Exposome and Immunity Training: How Pathogen Exposure Order Influences Innate Immune Cell Lineage Commitment and Function
Abstract
1. Introduction
2. Influence of Pathogen Exposure on Innate Immune Cell Development and Function
2.1. Pathogen Exposure, Epigenetic Modifications, and Lineage Determination
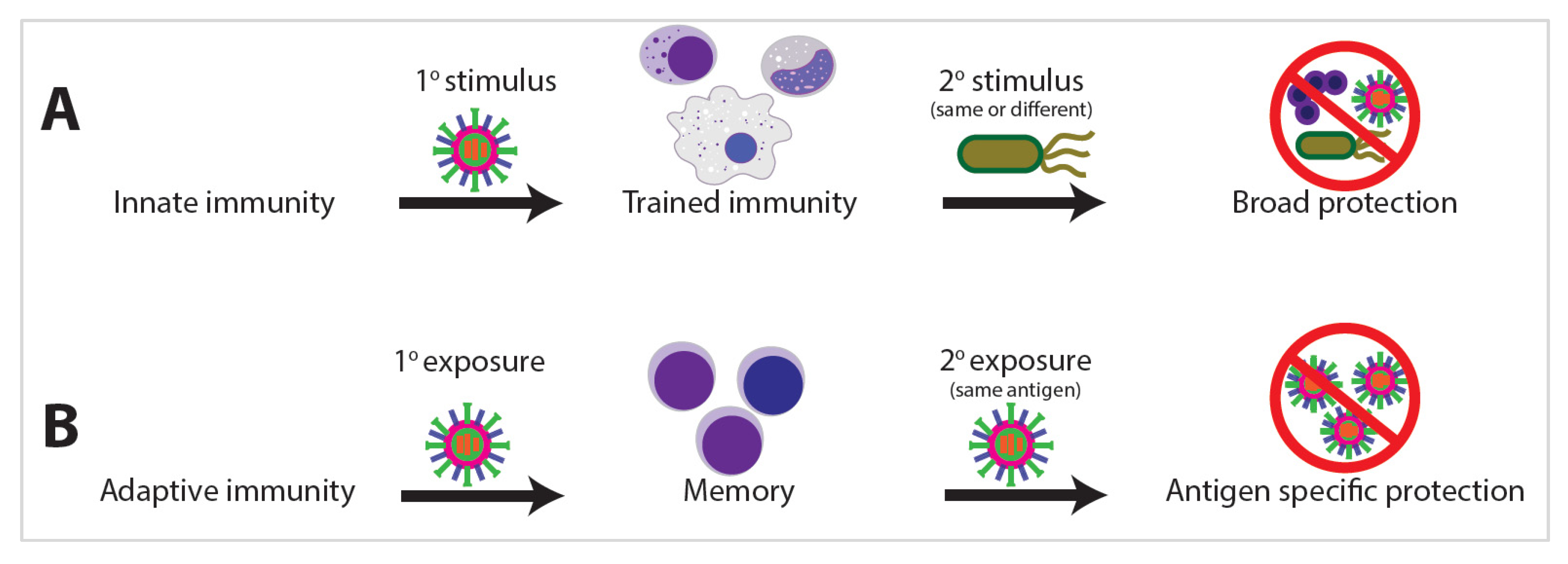
2.2. Discovery of Trained Immunity
2.3. Natural Killer Cells
2.4. Dendritic Cells
2.5. Monocytes and Macrophages
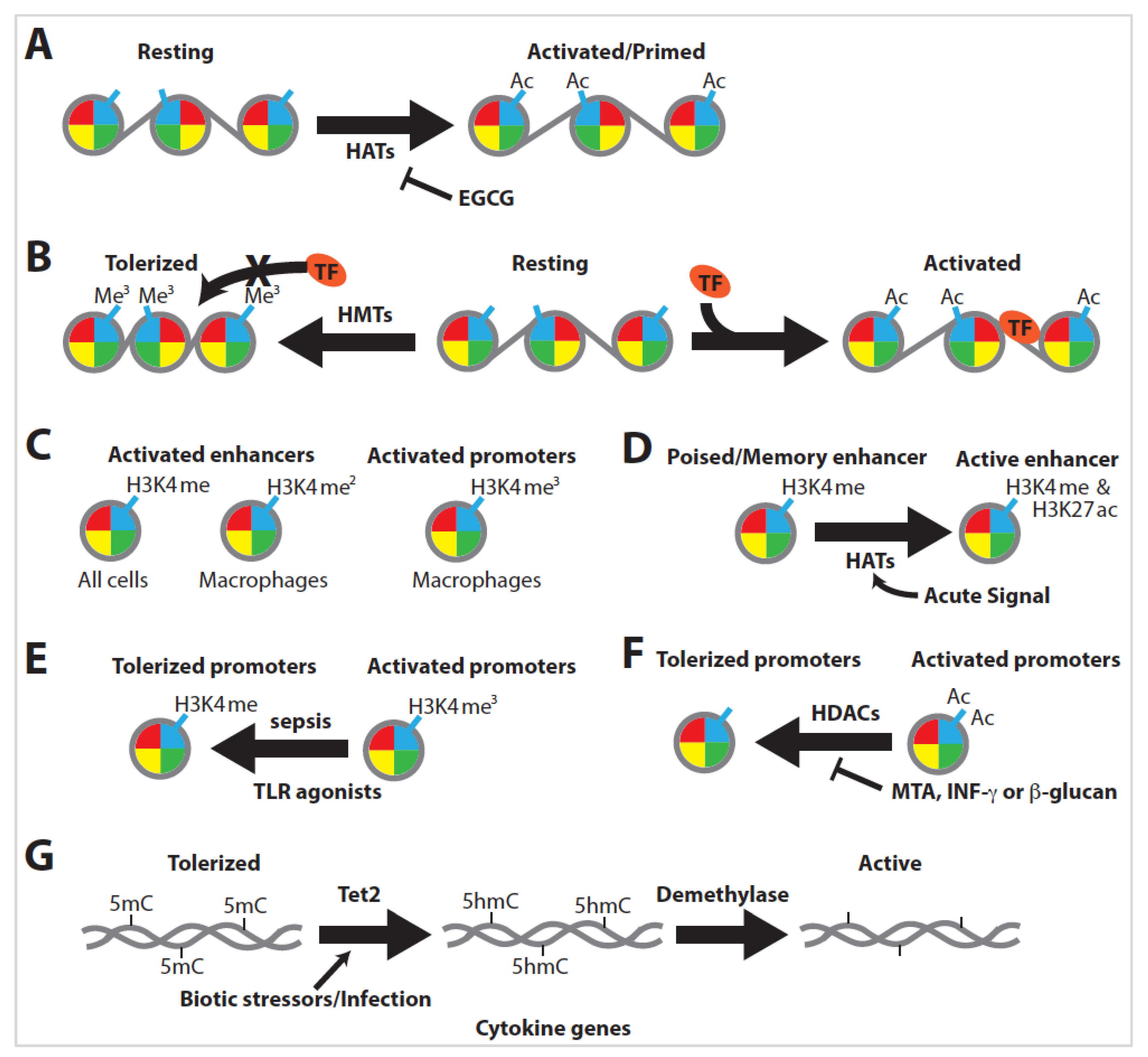
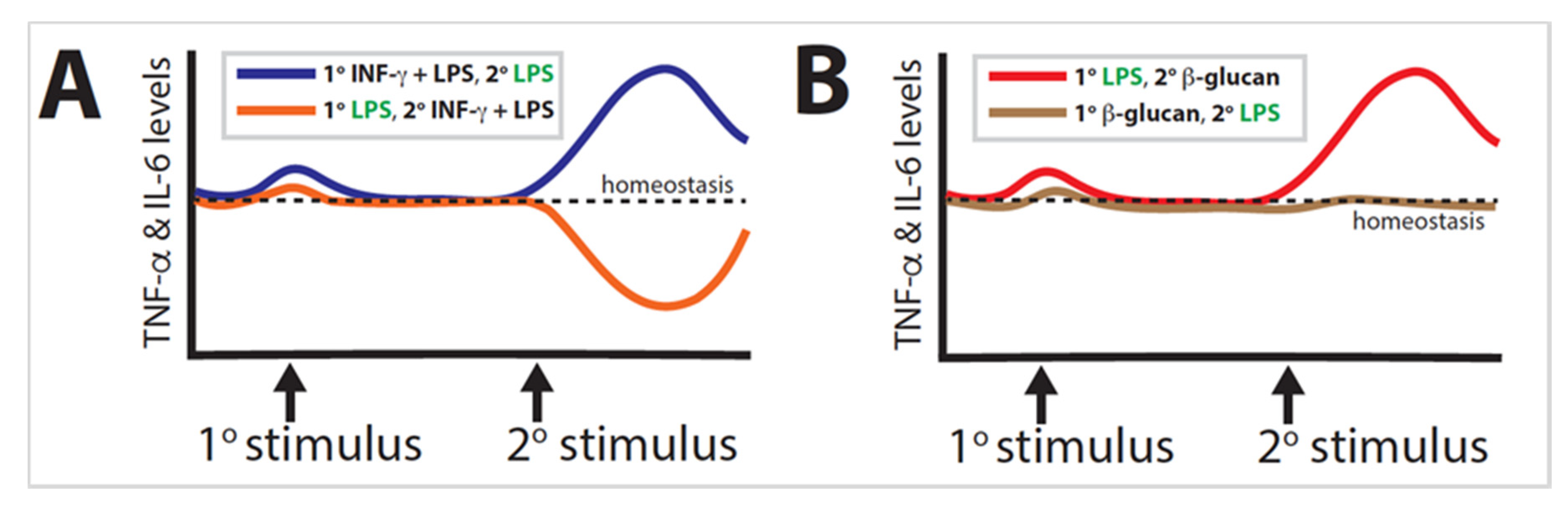
2.6. Macrophage Tolerance
2.7. Macrophage Training
2.8. Granulocytes
3. Epigenetic Regulation of Trained Immunity
3.1. Epigenetic Regulation
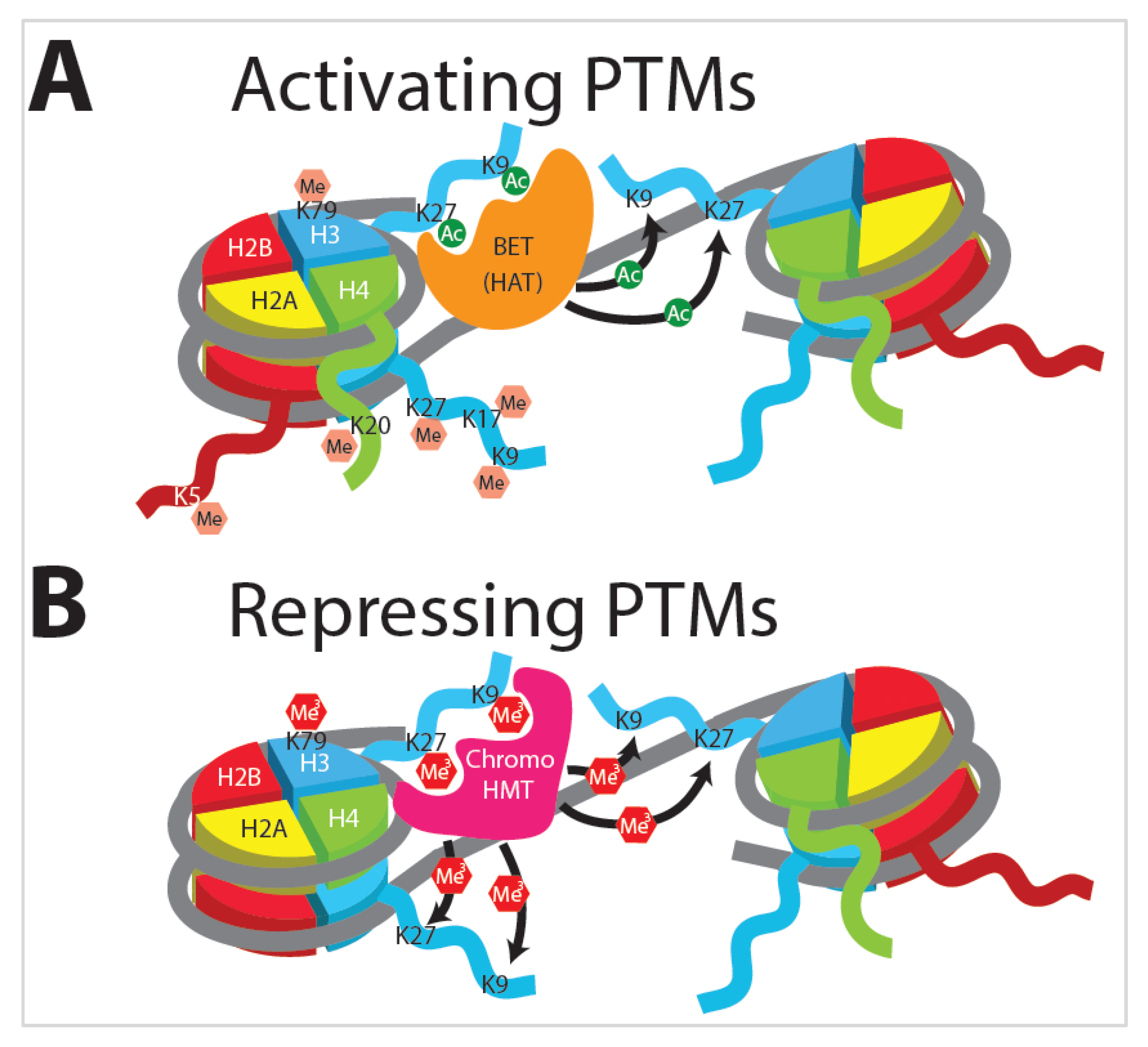
3.2. Histone Modifications
3.3. Histone Acetylation
3.4. Histone Methylation
3.5. Innate Immune Training Is Dependent on Histone Modification
3.6. DNA Methylation
3.7. Transcription Factors and Enhancers
3.8. Transcriptomics
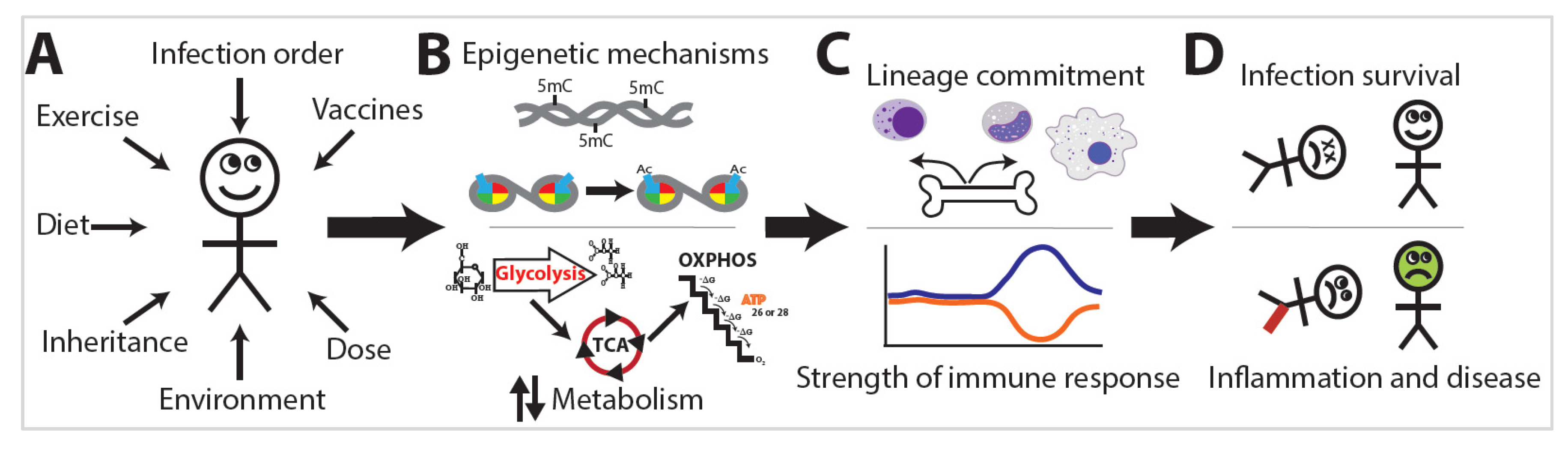
4. Exposome Alterations of Immune Function
4.1. Metabolism
4.2. Infection Survival
4.3. Inflammatory/Disease Phenotypes
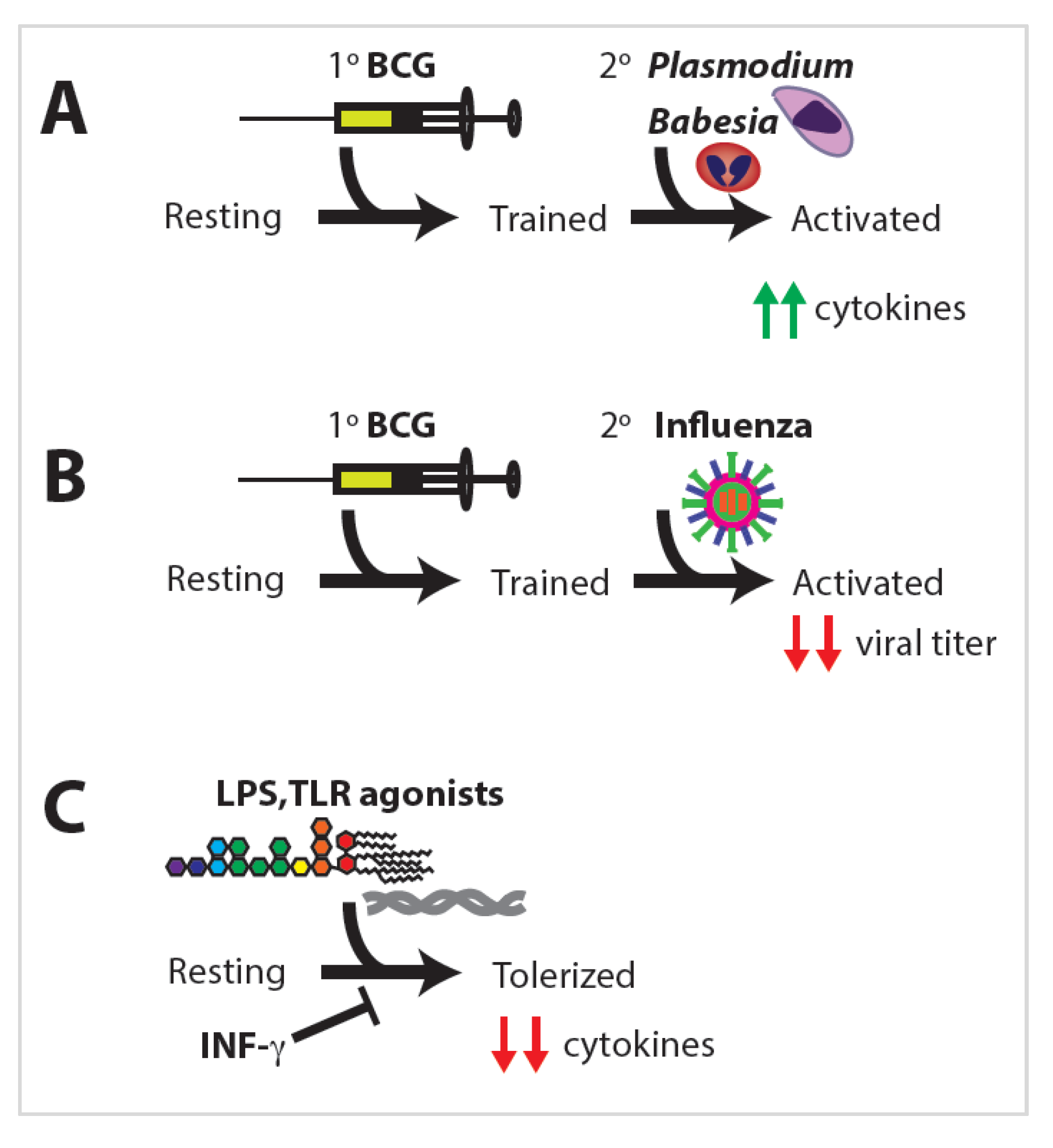
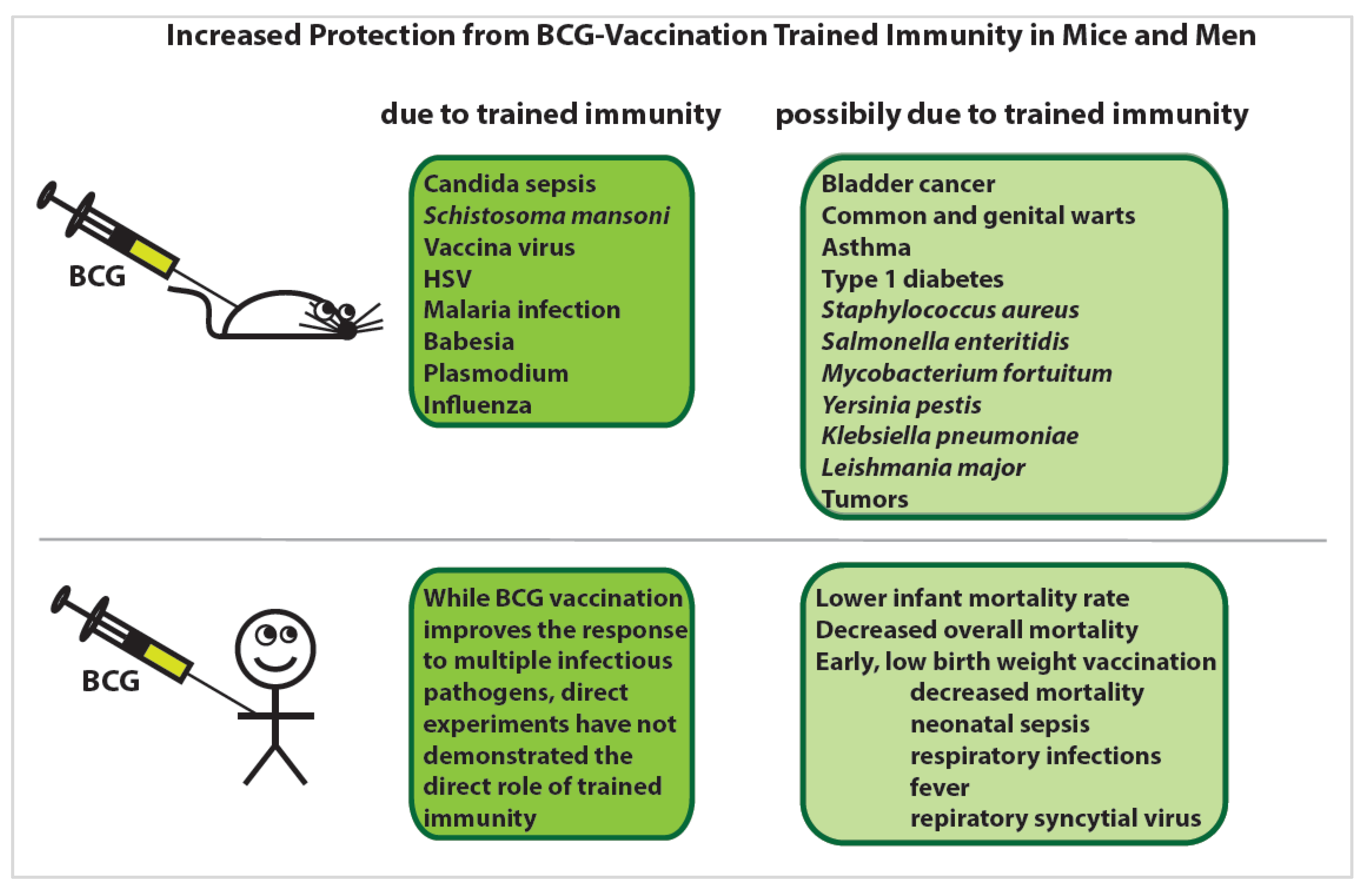
4.4. Heterologous Immune Protection
4.5. Sex-Dependent Differences
4.6. Trained Immunity Mechanisms
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BCG | Bacillus Calmette–Guerin |
| HELIX | The Human Early Life Exposome |
| DC | Dendritic cell |
| NK | Natural killer cells |
| LPS | Lipopolysaccharide |
| TLR | Toll-like receptor |
| HSC | Hematopoietic stem cells |
| MPP | Multipotent progenitors |
| MTB | Mycobacterium tuberculosis |
| HFD | High-fat diet |
| MCMV | Mouse cytomegalovirus |
| IL | Interleukin |
| oxLDL | Oxidized low-density lipoprotein |
| HAT | Histone acetyltransferase |
| HMT | Histone methyltransferase |
| MTA | Methylthioadenosine |
| EGCG | Epigallocatechin-3-gallate |
| ROS | Reactive oxygen species |
| TF | Transcription factor |
| DPT | Diphtheria, pertussis, and tetanus |
| MV | Measles vaccine |
| 5mC | 5-methylcytosine |
| 5hmc | 5-hydroxymethylcytosine |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| COVID-19 | Coronavirus Disease 2019 |
References
- Netea, M.G.; Joosten, L.A.B.; Latz, E.; Mills, K.H.G.; Natoli, G.; Stunnenberg, H.G.; O’Neill, L.A.J.; Xavier, R.J. Trained immunity: A program of innate immune memory in health and disease. Science 2016, 352, aaf1098. [Google Scholar] [CrossRef]
- Netea, M.G.; Domínguez-Andrés, J.; Barreiro, L.B.; Chavakis, T.; Divangahi, M.; Fuchs, E.; Joosten, L.A.B.; Van Der Meer, J.W.M.; Mhlanga, M.M.; Mulder, W.J.M.; et al. Defining trained immunity and its role in health and disease. Nat. Rev. Immunol. 2020, 20, 375–388. [Google Scholar] [CrossRef]
- Netea, M.G.; Quintin, J.; Van Der Meer, J.W. Trained Immunity: A Memory for Innate Host Defense. Cell Host Microbe 2011, 9, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Arts, R.J.; Moorlag, S.J.; Novakovic, B.; Li, Y.; Wang, S.-Y.; Oosting, M.; Kumar, V.; Xavier, R.J.; Wijmenga, C.; Joosten, L.A.; et al. BCG Vaccination Protects against Experimental Viral Infection in Humans through the Induction of Cytokines Associated with Trained Immunity. Cell Host Microbe 2018, 23, 89–100.e5. [Google Scholar] [CrossRef] [PubMed]
- Kleinnijenhuis, J.; Quintin, J.; Preijers, F.; Joosten, L.A.B.; Ifrim, D.C.; Saeed, S.; Jacobs, C.; Van Loenhout, J.; De Jong, D.; Stunnenberg, H.G.; et al. Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc. Natl. Acad. Sci. USA 2012, 109, 17537–17542. [Google Scholar] [CrossRef]
- Mantovani, A.; Netea, M.G. Trained Innate Immunity, Epigenetics, and Covid-19. N. Engl. J. Med. 2020, 383, 1078–1080. [Google Scholar] [CrossRef] [PubMed]
- Wild, C.P. Complementing the genome with an “Exposome”: The outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol. Biomark. Prev. 2005, 14, 1847–1850. [Google Scholar] [CrossRef]
- Jiang, C.; Wang, X.; Li, X.; Inlora, J.; Wang, T.; Liu, Q.; Snyder, M. Dynamic Human Environmental Exposome Revealed by Longitudinal Personal Monitoring. Cell 2018, 175, 277–291.e31. [Google Scholar] [CrossRef]
- Vrijheid, M.; Slama, R.; Robinson, O.; Chatzi, L.; Coen, M.; Hazel, P.V.D.; Thomsen, C.; Wright, J.; Athersuch, T.J.; Avellana, N.; et al. The Human Early-Life Exposome (HELIX): Project Rationale and Design. Environ. Health Perspect. 2014, 122, 535–544. [Google Scholar] [CrossRef]
- Vineis, P.; Chadeau-Hyam, M.; Gmuender, H.; Gulliver, J.; Herceg, Z.; Kleinjans, J.; Kogevinas, M.; Kyrtopoulos, S.; Nieuwenhuijsen, M.J.; Phillips, D.; et al. The exposome in practice: Design of the EXPOsOMICS project. Int. J. Hyg. Environ. Health 2017, 220, 142–151. [Google Scholar] [CrossRef]
- Schutsky, E.K.; DeNizio, J.E.; Hu, P.; Liu, M.Y.; Nabel, C.S.; Fabyanic, E.B.; Hwang, Y.; Bushman, F.D.; Wu, H.; Kohli, R.M. Nondestructive, base-resolution sequencing of 5-hydroxymethylcytosine using a DNA deaminase. Nat. Biotechnol. 2018, 36, 1083–1090. [Google Scholar] [CrossRef] [PubMed]
- Meadow, J.F.; Altrichter, A.E.; Bateman, A.C.; Stenson, J.; Brown, G.Z.; Green, J.L.; Bohannan, B.J. Humans differ in their personal microbial cloud. PeerJ 2015, 3, e1258. [Google Scholar] [CrossRef] [PubMed]
- Chung, M.K.; Kannan, K.; Louis, G.M.; Patel, C.J. Toward Capturing the Exposome: Exposure Biomarker Variability and Coexposure Patterns in the Shared Environment. Environ. Sci. Technol. 2018, 52, 8801–8810. [Google Scholar] [CrossRef] [PubMed]
- Laker, R.C.; Garde, C.; Camera, D.M.; Smiles, W.J.; Zierath, J.R.; Hawley, J.A.; Barrès, R. Transcriptomic and epigenetic responses to short-term nutrient-exercise stress in humans. Sci. Rep. 2017, 7, 15134. [Google Scholar] [CrossRef]
- Schloissnig, S.; Arumugam, M.; Sunagawa, S.; Mitreva, M.; Tap, J.; Zhu, A.; Waller, A.S.; Mende, D.R.; Kultima, J.R.; Martin, J.; et al. Genomic variation landscape of the human gut microbiome. Nature 2013, 493, 45–50. [Google Scholar] [CrossRef]
- Lax, S.; Smith, D.P.; Hampton-Marcell, J.; Owens, S.M.; Handley, K.M.; Scott, N.M.; Gibbons, S.M.; Larsen, P.; Shogan, B.D.; Weiss, S.; et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 2014, 345, 1048–1052. [Google Scholar] [CrossRef]
- Pfeifer, G.P. Environmental exposures and mutational patterns of cancer genomes. Genome Med. 2010, 2, 54. [Google Scholar] [CrossRef]
- Yuan, J.; Liu, Y. Progress of Genomics in Atherosclerosis-Coronary Heart Disease and Myocardial Infarction. Appl. Clin. Bioinform. 2018, 16, 219–240. [Google Scholar]
- McCreanor, J.; Cullinan, P.; Nieuwenhuijsen, M.J.; Stewart-Evans, J.; Malliarou, E.; Järup, L.; Harrington, R.; Svartengren, M.; Han, I.-K.; Ohman-Strickland, P.; et al. Respiratory effects of exposure to diesel traffic in persons with asthma. N. Engl. J. Med. 2007, 357, 2348–2358. [Google Scholar] [CrossRef]
- Frellstedt, L.; Waldschmidt, I.; Gosset, P.; Desmet, C.; Pirottin, D.; Bureau, F.; Farnir, F.; Franck, T.; Dupuis-Tricaud, M.-C.; Lekeux, P.; et al. Training Modifies Innate Immune Responses in Blood Monocytes and in Pulmonary Alveolar Macrophages. Am. J. Respir. Cell Mol. Biol. 2014, 51, 135–142. [Google Scholar] [CrossRef]
- Fujimura, K.E.; Demoor, T.; Rauch, M.; Faruqi, A.A.; Jang, S.; Johnson, C.C.; Boushey, H.A.; Zoratti, E.; Ownby, D.; Lukacs, N.W.; et al. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc. Natl. Acad. Sci. USA 2013, 111, 805–810. [Google Scholar] [CrossRef] [PubMed]
- Sørup, S.; Benn, C.S.; Poulsen, A.; Krause, T.G.; Aaby, P.; Ravn, H. Live Vaccine Against Measles, Mumps, and Rubella and the Risk of Hospital Admissions for Nontargeted Infections. JAMA 2014, 311, 826. [Google Scholar] [CrossRef] [PubMed]
- Bekkering, S.; Arts, R.J.; Novakovic, B.; Kourtzelis, I.; Van Der Heijden, C.D.; Li, Y.; Popa, C.D.; Ter Horst, R.; Van Tuijl, J.; Netea-Maier, R.T.; et al. Metabolic Induction of Trained Immunity through the Mevalonate Pathway. Cell 2018, 172, 135–146.e9. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Ehrlich, L.I.R.; Seita, J.; Murakami, P.; Doi, A.; Lindau, P.; Lee, H.; Aryee, M.J.; Irizarry, R.A.; Kim, K.; et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature 2010, 467, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Mitroulis, I.; Chen, L.-S.; Singh, R.P.; Kourtzelis, I.; Economopoulou, M.; Kajikawa, T.; Troullinaki, M.; Ziogas, A.; Ruppova, K.; Hosur, K.; et al. Secreted protein Del-1 regulates myelopoiesis in the hematopoietic stem cell niche. J. Clin. Investig. 2017, 127, 3624–3639. [Google Scholar] [CrossRef] [PubMed]
- Brasacchio, D.; Okabe, J.; Tikellis, C.; Balcerczyk, A.; George, P.; Baker, E.K.; Calkin, A.C.; Brownlee, M.; Cooper, M.E.; El-Osta, A. Hyperglycemia Induces a Dynamic Cooperativity of Histone Methylase and Demethylase Enzymes Associated With Gene-Activating Epigenetic Marks That Coexist on the Lysine Tail. Diabetes 2009, 58, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Seijkens, T.; Hoeksema, M.A.; Beckers, L.; Smeets, E.; Meiler, S.; Levels, J.; Tjwa, M.; De Winther, M.P.J.; Lutgens, E. Hypercholesterolemia-induced priming of hematopoietic stem and progenitor cells aggravates atherosclerosis. FASEB J. 2014, 28, 2202–2213. [Google Scholar] [CrossRef]
- Romee, R.; Schneider, S.E.; Leong, J.W.; Chase, J.M.; Keppel, C.R.; Sullivan, R.P.; Cooper, M.A.; Fehniger, T.A. Cytokine activation induces human memory-like NK cells. Blood 2012, 120, 4751–4760. [Google Scholar] [CrossRef]
- Hong, M.; Sandalova, E.; Low, D.; Gehring, A.J.; Fieni, S.; Amadei, B.; Urbani, S.; Chong, Y.-S.; Guccione, E.; Bertoletti, A. Trained immunity in newborn infants of HBV-infected mothers. Nat. Commun. 2015, 6, 6588. [Google Scholar] [CrossRef]
- Kleinnijenhuis, J.; Quintin, J.; Preijers, F.; Benn, C.S.; Joosten, L.A.B.; Jacobs, C.; van Loenhout, J.; Xavier, R.J.; Aaby, P.; van der Meer, J.W.M.; et al. Long-Lasting Effects of BCG Vaccination on Both Heterologous Th1/Th17 Responses and Innate Trained Immunity. J. Innate Immun. 2014, 6, 152–158. [Google Scholar] [CrossRef]
- Kaufmann, E.; Sanz, J.; Dunn, J.L.; Khan, N.; Mendonça, L.E.; Pacis, A.; Tzelepis, F.; Pernet, E.; Dumaine, A.; Grenier, J.-C.; et al. BCG Educates Hematopoietic Stem Cells to Generate Protective Innate Immunity against Tuberculosis. Cell 2018, 172, 176–190.e19. [Google Scholar] [CrossRef] [PubMed]
- Cenci, E.; Bartocci, A.; Puccetti, P.; Mocci, S.; Stanley, E.R.; Bistoni, F. Macrophage colony-stimulating factor in murine candidiasis: Serum and tissue levels during infection and protective effect of exogenous administration. Infect. Immun. 1991, 59, 868–872. [Google Scholar] [CrossRef] [PubMed]
- Mitroulis, I.; Ruppova, K.; Wang, B.; Chen, L.-S.; Grzybek, M.; Grinenko, T.; Eugster, A.; Troullinaki, M.; Palladini, A.; Kourtzelis, I.; et al. Modulation of Myelopoiesis Progenitors Is an Integral Component of Trained Immunity. Cell 2018, 172, 147–161.e12. [Google Scholar] [CrossRef] [PubMed]
- Martínez, A.; Bono, C.; Megías, J.; Yáñez, A.; Gozalbo, D.; Gil, M.L. Systemic Candidiasis and TLR2 Agonist Exposure Impact the Antifungal Response of Hematopoietic Stem and Progenitor Cells. Front. Cell. Infect. Microbiol. 2018, 8, 309. [Google Scholar] [CrossRef]
- Christ, A.; Günther, P.; Lauterbach, M.A.; Duewell, P.; Biswas, D.; Pelka, K.; Scholz, C.J.; Oosting, M.; Haendler, K.; Baßler, K.; et al. Western Diet Triggers NLRP3-Dependent Innate Immune Reprogramming. Cell 2018, 172, 162–175.e14. [Google Scholar] [CrossRef]
- van Kampen, E.; Jaminon, A.; van Berkel, T.J.C.; van Eck, M. Diet-induced (epigenetic) changes in bone marrow augment atherosclerosis. J. Leukoc. Biol. 2014, 96, 833–841. [Google Scholar] [CrossRef]
- Nagareddy, P.R.; Kraakman, M.; Masters, S.L.; Stirzaker, R.A.; Gorman, D.J.; Grant, R.W.; Dragoljevic, D.; Hong, E.S.; Abdel-Latif, A.; Smyth, S.S.; et al. Adipose Tissue Macrophages Promote Myelopoiesis and Monocytosis in Obesity. Cell Metab. 2014, 19, 821–835. [Google Scholar] [CrossRef]
- Singer, K.; DelProposto, J.; Morris, D.L.; Zamarron, B.; Mergian, T.; Maley, N.; Cho, K.W.; Geletka, L.; Subbaiah, P.; Muir, L.; et al. Diet-induced obesity promotes myelopoiesis in hematopoietic stem cells. Mol. Metab. 2014, 3, 664–675. [Google Scholar] [CrossRef]
- Murphy, A.J.; Akhtari, M.; Tolani, S.; Pagler, T.; Bijl, N.; Kuo, C.-L.; Wang, M.; Sanson, M.; Abramowicz, S.; Welch, C.; et al. ApoE regulates hematopoietic stem cell proliferation, monocytosis, and monocyte accumulation in atherosclerotic lesions in mice. J. Clin. Investig. 2011, 121, 4138–4149. [Google Scholar] [CrossRef]
- Clark, I.A.; Allison, A.C.; Cox, F.E.; Clark, A.C.A.I.A. Protection of mice against Babesia, and Plasmodium with BCG. Nat. Cell Biol. 1976, 259, 309–311. [Google Scholar] [CrossRef]
- Spencer, J.C.; Ganguly, R.; Waldman, R.H. Nonspecific protection of mice against influenza virus infection by local or systemic immunization with Bacille Calmette-Guerin. J. Infect. Dis. 1977, 136, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Blok, B.A.; Arts, R.J.W.; Van Crevel, R.; Benn, C.S.; Netea, M. Trained innate immunity as underlying mechanism for the long-term, nonspecific effects of vaccines. J. Leukoc. Biol. 2015, 98, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Lau, C.M.; Adams, N.M.; Geary, C.D.; Weizman, O.-E.; Rapp, M.; Pritykin, Y.; Leslie, C.S.; Sun, J.C. Epigenetic control of innate and adaptive immune memory. Nat. Immunol. 2018, 19, 963–972. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, J.G.; Goodarzi, M.; Drayton, D.L.; Von Andrian, U.H. T cell- and B cell-independent adaptive immunity mediated by natural killer cells. Nat. Immunol. 2006, 7, 507–516. [Google Scholar] [CrossRef]
- Sun, J.C.; Beilke, J.N.; Lanier, L.L. Adaptive immune features of natural killer cells. Nature 2009, 457, 557–561. [Google Scholar] [CrossRef]
- Keppel, M.P.; Yang, L.; Cooper, M.A. Murine NK Cell Intrinsic Cytokine-Induced Memory-like Responses Are Maintained following Homeostatic Proliferation. J. Immunol. 2013, 190, 4754–4762. [Google Scholar] [CrossRef]
- Romee, R.; Rosario, M.; Berrien-Elliott, M.M.; Wagner, J.A.; Jewell, B.A.; Schappe, T.; Leong, J.W.; Abdel-Latif, S.; Schneider, S.E.; Willey, S.; et al. Cytokine-induced memory-like natural killer cells exhibit enhanced responses against myeloid leukemia. Sci. Transl. Med. 2016, 8, 357ra123. [Google Scholar] [CrossRef]
- Kleinnijenhuis, J.; Quintin, J.; Preijers, F.; Joosten, L.A.; Jacobs, C.; Xavier, R.J.; Van Der Meer, J.W.; Van Crevel, R.; Netea, M.G. BCG-induced trained immunity in NK cells: Role for non-specific protection to infection. Clin. Immunol. 2014, 155, 213–219. [Google Scholar] [CrossRef]
- Askenase, M.H.; Han, S.-J.; Byrd, A.L.; Da Fonseca, D.M.; Bouladoux, N.; Wilhelm, C.; Konkel, J.E.; Hand, T.W.; Lacerda-Queiroz, N.; Su, X.-Z.; et al. Bone-Marrow-Resident NK Cells Prime Monocytes for Regulatory Function during Infection. Immunity 2015, 42, 1130–1142. [Google Scholar] [CrossRef]
- Sun, J.C.; Madera, S.; Bezman, N.A.; Beilke, J.N.; Kaplan, M.H.; Lanier, L.L. Proinflammatory cytokine signaling required for the generation of natural killer cell memory. J. Exp. Med. 2012, 209, 947–954. [Google Scholar] [CrossRef]
- Pickl, W.F.; Majdic, O.; Kohl, P.; Stöckl, J.; Riedl, E.; Scheinecker, C.; Bello-Fernandez, C.; Knapp, W. Molecular and functional characteristics of dendritic cells generated from highly purified CD14+ peripheral blood monocytes. J. Immunol. 1996, 157, 3850–3859. [Google Scholar] [PubMed]
- Bonifazi, P.; Zelante, T.; D’Angelo, C.; De Luca, A.; Moretti, S.; Bozza, S.; Perruccio, K.; Iannitti, R.G.; Giovannini, G.; Volpi, C.; et al. Balancing inflammation and tolerance in vivo through dendritic cells by the commensal Candida albicans. Mucosal Immunol. 2009, 2, 362–374. [Google Scholar] [CrossRef]
- Wen, H.; Dou, H.W.Y.; Hogaboam, C.M.; Kunkel, S.L. Epigenetic regulation of dendritic cell–derived interleukin-12 facilitates immunosuppression after a severe innate immune response. Blood 2008, 111, 1797–1804. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.L.; Buonomo, E.; Carey, M.; Cowardin, C.; Naylor, C.; Noor, Z.; Wills-Karp, M.; Petri, W.A., Jr. Bone Marrow Dendritic Cells from Mice with an Altered Microbiota Provide Interleukin 17A-Dependent Protection against Entamoeba histolytica Colitis. mBio 2014, 5, e01817-14. [Google Scholar] [CrossRef] [PubMed]
- Iwanowycz, S.; Wang, J.; Altomare, D.; Hui, Y.; Fan, D. Emodin Bidirectionally Modulates Macrophage Polarization and Epigenetically Regulates Macrophage Memory. J. Biol. Chem. 2016, 291, 11491–11503. [Google Scholar] [CrossRef]
- McCall, M.B.; Netea, M.G.; Hermsen, C.C.; Jansen, T.; Jacobs, L.; Golenbock, U.; Van Der Ven, A.J.A.M.; Sauerwein, R.W. Plasmodium falciparum infection causes proinflammatory priming of human TLR responses. J. Immunol. 2007, 179, 162–171. [Google Scholar] [CrossRef]
- Ifrim, D.C.; Quintin, J.; Joosten, L.A.B.; Jacobs, C.; Jansen, T.; Jacobs, L.; Gow, N.A.R.; Williams, D.L.; Van Der Meer, J.W.M.; Netea, M.G. Trained Immunity or Tolerance: Opposing Functional Programs Induced in Human Monocytes after Engagement of Various Pattern Recognition Receptors. Clin. Vaccine Immunol. 2014, 21, 534–545. [Google Scholar] [CrossRef]
- Strunk, T.; Prosser, A.; Levy, O.; Philbin, V.; Simmer, K.; Doherty, D.; Charles, A.; Richmond, P.; Burgner, D.; Currie, A. Responsiveness of human monocytes to the commensal bacterium Staphylococcus epidermidis develops late in gestation. Pediatr. Res. 2012, 72, 10–18. [Google Scholar] [CrossRef]
- Tannahill, G.M.; Curtis, A.M.; Adamik, J.; Palsson-McDermott, E.M.; McGettrick, A.F.; Goel, G.; Frezza, C.; Bernard, N.J.; Kelly, B.; Foley, N.H.; et al. Succinate is an inflammatory signal that induces IL-1 beta through HIF-1 alpha. Nature 2013, 496, 238–242. [Google Scholar] [CrossRef]
- Yáñez, A.; Hassanzadeh-Kiabi, N.; Ng, M.Y.; Megías, J.; Subramanian, A.; Liu, G.Y.; Underhill, D.M.; Gil, M.L.; Goodridge, H.S. Detection of a TLR2 agonist by hematopoietic stem and progenitor cells impacts the function of the macrophages they produce. Eur. J. Immunol. 2013, 43, 2114–2125. [Google Scholar] [CrossRef]
- Deng, H.; Maitra, U.; Morris, M.; Li, L. Molecular Mechanism Responsible for the Priming of Macrophage Activation. J. Biol. Chem. 2012, 288, 3897–3906. [Google Scholar] [CrossRef] [PubMed]
- Foster, S.L.; Hargreaves, D.C.; Medzhitov, R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 2007, 447, 972–978. [Google Scholar] [CrossRef] [PubMed]
- Nicodeme, E.; Jeffrey, K.L.; Schaefer, U.; Beinke, S.; Dewell, S.; Chung, C.-W.; Chandwani, R.; Marazzi, I.; Wilson, P.; Coste, H.; et al. Suppression of inflammation by a synthetic histone mimic. Nature 2010, 468, 1119–1123. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ivashkiv, L.B. IFN-gamma abrogates endotoxin tolerance by facilitating Toll-like receptor-induced chromatin remodeling. Proc. Natl. Acad. Sci. USA 2010, 107, 19438–19443. [Google Scholar] [CrossRef]
- Bekkering, S.; Blok, B.A.; Joosten, L.A.B.; Riksen, N.P.; Van Crevel, R.; Netea, M.G. In Vitro Experimental Model of Trained Innate Immunity in Human Primary Monocytes. Clin. Vaccine Immunol. 2016, 23, 926–933. [Google Scholar] [CrossRef]
- Jensen, K.J.; Larsen, N.; Biering-Sørensen, S.; Andersen, A.; Eriksen, H.B.; Monteiro, I.; Hougaard, D.; Aaby, P.; Netea, M.G.; Flanagan, K.L.; et al. Heterologous Immunological Effects of Early BCG Vaccination in Low-Birth-Weight Infants in Guinea-Bissau: A Randomized-controlled Trial. J. Infect. Dis. 2015, 211, 956–967. [Google Scholar] [CrossRef]
- Garcia-Valtanen, P.; Guzman-Genuino, R.M.; Williams, D.L.; Hayball, J.D.; Diener, K.R. Evaluation of trained immunity by beta-1, 3 (D)-glucan on murine monocytes in vitro and duration of response in vivo. Immunol. Cell Biol. 2017, 95, 601–610. [Google Scholar] [CrossRef]
- Bistoni, F.; Verducci, G.; Perito, S.; Vecchiarelli, A.; Puccetti, P.; Marconi, P.; Cassone, A. Immunomodulation by a low-virulence, agerminative variant of Candida albicans. Further evidence for macrophage activation as one of the effector mechanisms of nonspecific anti-infectious protection. Med Mycol. 1988, 26, 285–299. [Google Scholar] [CrossRef]
- Quintin, J.; Saeed, S.; Martens, J.H.; Giamarellos-Bourboulis, E.J.; Ifrim, D.C.; Logie, C.; Jacobs, L.; Jansen, T.; Kullberg, B.-J.; Wijmenga, C.; et al. Candida albicans Infection Affords Protection against Reinfection via Functional Reprogramming of Monocytes. Cell Host Microbe 2012, 12, 223–232. [Google Scholar] [CrossRef]
- Luty, A.J.F.; Lell, B.; Schmidt-Ott, R.; Lehman, L.G.; Luckner, D.; Greve, B.; Matousek, P.; Herbich, K.; Schmid, D.; Migot-Nabias, F.; et al. Interferon-gamma responses are associated with resistance to reinfection with Plasmodium falciparum in young African children. J. Infect. Dis. 1999, 179, 980–988. [Google Scholar] [CrossRef]
- Barton, E.S.; White, D.W.; Cathelyn, J.S.; Brett-McClellan, K.A.; Engle, M.; Diamond, M.S.; Miller, V.L.; Iv, H.W.V. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature 2007, 447, 326–329. [Google Scholar] [CrossRef] [PubMed]
- Marr, A.K.; MacIsaac, J.L.; Jiang, R.; Airo, A.M.; Kobor, M.S.; McMaster, W.R. Leishmania donovani Infection Causes Distinct Epigenetic DNA Methylation Changes in Host Macrophages. PLoS Pathog. 2014, 10, e1004419. [Google Scholar] [CrossRef] [PubMed]
- Bekkering, S.; Quintin, J.; Joosten, L.A.; Van Der Meer, J.W.; Netea, M.G.; Riksen, N.P. Oxidized Low-Density Lipoprotein Induces Long-Term Proinflammatory Cytokine Production and Foam Cell Formation via Epigenetic Reprogramming of Monocytes. Arter. Thromb. Vasc. Biol. 2014, 34, 1731–1738. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Xia, C.-Q.; Butfiloski, E.; Clare-Salzler, M. Effect of high glucose on cytokine production by human peripheral blood immune cells and type I interferon signaling in monocytes: Implications for the role of hyperglycemia in the diabetes inflammatory process and host defense against infection. Clin. Immunol. 2018, 195, 139–148. [Google Scholar] [CrossRef]
- Arts, R.J.; Novakovic, B.; Ter Horst, R.; Carvalho, A.; Bekkering, S.; Lachmandas, E.; Rodrigues, F.; Silvestre, R.; Cheng, S.-C.; Wang, S.-Y.; et al. Glutaminolysis and Fumarate Accumulation Integrate Immunometabolic and Epigenetic Programs in Trained Immunity. Cell Metab. 2016, 24, 807–819. [Google Scholar] [CrossRef]
- Crişan, T.O.; Cleophas, M.C.P.; Novakovic, B.; Erler, K.; Van De Veerdonk, F.L.; Stunnenberg, H.G.; Netea, M.G.; Dinarello, C.A.; Joosten, L.A. Uric acid priming in human monocytes is driven by the AKT–PRAS40 autophagy pathway. Proc. Natl. Acad. Sci. USA 2017, 114, 5485–5490. [Google Scholar] [CrossRef]
- Cui, H.; Banerjee, S.; Guo, S.; Xie, N.; Liu, G. IFN Regulatory Factor 2 Inhibits Expression of Glycolytic Genes and Lipopolysaccharide-Induced Proinflammatory Responses in Macrophages. J. Immunol. 2018, 200, 3218–3230. [Google Scholar] [CrossRef]
- Domínguez-Andrés, J.; Arts, R.J.W.; Ter Horst, R.; Gresnigt, M.S.; Smeekens, S.P.; Ratter, J.M.; Lachmandas, E.; Boutens, L.; Van De Veerdonk, F.L.; Joosten, L.A.B.; et al. Rewiring monocyte glucose metabolism via C-type lectin signaling protects against disseminated candidiasis. PLoS Pathog. 2017, 13, e1006632. [Google Scholar] [CrossRef]
- Chen, F.; Wu, W.; Millman, A.; Craft, J.F.; Chen, E.; Patel, N.; Boucher, J.L.; Urban, J.F., Jr.; Kim, C.C.; Gause, W.C. Neutrophils prime a long-lived effector macrophage phenotype that mediates accelerated helminth expulsion. Nat. Immunol. 2014, 15, 938–946. [Google Scholar] [CrossRef]
- Bruniquel, D.; Schwartz, R.H. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat. Immunol. 2003, 4, 235–240. [Google Scholar] [CrossRef]
- Pique-Regi, R.; Degner, J.F.; Pai, A.A.; Gaffney, D.J.; Gilad, Y.; Pritchard, J.K. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2010, 21, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Yadav, T.; Quivy, J.-P.; Almouzni, G. Chromatin plasticity: A versatile landscape that underlies cell fate and identity. Science 2018, 361, 1332–1336. [Google Scholar] [CrossRef] [PubMed]
- Freyne, B.; Donath, S.; Germano, S.; Gardiner, K.; Casalaz, D.; Robins-Browne, R.M.; Amenyogbe, N.; Messina, N.L.; Netea, M.G.; Flanagan, K.L.; et al. Neonatal BCG Vaccination Influences Cytokine Responses to Toll-like Receptor Ligands and Heterologous Antigens. J. Infect. Dis. 2018, 217, 1798–1808. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Maekawa, T.; Zhu, Y.; Renard-Guillet, C.; Chatton, B.; Inoue, K.; Uchiyama, T.; Ishibashi, K.-I.; Yamada, T.; Ohno, N.; et al. The transcription factor ATF7 mediates lipopolysaccharide-induced epigenetic changes in macrophages involved in innate immunological memory. Nat. Immunol. 2015, 16, 1034–1043. [Google Scholar] [CrossRef] [PubMed]
- Novakovic, B.; Habibi, E.; Wang, S.-Y.; Arts, R.J.W.; Davar, R.; Megchelenbrink, W.; Kim, B.; Kuznetsova, T.; Kox, M.; Zwaag, J.; et al. Beta-Glucan Reverses the Epigenetic State of LPS-Induced Immunological Tolerance. Cell 2016, 167, 1354–1368.e14. [Google Scholar] [CrossRef]
- Saeed, S.; Quintin, J.; Kerstens, H.H.D.; Rao, N.A.; Aghajanirefah, A.; Matarese, F.; Cheng, S.-C.; Ratter, J.; Berentsen, K.; Van Der Ent, M.A.; et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 2014, 345, 1251086. [Google Scholar] [CrossRef]
- Ostuni, R.; Piccolo, V.; Barozzi, I.; Polletti, S.; Termanini, A.; Bonifacio, S.; Curina, A.; Prosperini, E.; Ghisletti, S.; Natoli, G. Latent Enhancers Activated by Stimulation in Differentiated Cells. Cell 2013, 152, 157–171. [Google Scholar] [CrossRef]
- Finch, J.T.; Lutter, L.C.; Rhodes, D.; Brown, R.S.; Rushton, B.; Levitt, M.; Klug, A. Structure of nucleosome core particles of chromatin. Nature 1977, 269, 29–36. [Google Scholar] [CrossRef]
- Becker, P.B.; Workman, J.L. Nucleosome Remodeling and Epigenetics. Cold Spring Harb. Perspect. Biol. 2013, 5, a017905. [Google Scholar] [CrossRef]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.G.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef]
- Visel, A.; Blow, M.J.; Li, Z.; Zhang, T.; Akiyama, J.A.; Holt, A.; Plajzer-Frick, I.; Shoukry, M.; Wright, C.; Chen, F.; et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 2009, 457, 854–858. [Google Scholar] [CrossRef] [PubMed]
- Rada-Iglesias, A.; Bajpai, R.; Swigut, T.; Brugmann, S.A.; Flynn, R.A.; Wysocka, J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 2011, 470, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.-Y.; Schones, D.E.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-resolution profiling of histone methylations in the human genome. Cell 2007, 129, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, B.E.; Kamal, M.; Lindblad-Toh, K.; Bekiranov, S.; Bailey, D.K.; Huebert, D.J.; McMahon, S.; Karlsson, E.K.; Kulbokas, E.J.; Gingeras, T.R.; et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 2005, 120, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Pokholok, D.K.; Harbison, C.T.; Levine, S.; Cole, M.; Hannett, N.M.; Lee, T.I.; Bell, G.W.; Walker, K.; Rolfe, P.A.; Herbolsheimer, E.; et al. Genome-wide map of nucleosorne acetylation and methylation in yeast. Cell 2005, 122, 517–527. [Google Scholar] [CrossRef]
- Buro, L.J.; Chipumuro, E.; Henriksen, M.A. Menin and RNF20 recruitment is associated with dynamic histone modifications that regulate signal transducer and activator of transcription 1 (STAT1)-activated transcription of the interferon regulatory factor 1 gene (IRF1). Epigenetics Chromatin 2010, 3, 16. [Google Scholar] [CrossRef]
- Fang, T.C.; Schaefer, U.; Mecklenbrauker, I.; Stienen, A.; Dewell, S.; Chen, M.S.; Rioja, I.; Parravicini, V.; Prinjha, R.K.; Chandwani, R.; et al. Histone H3 lysine 9 di-methylation as an epigenetic signature of the interferon response. J. Exp. Med. 2012, 209, 661–669. [Google Scholar] [CrossRef]
- Ghisletti, S.; Barozzi, I.; Mietton, F.; Polletti, S.; De Santa, F.; Venturini, E.; Gregory, L.; Lonie, L.; Chew, A.; Wei, C.-L.; et al. Identification and Characterization of Enhancers Controlling the Inflammatory Gene Expression Program in Macrophages. Immunity 2010, 32, 317–328. [Google Scholar] [CrossRef]
- Saccani, S. Dynamic changes in histone H3 Lys 9 methylation occurring at tightly regulated inducible inflammatory genes. Genes Dev. 2002, 16, 2219–2224. [Google Scholar] [CrossRef]
- Heintzman, N.D.; Stuart, R.K.; Hon, G.; Fu, Y.; Ching, C.W.; Hawkins, R.D.; O Barrera, L.; Van Calcar, S.; Qu, C.; A Ching, K.; et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007, 39, 311–318. [Google Scholar] [CrossRef]
- Kaikkonen, M.U.; Spann, N.J.; Heinz, S.; Romanoski, C.E.; Allison, K.A.; Stender, J.D.; Chun, H.B.; Tough, D.F.; Prinjha, R.K.; Benner, C.; et al. Remodeling of the Enhancer Landscape during Macrophage Activation Is Coupled to Enhancer Transcription. Mol. Cell 2013, 51, 310–325. [Google Scholar] [CrossRef] [PubMed]
- Villagra, A.; Cheng, F.; Wang, H.-W.; Suarez, I.; Glozak, M.; Maurin, M.; Nguyen, D.; Wright, K.L.; Atadja, P.W.; Bhalla, K.N.; et al. The histone deacetylase HDAC11 regulates the expression of interleukin 10 and immune tolerance. Nat. Immunol. 2009, 10, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Giannopoulou, E.G.; Chan, C.H.; Park, S.; Gong, S.; Chen, J.; Hu, X.; Elemento, O.; Ivashkiv, L.B. Synergistic Activation of Inflammatory Cytokine Genes by Interferon-gamma-Induced Chromatin Remodeling and Toll-like Receptor Signaling. Immunity 2013, 39, 454–469. [Google Scholar] [CrossRef] [PubMed]
- Arts, R.J.W.; Blok, B.A.; Aaby, P.; Joosten, L.A.B.; de Jong, D.; van der Meer, J.W.M.; Benn, C.S.; van Crevel, R.; Netea, M.G. Long-term in vitro and in vivo effects of gamma-irradiated BCG on innate and adaptive immunity. J. Leukoc. Biol. 2015, 98, 995–1001. [Google Scholar] [CrossRef] [PubMed]
- Arts, J.W.R.; Carvalho, A.A.R.; La Rocca, C.; Palma, C.; Rodrigues, F.J.D.S.; Silvestre, R.; Kleinnijenhuis, J.; Lachmandas, E.; Gonçalves, L.G.; Belinha, A.; et al. Immunometabolic Pathways in BCG-Induced Trained Immunity. Cell Rep. 2016, 17, 2562–2571. [Google Scholar] [CrossRef] [PubMed]
- Schrum, J.E.; Crabtree, J.N.; Dobbs, K.R.; Kiritsy, M.C.; Reed, G.W.; Gazzinelli, R.T.; Netea, M.G.; Kazura, J.W.; Dent, A.E.; Fitzgerald, K.A.; et al. Cutting Edge: Plasmodium falciparum Induces Trained Innate Immunity. J. Immunol. 2018, 200, 1243–1248. [Google Scholar] [CrossRef]
- Klug, M.; Heinz, S.; Gebhard, C.; Schwarzfischer, L.; Krause, S.; Andreesen, R.; Rehli, M. Active DNA demethylation in human postmitotic cells correlates with activating histone modifications, but not transcription levels. Genome Biol. 2010, 11, R63. [Google Scholar] [CrossRef]
- Dowen, R.H.; Pelizzola, M.; Schmitz, R.J.; Lister, R.; Dowen, J.M.; Nery, J.R.; Dixon, J.E.; Ecker, J.R. Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. USA 2012, 109, E2183–E2191. [Google Scholar] [CrossRef]
- Pacis, A.; Tailleux, L.; Morin, A.M.; Lambourne, J.; MacIsaac, J.L.; Yotova, V.; Dumaine, A.; Danckaert, A.; Luca, F.; Grenier, J.-C.; et al. Bacterial infection remodels the DNA methylation landscape of human dendritic cells. Genome Res. 2015, 25, 1801–1811. [Google Scholar] [CrossRef]
- Klug, M.; Schmidhofer, S.; Gebhard, C.; Andreesen, R.; Rehli, M. 5-Hydroxymethylcytosine is an essential intermediate of active DNA demethylation processes in primary human monocytes. Genome Biol. 2013, 14, R46. [Google Scholar] [CrossRef]
- Ziller, M.J.; Gu, H.; Müller, F.; Donaghey, J.; Tsai, L.T.-Y.; Kohlbacher, O.; De Jager, P.L.; Rosen, E.D.; Bennett, D.A.; Bernstein, B.E.; et al. Charting a dynamic DNA methylation landscape of the human genome. Nature 2013, 500, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Ichiyama, K.; Chen, T.; Wang, X.; Yan, X.; Kim, B.; Tanaka, S.; Ndiaye-Lobry, D.; Deng, Y.; Zou, Y.; Zheng, P.; et al. The Methylcytosine Dioxygenase Tet2 Promotes DNA Demethylation and Activation of Cytokine Gene Expression in T Cells. Immunity 2015, 42, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Kozlenkov, A.; Li, J.; Apontes, P.; Hurd, Y.L.; Byne, W.M.; Koonin, E.V.; Wegner, M.; Mukamel, E.A.; Dracheva, S. A unique role for DNA (hydroxy)methylation in epigenetic regulation of human inhibitory neurons. Sci. Adv. 2018, 4, eaau6190. [Google Scholar] [CrossRef] [PubMed]
- Stadler, M.B.; Murr, R.; Burger, L.; Ivanek, R.; Lienert, F.; Schöler, A.; Van Nimwegen, E.; Wirbelauer, C.; Oakeley, E.J.; Gaidatzis, D.; et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 2011, 480, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Garcia-Bassets, I.; Benner, C.; Li, W.; Su, X.; Zhou, Y.; Qiu, J.; Liu, W.; Kaikkonen, M.U.; Ohgi, K.A.; et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 2011, 474, 390–394. [Google Scholar] [CrossRef]
- Ifrim, D.C.; Joosten, L.A.B.; Kullberg, B.; Jacobs, L.; Jansen, T.; Williams, D.L.; Gow, N.A.R.; van der Meer, J.W.M.; Netea, M.G.; Quintin, J. Candida albicans Primes TLR Cytokine Responses through a Dectin-1/Raf-1-Mediated Pathway. J. Immunol. 2013, 190, 4129–4135. [Google Scholar] [CrossRef]
- Rodríguez-Prados, J.-C.; Través, P.G.; Cuenca, J.; Rico, D.; Aragonés, J.; Martín-Sanz, P.; Cascante, M.; Boscá, L. Substrate Fate in Activated Macrophages: A Comparison between Innate, Classic, and Alternative Activation. J. Immunol. 2010, 185, 605–614. [Google Scholar] [CrossRef]
- Ifrim, D.C.; Quintin, J.; Meerstein-Kessel, L.; Plantinga, T.S.; Joosten, L.A.B.; Van Der Meer, J.W.M.; Van De Veerdonk, F.L.; Netea, M. Defective trained immunity in patients with STAT-1-dependent chronic mucocutaneaous candidiasis. Clin. Exp. Immunol. 2015, 181, 434–440. [Google Scholar] [CrossRef]
- Vakoc, C.R.; Sachdeva, M.M.; Wang, H.; Blobel, G.A. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol. Cell. Biol. 2006, 26, 9185–9195. [Google Scholar] [CrossRef]
- Rizzetto, L.; Ifrim, D.C.; Moretti, S.; Tocci, N.; Cheng, S.-C.; Quintin, J.; Renga, G.; Oikonomou, V.; De Filippo, C.; Weil, T.; et al. Fungal Chitin Induces Trained Immunity in Human Monocytes during Cross-talk of the Host with Saccharomyces cerevisiae. J. Biol. Chem. 2016, 291, 7961–7972. [Google Scholar] [CrossRef]
- Cheng, S.C.; Quintin, J.; Cramer, R.A.; Shepardson, K.M.; Saeed, S.; Kumar, V.; Giamarellos-Bourboulis, E.J.; Martens, J.H.A.; Rao, N.A.; Aghajanirefah, A.; et al. mTOR- and HIF-1 alpha-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 2014, 345, 1579. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O.; Wind, F.; Negelein, E. The metabolism of tumors in the body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.; Brayner, F.A.; Alves, L.C.; Dixit, R.; Barillas-Mury, C. Hemocyte Differentiation Mediates Innate Immune Memory in Anopheles gambiae Mosquitoes. Science 2010, 329, 1353–1355. [Google Scholar] [CrossRef] [PubMed]
- Pham, L.N.; Dionne, M.S.; Shirasu-Hiza, M.; Schneider, D. A Specific Primed Immune Response in Drosophila is Dependent on Phagocytes. PLoS Pathog. 2007, 3, e26. [Google Scholar] [CrossRef]
- Bistoni, F.; Vecchiarelli, A.; Cenci, E.; Puccetti, P.; Marconi, P.; Cassone, A. Evidence for Macrophage-Mediated Protection Against Lethal Candida albicans Infection. Infect. Immun. 1986, 51, 668–674. [Google Scholar] [CrossRef]
- Aaby, P.; Roth, A.; Ravn, H.; Napirna, B.M.; Rodrigues, A.; Lisse, I.M.; Stensballe, L.; Diness, B.R.; Lausch, K.R.; Lund, N.; et al. Randomized Trial of BCG Vaccination at Birth to Low-Birth-Weight Children: Beneficial Nonspecific Effects in the Neonatal Period? J. Infect. Dis. 2011, 204, 245–252. [Google Scholar] [CrossRef]
- Biering-Sorensen, S.; Aaby, P.; Napirna, B.M.; Roth, A.; Ravn, H.; Rodrigues, A.; Whittle, H.; Benn, C.S. Small Randomized Trial Among Low-Birth-Weight Children Receiving Bacillus Calmette-Guerin Vaccination at First Health Center Contact. Pediatric Infect. Dis. J. 2012, 31, 306–308. [Google Scholar] [CrossRef]
- Garly, M.-L.; Martins, C.L.; Balé, C.; Baldé, M.A.; Hedegaard, K.L.; Gustafson, P.; Lisse, I.; Whittle, H.; Aaby, P. BCG scar and positive tuberculin reaction associated with reduced child mortality in West Africa—A non-specific beneficial effect of BCG? Vaccine 2003, 21, 2782–2790. [Google Scholar] [CrossRef]
- Van’t Wout, J.W.; Poell, R.; Van Furth, R. The Role of BCG/PPD-Activated Macrophages in Resistance against Systemic Candidiasis in Mice. Scand. J. Immunol. 1992, 36, 713–720. [Google Scholar] [CrossRef]
- Sakuma, T.; Suenaga, T.; Yoshida, I.; Azuma, M. Mechanisms of Enhanced Resistance of Mycobacterium Bovis BCG-treated Mice to Ectromelia Virus Infection. Infect. Immun. 1983, 42, 567–573. [Google Scholar] [CrossRef]
- Basso, B.; Marini, V. Experimental Chagas disease. Innate immune response in Balb/c mice previously vaccinated with Trypanosoma rangeli. I. The macrophage shows immunological memory: Reality or fiction? Immunobiology 2014, 219, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Dodoo, D.; Omer, F.M.; Todd, J.; Akanmori, B.D.; Koram, K.A.; Riley, E.M. Absolute levels and ratios of proinflammatory and anti-inflammatory cytokine production in vitro predict clinical immunity to Plasmodium falciparum malaria. J. Infect. Dis. 2002, 185, 971–979. [Google Scholar] [CrossRef] [PubMed]
- Pfahlberg, A.; Schneider, D.; Uter, W.; Gefeller, O.; Kölmel, K.F.; Grange, J.M.; Mastrangelo, G.; Krone, B.; Botev, I.N.; Niin, M.; et al. Inverse association between melanoma and previous vaccinations against tuberculosis and smallpox: Results of the FEBIM study. J. Investig. Dermatol. 2002, 119, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Aaby, P.; Benn, C.; Nielsen, J.; Lisse, I.M.; Rodrigues, A.; Ravn, H. Testing the hypothesis that diphtheria–tetanus–pertussis vaccine has negative non-specific and sex-differential effects on child survival in high-mortality countries. BMJ Open 2012, 2, e000707. [Google Scholar] [CrossRef] [PubMed]
- Kronforst, K.D.; Mancuso, C.J.; Pettengill, M.; Ninkovic, J.; Coombs, M.R.P.; Stevens, C.; Otto, M.; Mallard, C.; Wang, X.; Goldmann, D.; et al. A Neonatal Model of Intravenous Staphylococcus epidermidis Infection in Mice <24 h Old Enables Characterization of Early Innate Immune Responses. PLoS ONE 2012, 7, e43897. [Google Scholar]
- De Castro, M.J.; Pardo-Seco, J.J.; Martinón-Torres, F. Nonspecific (Heterologous) Protection of Neonatal BCG Vaccination against Hospitalization Due to Respiratory Infection and Sepsis. Clin. Infect. Dis. 2015, 60, 1611–1619. [Google Scholar] [CrossRef]
- Leentjens, J.; Kox, M.; Stokman, R.; Gerretsen, J.; Diavatopoulos, D.A.; Van Crevel, R.; Rimmelzwaan, G.F.; Pickkers, P.; Netea, M.G. BCG Vaccination Enhances the Immunogenicity of Subsequent Influenza Vaccination in Healthy Volunteers: A Randomized, Placebo-Controlled Pilot Study. J. Infect. Dis. 2015, 212, 1930–1938. [Google Scholar] [CrossRef]
- de Bree, C.; Marijnissen, R.J.; Kel, J.M.; Huber, S.K.R.; Aaby, P.; Benn, C.S.; Wijnands, M.V.W.; Diavatopoulos, D.A.; van Crevel, R.; van Crevel, R.; et al. Bacillus Calmette-Guerin-Induced Trained Immunity Is Not Protective for Experimental Influenza A/Anhui/1/2013 (H7N9) Infection in Mice. Front. Immunol. 2018, 9, 869. [Google Scholar] [CrossRef]
- Chedid, L.; Parant, M.; Parant, F.; Lefrancher, P.; Choay, J.; Lederer, E. Enhancement of Nonspecific Immunity to Klebsiella pneumoniae Infection by a Synthetic Immunoadjuvant (N-Acetylmuramyl-L-alanyl-D-Isoglutamine) and Several Analogs. Proc. Natl. Acad. Sci. USA 1977, 74, 2089–2093. [Google Scholar] [CrossRef]
- De Bree, C.L.; Janssen, R.; Aaby, P.; Van Crevel, R.; Joosten, L.A.; Benn, C.S.; Netea, M.G. The impact of sex hormones on BCG-induced trained immunity. J. Leukoc. Biol. 2018, 104, 573–578. [Google Scholar] [CrossRef]
- Lathrop, S.K.; Bloom, S.M.; Rao, S.M.; Nutsch, K.; Lio, C.-W.; A Santacruz, N.; Peterson, D.A.; Stappenbeck, T.S.; Hsieh, C.-S. Peripheral education of the immune system by colonic commensal microbiota. Nature 2011, 478, 250–254. [Google Scholar] [CrossRef] [PubMed]
- Weavers, H.; Evans, I.R.; Martin, P.; Wood, W. Corpse Engulfment Generates a Molecular Memory that Primes the Macrophage Inflammatory Response. Cell 2016, 165, 1658–1671. [Google Scholar] [CrossRef] [PubMed]
- Yona, S.; Kim, K.-W.; Wolf, Y.; Mildner, A.; Varol, D.; Breker, M.; Strauss-Ayali, D.; Viukov, S.; Guilliams, M.; Misharin, A.; et al. Fate Mapping Reveals Origins and Dynamics of Monocytes and Tissue Macrophages under Homeostasis. Immunity 2013, 38, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Jeyanathan, M.; Afkhami, S.; Smaill, F.; Miller, M.S.; Lichty, B.D.; Xing, Z. Immunological considerations for COVID-19 vaccine strategies. Nat. Rev. Immunol. 2020, 20, 615–632. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adams, K.; Weber, K.S.; Johnson, S.M. Exposome and Immunity Training: How Pathogen Exposure Order Influences Innate Immune Cell Lineage Commitment and Function. Int. J. Mol. Sci. 2020, 21, 8462. https://doi.org/10.3390/ijms21228462
Adams K, Weber KS, Johnson SM. Exposome and Immunity Training: How Pathogen Exposure Order Influences Innate Immune Cell Lineage Commitment and Function. International Journal of Molecular Sciences. 2020; 21(22):8462. https://doi.org/10.3390/ijms21228462
Chicago/Turabian StyleAdams, Kevin, K. Scott Weber, and Steven M. Johnson. 2020. "Exposome and Immunity Training: How Pathogen Exposure Order Influences Innate Immune Cell Lineage Commitment and Function" International Journal of Molecular Sciences 21, no. 22: 8462. https://doi.org/10.3390/ijms21228462
APA StyleAdams, K., Weber, K. S., & Johnson, S. M. (2020). Exposome and Immunity Training: How Pathogen Exposure Order Influences Innate Immune Cell Lineage Commitment and Function. International Journal of Molecular Sciences, 21(22), 8462. https://doi.org/10.3390/ijms21228462





