Comparative Ubiquitome Analysis under Heat Stress Reveals Diverse Functions of Ubiquitination in Saccharina japonica
Abstract
:1. Introduction
2. Results
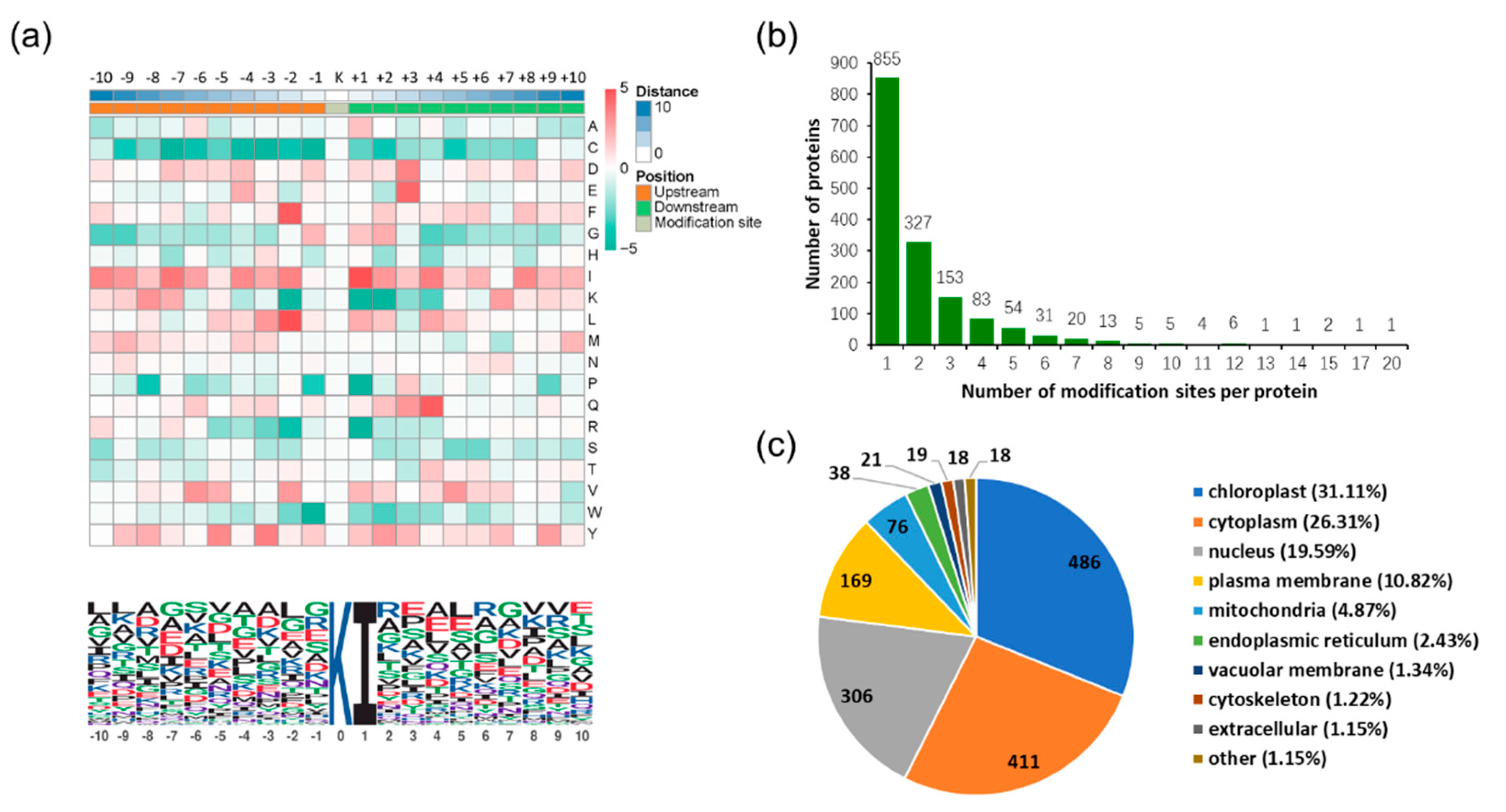
2.1. Overview of the Ubiquitomic Analysis
2.2. Properties of Ubiquitination Sites and Motifs
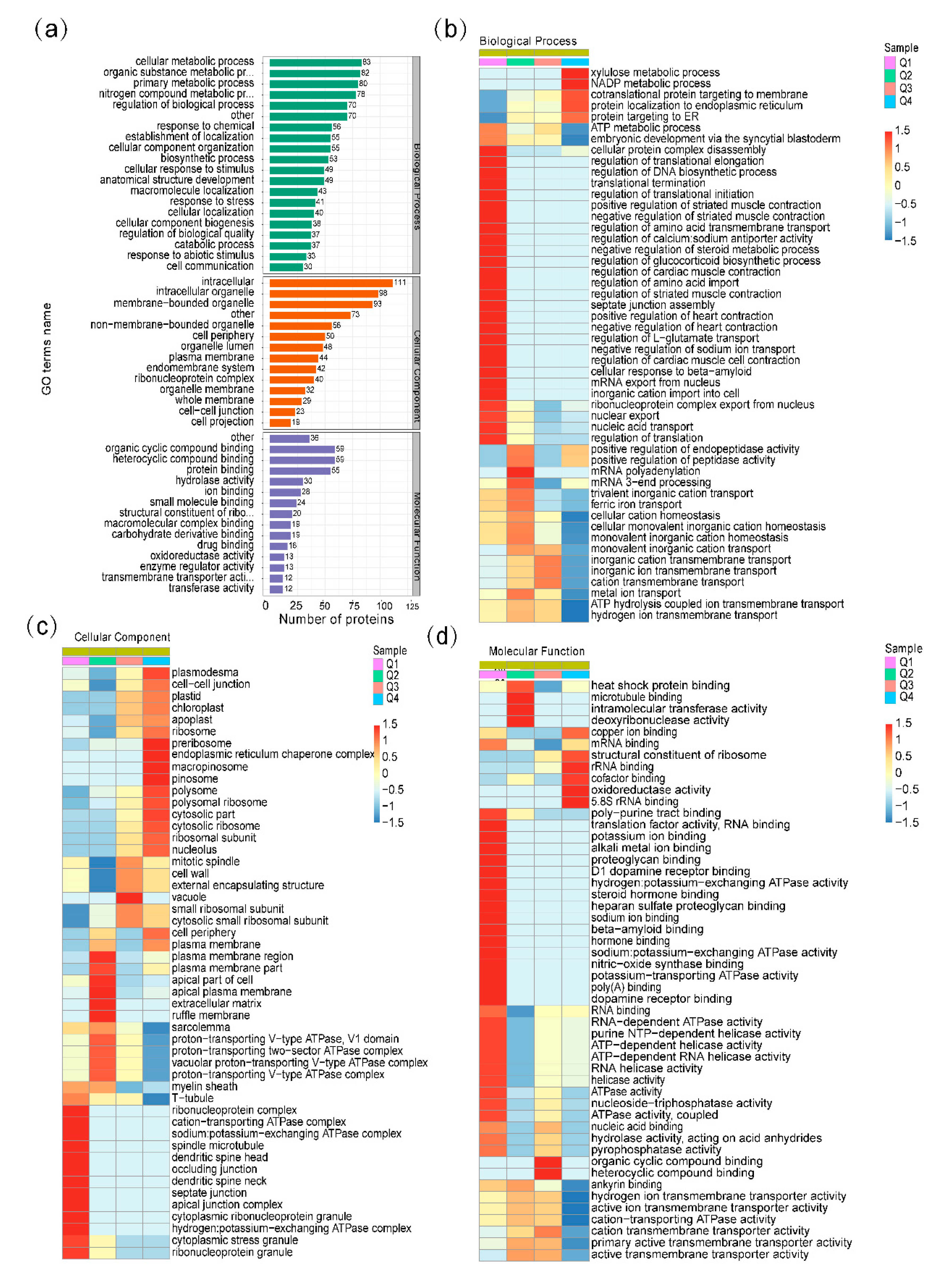
2.3. Gene Ontology (GO) Functional Classification and GO Enrichment-Based Clustering Analysis of the Differentially Expressed Ubiquitinated Proteins
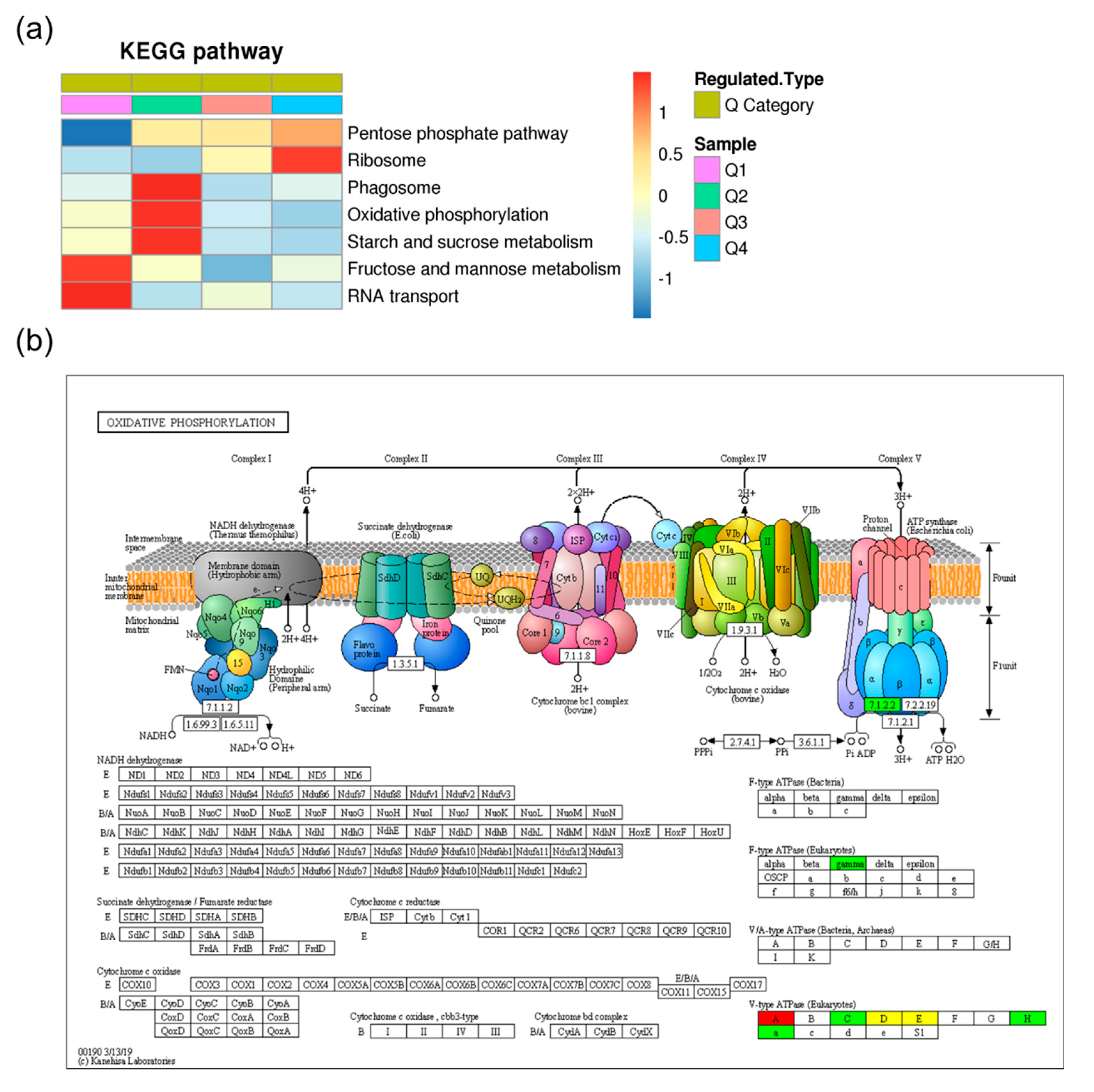
2.4. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis of the Differentially Expressed Ubiquitinated Proteins
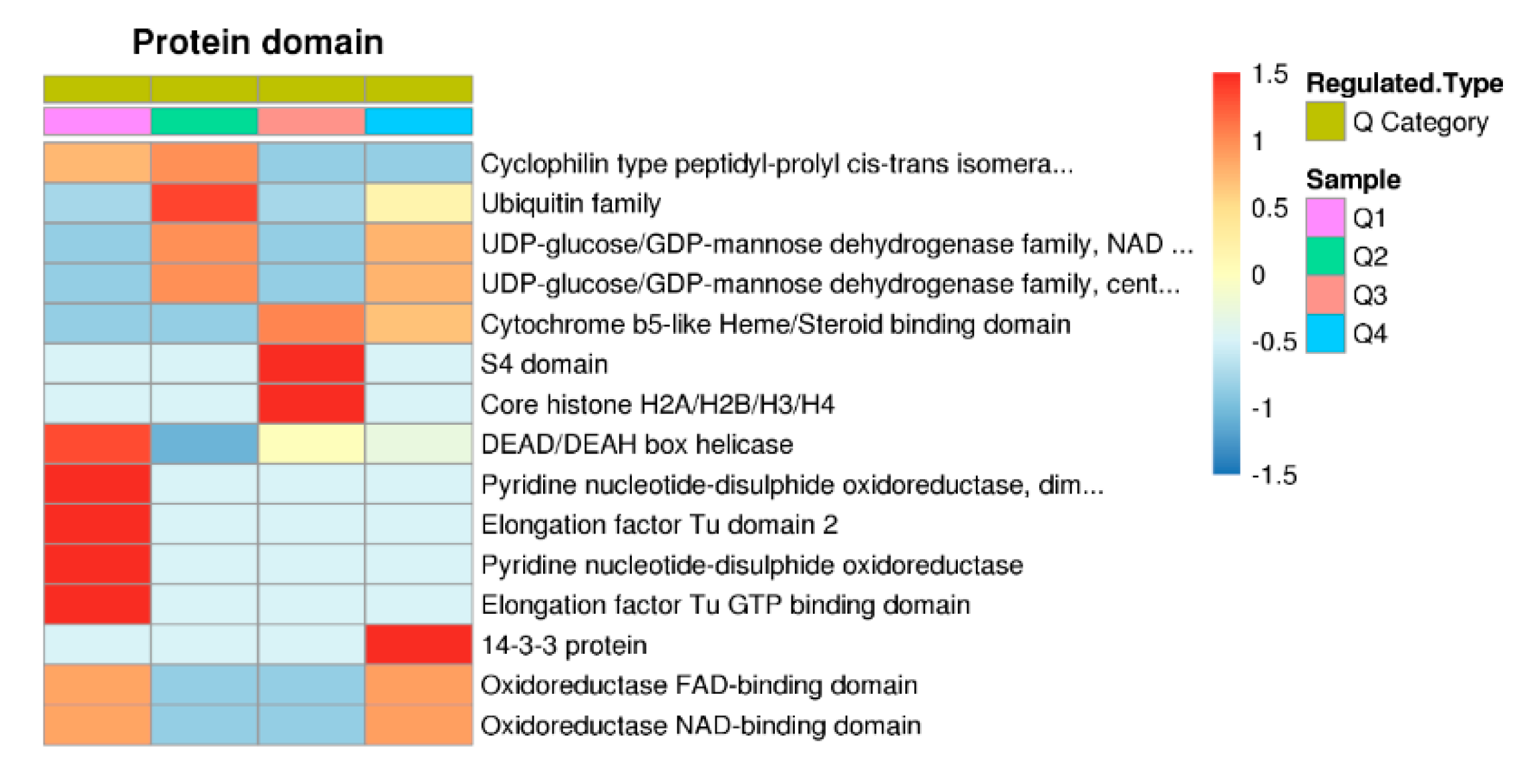
2.5. Protein Domain Based Enrichment Analysis of the Differentially Expressed Lysine Ubiquitinated Proteins
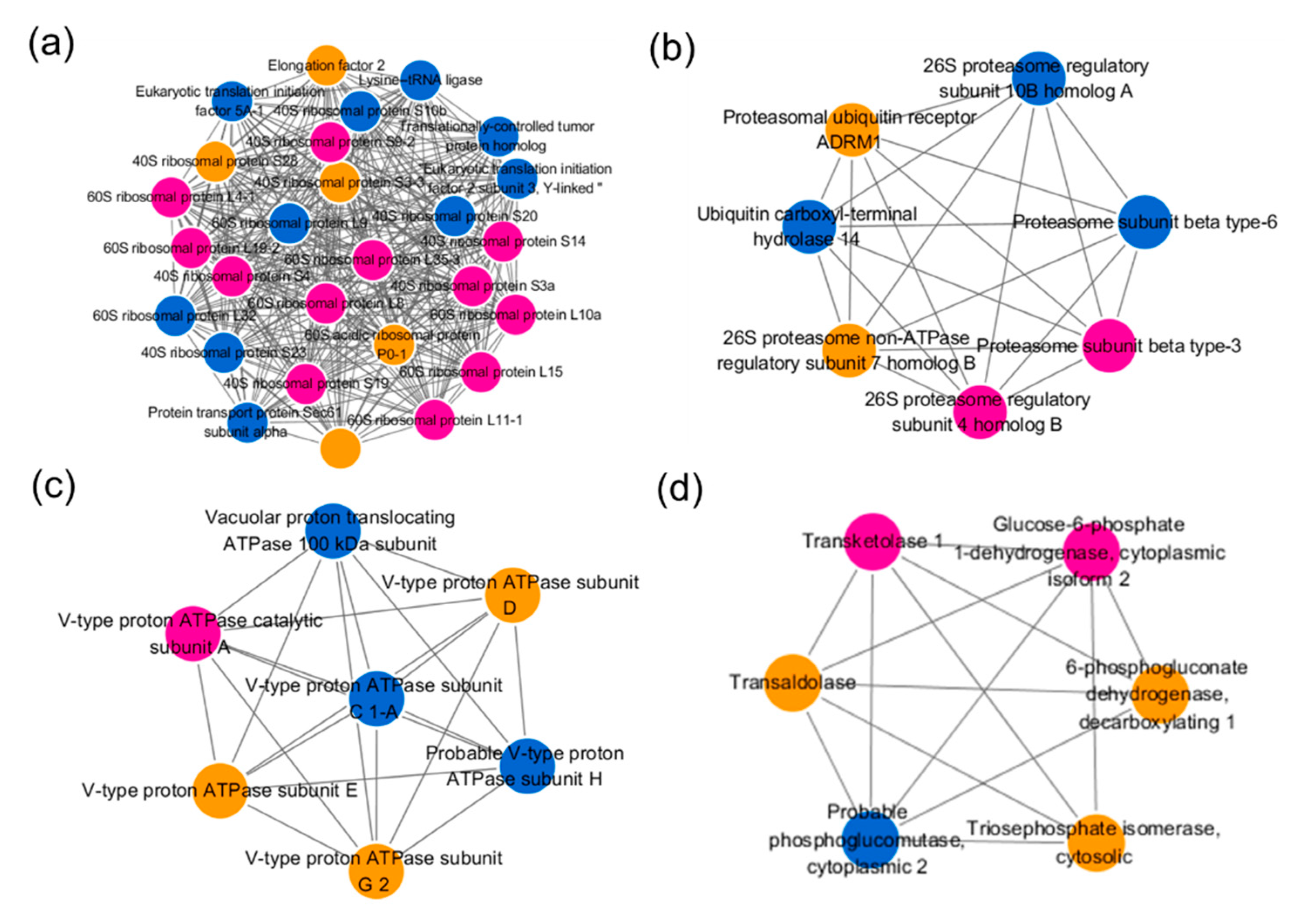
2.6. Protein–Protein Interaction (PPI) Network Analysis of Differentially Expressed Ubiquitinated Proteins
3. Discussion
3.1. Ubiquitination Involved in the Heat Response of S. japonica
3.2. The UPS-Related Protein Showed Differential Ubiquitination
3.3. Key Biological Processes are Modified by Ubiquitination under Heat Stress
4. Materials and Methods
4.1. Sample Preparation
4.2. Protein Extraction
4.3. Trypsin Digestion
4.4. High Performance Liquid Chromatography (HPLC) Fractionation
4.5. Affinity Enrichment of Ubiquitinated Peptides
4.6. Liquid Chromatography (LC)-Tandem Mass Spectroscopy (MS/MS) Analysis
4.7. Database Search and Quantification
4.8. Bioinformatics Methods
4.9. Western Blot Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, F.; Pang, S. Performances of growth, photochemical efficiency, and stress tolerance of young sporophytes from seven populations of Saccharina japonica (Phaeophyta) under short-term heat stress. J. Appl. Phycol. 2009, 22, 221–229. [Google Scholar] [CrossRef]
- Liu, F.; Wang, W.; Sun, X.; Liang, Z.; Wang, F. RNA-Seq revealed complex response to heat stress on transcriptomic level in Saccharina japonica (Laminariales, Phaeophyta). J. Appl. Phycol. 2013, 26, 1585–1596. [Google Scholar] [CrossRef]
- Pang, S.; Jin, Z.H.; Sun, J.Z.; Gao, S.Q. Temperature tolerance of young sporophytes from two populations of Laminaria japonica revealed by chlorophyll fluorescence measurements and short-term growth and survival performances in tank culture. Aquaculture 2007, 262, 493–503. [Google Scholar] [CrossRef]
- Liu, F.; Wang, W.; Sun, X.; Liang, Z.; Wang, F. Conserved and novel heat stress-responsive microRNAs were identified by deep sequencing in Saccharina japonica (Laminariales, Phaeophyta). Plant Cell Environ. 2015, 38, 1357–1367. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhang, P.; Liang, Z.; Wang, W.; Sun, X.; Wang, F. Dynamic profile of proteome revealed multiple levels of regulation under heat stress in Saccharina japonica. J. Appl. Phycol. 2019, 31, 3077–3089. [Google Scholar] [CrossRef]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulman, B.A.; Harper, J.W. Ubiquitin-like protein activation by E1 enzymes: The apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 2009, 10, 319–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, Y.; Rape, M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 2009, 10, 755–764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morreale, F.E.; Walden, H. Types of ubiquitin ligases. Cell 2016, 165, 248–248.e1. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Shabek, N. Ubiquitin ligases: Structure, function, and regulation. Annu. Rev. Biochem. 2017, 86, 129–157. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Paige, J.S.; Jaffrey, S.R. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat. Biotechnol. 2010, 28, 868–873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haglund, K.; Dikic, I. Ubiquitylation and cell signaling. EMBO J. 2005, 24, 3353–3359. [Google Scholar] [CrossRef] [Green Version]
- Mukhopadhyay, D.; Riezman, H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 2007, 315, 201–205. [Google Scholar] [CrossRef] [Green Version]
- Shih, S.C.; Prag, G.; Francis, S.A.; Sutanto, M.A.; Hurley, J.H.; Hicke, L. A ubiquitin-binding motif required for intramolecular monoubiquitylation, the CUE domain. EMBO J. 2003, 22, 1273–1281. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Shi, X.; Zou, Y.; Guo, J.; Huo, N.; Chen, S.; Zhao, C.; Li, H.; Wu, G.; Peng, Y. Proteome-wide identification and functional analysis of ubiquitinated proteins in peach leaves. Sci. Rep. 2020, 10, 2447. [Google Scholar] [CrossRef] [Green Version]
- Vierstra, R.D. The ubiquitin–26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Liu, H.; Chong, K.; Xu, Y. Roles of ubiquitination-mediated protein degradation in plant responses to abiotic stresses. Environ. Exp. Bot. 2015, 114, 92–103. [Google Scholar] [CrossRef]
- Chen, Z.J.; Sun, L.J. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 2009, 33, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, C.; Wei, C.; Liu, X.; Wang, M.; Yu, F.; Xie, Q.; Tu, J. The RING finger ubiquitin E3 ligase OsHTAS enhances heat tolerance by promoting H2O2-induced stomatal closure in rice. Plant Physiol. 2016, 170, 429–443. [Google Scholar] [CrossRef] [Green Version]
- Serrano, I.; Campos, L.; Rivas, S. Roles of E3 ubiquitin-ligases in nuclear protein homeostasis during plant stress responses. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Lim, S.D.; Cho, H.Y.; Park, Y.C.; Ham, D.J.; Lee, J.K.; Jang, C.S. The rice RING finger E3 ligase, OsHCI1, drives nuclear export of multiple substrate proteins and its heterogeneous overexpression enhances acquired thermotolerance. J. Exp. Bot. 2013, 64, 2899–2914. [Google Scholar] [CrossRef]
- Kim, J.H.; Lim, S.D.; Jang, C.S. Oryza sativa heat-induced RING finger protein 1 (OsHIRP1) positively regulates plant response to heat stress. Plant Mol. Biol. 2019, 99, 545–559. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, W.; Wang, W.; Zhang, G.; Liu, Y.; Wang, Y.; Wang, W. Wheat f-box protein gene TaFBA1 Is involved in plant tolerance to heat stress. Front. Plant Sci. 2018, 9, 521. [Google Scholar] [CrossRef] [Green Version]
- Morimoto, K.; Ohama, N.; Kidokoro, S.; Mizoi, J.; Takahashi, F.; Todaka, D.; Mogami, J.; Sato, H.; Qin, F.; Kim, J.S.; et al. BPM-CUL3 E3 ligase modulates thermotolerance by facilitating negative regulatory domain-mediated degradation of DREB2A in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E8528–E8536. [Google Scholar] [CrossRef] [Green Version]
- Von Kampen, J.; Nieländer, U.; Wettern, M. Expression of ubiquitin genes in Chlamydomonas reinhardtii: Involvement in stress response and cell cycle. Planta 1995, 197, 528–534. [Google Scholar] [CrossRef]
- Li, G.-Q.; Zang, X.; Zhang, X.-C.; Lu, N.; Ding, Y.; Gong, L.; Chen, W.-C. Cloning of ubiquitin-activating enzyme and ubiquitin-conjugating enzyme genes from Gracilaria lemaneiformis and their activity under heat shock. Gene 2014, 538, 155–163. [Google Scholar] [CrossRef]
- Penna, A.; Crinelli, R.; Magnani, M. Modulation of the heat shock ubiquitin pool in Skeletonema costatum (bacillariophyceae). J. Phycol. 1996, 32, 409–415. [Google Scholar] [CrossRef]
- Schiedlmeier, B. Repetitious structure and transcription control of a polyubiquitin gene in Volvox carteri. Curr. Genet. 1994, 25, 169–177. [Google Scholar] [CrossRef]
- Vayda, M.E.; Yuan, M.L. The heat shock response of an antarctic alga is evident at 5 °C. Plant Mol. Biol. 1994, 24, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Kang, H.; Liu, W.; Wang, G.L. Comprehensive profiling of the rice ubiquitome reveals the significance of lysine ubiquitination in young leaves. J. Proteome Res. 2015, 14, 2017–2025. [Google Scholar] [CrossRef]
- Zhang, N.; Zhang, L.; Shi, C.; Tian, Q.; Lv, G.; Wang, Y.; Cui, D.; Chen, F. Comprehensive profiling of lysine ubiquitome reveals diverse functions of lysine ubiquitination in common wheat. Sci. Rep. 2017, 7, 13601. [Google Scholar] [CrossRef] [Green Version]
- Walton, A.; Stes, E.; Cybulski, N.; Van Bel, M.; Iñigo, S.; Durand, A.N.; Timmerman, E.; Heyman, J.; Pauwels, L.; De Veylder, L.; et al. It’s time for some “site”-seeing: Novel tools to monitor the ubiquitin landscape in Arabidopsis thaliana. Plant Cell 2016, 28, 6–16. [Google Scholar] [CrossRef] [Green Version]
- Ferguson, D.L.; Guikema, J.A.; Paulsen, G.M. Ubiquitin pool modulation and protein degradation in wheat roots during high temperature stress. Plant Physiol. 1990, 92, 740–746. [Google Scholar] [CrossRef] [Green Version]
- Shimogawara, K.; Muto, S. Heat Shock Induced Change in Protein Ubiquitination in Chlamydomonas. Plant Cell Physiol. 1989, 30, 9–16. [Google Scholar] [CrossRef]
- Wettern, M.; Parag, H.A.; Pollmann, L.; Ohad, I.; Kulka, R.G. Ubiquitin in Chlamydomonas reinhardii. Distribution in the cell and effect of heat shock and photoinhibition on its conjugate pattern. J. Biol. Inorg. Chem. 1990, 191, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, I.B.; Bowen, L.J.H. Protein synthesis and breakdown during heat shock of cultured pear (Pyrus communis L.) cells. Plant Physiol. 1994, 104, 1429–1437. [Google Scholar] [CrossRef] [Green Version]
- Pickart, C.M.; Fushman, D. Polyubiquitin chains: Polymeric protein signals. Curr. Opin. Chem. Biol. 2004, 8, 610–616. [Google Scholar] [CrossRef]
- Sadanandom, A.; Bailey, M.; Ewan, R.; Lee, J.; Nelis, S. The ubiquitin-proteasome system: Central modifier of plant signalling. New Phytol. 2012, 196, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Smalle, J.; Vierstra, R.D. The ubiquitin 26s proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004, 55, 555–590. [Google Scholar] [CrossRef]
- Gardner, R.G.; Shearer, A.G.; Hampton, R.Y. In vivo action of the HRD ubiquitin ligase complex: Mechanisms of endoplasmic reticulum quality control and sterol regulation. Mol. Cell. Biol. 2001, 21, 4276–4291. [Google Scholar] [CrossRef] [Green Version]
- Brodsky, J.L. The protective and destructive roles played by molecular chaperones during ERAD (endoplasmic-reticulum-associated degradation). Biochem. J. 2007, 404, 353–363. [Google Scholar] [CrossRef]
- Saracco, S.A.; Hansson, M.; Scalf, M.; Walker, J.M.; Smith, L.M.; Vierstra, R.D. Tandem affinity purification and mass spectrometric analysis of ubiquitylated proteins in Arabidopsis. Plant J. 2009, 59, 344–358. [Google Scholar] [CrossRef] [Green Version]
- Spence, J.; Gali, R.R.; Dittmar, G.; Sherman, F.; Karin, M.; Finley, D. Cell cycle–regulated modification of the ribosome by a variant multiubiquitin chain. Cell 2000, 102, 67–76. [Google Scholar] [CrossRef] [Green Version]
- Kraft, C.; Deplazes, A.; Sohrmann, M.; Peter, M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat. Cell Biol. 2008, 10, 602–610. [Google Scholar] [CrossRef]
- Matsuo, Y.; Ikeuchi, K.; Saeki, Y.; Iwasaki, S.; Schmidt, C.; Udagawa, T.; Sato, F.; Tsuchiya, H.; Becker, T.; Tanaka, K.; et al. Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Sung, M.K.; Porras-Yakushi, T.R.; Reitsma, J.M.; Huber, F.M.; Sweredoski, M.J.; Hoelz, A.; Hess, S.; Deshaies, R.J. A conserved quality-control pathway that mediates degradation of unassembled ribosomal proteins. eLife 2016, 5. [Google Scholar] [CrossRef]
- Wu, C.C.C.; Peterson, A.; Zinshteyn, B.; Regot, S.; Green, R. Ribosome collisions trigger general stress responses to regulate cell fate. Cell 2020, 182, 404–416.e14. [Google Scholar] [CrossRef]
- Marshansky, V.; Rubinstein, J.L.; Grüber, G. Eukaryotic V-ATPase: Novel structural findings and functional insights. Biochim. Biophys. Acta 2014, 1837, 857–879. [Google Scholar] [CrossRef] [Green Version]
- Stevens, T.H.; Forgac, M. Structure, function and regulation of the vacuolar (H+)-ATPase. Annu. Rev. Cell Dev. Biol. 1997, 13, 779–808. [Google Scholar] [CrossRef]
- Kawasaki-Nishi, S.; Nishi, T.; Forgac, M. Arg-735 of the 100-kDa subunit a of the yeast V-ATPase is essential for proton translocation. Proc. Natl. Acad. Sci. USA 2001, 98, 2397–12402. [Google Scholar] [CrossRef] [Green Version]
- Seol, J.H.; Shevchenko, A.; Shevchenko, A.; Deshaies, R.J. Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat. Cell Biol. 2001, 3, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Landi, S.; Nurcato, R.; De Lillo, A.; Lentini, M.; Grillo, S.; Esposito, S. Glucose-6-phosphate dehydrogenase plays a central role in the response of tomato (Solanum lycopersicum) plants to short and long-term drought. Plant Physiol. Biochem. 2016, 105, 79–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Wu, R.; Wan, Q.; Xie, G.; Bi, Y. Glucose-6-phosphate dehydrogenase plays a pivotal role in nitric oxide-involved defense against oxidative stress under salt stress in red kidney bean. Plant Cell Physiol. 2007, 48, 511–522. [Google Scholar] [CrossRef] [Green Version]
- Ye, N.; Zhang, X.; Miao, M.; Fan, X.; Zheng, Y.; Xu, D.; Wang, J.; Zhou, L.; Wang, D.; Gao, Y.; et al. Saccharina genomes provide novel insight into kelp biology. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Michel, G.; Tonon, T.; Scornet, D.; Cock, J.M.; Kloareg, B. The cell wall polysaccharide metabolism of the brown alga Ectocarpus siliculosus. Insights into the evolution of extracellular matrix polysaccharides in Eukaryotes. New Phytol. 2010, 188, 82–97. [Google Scholar] [CrossRef]
- Zhang, P.; Shao, Z.; Jin, W.; Duan, D. Comparative characterization of two GDP-mannose dehydrogenase genes from Saccharina japonica (Laminariales, Phaeophyceae). BMC Plant Biol. 2016, 16. [Google Scholar] [CrossRef] [Green Version]
- Gimmler, H. Algal Adaptation to Environmental Stresses; Springer: Berlin/Heidelberg, Germany, 2001. [Google Scholar]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]

| Name | Upregulated (>1.5) | Downregulated (<0.67) |
|---|---|---|
| Sites | 152 | 208 |
| Proteins | 106 | 131 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pengyan, Z.; Fuli, L.; Siqing, C.; Zhourui, L.; Wenjun, W.; Xiutao, S. Comparative Ubiquitome Analysis under Heat Stress Reveals Diverse Functions of Ubiquitination in Saccharina japonica. Int. J. Mol. Sci. 2020, 21, 8210. https://doi.org/10.3390/ijms21218210
Pengyan Z, Fuli L, Siqing C, Zhourui L, Wenjun W, Xiutao S. Comparative Ubiquitome Analysis under Heat Stress Reveals Diverse Functions of Ubiquitination in Saccharina japonica. International Journal of Molecular Sciences. 2020; 21(21):8210. https://doi.org/10.3390/ijms21218210
Chicago/Turabian StylePengyan, Zhang, Liu Fuli, Chen Siqing, Liang Zhourui, Wang Wenjun, and Sun Xiutao. 2020. "Comparative Ubiquitome Analysis under Heat Stress Reveals Diverse Functions of Ubiquitination in Saccharina japonica" International Journal of Molecular Sciences 21, no. 21: 8210. https://doi.org/10.3390/ijms21218210
APA StylePengyan, Z., Fuli, L., Siqing, C., Zhourui, L., Wenjun, W., & Xiutao, S. (2020). Comparative Ubiquitome Analysis under Heat Stress Reveals Diverse Functions of Ubiquitination in Saccharina japonica. International Journal of Molecular Sciences, 21(21), 8210. https://doi.org/10.3390/ijms21218210






