Structural and Functional Enrichment Analyses for Antimicrobial Peptides
Abstract
1. Introduction
2. Results
2.1. Enriched AMP Functions
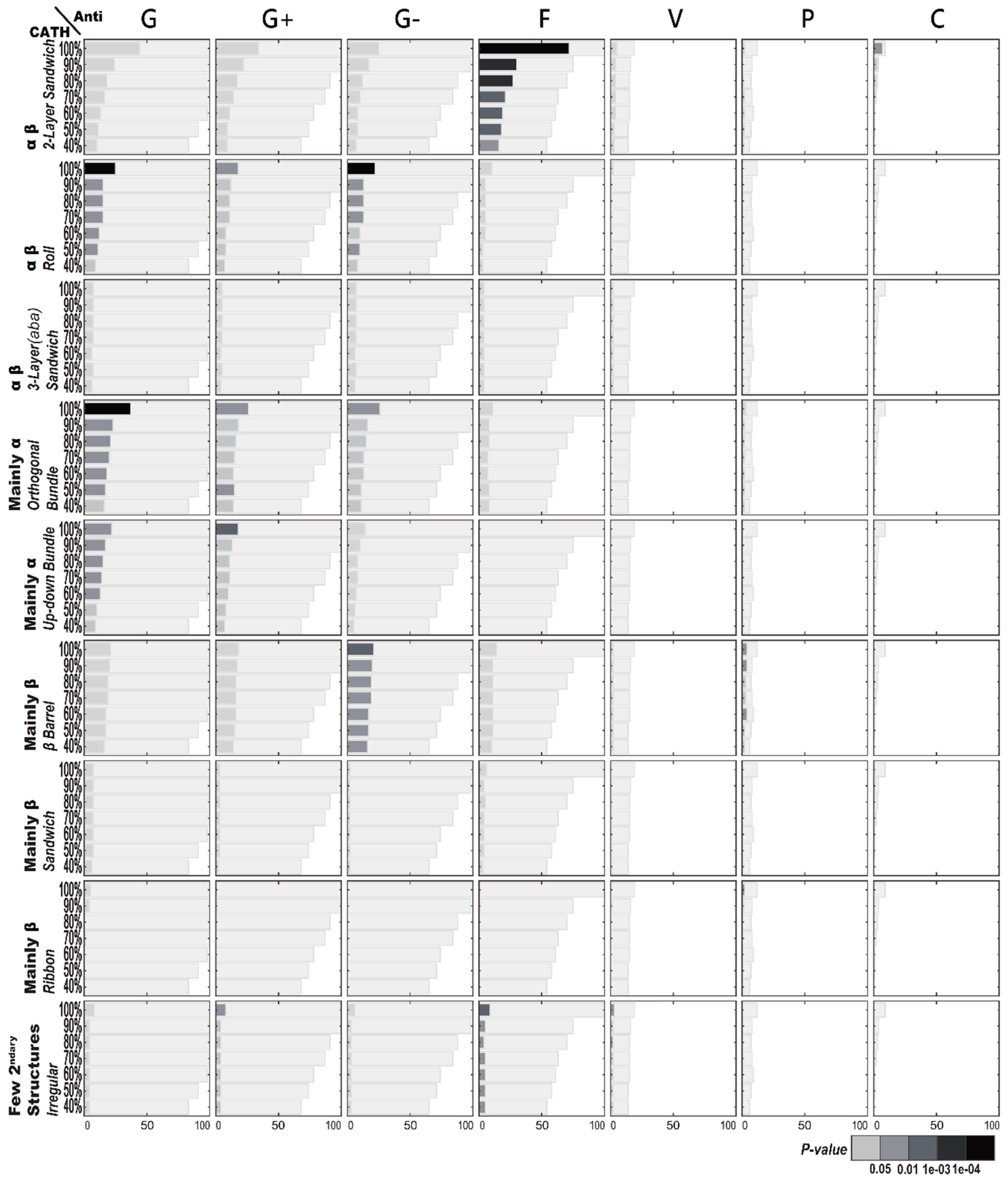
2.1.1. Enriched AMP Functions in Terms of CATH Structures
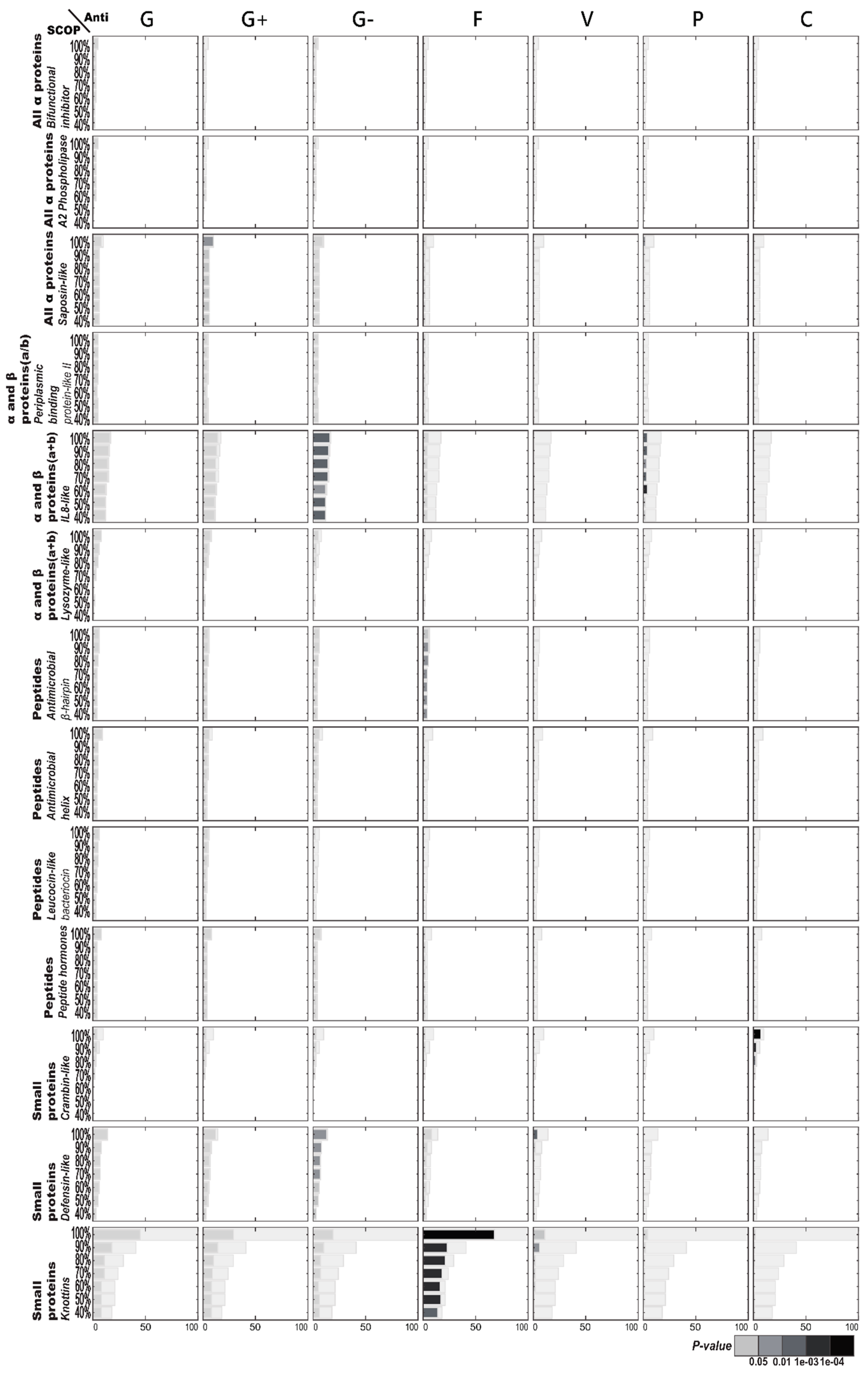
2.1.2. Enriched AMP Functions in Terms of SCOP Structures
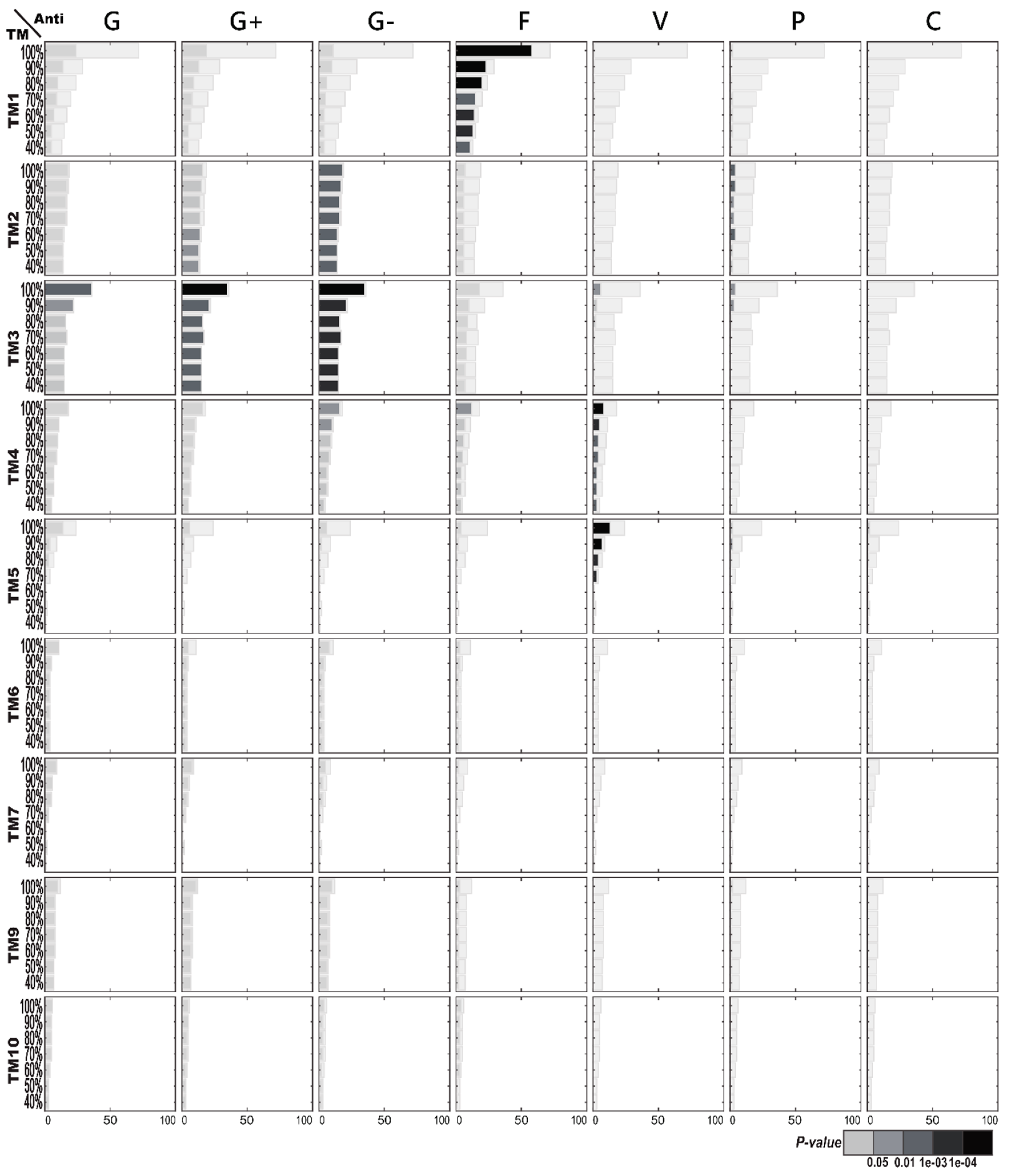
2.1.3. Enriched AMP Functions in Terms of TM Structures
2.2. Enriched AMP Structures
2.2.1. Enriched CATH Structures in Terms of AMP Functions
2.2.2. Enriched SCOP Structures in Terms of AMP Functions
2.2.3. Enriched TM Structures in Terms of AMP Functions
2.3. AMP Structure–Function Enrichment Analysis
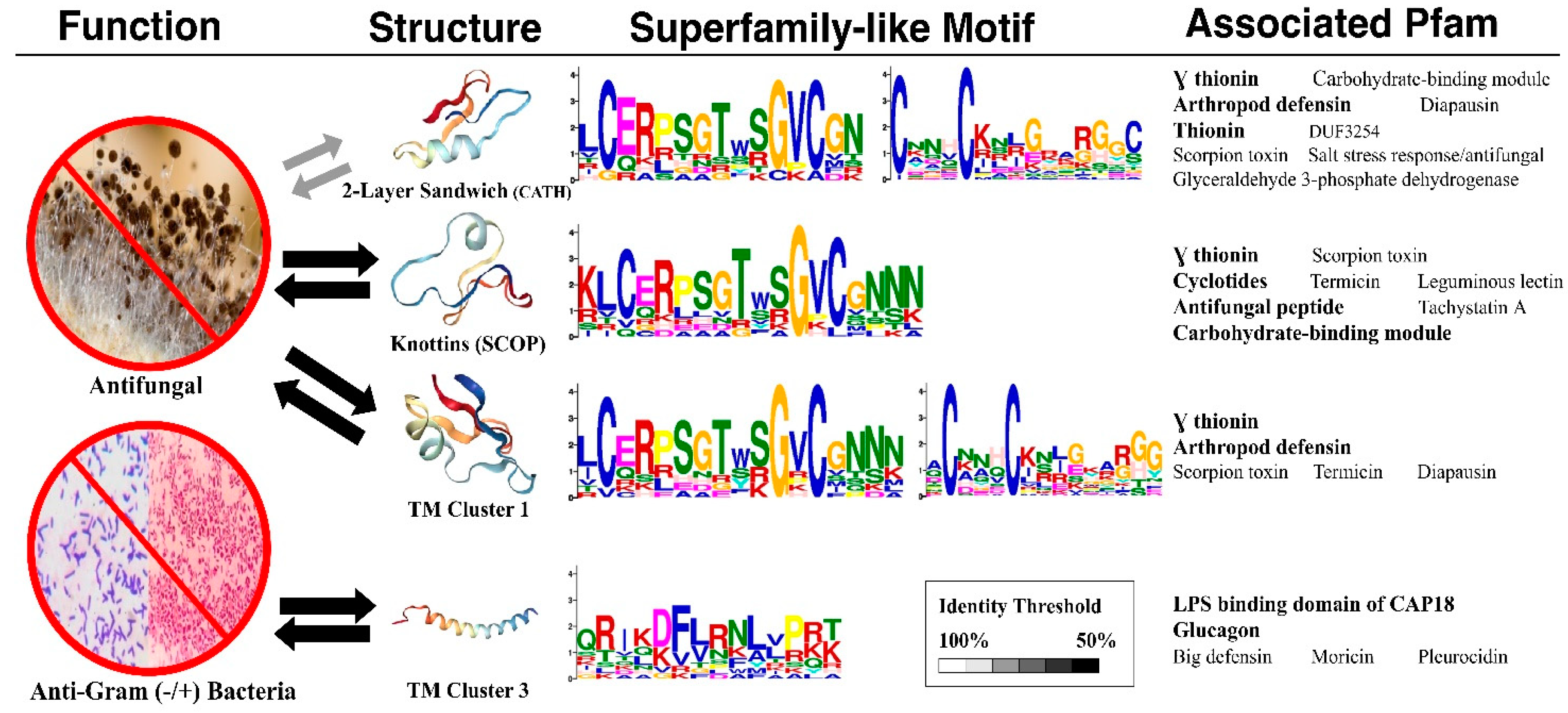
2.4. AMP Structure–Function Enrichments Associated with Sequences
2.4.1. Superfamily-Like Sequence Motifs
2.4.2. Associated Pfam Families or Domains
3. Discussion
4. Materials and Methods
4.1. Antimicrobial Peptides
4.2. Antimicrobial Peptide Structures
4.3. Representative Antimicrobial Peptides
4.4. Structure Classification Methods
4.4.1. CATH Structural Classification
4.4.2. SCOP Structural Classification
4.4.3. TM Structural Classification
4.5. Enrichment Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ADAM | A Database of Anti-Microbial Peptides |
| AMP | Antimicrobial peptide |
| CATH | Class, Architecture, Topology, Homology |
| SCOP | Structural Classification of Proteins |
| TM | Template Modeling |
References
- Mahlapuu, M.; Hakansson, J.; Ringstad, L.; Bjorn, C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.T.; Lee, C.C.; Yang, J.R.; Lai, J.Z.; Chang, K.Y. A large-scale structural classification of antimicrobial peptides. BioMed Res. Int. 2015, 2015, 475062. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.D.T.; Pedron, C.N.; Higashikuni, Y.; Kramer, R.M.; Cardoso, M.H.; Oshiro, K.G.N.; Franco, O.L.; Silva Junior, P.I.; Silva, F.D.; Oliveira Junior, V.X.; et al. Structure-function-guided exploration of the antimicrobial peptide polybia-CP identifies activity determinants and generates synthetic therapeutic candidates. Commun. Biol. 2018, 1, 221. [Google Scholar] [CrossRef] [PubMed]
- Braff, M.H.; Hawkins, M.A.; Di Nardo, A.; Lopez-Garcia, B.; Howell, M.D.; Wong, C.; Lin, K.; Streib, J.E.; Dorschner, R.; Leung, D.Y.; et al. Structure-function relationships among human cathelicidin peptides: Dissociation of antimicrobial properties from host immunostimulatory activities. J. Immunol. 2005, 174, 4271–4278. [Google Scholar] [CrossRef]
- Wu, P.-S.; Lai, S.-J.; Fung, K.-M.; Tseng, T.-S. Characterization of the structure–function relationship of a novel salt-resistant antimicrobial peptide, RR12. RSC Adv. 2020, 10, 23624–23631. [Google Scholar] [CrossRef]
- Lacerda, A.F.; Vasconcelos, E.A.; Pelegrini, P.B.; Grossi de Sa, M.F. Antifungal defensins and their role in plant defense. Front. Microbiol. 2014, 5, 116. [Google Scholar] [CrossRef]
- Tam, J.P.; Wang, S.; Wong, K.H.; Tan, W.L. Antimicrobial Peptides from Plants. Pharm. Basel 2015, 8, 711–757. [Google Scholar] [CrossRef]
- Dawson, N.L.; Lewis, T.E.; Das, S.; Lees, J.G.; Lee, D.; Ashford, P.; Orengo, C.A.; Sillitoe, I. CATH: An expanded resource to predict protein function through structure and sequence. Nucleic Acids Res. 2017, 45, D289–D295. [Google Scholar] [CrossRef]
- Chandonia, J.M.; Fox, N.K.; Brenner, S.E. SCOPe: Classification of large macromolecular structures in the structural classification of proteins-extended database. Nucleic Acids Res. 2019, 47, D475–D481. [Google Scholar] [CrossRef]
- Zhang, Y.; Skolnick, J. TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005, 33, 2302–2309. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful pproach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Molesini, B.; Treggiari, D.; Dalbeni, A.; Minuz, P.; Pandolfini, T. Plant cystine-knot peptides: Pharmacological perspectives. Br. J. Clin. Pharmacol. 2017, 83, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Postic, G.; Gracy, J.; Perin, C.; Chiche, L.; Gelly, J.C. KNOTTIN: The database of inhibitor cystine knot scaffold after 10 years, toward a systematic structure modeling. Nucleic Acids Res. 2018, 46, D454–D458. [Google Scholar] [CrossRef]
- Schneider, G.; Clark, D.E. Automated De Novo Drug Design: Are We Nearly There Yet? Angew. Chem. Int. Ed. Engl. 2019, 58, 10792–10803. [Google Scholar] [CrossRef]
- Torres, M.D.T.; Sothiselvam, S.; Lu, T.K.; de la Fuente-Nunez, C. Peptide Design Principles for Antimicrobial Applications. J. Mol. Biol. 2019, 431, 3547–3567. [Google Scholar] [CrossRef]
- Wang, C.K.; Shih, L.Y.; Chang, K.Y. Large-Scale Analysis of Antimicrobial Activities in Relation to Amphipathicity and Charge Reveals Novel Characterization of Antimicrobial Peptides. Molecules 2017, 22, 2037. [Google Scholar] [CrossRef]
- Waghu, F.H.; Barai, R.S.; Gurung, P.; Idicula-Thomas, S. CAMPR3: A database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res. 2016, 44, D1094–D1097. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef]
- Burley, S.K.; Berman, H.M.; Bhikadiya, C.; Bi, C.; Chen, L.; Di Costanzo, L.; Christie, C.; Dalenberg, K.; Duarte, J.M.; Dutta, S.; et al. RCSB Protein Data Bank: Biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019, 47, D464–D474. [Google Scholar] [CrossRef]
- Mirdita, M.; Steinegger, M.; Söding, J. MMseqs2 desktop and local web server app for fast, interactive sequence searches. Bioinformatics 2019, 35, 2856–2858. [Google Scholar] [CrossRef] [PubMed]
- Michie, A.D.; Orengo, C.A.; Thornton, J.M. Analysis of Domain Structural Class Using an Automated Class Assignment Protocol. J. Mol. Biol. 1996, 262, 168–185. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics 2010, 26, 889–895. [Google Scholar] [CrossRef] [PubMed]



| Type | Structure | Sequence Identity Threshold (%) | F(x) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1st Level | 2nd Level | 100 | 90 | 80 | 70 | 60 | 50 | 40 | ||
| CATH | α β | * | ⟷ | F | ||||||
| 2-Layer Sandwich | ⟷ | ⟷ | ⟷ | → | F | |||||
| Roll | ← | G | ||||||||
| Roll | ← | G- | ||||||||
| Mainly α | * | ← | ← | ← | ← | G | ||||
| * | ← | G+ | ||||||||
| Orthogonal Bundle | ← | G | ||||||||
| Mainly β | * | → | P | |||||||
| Beta Barrel | ← | G- | ||||||||
| SCOP | α and β (α + β) | * | ← | G | ||||||
| * | ← | G+ | ||||||||
| IL8-like | ⟷ | ← | ← | ← | G- | |||||
| Peptides | * | → | G | |||||||
| * | → | G+ | ||||||||
| Small proteins | * | ⟷ | ⟷ | ⟷ | ⟷ | ⟷ | ⟷ | ⟷ | F | |
| Knottins | ⟷ | ⟷ | ⟷ | ⟷ | → | ⟷ | → | F | ||
| Crambin-like | ⟷ | ⟷ | ⟷ | C | ||||||
| Defensin-like | ← | G- | ||||||||
| TM | TM1 | ⟷ | ⟷ | ⟷ | → | ⟷ | ⟷ | → | F | |
| TM2 | ⟷ | ⟷ | ⟷ | ⟷ | → | ⟷ | ⟷ | G- | ||
| TM3 | ⟷ | G | ||||||||
| TM3 | ⟷ | ⟷ | ⟷ | ⟷ | ← | ⟷ | → | G+ | ||
| TM3 | ⟷ | ⟷ | ⟷ | ⟷ | ⟷ | ⟷ | ⟷ | G- | ||
| TM4 | ← | G | ||||||||
| TM4 | ← | G+ | ||||||||
| TM4 | ← | G- | ||||||||
| TM4 | ⟷ | → | → | → | → | V | ||||
| TM5 | ⟷ | ⟷ | → | → | V | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lo, S.C.; Xie, Z.-R.; Chang, K.Y. Structural and Functional Enrichment Analyses for Antimicrobial Peptides. Int. J. Mol. Sci. 2020, 21, 8783. https://doi.org/10.3390/ijms21228783
Lo SC, Xie Z-R, Chang KY. Structural and Functional Enrichment Analyses for Antimicrobial Peptides. International Journal of Molecular Sciences. 2020; 21(22):8783. https://doi.org/10.3390/ijms21228783
Chicago/Turabian StyleLo, Sheng C., Zhong-Ru Xie, and Kuan Y. Chang. 2020. "Structural and Functional Enrichment Analyses for Antimicrobial Peptides" International Journal of Molecular Sciences 21, no. 22: 8783. https://doi.org/10.3390/ijms21228783
APA StyleLo, S. C., Xie, Z.-R., & Chang, K. Y. (2020). Structural and Functional Enrichment Analyses for Antimicrobial Peptides. International Journal of Molecular Sciences, 21(22), 8783. https://doi.org/10.3390/ijms21228783







