CaHsfA1d Improves Plant Thermotolerance via Regulating the Expression of Stress- and Antioxidant-Related Genes
Abstract
:1. Introduction
2. Results
2.1. Isolation and Characterization Analyses of CaHsfA1d
2.2. Transactivation Activity Analysis of the CaHsfA1d Protein
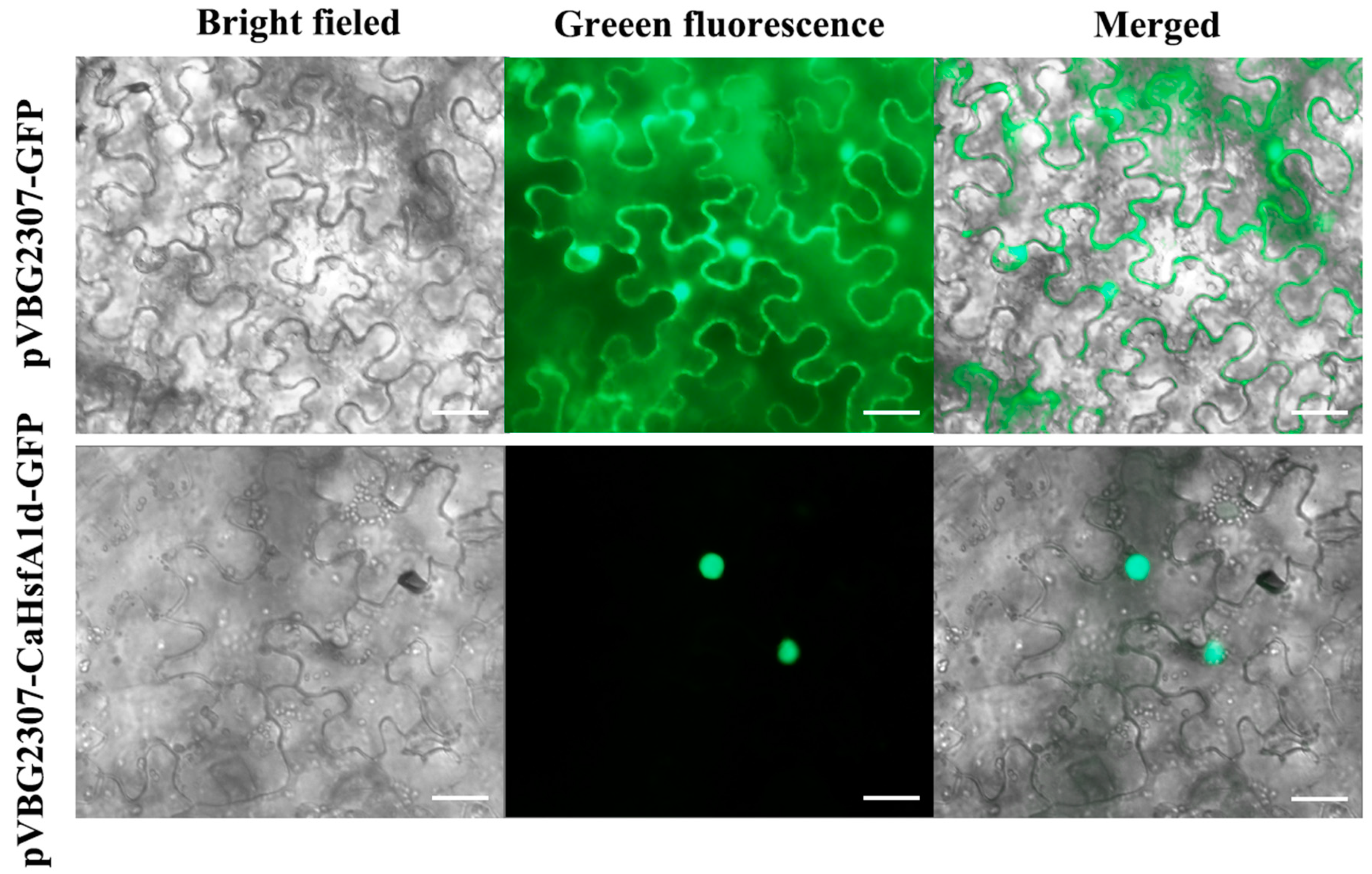
2.3. Subcellular Location of the CaHsfA1d Protein in Tobacco
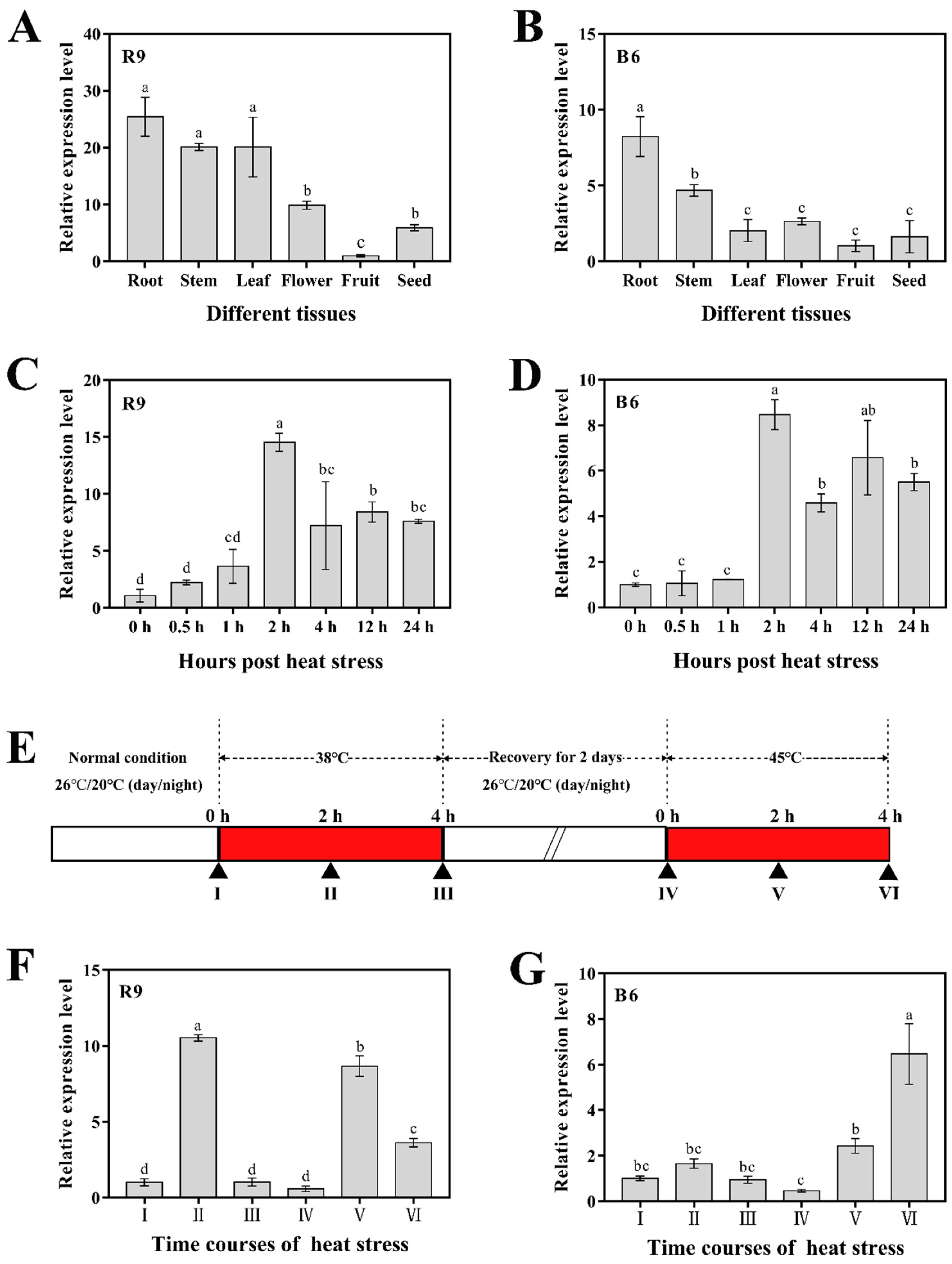
2.4. Expression Analyses of CaHsfA1d in Different Tissues and Response to Heat Stress
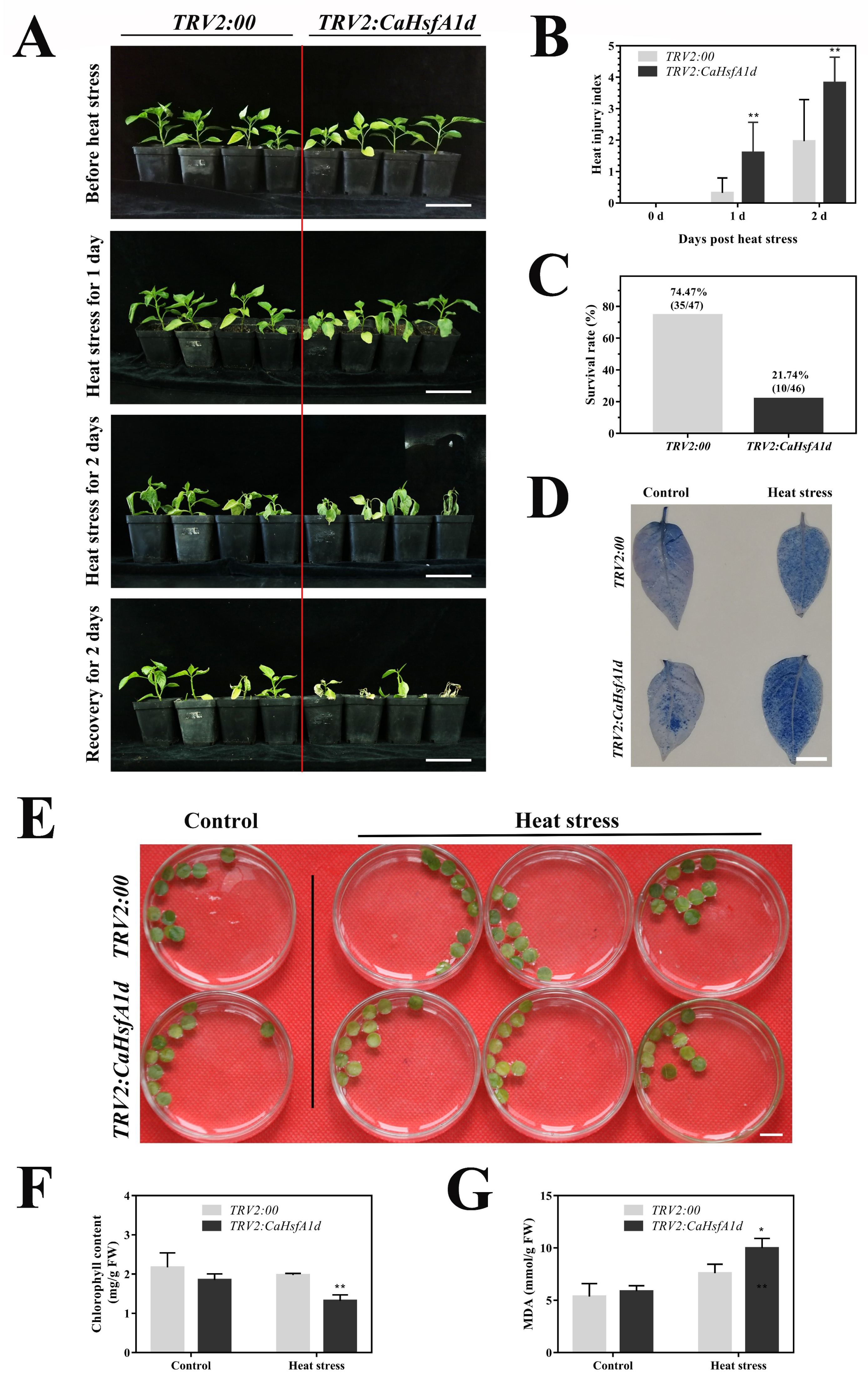
2.5. Performance of the CaHsfA1d-Silenced Pepper under Heat Stress
2.6. Thermotolerance Analyses of the CaHsfA1d-Overexpression Arabidopsis
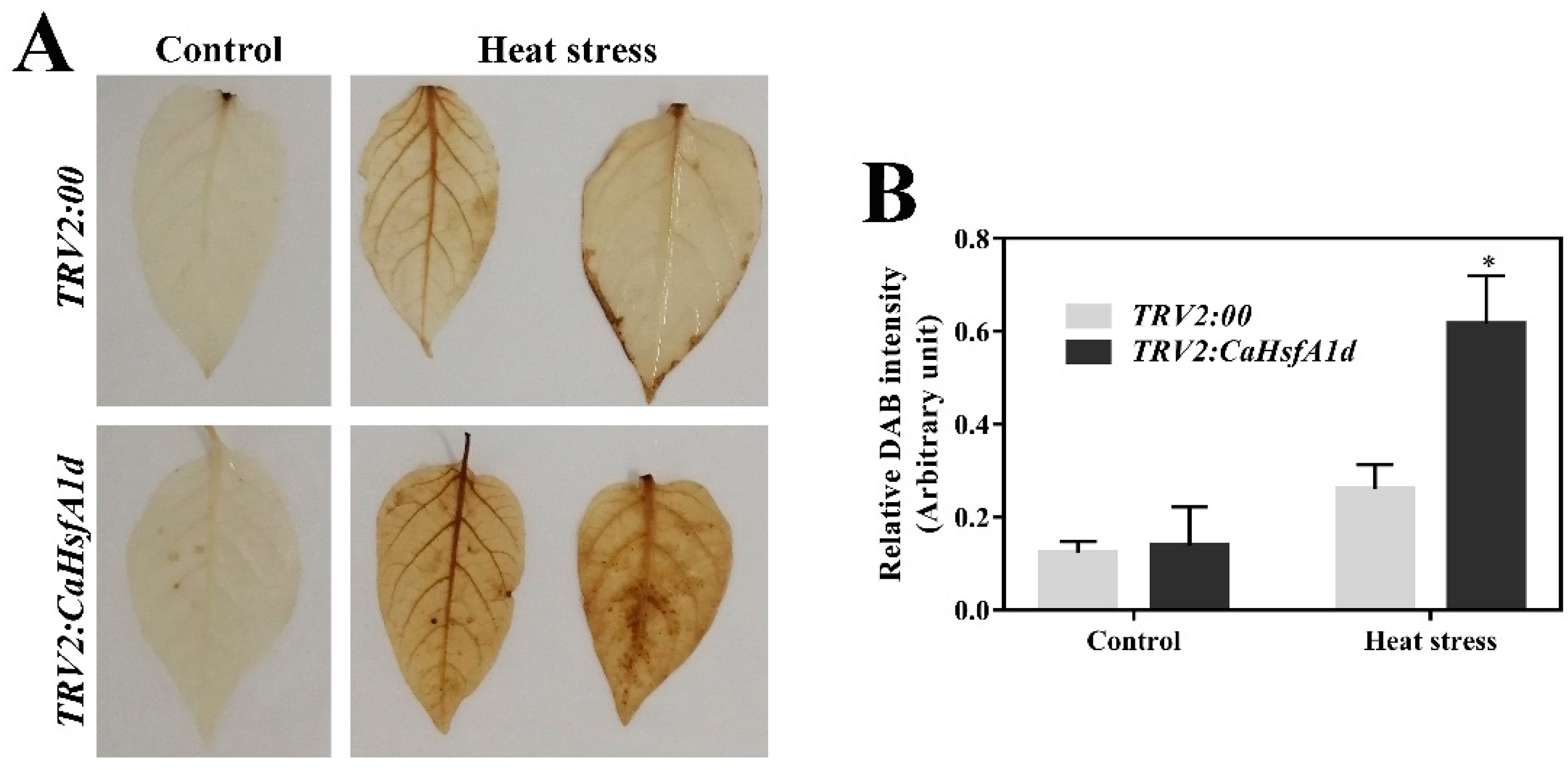
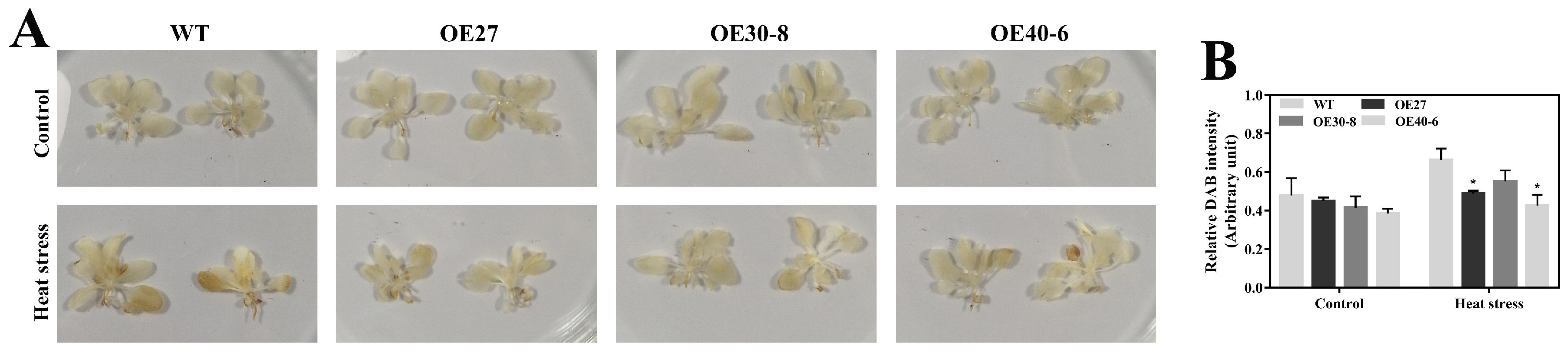
2.7. Association between CaHsfA1d and H2O2 Accumulation in Heat Stress
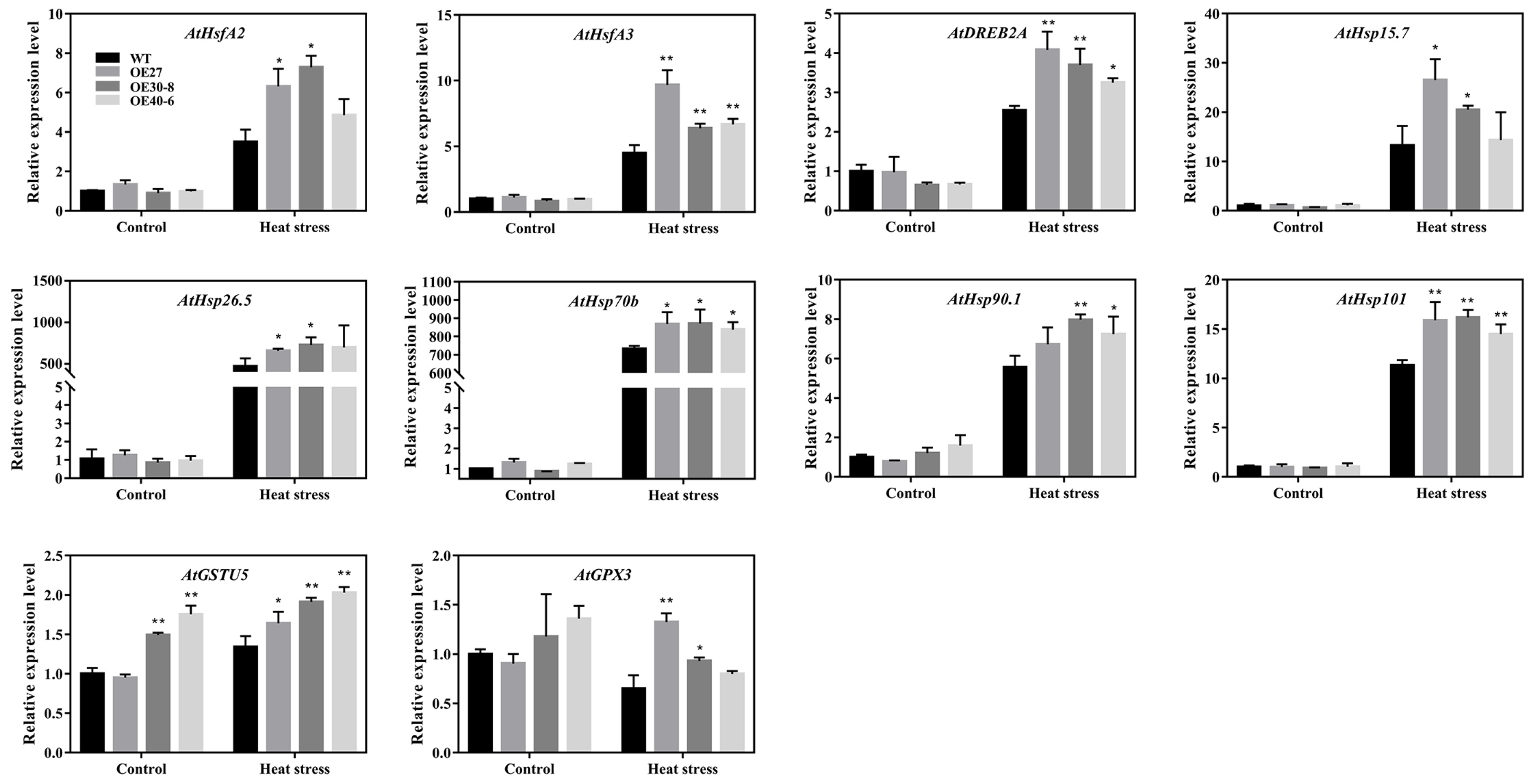
2.8. Expression Analyses of the Stress-Related Genes in Transgenic Arabidopsis
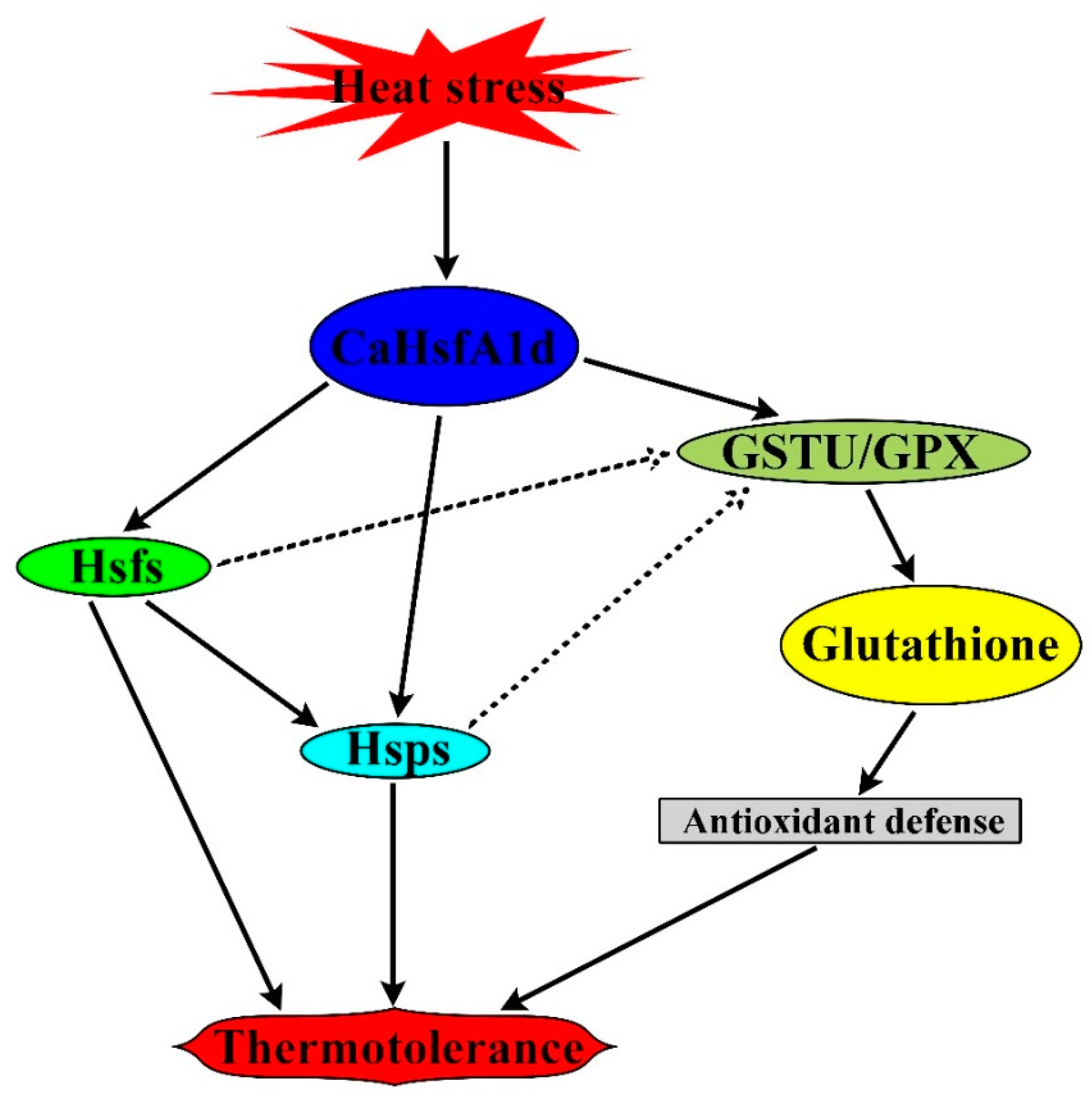
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Cloning and Expression Pattern of the CaHsfA1d Gene
4.3. Transactivation Activity Assay in Yeast
4.4. Subcellular Localization in Tobacco
4.5. The Thermotolerance Analyses in the CaHsfA1d-Silenced Pepper
4.6. The Thermotolerance Analyses in the CaHsfA1d-Overexpression Arabidopsis
4.7. RNA Isolation and qRT-PCR Analyses
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhao, C.; Liu, B.; Piao, S.; Wang, X.; Lobell, D.B.; Huang, Y.; Huang, M.; Yao, Y.; Bassu, S.; Ciais, P.; et al. Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl. Acad. Sci. USA 2017, 114, 9326–9331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foyer, C.H.; Descourvieres, P.; Kunert, K.J. Protection against oxygen radicals—An important defense-mechanism studied in transgenic plants. Plant Cell Environ. 1994, 17, 507–523. [Google Scholar] [CrossRef]
- Von Koskull-Doring, P.; Scharf, K.D.; Nover, L. The diversity of plant heat stress transcription factors. Trends Plant Sci. 2007, 12, 452–457. [Google Scholar] [CrossRef]
- Guo, M.; Liu, J.H.; Ma, X.; Luo, D.X.; Gong, Z.H.; Lu, M.H. The plant heat stress transcription factors (HSFs): Structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 2016, 7, 114. [Google Scholar] [CrossRef] [Green Version]
- Mittler, R.; Finka, A.; Goloubinoff, P. How do plants feel the heat? Trends Biochem. Sci. 2012, 37, 118–125. [Google Scholar] [CrossRef]
- Scharf, K.-D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2012, 1819, 104–119. [Google Scholar] [CrossRef]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Li, G.; Zhang, Y.; Zhang, H.; Zhang, Y.; Zhao, L.; Liu, Z.; Guo, X. Characteristics and regulating role in thermotolerance of the heat shock transcription factor ZmHsf12 from Zea mays L. J. Plant Biol. 2019, 62, 329–341. [Google Scholar] [CrossRef]
- Nishizawa-Yokoi, A.; Nosaka, R.; Hayashi, H.; Tainaka, H.; Maruta, T.; Tamoi, M.; Ikeda, M.; Ohme-Takagi, M.; Yoshimura, K.; Yabuta, Y.; et al. HsfA1d and HsfA1e involved in the transcriptional regulation of HsfA2 function as key regulators for the Hsf signaling network in response to environmental stress. Plant Cell Physiol. 2011, 52, 933–945. [Google Scholar] [CrossRef]
- Pirkkala, L.; Nykanen, P.; Sistonen, L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001, 15, 1118–1131. [Google Scholar] [CrossRef] [Green Version]
- Jaiswal, L.; De, S.; Singh, R.K.; Baithalu, R.K. Molecular characterization and protein structure prediction of heat shock transcriptional factors in goat (Capra hircus) and sheep (Ovis aries). Anim. Biotechnol. 2020, 31, 432–439. [Google Scholar] [CrossRef]
- Akerfelt, M.; Morimoto, R.I.; Sistonen, L. Heat shock factors: integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 2010, 11, 545–555. [Google Scholar] [CrossRef]
- Guo, J.K.; Wu, J.; Ji, Q.; Wang, C.; Luo, L.; Yuan, Y.; Wang, Y.H.; Wang, J. Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J. Genet. Genom. 2008, 35, 105–118. [Google Scholar] [CrossRef]
- Berz, J.; Simm, S.; Schuster, S.; Scharf, K.D.; Schleiff, E.; Ebersberger, I. HEATSTER: A database and web server for identification and classification of heat stress transcription factors in plants. Bioinform. Biol. Insights 2019, 13. [Google Scholar] [CrossRef] [Green Version]
- Xue, G.P.; Sadat, S.; Drenth, J.; McIntyre, C.L. The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes. J. Exp. Bot. 2014, 65, 539–557. [Google Scholar] [CrossRef] [Green Version]
- Li, G.L.; Zhang, H.N.; Shao, H.; Wang, G.Y.; Zhang, Y.Y.; Zhang, Y.J.; Zhao, L.N.; Guo, X.L.; Sheteiwy, M.S. ZmHsf05, a new heat shock transcription factor from Zea mays L. improves thermotolerance in Arabidopsis thaliana and rescues thermotolerance defects of the athsfa2 mutant. Plant Sci. 2019, 283, 375–384. [Google Scholar] [CrossRef]
- Nover, L.; Bharti, K.; Doring, P.; Mishra, S.K.; Ganguli, A.; Scharf, K.D. Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperon 2001, 6, 177–189. [Google Scholar] [CrossRef]
- Guo, M.; Lu, J.P.; Zhai, Y.F.; Chai, W.G.; Gong, Z.H.; Lu, M.H. Genome-wide analysis, expression profile of heat shock factor gene family (CaHsfs) and characterisation of CaHsfA2 in pepper (Capsicum annuum L.). BMC Plant Biol. 2015, 15, 151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuang, L.; Cao, W.; Wang, J.; Yu, J.; Yang, Z.; Huang, B. Characterization and functional analysis of FaHsfC1b from festuca arundinacea conferring heat tolerance in arabidopsis. Int. J. Mol. Sci. 2018, 19, 2702. [Google Scholar] [CrossRef] [Green Version]
- Lohmann, C.; Eggers-Schumacher, G.; Wunderlich, M.; Schoffl, F. Two different heat shock transcription factors regulate immediate early expression of stress genes in Arabidopsis. Mol. Genet. Genom. 2004, 271, 11–21. [Google Scholar] [CrossRef]
- Lee, J.H.; Hubel, A.; Schoffl, F. Derepression of the activity of genetically-engineered heat-shock factor causes constitutive synthesis of heat-shock proteins and increased thermotolerance in transgenic Arabidopsis. Plant J. 1995, 8, 603–612. [Google Scholar] [CrossRef]
- Prandl, R.; Hinderhofer, K.; Eggers-Schumacher, G.; Schoffl, F. HSF3, a new heat shock factor from Arabidopsis thaliana, derepresses the heat shock response and confers thermotolerance when overexpressed in transgenic plants. Mol. Gen. Genet. 1998, 258, 269–278. [Google Scholar] [CrossRef]
- Yoshida, T.; Ohama, N.; Nakajima, J.; Kidokoro, S.; Mizoi, J.; Nakashima, K.; Maruyama, K.; Kim, J.M.; Seki, M.; Todaka, D.; et al. Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol. Genet. Genom. 2011, 286, 321–332. [Google Scholar] [CrossRef]
- Liu, H.C.; Liao, H.T.; Charng, Y.Y. The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 2011, 34, 738–751. [Google Scholar] [CrossRef]
- Shi, H.T.; Tan, D.X.; Reiter, R.J.; Ye, T.T.; Yang, F.; Chan, Z.L. Melatonin induces class A1 heat-shock factors (HSFA1s) and their possible involvement of thermotolerance in Arabidopsis. J. Pineal Res. 2015, 58, 335–342. [Google Scholar] [CrossRef]
- Liu, H.C.; Charng, Y.Y. Common and distinct functions of Arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development. Plant Physiol. 2013, 163, 276–290. [Google Scholar] [CrossRef] [Green Version]
- Mishra, S.K. In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev. 2002, 16, 1555–1567. [Google Scholar] [CrossRef] [Green Version]
- El-shershaby, A.; Ullrich, S.; Simm, S.; Scharf, K.D.; Schleiff, E.; Fragkostefanakis, S. Functional diversification of tomato HsfA1 factors is based on DNA binding domain properties. Gene 2019, 714. [Google Scholar] [CrossRef]
- Charng, Y.Y.; Liu, H.C.; Liu, N.Y.; Chi, W.T.; Wang, C.N.; Chang, S.H.; Wang, T.T. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 2007, 143, 251–262. [Google Scholar] [CrossRef] [Green Version]
- Chan-Schaminet, K.Y.; Baniwal, S.K.; Bublak, D.; Nover, L.; Scharf, K.D. Specific interaction between tomato HsfA1 and HsfA2 creates hetero-oligomeric superactivator complexes for synergistic activation of heat stress gene expression. J. Biol. Chem. 2009, 284, 20848–20857. [Google Scholar] [CrossRef] [Green Version]
- Gong, B.; Yi, J.; Wu, J.; Sui, J.; Khan, M.A.; Wu, Z.; Zhong, X.; Seng, S.; He, J.; Yi, M. LlHSFA1, a novel heat stress transcription factor in lily (Lilium longiflorum), can interact with LlHSFA2 and enhance the thermotolerance of transgenic Arabidopsis thaliana. Plant Cell Rep. 2014, 33, 1519–1533. [Google Scholar] [CrossRef]
- Lin, Y.X.; Jiang, H.Y.; Chu, Z.X.; Tang, X.L.; Zhu, S.W.; Cheng, B.J. Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genom. 2011, 12, 76. [Google Scholar] [CrossRef] [Green Version]
- Li, H.C.; Zhang, H.N.; Li, G.L.; Liu, Z.H.; Zhang, Y.M.; Zhang, H.M.; Guo, X.L. Expression of maize heat shock transcription factor gene ZmHsf06 enhances the thermotolerance and drought-stress tolerance of transgenic Arabidopsis. Funct. Plant Biol. 2015, 42, 1080–1091. [Google Scholar] [CrossRef]
- Schramm, F.; Larkindale, J.; Kiehlmann, E.; Ganguli, A.; Englich, G.; Vierling, E.; von Koskull-Doring, P. A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. 2008, 53, 264–274. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, T.; Sakuma, Y.; Todaka, D.; Maruyama, K.; Qin, F.; Mizoi, J.; Kidokoro, S.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis heat-shock transcription factor HsfA3 in the transcriptional cascade downstream of the DREB2A stress-regulatory system. Biochem. Biophys. Res. Commun. 2008, 368, 515–521. [Google Scholar] [CrossRef]
- Qin, F.; Kakimoto, M.; Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Tran, L.S.P.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J. 2007, 50, 54–69. [Google Scholar] [CrossRef]
- Baniwal, S.K.; Chan, K.Y.; Scharf, K.D.; Nover, L. Role of heat stress transcription factor HsfA5 as specific repressor of HsfA4. J. Biol. Chem. 2007, 282, 3605–3613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.C.; Niu, C.Y.; Yang, C.R.; Jinn, T.L. The heat stress factor HSFA6b connects ABA signaling and ABA-mediated heat responses. Plant Physiol. 2016, 172, 1182–1199. [Google Scholar] [CrossRef]
- Guo, M.; Yin, Y.X.; Ji, J.J.; Ma, B.P.; Lu, M.H.; Gong, Z.H. Cloning and expression analysis of heat-shock transcription factor gene CaHsfA2 from pepper (Capsicum annuum L.). Genet. Mol. Res. 2014, 13, 1865–1875. [Google Scholar] [CrossRef]
- Zhai, Y.; Guo, M.; Wang, H.; Lu, J.; Liu, J.; Zhang, C.; Gong, Z.; Lu, M. Autophagy, a conserved mechanism for protein degradation, responds to heat, and other abiotic stresses in Capsicum annuum L. Front. Plant Sci. 2016, 7, 131. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.L.; Schiff, M.; Dinesh-Kumar, S.P. Virus-induced gene silencing in tomato. Plant J. 2002, 31, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Gai, W.X.; Ma, X.; Qiao, Y.M.; Shi, B.H.; Ul Haq, S.; Li, Q.H.; Wei, A.M.; Liu, K.K.; Gong, Z.H. Characterization of the bZIP transcription factor family in pepper (Capsicum annuum L.): CabZIP25 positively modulates the salt tolerance. Front. Plant Sci. 2020, 11, 139. [Google Scholar] [CrossRef]
- Uzilday, B.; Turkan, I.; Sekmen, A.H.; Ozgur, R.; Karakaya, H.C. Comparison of ROS formation and antioxidant enzymes in Cleome gynandra (C4) and Cleome spinosa (C3) under drought stress. Plant. Sci. 2012, 182, 59–70. [Google Scholar] [CrossRef] [Green Version]
- Feng, X.H.; Zhang, H.X.; Ali, M.; Gai, W.X.; Cheng, G.X.; Yu, Q.H.; Yang, S.B.; Li, X.X.; Gong, Z.H. A small heat shock protein CaHsp25.9 positively regulates heat, salt, and drought stress tolerance in pepper (Capsicum annuum L.). Plant Physiol. Biochem. 2019, 142, 151–162. [Google Scholar] [CrossRef]
- Muthuramalingam, P.; Jeyasri, R.; Bharathi, R.; Suba, V.; Pandian, S.T.K.; Ramesh, M. Global integrated omics expression analyses of abiotic stress signaling HSF transcription factor genes in Oryza sativa L.: An in silico approach. Genomics 2020, 112, 908–918. [Google Scholar] [CrossRef]
- Zhou, J.; Xu, X.C.; Cao, J.J.; Yin, L.L.; Xia, X.J.; Shi, K.; Zhou, Y.H.; Yu, J.Q. Heat shock factor HsfA1a is essential for R gene-mediated nematode resistance and triggers H2O2 production. Plant Physiol. 2018, 176, 2456–2471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotak, S.; Port, M.; Ganguli, A.; Bicker, F.; von Koskull-Doring, P. Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J. 2004, 39, 98–112. [Google Scholar] [CrossRef]
- Xin, H.; Zhang, H.; Zhong, X.; Lian, Q.; Dong, A.; Cao, L.; Yi, M.; Cong, R. Over-expression of LlHsfA2b, a lily heat shock transcription factor lacking trans-activation activity in yeast, can enhance tolerance to heat and oxidative stress in transgenic Arabidopsis seedlings. Plant Cell Tissue Organ Cult. (PCTOC) 2017, 130, 617–629. [Google Scholar] [CrossRef]
- Lyck, R.; Harmening, U.; Hohfeld, I.; Treuter, E.; Scharf, K.D.; Nover, L. Intracellular distribution and identification of the nuclear localization signals of two plant heat-stress transcription factors. Planta 1997, 202, 117–125. [Google Scholar] [CrossRef]
- Whiteside, S.T.; Goodbourn, S. Signal transduction and nuclear targeting: regulation of transcription factor activity by subcellular localization. J. Cell Sci. 1993, 104, 949–955. [Google Scholar]
- Zhu, B.; Ye, C.; Lu, H.; Chen, X.; Chai, G.; Chen, J.; Wang, C. Identification and characterization of a novel heat shock transcription factor gene, GmHsfA1, in soybeans (Glycine max). J. Plant Res. 2006, 119, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Jiang, T.; Zhang, C.; Li, X.; Wang, C.; Zhang, Y.; Li, T.; Dirk, L.M.A.; Downie, A.B.; Zhao, T. Maize HSFA2 and HSBP2 antagonistically modulate raffinose biosynthesis and heat tolerance in Arabidopsis. Plant J. 2019, 100, 128–142. [Google Scholar] [CrossRef]
- Li, Z.J.; Zhang, L.L.; Wang, A.X.; Xu, X.Y.; Li, J.F. Ectopic overexpression of SlHsfA3, a heat stress transcription factor from tomato, confers increased thermotolerance and salt yypersensitivity in germination in transgenic Arabidopsis. PLoS ONE 2013, 8, e54880. [Google Scholar] [CrossRef] [Green Version]
- Li, X.-D.; Wang, X.-L.; Cai, Y.-M.; Wu, J.-H.; Mo, B.-T.; Yu, E.-R. Arabidopsis heat stress transcription factors A2 (HSFA2) and A3 (HSFA3) function in the same heat regulation pathway. Acta Physiol. Plant. 2017, 39. [Google Scholar] [CrossRef]
- Guo, M.; Liu, J.H.; Lu, J.P.; Zhai, Y.F.; Wang, H.; Gong, Z.H.; Wang, S.B.; Lu, M.H. Genome-wide analysis of the CaHsp20 gene family in pepper: comprehensive sequence and expression profile analysis under heat stress. Front. Plant Sci. 2015, 6, 806. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.J.; Cheng, G.X.; Khan, A.; Wei, A.M.; Yu, Q.H.; Yang, S.B.; Luo, D.X.; Gong, Z.H. CaHSP16.4, a small heat shock protein gene in pepper, is involved in heat and drought tolerance. Protoplasma 2019, 256, 39–51. [Google Scholar] [CrossRef]
- Kim, D.H.; Xu, Z.Y.; Hwang, I. AtHSP17.8 overexpression in transgenic lettuce gives rise to dehydration and salt stress resistance phenotypes through modulation of ABA-mediated signaling. Plant Cell Rep. 2013, 32, 1953–1963. [Google Scholar] [CrossRef]
- Sun, J.-T.; Cheng, G.-X.; Huang, L.-J.; Liu, S.; Ali, M.; Khan, A.; Yu, Q.-H.; Yang, S.-B.; Luo, D.-X.; Gong, Z.-H. Modified expression of a heat shock protein gene, CaHSP22.0, results in high sensitivity to heat and salt stress in pepper (Capsicum annuum L.). Sci. Hortic. 2019, 249, 364–373. [Google Scholar] [CrossRef]
- Wu, T.Y.; Juan, Y.T.; Hsu, Y.H.; Wu, S.H.; Liao, H.T.; Fung, R.W.; Charng, Y.Y. Interplay between heat shock proteins HSP101 and HSA32 prolongs heat acclimation memory posttranscriptionally in Arabidopsis. Plant Physiol. 2013, 161, 2075–2084. [Google Scholar] [CrossRef] [Green Version]
- Guo, M.; Liu, J.H.; Ma, X.; Zhai, Y.F.; Gong, Z.H.; Lu, M.H. Genome-wide analysis of the Hsp70 family genes in pepper (Capsicum annuum L.) and functional identification of CaHsp70-2 involvement in heat stress. Plant Sci. 2016, 252, 246–256. [Google Scholar] [CrossRef]
- Guo, M.; Zhai, Y.F.; Lu, J.P.; Chai, L.; Chai, W.G.; Gong, Z.H.; Lu, M.H. Characterization of CaHsp70-1, a pepper heat-shock protein gene in response to heat stress and some regulation exogenous substances in Capsicum annuum L. Int. J. Mol. Sci. 2014, 15, 19741–19759. [Google Scholar] [CrossRef] [Green Version]
- Burke, J.J.; Chen, J.P. Enhancement of reproductive heat tolerance in plants. PLoS ONE 2015, 10, e0122933. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Zhang, H.; Zhao, H.; Gao, T.; Song, A.; Jiang, J.; Chen, F.; Chen, S. Chrysanthemum CmHSFA4 gene positively regulates salt stress tolerance in transgenic chrysanthemum. Plant Biotechnol. J. 2018, 16, 1311–1321. [Google Scholar] [CrossRef] [Green Version]
- Huang, P.; Chung, M.S.; Ju, H.W.; Na, H.S.; Lee, D.J.; Cheong, H.S.; Kim, C.S. Physiological characterization of the Arabidopsis thaliana oxidation-related zinc finger 1, a plasma membrane protein involved in oxidative stress. J. Plant Res. 2011, 124, 699–705. [Google Scholar] [CrossRef]
- Chen, S.; Yu, M.; Li, H.; Wang, Y.; Lu, Z.; Zhang, Y.; Liu, M.; Qiao, G.; Wu, L.; Han, X.; et al. SaHsfA4c from Sedum alfredii hance enhances cadmium tolerance by regulating ROS-scavenger activities and heat shock proteins expression. Front. Plant Sci. 2020, 11, 142. [Google Scholar] [CrossRef] [Green Version]
- Shen, Z.D.; Ding, M.Q.; Sun, J.; Deng, S.R.; Zhao, R.; Wang, M.J.; Ma, X.J.; Wang, F.F.; Zhang, H.L.; Qian, Z.Y.; et al. Overexpression of PeHSF mediates leaf ROS homeostasis in transgenic tobacco lines grown under salt stress conditions. Plant Cell Tissue Organ 2013, 115, 299–308. [Google Scholar] [CrossRef]
- Panchuk, I.I.; Volkov, R.A.; Schoffl, F. Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol. 2002, 129, 838–853. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Yang, J.; Li, W.; Chen, Y.; Lu, H.; Zhao, S.; Li, D.; Wei, M.; Li, C. PuHSFA4a enhances tolerance to excess zinc by regulating reactive oxygen species production and root development in Populus. Plant Physiol. 2019, 180, 2254–2271. [Google Scholar] [CrossRef] [Green Version]
- Miao, Y.; Lv, D.; Wang, P.; Wang, X.C.; Chen, J.; Miao, C.; Song, C.P. An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 2006, 18, 2749–2766. [Google Scholar] [CrossRef] [Green Version]
- Wagner, U.; Edwards, R.; Dixon, D.P.; Mauch, F. Probing the diversity of the arabidopsis glutathione S-transferase gene family. Plant Mol. Biol. 2002, 49, 515–532. [Google Scholar] [CrossRef]
- Wydro, M.; Kozubek, E.; Lehmann, P. Optimization of transient Agrobacterium-mediated gene expression system in leaves of Nicotiana benthamiana. Acta Biochim. Pol. 2006, 53, 289–298. [Google Scholar] [CrossRef]
- Chakraborty, J.; Sen, S.; Ghosh, P.; Jain, A.; Das, S. Inhibition of multiple defense responsive pathways by CaWRKY70 transcription factor promotes susceptibility in chickpea under Fusarium oxysporum stress condition. BMC Plant Biol. 2020, 20, 319. [Google Scholar] [CrossRef] [PubMed]
- ThordalChristensen, H.; Zhang, Z.G.; Wei, Y.D.; Collinge, D.B. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 1997, 11, 1187–1194. [Google Scholar] [CrossRef]
- Cai, S.Y.; Zhang, Y.; Xu, Y.P.; Qi, Z.Y.; Li, M.Q.; Ahammed, G.J.; Xia, X.J.; Shi, K.; Zhou, Y.H.; Reiter, R.J.; et al. HsfA1a upregulates melatonin biosynthesis to confer cadmium tolerance in tomato plants. J. Pineal Res. 2017, 62. [Google Scholar] [CrossRef]
- Stewart, R.R.; Bewley, J.D. Lipid peroxidation associated with accelerated aging of soybean axes. Plant Physiol. 1980, 65, 245–248. [Google Scholar] [CrossRef] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]



| Gene | Plant | Function | Reference |
|---|---|---|---|
| HsfA1a/HsfA1b | Arabidopsis | The thermotolerance of HsfA1a and HsfA1b double-knockout mutant are significantly impaired. Overexpression of HsfA1a or HsfA1b enhanced the thermotolerance. | [20,21,22] |
| HsfA1 | Arabidopsis | Arabidopsis Hsp90 can interact with HsfA1 to inhibit the accumulation of HsfA1 protein in the nucleus. | [23] |
| HsfA1a/HsfA1b/HsfA1d/HsfA1e | Arabidopsis | The quadruple-knockout Arabidopsis mutant display extremely weakened BT and AT. HsfA1s may be involved in melatonin-mediated heat tolerance. | [24,25] |
| HsfA1a/HsfA1b/HsfA1d | Arabidopsis | The HsfA1a/HsfA1b/HsfA1d are also involved in thermotolerance to mild heat stress. | [26] |
| HsfA1a | Solanum lycopersicum | The expression of HsfA1a is constitutive under control and stress conditions. | [28] |
| HsfA1 | Solanum lycopersicum | The physical interaction between HsfA1 and heat stress-inducible HsfA2 can form activator heterodimers, resulting in the transactivation activity of target heat stress genes expression. | [30] |
| LiHsfA1 | Lilium longiflorum | Overexpression of LiHsfA1 gene improves the thermotolerance of transgenic. Arabidopsis and up-regulated the expression of putative stress-response genes. | [31] |
| GmHsfA1 | Glycine max | The transgenic soybeans with its overexpression showed obviously enhance thermotolerance under heat stress. | [1] |
| ZmHsf06/ZmHsf12 | Zea mays | The heat tolerance of the Arabidopsis seedlings overexpressed with ZmHsf06 or ZmHsf12 was increased. | [8,33] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gai, W.-X.; Ma, X.; Li, Y.; Xiao, J.-J.; Khan, A.; Li, Q.-H.; Gong, Z.-H. CaHsfA1d Improves Plant Thermotolerance via Regulating the Expression of Stress- and Antioxidant-Related Genes. Int. J. Mol. Sci. 2020, 21, 8374. https://doi.org/10.3390/ijms21218374
Gai W-X, Ma X, Li Y, Xiao J-J, Khan A, Li Q-H, Gong Z-H. CaHsfA1d Improves Plant Thermotolerance via Regulating the Expression of Stress- and Antioxidant-Related Genes. International Journal of Molecular Sciences. 2020; 21(21):8374. https://doi.org/10.3390/ijms21218374
Chicago/Turabian StyleGai, Wen-Xian, Xiao Ma, Yang Li, Jing-Jing Xiao, Abid Khan, Quan-Hui Li, and Zhen-Hui Gong. 2020. "CaHsfA1d Improves Plant Thermotolerance via Regulating the Expression of Stress- and Antioxidant-Related Genes" International Journal of Molecular Sciences 21, no. 21: 8374. https://doi.org/10.3390/ijms21218374
APA StyleGai, W.-X., Ma, X., Li, Y., Xiao, J.-J., Khan, A., Li, Q.-H., & Gong, Z.-H. (2020). CaHsfA1d Improves Plant Thermotolerance via Regulating the Expression of Stress- and Antioxidant-Related Genes. International Journal of Molecular Sciences, 21(21), 8374. https://doi.org/10.3390/ijms21218374






