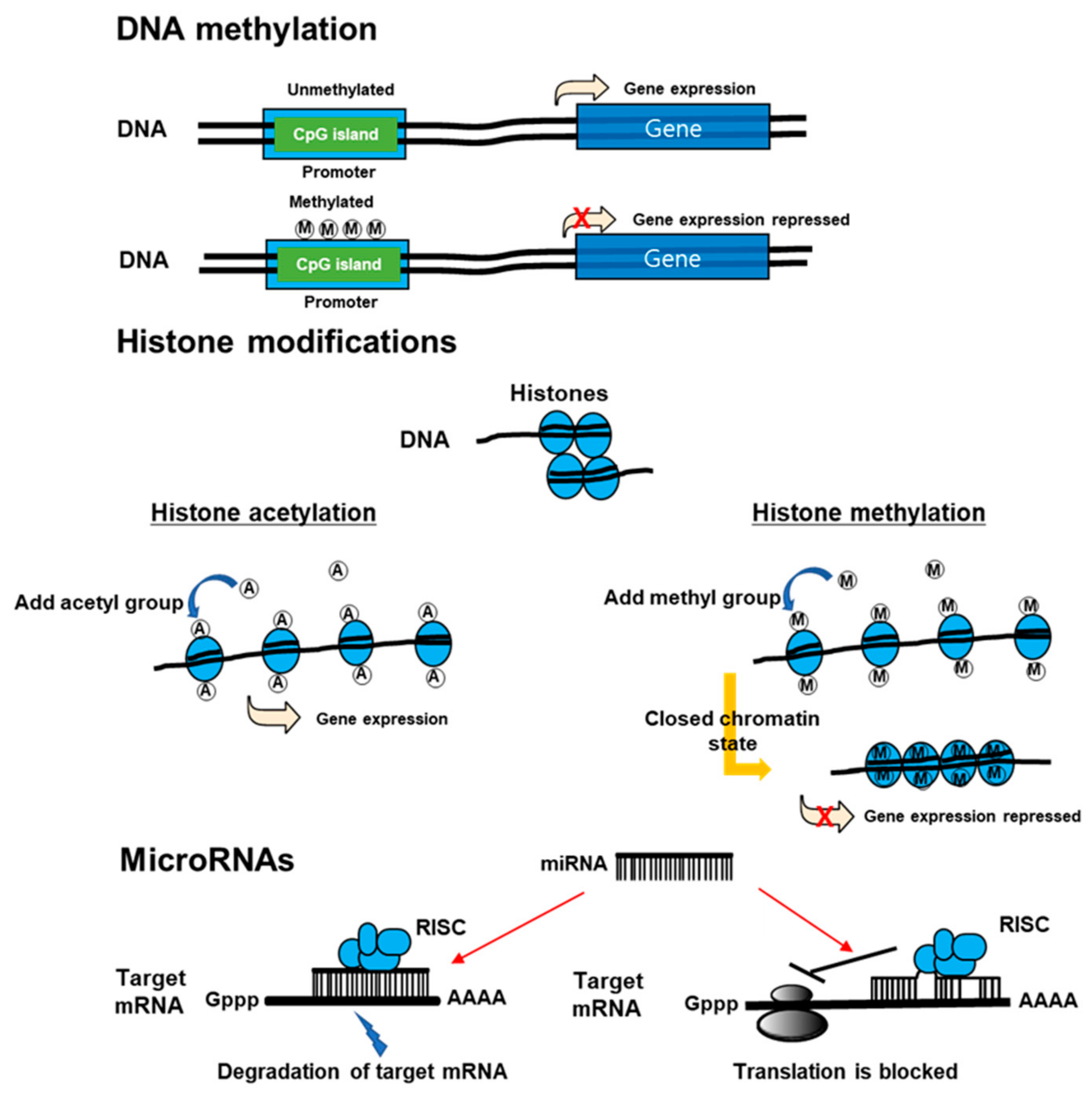
3. DNA Methylation
4. Histone Modification
5. Noncoding RNA
5.1. MicroRNAs (miRNAs)
5.2. Long Non-Coding (lnc) RNAs
6. The Possibility of Epigenetics in Treatment of Osteoporosis and OVF
7. Future Perspectives
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ballane, G.; Cauley, J.A.; Luckey, M.M.; Fuleihan, G.E.-H. Worldwide prevalence and incidence of osteoporotic vertebral fractures. Osteoporos. Int. 2017, 28, 1531–1542. [Google Scholar] [CrossRef] [PubMed]
- Cauley, J. Epidemiology of ageing, fracture and falls. Geographic and ethnic disparities in osteoporotic fractures. Bone Abstr. 2014, 10, 338–351. [Google Scholar] [CrossRef]
- Richards, J.B.; Kavvoura, F.K.; Rivadeneira, F.; Styrkarsdottir, U.; Estrada, K.; Halldórsson, B.; Hsu, Y.-H.; Zillikens, M.C.; Wilson, S.G.; Mullin, B.H.; et al. Collaborative meta-analysis: Associations of 150 candidate genes with osteoporosis and osteoporotic fracture. Ann. Intern. Med. 2009, 151, 528–537. [Google Scholar] [CrossRef] [PubMed]
- Ahn, T.-K.; Kim, J.-O.; Han, I.-B.; Choi, H.; Jo, M.-J.; Sohn, S.; Ropper, A.E.; Kim, N.K.; Han, I.-B. Polymorphisms of miR-146a, miR-149, miR-196a2, and miR-499 are associated with osteoporotic vertebral compression fractures in Korean postmenopausal women. J. Orthop. Res. 2017, 36, 244–253. [Google Scholar] [CrossRef]
- Ahn, T.-K.; Kim, J.O.; Kim, H.W.; Park, H.S.; Shim, J.H.; Ropper, A.E.; Han, I.; Kim, N.K. 3′-UTR Polymorphisms of MTHFR and TS Associated with Osteoporotic Vertebral Compression Fracture Susceptibility in Postmenopausal Women. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef]
- Cheng, C.; Wentworth, K.; Shoback, D.M. New Frontiers in Osteoporosis Therapy. Annu. Rev. Med. 2020, 71, 277–288. [Google Scholar] [CrossRef]
- Hu, L.; Yin, C.; Zhao, F.; Ali, A.; Ma, J.; Qian, A. Mesenchymal Stem Cells: Cell Fate Decision to Osteoblast or Adipocyte and Application in Osteoporosis Treatment. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef]
- Macías, I.; Alcorta-Sevillano, N.; Rodríguez, C.I.; Infante, A. Osteoporosis and the Potential of Cell-Based Therapeutic Strategies. Int. J. Mol. Sci. 2020, 21. [Google Scholar] [CrossRef]
- Sanghani-Kerai, A.; McCreary, D.; Lancashire, H.; Osagie-Clouard, L.; Coathup, M.; Blunn, G. Stem Cell Interventions for Bone Healing: Fractures and Osteoporosis. Curr. Stem Cell Res. Ther. 2018, 13, 369–377. [Google Scholar] [CrossRef]
- Schindeler, A.; McDonald, M.M.; Bokko, P.; Little, D.G. Bone remodeling during fracture repair: The cellular picture. Semin. Cell Dev. Biol. 2008, 19, 459–466. [Google Scholar] [CrossRef]
- Bolander, M.E. Regulation of Fracture Repair by Growth Factors. Exp. Biol. Med. 1992, 200, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Einhorn, T.A. The Cell and Molecular Biology of Fracture Healing. Clin. Orthop. Relat. Res. 1998, 355, S7–S21. [Google Scholar] [CrossRef] [PubMed]
- Gerstenfeld, L.C.; Cullinane, D.M.; Barnes, G.L.; Graves, D.T.; Einhorn, T.A. Fracture healing as a post-natal developmental process: Molecular, spatial, and temporal aspects of its regulation. J. Cell. Biochem. 2003, 88, 873–884. [Google Scholar] [CrossRef] [PubMed]
- Ai-Aql, Z.S.; Alagl, A.S.; Graves, D.T.; Gerstenfeld, L.C.; Einhorn, T.A. Molecular mechanisms controlling bone formation during fracture healing and distraction osteogenesis. J. Dent. Res. 2008, 87, 107–118. [Google Scholar] [CrossRef]
- Giannoudis, P.V.; Einhorn, T.A.; Marsh, D. Fracture healing: The diamond concept. Injury 2007, 38, S3–S6. [Google Scholar] [CrossRef]
- Stone, C. A molecular approach to bone regeneration. Br. J. Plast. Surg. 1997, 50, 369–373. [Google Scholar] [CrossRef]
- Barnes, G.L.; Kostenuik, P.J.; Gerstenfeld, L.C.; A Einhorn, T. Growth Factor Regulation of Fracture Repair. J. Bone Miner. Res. 1999, 14, 1805–1815. [Google Scholar] [CrossRef]
- Khosla, S.; Riggs, B.L. Pathophysiology of Age-Related Bone Loss and Osteoporosis. Endocrinol. Metab. Clin. N. Am. 2005, 34, 1015–1030. [Google Scholar] [CrossRef]
- Baccarelli, A.A.; Bollati, V. Epigenetics and environmental chemicals. Curr. Opin. Pediatr. 2009, 21, 243–251. [Google Scholar] [CrossRef]
- Yasui, T.; Hirose, J.; Aburatani, H.; Tanaka, S. Epigenetic regulation of osteoclast differentiation. Ann. N. Y. Acad. Sci. 2011, 1240, 7–13. [Google Scholar] [CrossRef]
- Kurotaki, D.; Yoshida, H.; Tamura, T. Epigenetic and transcriptional regulation of osteoclast differentiation. Bone 2020, 138. [Google Scholar] [CrossRef] [PubMed]
- Guil, S.; Esteller, M. DNA methylomes, histone codes and miRNAs: Tying it all together. Int. J. Biochem. Cell Biol. 2009, 41, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, A.P. The Key Role of Epigenetics in Human Disease Prevention and Mitigation. N. Engl. J. Med. 2018, 378, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Lyko, F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018, 19, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Ganser, L.R.; Kelly, M.L.; Herschlag, D.; Al-Hashimi, H.M. The roles of structural dynamics in the cellular functions of RNAs. Nat. Rev. Mol. Cell Biol. 2019, 20, 474–489. [Google Scholar] [CrossRef]
- Michalak, E.M.; Burr, M.L.; Bannister, A.J.; Dawson, M.A. The roles of DNA, RNA and histone methylation in ageing and cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 573–589. [Google Scholar] [CrossRef]
- Schübeler, D. Function and information content of DNA methylation. Nat. Cell Biol. 2015, 517, 321–326. [Google Scholar] [CrossRef]
- Pu, M.; Chen, J.; Tao, Z.; Miao, L.; Qi, X.; Wang, Y.; Ren, J. Regulatory network of miRNA on its target: Coordination between transcriptional and post-transcriptional regulation of gene expression. Cell. Mol. Life Sci. 2019, 76, 441–451. [Google Scholar] [CrossRef]
- Adams, B.D.; Parsons, C.; Walker, L.; Zhang, W.C.; Slack, F.J. Targeting noncoding RNAs in disease. J. Clin. Investig. 2017, 127, 761–771. [Google Scholar] [CrossRef]
- Mohammad, H.P.; Barbash, O.; Creasy, C.L. Targeting epigenetic modifications in cancer therapy: Erasing the roadmap to cancer. Nat. Med. 2019, 25, 403–418. [Google Scholar] [CrossRef]
- Piunti, A.; Shilatifard, A. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science 2016, 352, aad9780. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Roberts, C.W. Targeting EZH2 in cancer. Nat. Med. 2016, 22, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Sabari, B.R.; Zhang, D.; Allis, C.D.; Zhao, Y. Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol. 2017, 18, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.M.; et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009, 462, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.-P.; Shao, J.-Z.; Xiang, L. GADD45A Protein Plays an Essential Role in Active DNA Demethylation during Terminal Osteogenic Differentiation of Adipose-derived Mesenchymal Stem Cells. J. Biol. Chem. 2011, 286, 41083–41094. [Google Scholar] [CrossRef]
- Pei, M. Faculty Opinions recommendation of Epigenetic modifications and canonical wingless/int-1 class (WNT) signaling enable trans-differentiation of nonosteogenic cells into osteoblasts. Fac. Opin. Post-Publ. Peer Rev. Biomed. Lit. 2017, 289, 20120–20128. [Google Scholar] [CrossRef]
- Gong, Y.; Slee, R.B.; Fukai, N.; Rawadi, G.; Roman-Roman, S.; Reginato, A.M.; Wang, H.; Cundy, T.; Glorieux, F.H.; Lev, D.; et al. LDL Receptor-Related Protein 5 (LRP5) Affects Bone Accrual and Eye Development. Cell 2001, 107, 513–523. [Google Scholar] [CrossRef]
- Boyden, L.M.; Mao, J.; Belsky, J.; Mitzner, L.; Farhi, A.; Mitnick, M.A.; Wu, D.; Insogna, K.; Lifton, R.P. High bone density due to a mutation in LDL-receptor-related protein 5. N. Engl. J. Med. 2002, 346, 1513–1521. [Google Scholar] [CrossRef]
- Balemans, W.; Patel, N.; Ebeling, M.; Van Hul, E.; Wuyts, W.; Lacza, C.; Dioszegi, M.; Dikkers, F.; Hildering, P.; Willems, P.; et al. Identification of a 52 kb deletion downstream of the SOST gene in patients with van Buchem disease. J. Med. Genet. 2002, 39, 91–97. [Google Scholar] [CrossRef]
- Staehling-Hampton, K.; Proll, S.; Paeper, B.W.; Zhao, L.; Charmley, P.; Brown, A.; Gardner, J.C.; Galas, D.; Schatzman, R.C.; Beighton, P.; et al. A 52-kb deletion in theSOST-MEOX1 intergenic region on 17q12-q21 is associated with van Buchem disease in the Dutch population. Am. J. Med. Genet. 2002, 110, 144–152. [Google Scholar] [CrossRef]
- Agholme, F.; Li, X.; Isaksson, H.; Ke, H.Z.; Aspenberg, P. Sclerostin antibody treatment enhances metaphyseal bone healing in rats. J. Bone Miner. Res. 2010, 25, 2412–2418. [Google Scholar] [CrossRef]
- Ott, S. Faculty Opinions recommendation of Single-dose, placebo-controlled, randomized study of AMG 785, a sclerostin monoclonal antibody. Fac. Opin. Post-Publ. Peer Rev. Biomed. Lit. 2010, 26, 19–26. [Google Scholar] [CrossRef]
- Taylor, S.; Ominsky, M.S.; Hu, R.; Pacheco, E.; He, Y.D.; Brown, D.L.; Aguirre, J.I.; Wronski, T.J.; Buntich, S.; Afshari, C.A.; et al. Time-dependent cellular and transcriptional changes in the osteoblast lineage associated with sclerostin antibody treatment in ovariectomized rats. Bone 2016, 84, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yang, L.; Wang, H.; Chen, X.; Jiang, W.; Wang, Z.; Liu, S.; Liu, Y. Alterations in DNA methylation profiles in cancellous bone of postmenopausal women with osteoporosis. FEBS Open Bio 2020, 10, 1516–1531. [Google Scholar] [CrossRef] [PubMed]
- Harvey, N.; Sheppard, A.; Godfrey, K.M.; McLean, C.; Garratt, E.; Ntani, G.; Davies, L.; Murray, R.; Inskip, H.; Gluckman, P.D.; et al. Childhood bone mineral content is associated with methylation status of the RXRA promoter at birth. J. Bone Miner. Res. 2014, 29, 600–607. [Google Scholar] [CrossRef]
- Curtis, E.M.; Murray, R.; Titcombe, P.; Cook, E.; Clarke-Harris, R.; Costello, P.; Garratt, E.; Holbrook, J.D.; Barton, S.; Inskip, H.; et al. Perinatal DNA Methylation at CDKN2A Is Associated with Offspring Bone Mass: Findings From the Southampton Women’s Survey. J. Bone Miner. Res. 2017, 32, 2030–2040. [Google Scholar] [CrossRef]
- Harvey, N.; Lillycrop, K.A.; Garratt, E.; Sheppard, A.; Mc Lean, C.; Burdge, G.C.; Slater-Jefferies, J.; Rodford, J.; Crozier, S.R.; Inskip, H.; et al. Evaluation of Methylation Status of the eNOS Promoter at Birth in Relation to Childhood Bone Mineral Content. Calcif. Tissue Int. 2012, 90, 120–127. [Google Scholar] [CrossRef]
- Morris, J.A.; Tsai, P.-C.; Joehanes, R.; Zheng, J.; Trajanoska, K.; Soerensen, M.; Forgetta, V.; Castillo-Fernandez, J.E.; Frost, M.; Spector, T.D.; et al. Epigenome-wide Association of DNA Methylation in Whole Blood with Bone Mineral Density. J. Bone Miner. Res. 2017, 32, 1644–1650. [Google Scholar] [CrossRef]
- Jintaridth, P.; Tungtrongchitr, R.; Preutthipan, S.; Mutirangura, A. Hypomethylation of Alu Elements in Post-Menopausal Women with Osteoporosis. PLoS ONE 2013, 8, e70386. [Google Scholar] [CrossRef]
- Dieker, J.; Muller, S. Epigenetic Histone Code and Autoimmunity. Clin. Rev. Allergy Immunol. 2009, 39, 78–84. [Google Scholar] [CrossRef]
- Zhang, P.; Liu, Y.; Jin, C.; Zhang, M.; Lv, L.; Zhang, X.; Liu, H.; Zhou, Y. Histone H3K9 Acetyltransferase PCAF Is Essential for Osteogenic Differentiation Through Bone Morphogenetic Protein Signaling and May Be Involved in Osteoporosis. Stem Cells 2016, 34, 2332–2341. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Liu, Y.; Jin, C.; Zhang, M.; Tang, F.; Zhou, Y. Histone Acetyltransferase GCN5 Regulates Osteogenic Differentiation of Mesenchymal Stem Cells by Inhibiting NF-kappaB. J. Bone Miner. Res. 2016, 31, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.; Su, X.; Gao, B.; Shuai, Y.; Chen, J.; Deng, Z.; Liao, L.; Jin, Y. Epigenetic inhibition of Wnt pathway suppresses osteogenic differentiation of BMSCs during osteoporosis. Cell Death Dis. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Camilleri, E.T.; Dudakovic, A.; Riester, S.M.; Galeano-Garces, C.; Paradise, C.R.; Bradley, E.W.; McGee-Lawrence, M.E.; Im, H.-J.; Karperien, M.H.; Krych, A.J.; et al. Loss of histone methyltransferase Ezh2 stimulates an osteogenic transcriptional program in chondrocytes but does not affect cartilage development. J. Biol. Chem. 2018, 293, 19001–19011. [Google Scholar] [CrossRef] [PubMed]
- Lui, J.C.; Garrison, P.; Nguyen, Q.; Ad, M.; Keembiyehetty, C.; Chen, W.; Jee, Y.H.; Landman, E.; Nilsson, O.; Barnes, K.M.; et al. EZH1 and EZH2 promote skeletal growth by repressing inhibitors of chondrocyte proliferation and hypertrophy. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Dudakovic, A.; Camilleri, E.T.; Paradise, C.R.; Samsonraj, R.M.; Gluscevic, M.; Paggi, C.A.; Begun, D.L.; Khani, F.; Pichurin, O.; Ahmed, F.S.; et al. Enhancer of zeste homolog 2 (Ezh2) controls bone formation and cell cycle progression during osteogenesis in mice. J. Biol. Chem. 2018, 293, 12894–12907. [Google Scholar] [CrossRef]
- Prokopuk, L.; Stringer, J.M.; White, C.R.; Vossen, R.H.A.M.; White, S.J.; Cohen, A.S.A.; Gibson, W.T.; Western, P.S. Loss of maternal EED results in postnatal overgrowth. Clin. Epigenet. 2018, 10. [Google Scholar] [CrossRef]
- Wang, L.; Niu, N.; Li, L.; Shao, R.; Ouyang, H.; Zou, W. H3K36 trimethylation mediated by SETD2 regulates the fate of bone marrow mesenchymal stem cells. PLoS Biol. 2018, 16, e2006522. [Google Scholar] [CrossRef]
- Hemming, S.; Cakouros, D.; Isenmann, S.; Cooper, L.; Menicanin, D.; Zannettino, A.; Gronthos, S. EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cells 2014, 32, 802–815. [Google Scholar] [CrossRef]
- Ye, L.; Fan, Z.; Yu, B.; Chang, J.; Al Hezaimi, K.; Zhou, X.; Park, N.H.; Wang, C.Y. Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell 2012, 11, 50–61. [Google Scholar] [CrossRef]
- Lee, J.Y.; Mehrazarin, S.; Alshaikh, A.; Kim, S.; Chen, W.; Lux, R.; Gwack, Y.; Kim, R.H.; Kang, M.K. Histone Lys demethylase KDM3C demonstrates anti-inflammatory effects by suppressing NF-kappaB signaling and osteoclastogenesis. FASEB J. 2019, 33, 10515–10527. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, J.; Li, J.; Hu, G.; Shan, S.; Li, Q.; Zhang, X. KDM5A controls bone morphogenic protein 2-induced osteogenic differentiation of bone mesenchymal stem cells during osteoporosis. Cell Death Dis. 2016, 7, e2335. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.-S.; Lian, W.-S.; Lee, M.S.; Weng, W.-T.; Huang, Y.-H.; Chen, Y.-S.; Sun, Y.-C.; Wu, S.-L.; Chuang, P.-C.; Ko, J.-Y. Histone demethylase UTX counteracts glucocorticoid deregulation of osteogenesis by modulating histone-dependent and -independent pathways. J. Mol. Med. 2017, 95, 499–512. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.-T.; Li, H.-Q.; Liu, F. Selective histone deacetylase small molecule inhibitors: Recent progress and perspectives. Expert Opin. Ther. Patents 2016, 27, 621–636. [Google Scholar] [CrossRef] [PubMed]
- Cantley, M.; Fairlie, D.; Bartold, P.; Rainsford, K.; Le, G.; Lucke, A.; Holding, C.; Haynes, D. Inhibitors of histone deacetylases in class I and class II suppress human osteoclasts in vitro. J. Cell. Physiol. 2011, 226, 3233–3241. [Google Scholar] [CrossRef]
- Schroeder, T.M.; Westendorf, J.J. Histone Deacetylase Inhibitors Promote Osteoblast Maturation. J. Bone Miner. Res. 2005, 20, 2254–2263. [Google Scholar] [CrossRef]
- Lee, H.W.; Suh, J.H.; Kim, A.Y.; Lee, Y.S.; Park, S.Y.; Kim, J.B. Histone Deacetylase 1-Mediated Histone Modification Regulates Osteoblast Differentiation. Mol. Endocrinol. 2006, 20, 2432–2443. [Google Scholar] [CrossRef]
- Schroeder, T.M.; Kahler, R.A.; Li, X.; Westendorf, J.J. Histone Deacetylase 3 Interacts with Runx2 to Repress the Osteocalcin Promoter and Regulate Osteoblast Differentiation. J. Biol. Chem. 2004, 279, 41998–42007. [Google Scholar] [CrossRef]
- Wein, M.N.; Spatz, J.; Nishimori, S.; Doench, J.; Root, D.; Babij, P.; Nagano, K.; Baron, R.; Brooks, D.; Bouxsein, M.; et al. HDAC5 Controls MEF2C-Driven Sclerostin Expression in Osteocytes. J. Bone Miner. Res. 2015, 30, 400–411. [Google Scholar] [CrossRef]
- Li, H.; Xie, H.; Liu, W.; Hu, R.; Huang, B.; Tan, Y.F.; Xu, K.; Sheng, Z.F.; Zhou, H.D.; Wu, X.P.; et al. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J. Clin. Investig. 2009, 119, 3666–3677. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, K.; Youn, B.U.; Jin, H.M.; Kim, J.-Y.; Moon, J.B.; Ko, A.; Seo, S.-B.; Lee, K.-Y.; Kim, N. RANKL induces NFATc1 acetylation and stability via histone acetyltransferases during osteoclast differentiation. Biochem. J. 2011, 436, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Reedquist, K.; Grabiec, A.M. Faculty Opinions recommendation of Inhibition of HDAC activity by ITF2357 ameliorates joint inflammation and prevents cartilage and bone destruction in experimental arthritis. Fac. Opin. Post-Publ. Peer Rev. Biomed. Lit. 2011, 17, 391–396. [Google Scholar] [CrossRef]
- Jin, Z.; Wei, W.; Dechow, P.C.; Wan, Y. HDAC7 inhibits osteoclastogenesis by reversing RANKL-triggered beta-catenin switch. Mol. Endocrinol. 2013, 27, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Pham, L.; Kaiser, B.; Romsa, A.; Schwarz, T.; Gopalakrishnan, R.; Jensen, E.D.; Mansky, K.C. HDAC3 and HDAC7 Have Opposite Effects on Osteoclast Differentiation. J. Biol. Chem. 2011, 286, 12056–12065. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Wei, W.; Huynh, H.; Wan, Y. HDAC9 Inhibits Osteoclastogenesis via Mutual Suppression of PPARgamma/RANKL Signaling. Mol. Endocrinol. 2015, 29, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.S.; Yoon, S.-H.; Wein, M.N. Role of histone deacetylases in bone development and skeletal disorders. Bone 2020, 115606. [Google Scholar] [CrossRef] [PubMed]
- Backesjo, C.M.; Li, Y.; Lindgren, U.; Haldosen, L.A. Activation of Sirt1 decreases adipocyte formation during osteoblast differentiation of mesenchymal stem cells. J. Bone Miner. Res. 2006, 21, 993–1002. [Google Scholar] [CrossRef]
- Xiao, F.; Zhou, Y.; Liu, Y.; Xie, M.; Guo, G. Inhibitory Effect of Sirtuin6 (SIRT6) on Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells. Med. Sci. Monit. 2019, 25, 8412–8421. [Google Scholar] [CrossRef]
- Pratt, A.J.; Macrae, I.J. The RNA-induced Silencing Complex: A Versatile Gene-silencing Machine. J. Biol. Chem. 2009, 284, 17897–17901. [Google Scholar] [CrossRef]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 924–933. [Google Scholar] [CrossRef]
- Cohen, M.M., Jr. Perspectives on RUNX genes: An update. Am. J. Med. Genet. A 2009, 149, 2629–2646. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xie, R.L.; Croce, C.M.; Stein, J.L.; Lian, J.B.; van Wijnen, A.J.; Stein, G.S. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor RUNX2. Proc. Natl. Acad. Sci. USA 2011, 108, 9863–9868. [Google Scholar] [CrossRef] [PubMed]
- Gaur, T.; Hussain, S.; Mudhasani, R.; Parulkar, I.; Colby, J.L.; Frederick, D.; Kream, B.E.; Van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; et al. Dicer inactivation in osteoprogenitor cells compromises fetal survival and bone formation, while excision in differentiated osteoblasts increases bone mass in the adult mouse. Dev. Biol. 2010, 340, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Luzi, E.; Marini, F.; Sala, S.C.; Tognarini, I.; Galli, G.; Brandi, M.L. Osteogenic Differentiation of Human Adipose Tissue-Derived Stem Cells Is Modulated by the miR-26a Targeting of the SMAD1 Transcription Factor. J. Bone Miner. Res. 2007, 23, 287–295. [Google Scholar] [CrossRef]
- Schaap-Oziemlak, A.M.; Raymakers, R.A.; Bergevoet, S.M.; Gilissen, C.; Jansen, B.J.; Adema, G.J.; Kogler, G.; le Sage, C.; Agami, R.; van der Reijden, B.A.; et al. MicroRNA hsa-miR-135b regulates mineralization in osteogenic differentiation of human unrestricted somatic stem cells. Stem Cells Dev. 2010, 19, 877–885. [Google Scholar] [CrossRef]
- Mizuno, Y.; Yagi, K.; Tokuzawa, Y.; Kanesaki-Yatsuka, Y.; Suda, T.; Katagiri, T.; Fukuda, T.; Maruyama, M.; Okuda, A.; Amemiya, T.; et al. miR-125b inhibits osteoblastic differentiation by down-regulation of cell proliferation. Biochem. Biophys. Res. Commun. 2008, 368, 267–272. [Google Scholar] [CrossRef]
- Itoh, T.; Nozawa, Y.; Akao, Y. MicroRNA-141 and -200a Are Involved in Bone Morphogenetic Protein-2-induced Mouse Pre-osteoblast Differentiation by Targeting Distal-less Homeobox 5. J. Biol. Chem. 2009, 284, 19272–19279. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, L.; Xing, L.; Chen, D. MicroRNA-204 Regulates Runx2 Protein Expression and Mesenchymal Progenitor Cell Differentiation. Stem Cells 2009, 28, 357–364. [Google Scholar] [CrossRef]
- Hassan, M.Q.; Maeda, Y.; Taipaleenmaki, H.; Zhang, W.; Jafferji, M.; Gordon, J.A.R.; Li, Z.; Croce, C.M.; Van Wijnen, A.J.; Stein, J.L.; et al. miR-218 Directs a Wnt Signaling Circuit to Promote Differentiation of Osteoblasts and Osteomimicry of Metastatic Cancer Cells. J. Biol. Chem. 2012, 287, 42084–42092. [Google Scholar] [CrossRef]
- Zhang, W.B.; Zhong, W.J.; Wang, L. A signal-amplification circuit between miR-218 and Wnt/beta-catenin signal promotes human adipose tissue-derived stem cells osteogenic differentiation. Bone 2014, 58, 59–66. [Google Scholar] [CrossRef]
- Liu, T.; Hou, L.; Zhao, Y.; Huang, Y. Epigenetic silencing of HDAC1 by miR-449a upregulates Runx2 and promotes osteoblast differentiation. Int. J. Mol. Med. 2014, 35, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Cheng, P.; Xie, H.; Zhou, H.-D.; Wu, X.-P.; Liao, E.-Y.; Luo, X. MiR-503 Regulates Osteoclastogenesis via Targeting RANK. J. Bone Miner. Res. 2014, 29, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, H.; Nakasa, T.; Adachi, N.; Nagata, Y.; Ishikawa, M.; Deie, M.; Suzuki, O.; Ochi, M. Overexpression of microRNA-223 in rheumatoid arthritis synovium controls osteoclast differentiation. Mod. Rheumatol. 2013, 23, 674–685. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-T.; Chen, S.-Y.; Wang, C.-R.; Liu, M.-F.; Lin, C.-C.; Jou, I.-M.; Shiau, A.-L.; Wu, C.-L. Brief Report: Amelioration of collagen-induced arthritis in mice by lentivirus-mediated silencing of microRNA-223. Arthritis Rheum. 2012, 64, 3240–3245. [Google Scholar] [CrossRef] [PubMed]
- Zarecki, P.; Hackl, M.; Grillari, J.; Debono, M.; Eastell, R. Serum microRNAs as novel biomarkers for osteoporotic vertebral fractures. Bone 2019, 130. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Zhao, Q.; Guo, Z.; Chen, Z.; Hu, Y.; Su, J.; Chen, L.; He, Z.; Cai, X.; Chen, M.; et al. Hsa_Circ_0001275: A Potential Novel Diagnostic Biomarker for Postmenopausal Osteoporosis. Cell. Physiol. Biochem. 2018, 46, 2508–2516. [Google Scholar] [CrossRef] [PubMed]
- Huynh, N.P.T.; Anderson, B.A.; Guilak, F.; McAlinden, A. Emerging roles for long noncoding RNAs in skeletal biology and disease. Connect. Tissue Res. 2017, 58, 116–141. [Google Scholar] [CrossRef]
- Zhu, X.X.; Yan, Y.W.; Chen, D.; Ai, C.Z.; Lu, X.; Xu, S.S.; Jiang, S.; Zhong, G.S.; Chen, D.B.; Jiang, Y.Z. Long non-coding RNA HoxA-AS3 interacts with EZH2 to regulate lineage commitment of mesenchymal stem cells. Oncotarget 2016, 7, 63561–63570. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, P.; Sun, X.; Zhou, L.; Zhao, J. Long non-coding RNA DANCR regulates proliferation and apoptosis of chondrocytes in osteoarthritis via miR-216a-5p-JAK2-STAT3 axis. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef]
- Peng, S.; Cao, L.; He, S.; Zhong, Y.; Ma, H.; Zhang, Y.; Shuai, C. An Overview of Long Noncoding RNAs Involved in Bone Regeneration from Mesenchymal Stem Cells. Stem Cells Int. 2018, 2018, 1–11. [Google Scholar] [CrossRef]
- Jia, Q.; Jiang, W.; Ni, L. Down-regulated non-coding RNA (lncRNA-ANCR) promotes osteogenic differentiation of periodontal ligament stem cells. Arch. Oral Biol. 2015, 60, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.J.; Zhang, C.H.; Chen, Z.W.; Wang, Z.X.; Yang, D.C.; Zhang, F.L.; Feng, K.H. LncRNA HOTAIR inhibited osteogenic differentiation of BMSCs by regulating Wnt/beta-catenin pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7232–7246. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Tian, N.; Liu, H.; Tao, X.Z.; Wang, M.X.; Huang, W. LncRNAp21 promotes osteogenic differentiation of mesenchymal stem cells in the rat model of osteoporosis by the Wnt/beta-catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4303–4309. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Liu, J.; Zhao, J.; Ma, J.X.; Jia, H.B.; Zhang, Y.; Xing, G.S.; Ma, X.L. LncRNA-H19 Modulates Wnt/beta-catenin Signaling by Targeting Dkk4 in Hindlimb Unloaded Rat. Orthop. Surg. 2017, 9, 319–327. [Google Scholar] [CrossRef]
- Li, D.; Tian, Y.; Yin, C.; Huai, Y.; Zhao, Y.; Su, P.; Wang, X.; Pei, J.; Zhang, K.; Yang, C.; et al. Silencing of lncRNA AK045490 Promotes Osteoblast Differentiation and Bone Formation via beta-Catenin/TCF1/RUNX2 Signaling Axis. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef]
- Yin, C.; Tian, Y.; Yu, Y.; Wang, H.; Wu, Z.; Huang, Z.; Zhang, Y.; Li, D.; Yang, C.; Wang, X.; et al. A novel long noncoding RNA AK016739 inhibits osteoblast differentiation and bone formation. J. Cell. Physiol. 2019, 234, 11524–11536. [Google Scholar] [CrossRef]
- Zhang, R.-F.; Liu, J.-W.; Yu, S.-P.; Sun, D.; Wang, X.-H.; Fu, J.-S.; Xie, Z. LncRNA UCA1 affects osteoblast proliferation and differentiation by regulating BMP-2 expression. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 6774–6782. [Google Scholar] [CrossRef] [PubMed]
- Li, X.G.; Liu, S.C.; Qiao, X.F.; Kong, Y.; Liu, J.G.; Peng, X.M.; Wang, Y.X.; Abdulkarim Mohammed Al-Mohana, R.A. LncRNA MEG3 promotes proliferation and differentiation of osteoblasts through Wnt/beta-catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4521–4529. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, C.; Zhang, A.; Yin, S.; Wang, T.; Wang, Y.; Wang, M.; Liu, Y.; Ying, Q.; Sun, J.; et al. Down-regulation of long non-coding RNA MEG3 suppresses osteogenic differentiation of periodontal ligament stem cells (PDLSCs) through miR-27a-3p/IGF1 axis in periodontitis. Aging 2019, 11, 5334–5350. [Google Scholar] [CrossRef]
- Han, Y.; Liu, C.; Lei, M.; Sun, S.; Zheng, W.; Niu, Y.; Xia, X. LncRNA TUG1 was upregulated in osteoporosis and regulates the proliferation and apoptosis of osteoclasts. J. Orthop. Surg. Res. 2019, 14, 416. [Google Scholar] [CrossRef]
- Liu, C.; Cao, Z.; Bai, Y.; Dou, C.; Gong, X.; Liang, M.; Dong, R.; Quan, H.; Li, J.; Dai, J.; et al. LncRNA AK077216 promotes RANKL-induced osteoclastogenesis and bone resorption via NFATc1 by inhibition of NIP45. J. Cell. Physiol. 2019, 234, 1606–1617. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-Z.; Zhang, C.-Y.; Huang, L.-L.; Liu, W. Elevated expression of lncRNA SNHG15 in spinal tuberculosis: Preliminary results. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 9017–9024. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, X.Q.; Liu, X.T.; Guo, R.; Zhang, R.D. Integrative analysis reveals key mRNAs and lncRNAs in monocytes of osteoporotic patients. Math. Biosci. Eng. 2019, 16, 5947–5970. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.-S.; Zhang, X.-B.; Zhu, X.-T.; Wang, C.-S. LncRNA Bmncr alleviates the progression of osteoporosis by inhibiting RANML-induced osteoclast differentiation. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 9199–9206. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Yu, D.; Chu, W.; Liu, Z.; Li, H.; Zhai, Z. LncRNA expression profiles and the negative regulation of lncRNA-NOMMUT037835.2 in osteoclastogenesis. Bone 2020, 130. [Google Scholar] [CrossRef]
- Yang, Y.-S.; Xie, J.; Chaugule, S.; Wang, D.; Kim, J.-M.; Kim, J.; Tai, P.W.; Seo, S.-K.; Gravallese, E.; Gao, G.; et al. Bone-Targeting AAV-Mediated Gene Silencing in Osteoclasts for Osteoporosis Therapy. Mol. Ther. Methods Clin. Dev. 2020, 17, 922–935. [Google Scholar] [CrossRef]
- Lin, C.; Yu, S.; Jin, R.; Xiao, Y.; Pan, M.; Pei, F.; Zhu, X.; Huang, H.; Zhang, Z.; Chen, S.; et al. Circulating miR-338 Cluster activities on osteoblast differentiation: Potential Diagnostic and Therapeutic Targets for Postmenopausal Osteoporosis. Theranostics 2019, 9, 3780–3797. [Google Scholar] [CrossRef]
- Ahmad, N.; Kushwaha, P.; Karvande, A.; Tripathi, A.K.; Kothari, P.; Adhikary, S.; Khedgikar, V.; Mishra, V.K.; Trivedi, R. MicroRNA-672-5p Identified during Weaning Reverses Osteopenia and Sarcopenia in Ovariectomized Mice. Mol. Ther. Nucleic Acids 2019, 14, 536–549. [Google Scholar] [CrossRef]
- Gao, Y.; Ge, W. The histone methyltransferase DOT1L inhibits osteoclastogenesis and protects against osteoporosis. Cell Death Dis. 2018, 9, 1–16. [Google Scholar] [CrossRef]
- Jing, H.; Liao, L.; An, Y.; Su, X.; Liu, S.; Shuai, Y.; Zhang, X.; Jin, Y. Suppression of EZH2 Prevents the Shift of Osteoporotic MSC Fate to Adipocyte and Enhances Bone Formation During Osteoporosis. Mol. Ther. 2016, 24, 217–229. [Google Scholar] [CrossRef]
- Li, B.; Zhao, J.; Ma, J.-X.; Li, G.-M.; Zhang, Y.; Xing, G.-S.; Liu, J.; Ma, X.-L. Overexpression of DNMT1 leads to hypermethylation of H19 promoter and inhibition of Erk signaling pathway in disuse osteoporosis. Bone 2018, 111, 82–91. [Google Scholar] [CrossRef] [PubMed]
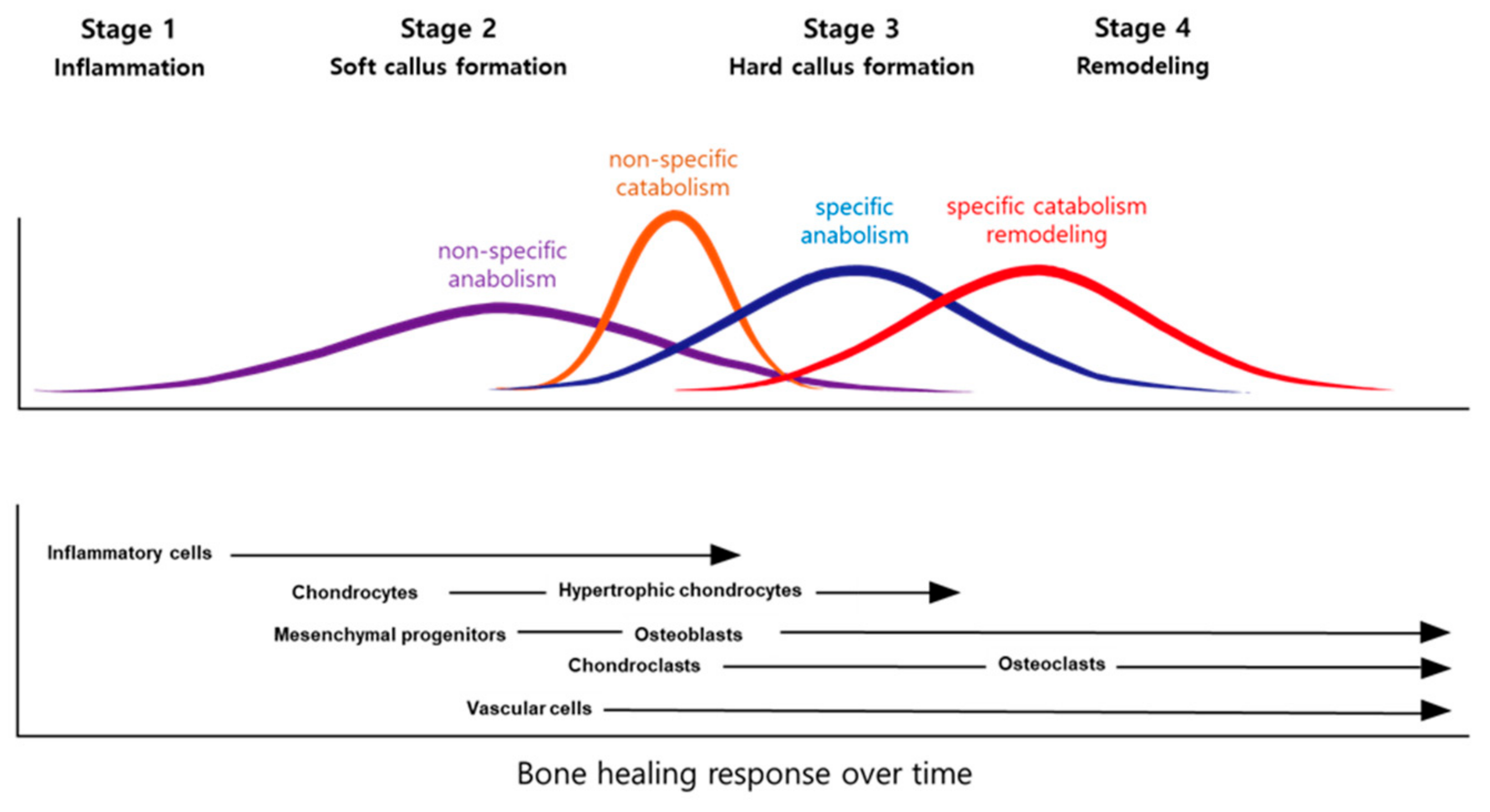
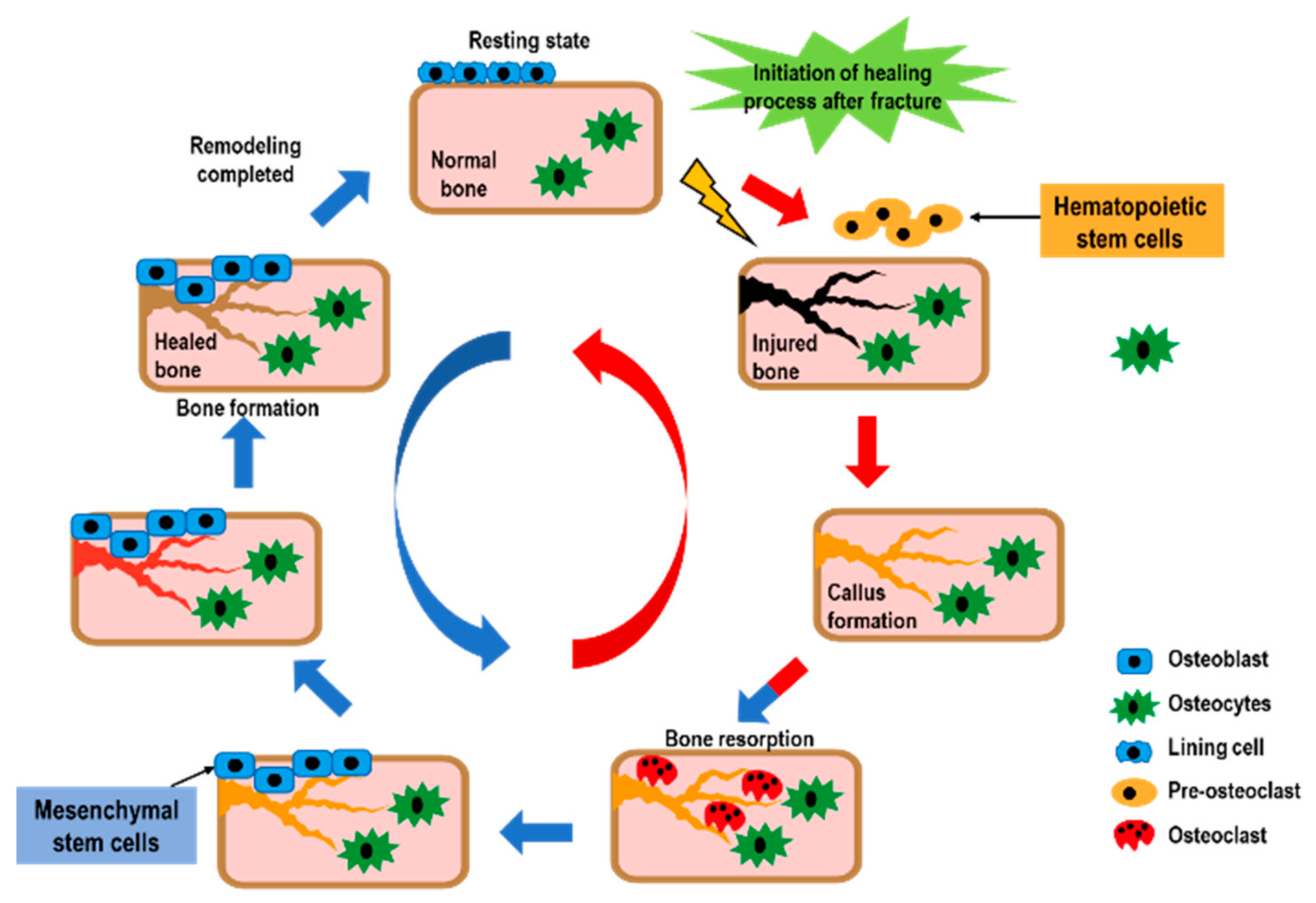

| HDAC | Class | Mechanism | Functions in the Bone |
|---|---|---|---|
| HDAC1 | I | Down-regulation of RUNX2 | Suppress the differentiation of osteoblasts |
| HDAC2 | I | Down-regulation of FoxO1 | Promote RANKL-induced osteoclastogenesis |
| HDAC3 | I | Down-regulation of RUNX2 | Maintain bone mass |
| HDAC4 | II | Down regulation of RUNX2 | Suppress endochondral ossification |
| HDAC5 | II | Down regulation of RUNX2 | Suppress the differentiation of osteoblasts |
| HDAC7 | II | Down regulation of RUNX2 | Regulate endochondral ossification |
| HDAC8 | I | Up-regulation of Homeobox transcription factors Otx2 and Lhx1 | Regulate the intramembranous ossification |
| HDAC9 | II | Down-regulation of RANKL | Suppress osteoclastogenesis |
| ncRNA | Length (nt) | Characteristics |
|---|---|---|
| MicroRNA(miRNA) | 20–24 |
|
| Small interfering RNA (siRNA) | 20–24 |
|
| Piwi-interacting RNA (piRNA) | 24–31 |
|
| Promoter-associated RNA (PAR) | 16–200 |
|
| Enhancer RNA (eRNA) | 100–9000 |
|
| Long non-coding RNA (lncRNA) | >200 |
|
| miRNA Family | Pathway or Affected Molecule | Effect on Osteoblasts and Osteoclasts |
|---|---|---|
| miR-33-5p | Hmga2 | Promote the differentiation of osteoblasts |
| miR-96 | EGFR, HB-EGF Wnt/β-catenin signaling pathway | Promote the differentiation of osteoblasts |
| miR-139-5p | NOTCH1, Wnt/β-catenin pathway | Promote the differentiation of osteoblasts |
| miR-194 | RUNX2 | Promote the differentiation of osteoblasts |
| miR-216a | PI3K/AKT pathway BMP/TGF-β signaling pathway | Promote the differentiation of osteoblasts |
| miRNA-433-3p | DKK1 | Promote the differentiation of osteoblasts |
| miR-542-3p | SFRP1 BMP-7/PI3K-surviving pathway NKIRAS2, NF-κB signaling pathway | Promote the differentiation of osteoblasts Inhibit the differentiation of osteoblasts |
| miR-26a | Smad1 | Inhibit the differentiation of osteoblasts |
| miR-100 | Smad1 | Inhibit the differentiation of osteoblasts |
| miR-124 | Dlx3, Dlx5, and Dlx2 GSK-3β, Wnt/β-catenin pathway | Inhibit the differentiation of osteoblasts |
| miR-125b | BMPR1b | Inhibit the differentiation of osteoblasts |
| miR-153 | BMPR2 | Inhibit the differentiation of osteoblasts |
| miR-203a-3p | Smad9, Wnt/β-catenin signaling pathway | Inhibit the differentiation of osteoblasts |
| miR-214-3p | ATF4 | Inhibit the differentiation of osteoblasts |
| miR-375 | RUNX2 | Inhibit the differentiation of osteoblasts |
| miR-19a | TWIST and RUNX2 | Promote the differentiation of osteoclasts |
| miR-21 | RANKL, PI3K/Akt signaling pathway, PDCD4 | Promote the differentiation of osteoclasts |
| miR-155 | TNF-α, IL-1β, RANKL, M-CSF, RANK, TRAP, Bcl-2, LEPR, AMPK, p-AMPK, OPG, Bax, TAB 1 | Promote the differentiation of osteoclasts |
| miR-182 | Foxo3, Maml1 | Promote the differentiation of osteoclasts |
| miR-183 | RANKL, HO-1 | Promote the differentiation of osteoclasts |
| miR-223 | TWIST and RUNX2 | Promote the differentiation of osteoclasts |
| miR-214 | Pten, PI3K/Akt pathway | Promote the differentiation of osteoclasts |
| miR-7b | DC-STAMP | Inhibit the differentiation of osteoclasts |
| miR-17 | RANKL | Inhibit the differentiation of osteoclasts |
| miR-26a | CTGF/CCN2 | Inhibit the differentiation of osteoclasts |
| miR-31 | RhoA | Inhibit the differentiation of osteoclasts |
| miR-126-5p | PTHrP and MMP-13 | Inhibit the differentiation of osteoclasts |
| miR-141 | Calcr, EphA2 | Inhibit the differentiation of osteoclasts |
| miR-503 | RANK | Inhibit the differentiation of osteoclasts |
| lncRNA | Pathway or Affected Molecule | Effect on Osteoblasts and Osteoclasts |
|---|---|---|
| lncRNA HIF1A-AS1 | SIRT1 | Promote the differentiation of osteoblasts |
| LncRNA HoxA-AS3 | EZH2, H3K27me3, RUNX2 | Promote the differentiation of osteoblasts |
| lncRNA MALAT1 | miR-143, miR-204 | Promote the differentiation of osteoblasts |
| lncRNA MODR | miR-454 | Promote the differentiation of osteoblasts |
| LncRNA KCNQ1OT1 | miR-214 | Promote the differentiation of osteoblasts |
| LncRNA NTF3-5 | miR-93-3p | Promote the differentiation of osteoblasts |
| LncRNA POIR | miR-182 | Promote the differentiation of osteoblasts |
| LncRNA Linc-ROR | miR-145 | Promote the differentiation of osteoblasts |
| LncRNA H19 | miR-675, miR-141, miR-22 Wnt/β-catenin pathway | Promote the differentiation of osteoblasts Inhibit the differentiation of osteoblasts |
| LncRNA-DANCR | EZH2, H3K27me3, RUNX2, p38 MAPK | Inhibit the differentiation of osteoblasts |
| LncRNA ANCR | Wnt/β-catenin pathway | Inhibit the differentiation of osteoblasts |
| LncRNA HOTAIR | Wnt/β-catenin pathway | Inhibit the differentiation of osteoblasts |
| lncRNA p21 | E2, Wnt/β-catenin pathway | Inhibit the differentiation of osteoblasts |
| Lnc-AK045490 | β-catenin, TCF1, LEF1, and RUNX2 | Inhibit the differentiation of osteoblasts |
| Lnc-AK016739 | osteoblastic TF | Inhibit the differentiation of osteoblasts |
| lncRNA UCA1 | BMP-2/(Smad1//5/8) | Inhibit the differentiation of osteoblasts |
| LncRNA MEG3 | Wnt/β-catenin signaling pathway IGF1 | Promote the differentiation of osteoclasts Inhibit the differentiation of osteoblasts |
| LncRNA HOTAIR | miR-17-5p, Smad7 | Inhibit the differentiation of osteoblasts |
| LncRNA MIAT | miR-150-5p | Inhibit the differentiation of osteoblasts |
| lncRNA-ORLNC1 | miR-296 | Inhibit the differentiation of osteoblasts |
| LncRNA MEG3 | miR-133a-3p | Inhibit the differentiation of osteoblasts |
| LncRNA TSIX | miR-30a-5p, and RUNX2 | Promote the apoptosis of osteoblasts |
| lncRNA TUG1 | PTEN | Promote the differentiation of osteoclasts |
| lncRNA AK077216 | NIP45, NFATc1 | Promote the differentiation of osteoclasts |
| lncRNA SNHG15 | RANK/RANKL pathway | Promote the differentiation of osteoclasts |
| LncRNA-Jak3 | NFATc1, CTSK | Promote the differentiation of osteoclasts |
| LncRNA LINC00311 | DDL3 | Promote the differentiation of osteoclasts |
| LncRNA RP11-498C9.17 | HDAC4 | Inhibit the differentiation of osteoclasts |
| LncRNA Bmncr | RANK | Inhibit the differentiation of osteoclasts |
| LncRNA NONMMUT037835.2 | RANK, NF-κB/MAPK signaling pathway | Inhibit the differentiation of osteoclasts |
| LncRNA-NEF | IL-6 | Inhibit the differentiation of osteoclasts |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.-T.; Lee, Y.-S.; Han, I. The Role of Epigenomics in Osteoporosis and Osteoporotic Vertebral Fracture. Int. J. Mol. Sci. 2020, 21, 9455. https://doi.org/10.3390/ijms21249455
Kim K-T, Lee Y-S, Han I. The Role of Epigenomics in Osteoporosis and Osteoporotic Vertebral Fracture. International Journal of Molecular Sciences. 2020; 21(24):9455. https://doi.org/10.3390/ijms21249455
Chicago/Turabian StyleKim, Kyoung-Tae, Young-Seok Lee, and Inbo Han. 2020. "The Role of Epigenomics in Osteoporosis and Osteoporotic Vertebral Fracture" International Journal of Molecular Sciences 21, no. 24: 9455. https://doi.org/10.3390/ijms21249455
APA StyleKim, K.-T., Lee, Y.-S., & Han, I. (2020). The Role of Epigenomics in Osteoporosis and Osteoporotic Vertebral Fracture. International Journal of Molecular Sciences, 21(24), 9455. https://doi.org/10.3390/ijms21249455






