Loss of Parkin Results in Altered Muscle Stem Cell Differentiation during Regeneration
Abstract
:1. Introduction
2. Results
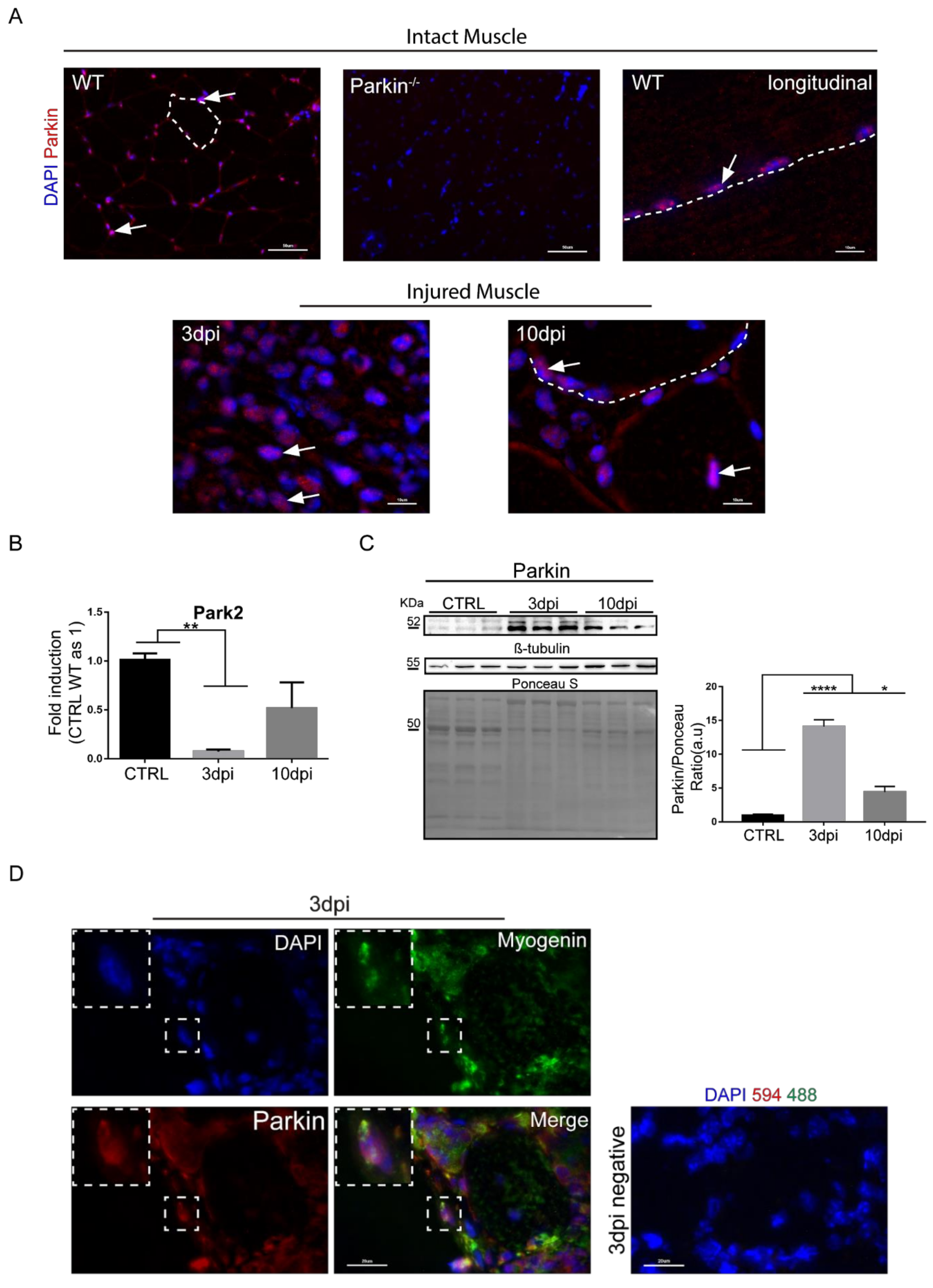
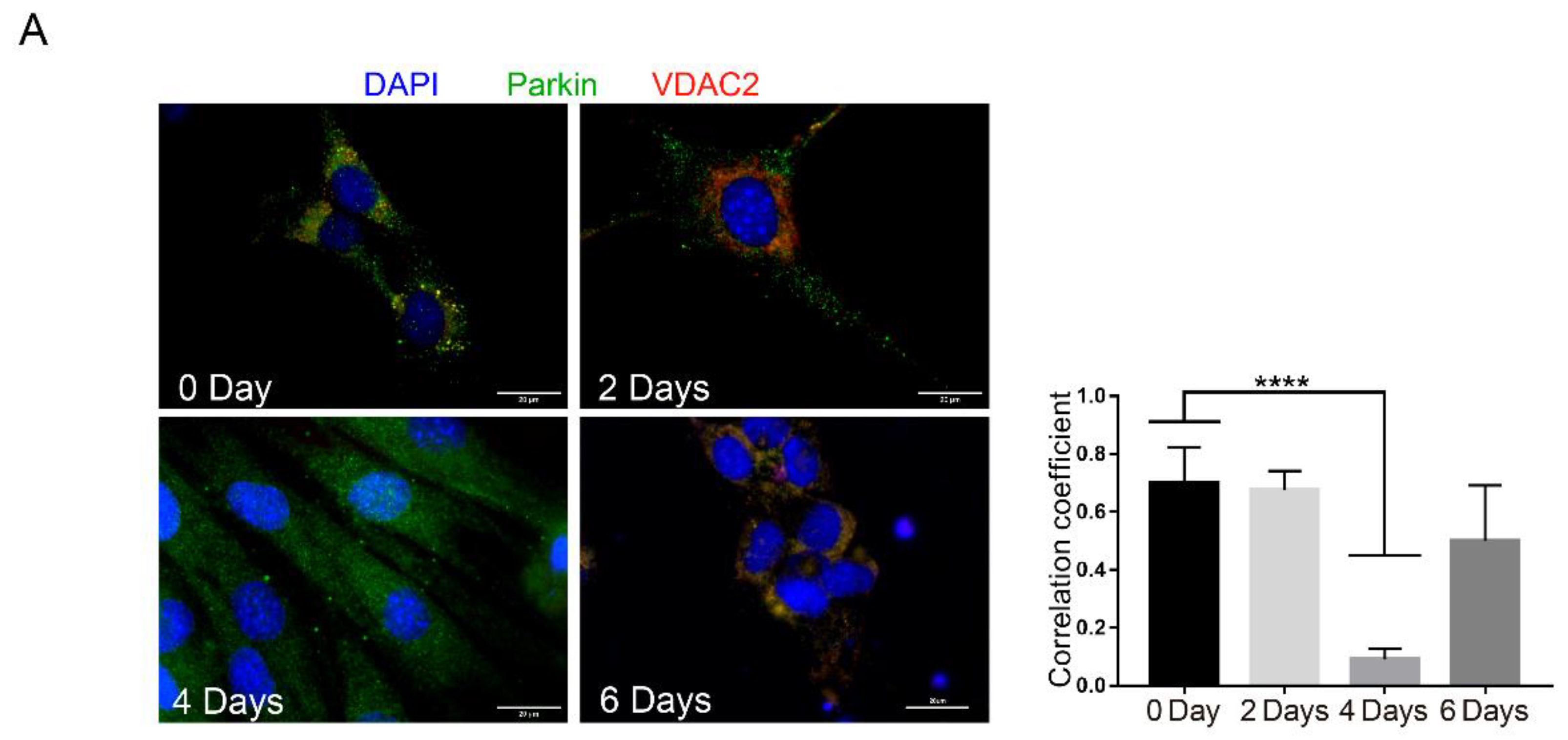
2.1. Parkin Is Expressed in MuSCs during Regeneration In Vivo and in Myoblast in Culture
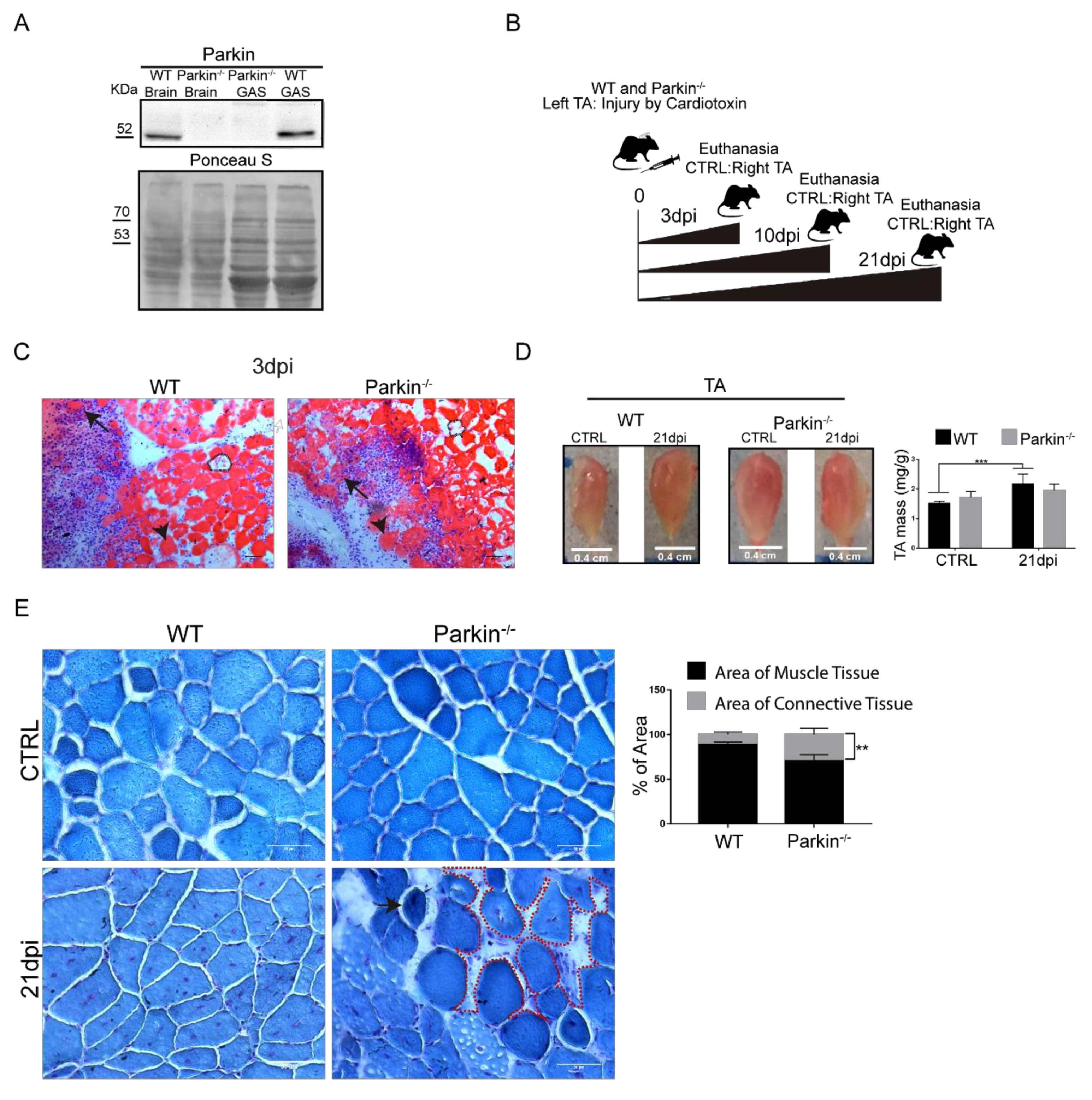
2.2. Parkin Knockout Mice Show Reduced Ability to Undergo Skeletal Muscle Repair after Injury
2.3. Parkin Deficiency Leads to Impaired Assembly of Myofibres after Skeletal Muscle Injury
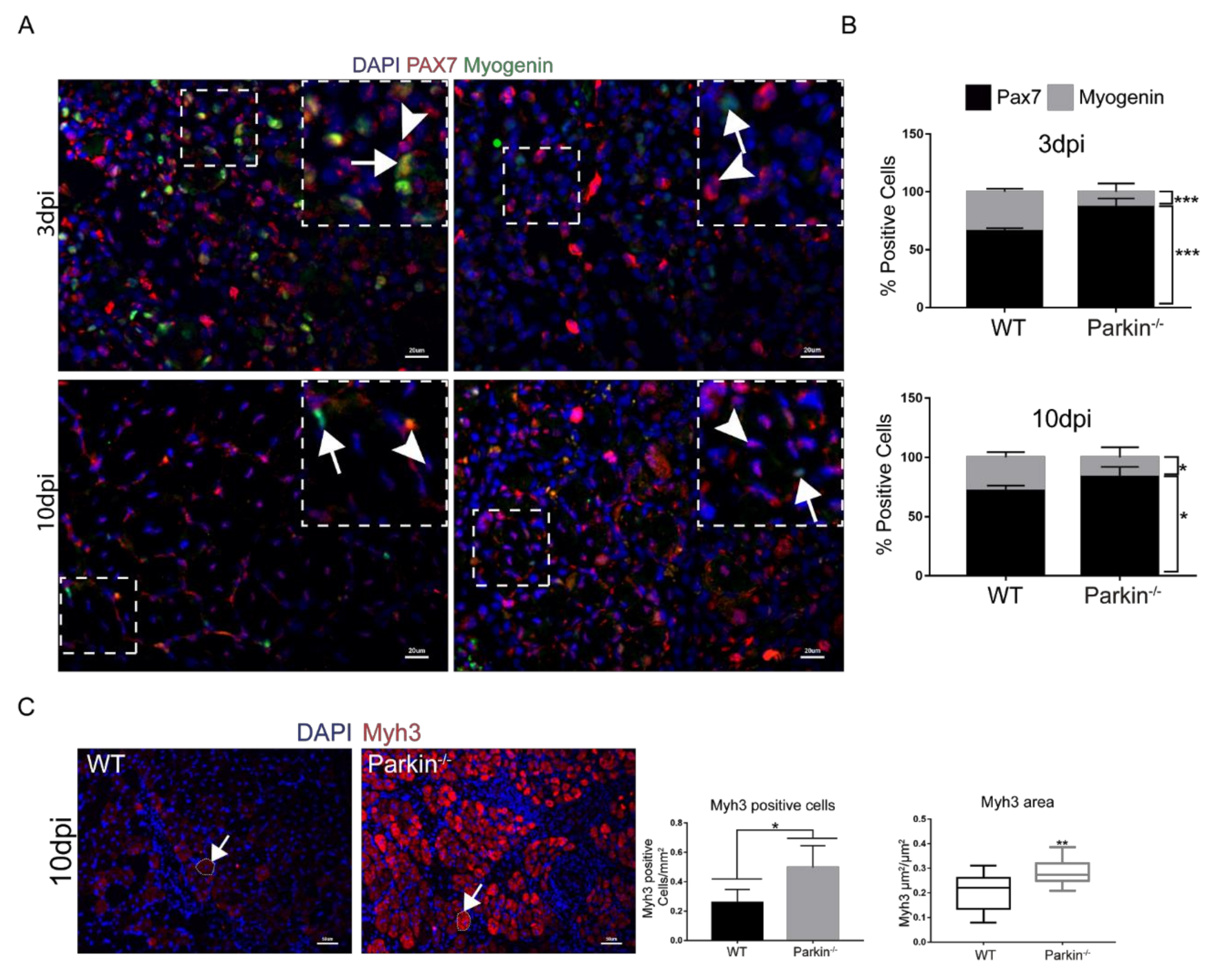
2.4. Parkin−/− MuSCs Display Reduced Differentiation Capacity and Increased Proliferation
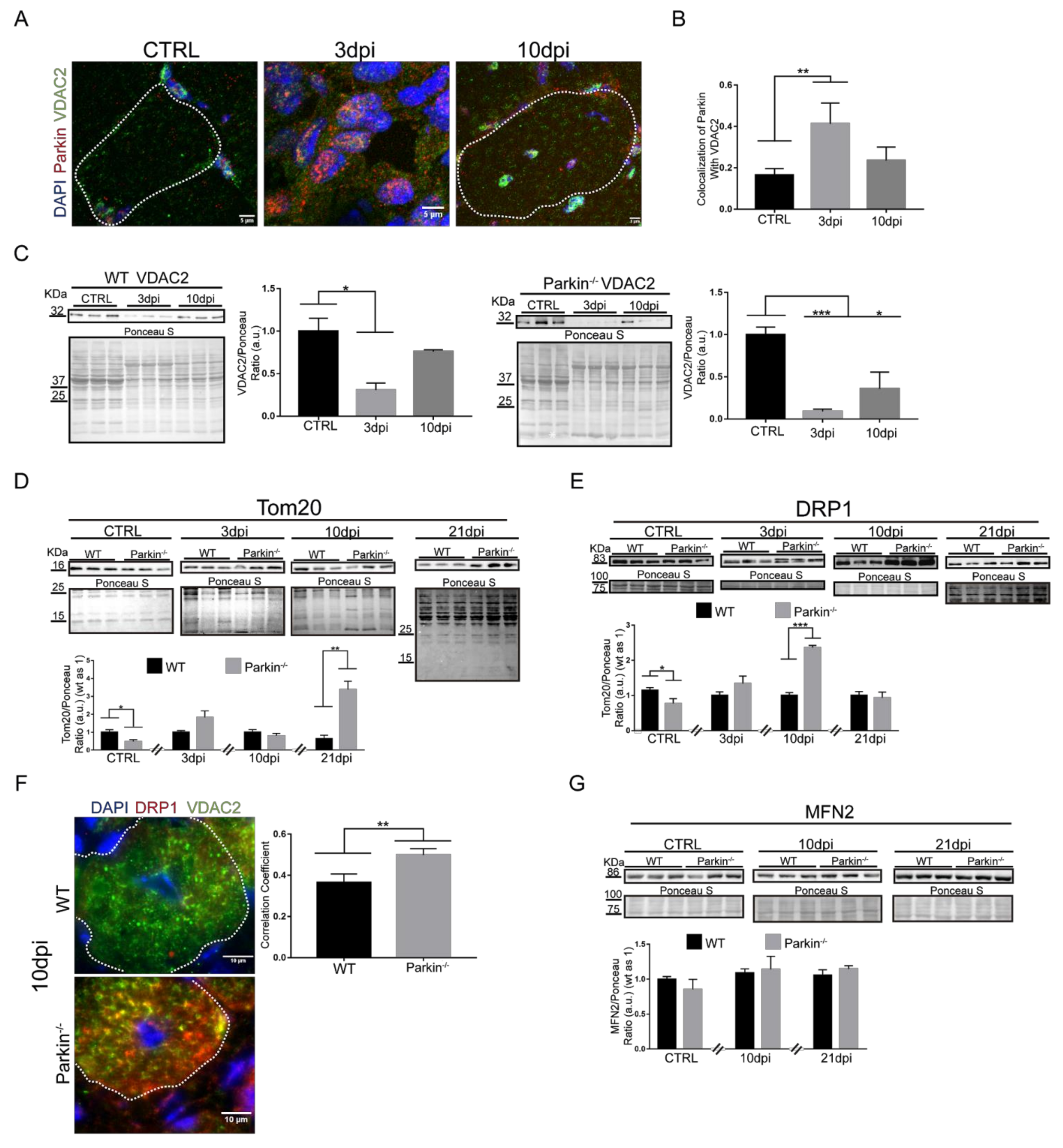
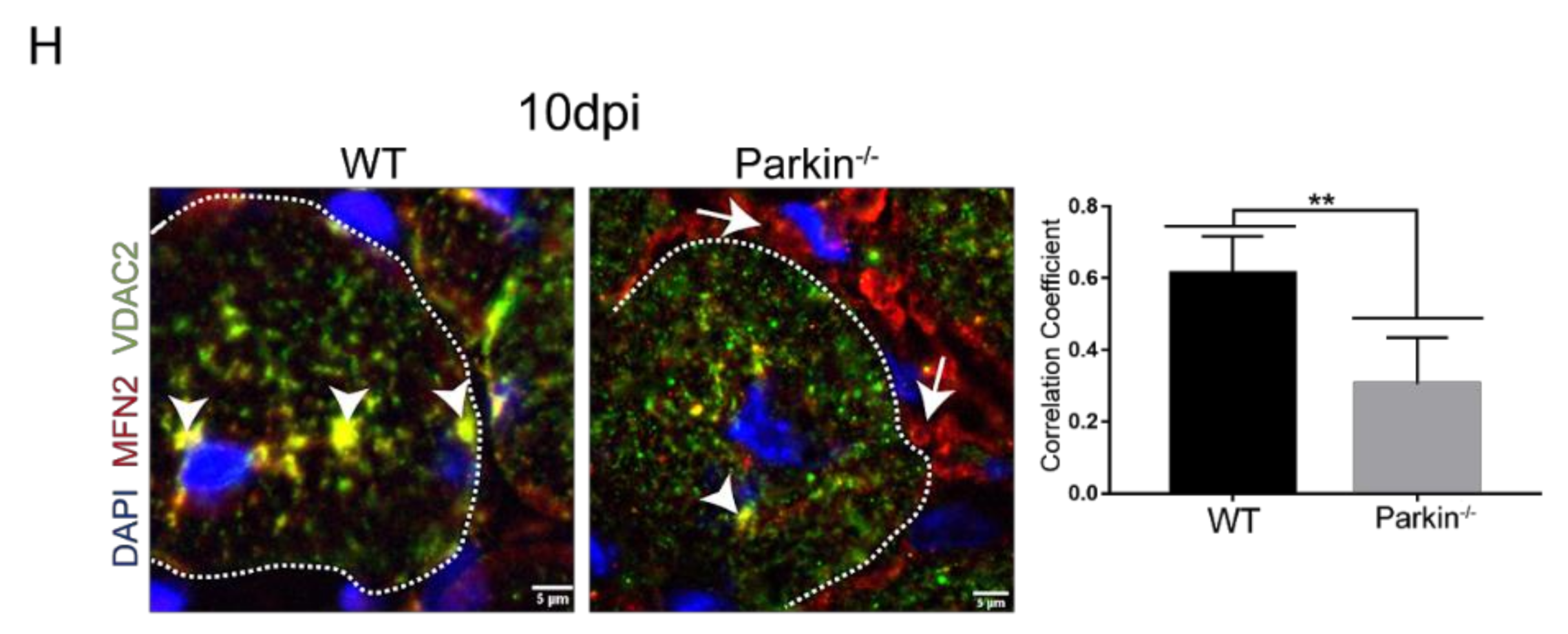
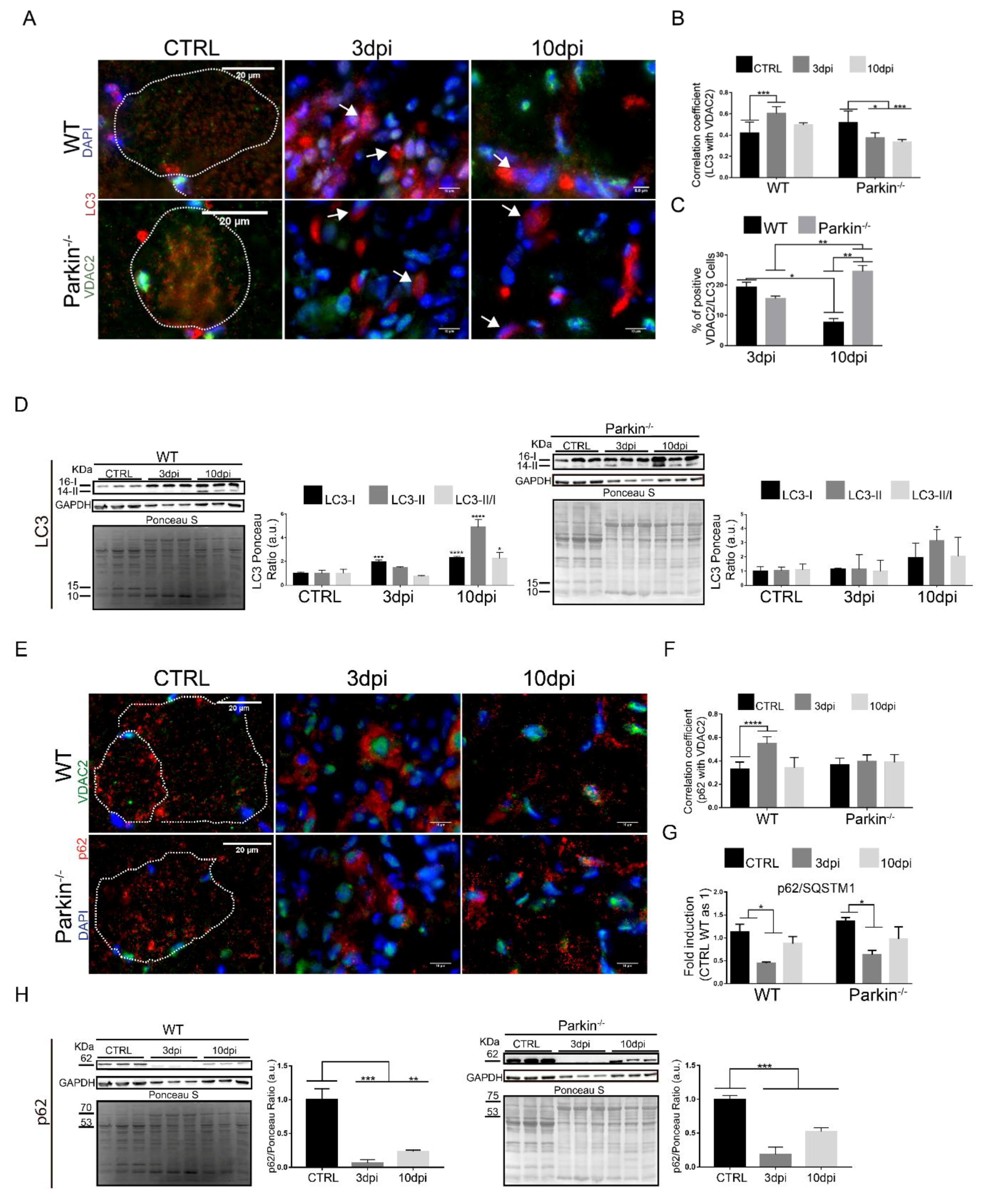
2.5. Parkin Deficiency Alters Mitochondrial Protein Levels during Skeletal Muscle Regeneration
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Cell Culture
4.3. Mitochondrial Fractionation
4.4. Mophological Analyzes
4.5. Immunofluorescence
4.6. Confocal Images
4.7. Western Blot
4.8. Real-Time PCR
4.9. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CSA | Cross-sectional area |
| CTRL | Control |
| CTX | Cardiotoxin |
| Dpi | Day post-injury |
| DRP1 | Dynamin-related protein 1 |
| Gas | Gastrocnemius |
| LC3-I | Microtubule-associated protein 1A/1B-light chain 3-I |
| LC3-II | Microtubule-associated protein 1A/1B-light chain 3-II |
| MFN2 | Mitofusin 2 |
| MuSCs | Muscle stem cells |
| Myh3 | Myosin heavy chain 3 |
| NCAM | Neural cell adhesion molecule |
| p62/SQSTM1 | Sequestosome 1 |
| Pax7 | Paired-box transcription factor 7 |
| TA | Tibialis Anterior |
| Tom20 | Translocase of outer membrane 20 |
| VDAC2 | Voltage dependent anion-selective channels 2 |
References
- Wen, Y.; Bi, P.; Liu, W.; Asakura, A.; Keller, C.; Kuang, S. Constitutive Notch activation upregulates Pax7 and promotes the self-renewal of skeletal muscle satellite cells. Mol. Cell. Biol. 2012, 32, 2300–2311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lepper, C.; Partridge, T.A.; Fan, C.-M. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development 2011, 138, 3639–3646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olguín, H.C.; Pisconti, A. Marking the tempo for myogenesis: Pax7 and the regulation of muscle stem cell fate decisions. J. Cell. Mol. Med. 2012, 16, 1013–1025. [Google Scholar] [CrossRef] [PubMed]
- Tidball, J.G. Regulation of muscle growth and regeneration by the immune system. Nat. Rev. Immunol. 2017, 17, 165. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, Y.; Gu, L.; Lan, M.; Liu, C.; Wang, M.; Su, Y.; Ge, M.; Wang, T.; Yu, Y. Islr regulates canonical Wnt signaling-mediated skeletal muscle regeneration by stabilizing Dishevelled-2 and preventing autophagy. Nat. Commun. 2018, 9, 5129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiaffino, S.; Rossi, A.C.; Smerdu, V.; Leinwand, L.A.; Reggiani, C. Developmental myosins: Expression patterns and functional significance. Skelet. Muscle 2015, 5, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baechler, B.L.; Bloemberg, D.; Quadrilatero, J. Mitophagy regulates mitochondrial network signaling, oxidative stress, and apoptosis during myoblast differentiation. Autophagy 2019, 15, 1606–1619. [Google Scholar] [CrossRef] [PubMed]
- Sin, J.; Andres, A.M.; Taylor, D.J.; Weston, T.; Hiraumi, Y.; Stotland, A.; Kim, B.J.; Huang, C.; Doran, K.S.; Gottlieb, R.A. Mitophagy is required for mitochondrial biogenesis and myogenic differentiation of C2C12 myoblasts. Autophagy 2016, 12, 369–380. [Google Scholar] [CrossRef] [Green Version]
- Lampert, M.A.; Orogo, A.M.; Najor, R.H.; Hammerling, B.C.; Leon, L.J.; Wang, B.J.; Kim, T.; Sussman, M.A.; Gustafsson, Å.B. BNIP3L/NIX and FUNDC1-mediated mitophagy is required for mitochondrial network remodeling during cardiac progenitor cell differentiation. Autophagy 2019, 15, 1182–1198. [Google Scholar] [CrossRef]
- Gustafsson, Å.B.; Dorn, G.W. Evolving and expanding the roles of mitophagy as a homeostatic and pathogenic process. Physiol. Rev. 2018, 99, 853–892. [Google Scholar] [CrossRef]
- Kubli, D.A.; Zhang, X.; Lee, Y.; Hanna, R.A.; Quinsay, M.N.; Nguyen, C.K.; Jimenez, R.; Petrosyan, S.; Murphy, A.N.; Gustafsson, Å.B. Parkin protein deficiency exacerbates cardiac injury and reduces survival following myocardial infarction. J. Biol. Chem. 2013, 288, 915–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panicker, N.; Dawson, V.L.; Dawson, T.M. Activation mechanisms of the E3 ubiquitin ligase parkin. Biochem. J. 2017, 474, 3075–3086. [Google Scholar] [CrossRef]
- Peker, N.; Donipadi, V.; Sharma, M.; McFarlane, C.; Kambadur, R. Loss of Parkin impairs mitochondrial function and leads to muscle atrophy. Am. J. Physiol. Cell Physiol. 2018, 315, C164–C185. [Google Scholar] [CrossRef] [Green Version]
- Mahdy, M.A.; Warita, K.; Hosaka, Y.Z. Early ultrastructural events of skeletal muscle damage following cardiotoxin-induced injury and glycerol-induced injury. Micron 2016, 91, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Gillon, A.; Sheard, P. Elderly mouse skeletal muscle fibres have a diminished capacity to upregulate NCAM production in response to denervation. Biogerontology 2015, 16, 811–823. [Google Scholar] [CrossRef]
- Ramadasan-Nair, R.; Gayathri, N.; Mishra, S.; Sunitha, B.; Mythri, R.B.; Nalini, A.; Subbannayya, Y.; Harsha, H.C.; Kolthur-Seetharam, U.; Bharath, M.M.S. Mitochondrial alterations and oxidative stress in an acute transient mouse model of muscle degeneration implications for muscular dystrophy and related muscle pathologies. J. Biol. Chem. 2014, 289, 485–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dogan, S.A.; Cerutti, R.; Beninca, C.; Brea-Calvo, G.; Jacobs, H.T.; Zeviani, M.; Szibor, M.; Viscomi, C. Perturbed redox signaling exacerbates a mitochondrial myopathy. Cell Metab. 2018, 28, 764–775.e765. [Google Scholar] [CrossRef] [Green Version]
- Servián-Morilla, E.; Cabrera-Serrano, M.; Rivas-Infante, E.; Carvajal, A.; Lamont, P.; Pelayo-Negro, A.; Ravenscroft, G.; Junckerstorff, R.; Dyke, J.; Fletcher, S. Altered myogenesis and premature senescence underlie human TRIM32-related myopathy. ActaNeuropathol. Commun. 2019, 7, 30. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, M.; Schüler, S.C.; Hüttner, S.S.; von Eyss, B.; von Maltzahn, J. Adult stem cells at work: Regenerating skeletal muscle. Cell. Mol. Life Sci. 2019, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Fadda, L.; Lombardi, R.; Soliveri, P.; Lauria, G.; Defazio, G.; Tagliavini, F. Skin nerve α-synuclein deposits in a parkinsonian patient with heterozygous parkin mutation. Parkinsonism Relat. Disord. 2018. [Google Scholar] [CrossRef]
- Pinto, M.; Nissanka, N.; Moraes, C.T. Lack of Parkin anticipates the phenotype and affects mitochondrial morphology and mtDNA levels in a mouse model of Parkinson’s Disease. J. Neurosci. 2018, 38, 1042–1053. [Google Scholar] [CrossRef] [PubMed]
- Romanello, V.; Sandri, M. Mitochondria quality control and muscle mass maintenance. Front. Physiol. 2016, 6, 422. [Google Scholar] [CrossRef] [PubMed]
- Garber, C.E.; Friedman, J.H. Effects of fatigue on physical activity and function in patients with Parkinson’s disease. Neurology 2003, 60, 1119–1124. [Google Scholar] [CrossRef]
- Stevens-Lapsley, J.; Kluger, B.M.; Schenkman, M. Quadriceps muscle weakness, activation deficits, and fatigue with Parkinson disease. Neurorehabil. Neural Repair. 2012, 26, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Gouspillou, G.; Godin, R.; Piquereau, J.; Picard, M.; Mofarrahi, M.; Mathew, J.; Purves-Smith, F.M.; Sgarioto, N.; Hepple, R.T.; Burelle, Y. Protective role of Parkin in skeletal muscle contractile and mitochondrial function. J. Physiol. 2018, 596, 2565–2579. [Google Scholar] [CrossRef]
- Yamamoto, M.; Legendre, N.P.; Biswas, A.A.; Lawton, A.; Yamamoto, S.; Tajbakhsh, S.; Kardon, G.; Goldhamer, D.J. Loss of MyoD and Myf5 in skeletal muscle stem cells results in altered myogenic programming and failed regeneration. Stem Cell Rep. 2018, 10, 956–969. [Google Scholar] [CrossRef] [Green Version]
- Murphy, M.M.; Keefe, A.C.; Lawson, J.A.; Flygare, S.D.; Yandell, M.; Kardon, G. Transiently active Wnt/β-catenin signaling is not required but must be silenced for stem cell function during muscle regeneration. Stem Cell Rep. 2014, 3, 475–488. [Google Scholar] [CrossRef] [Green Version]
- Rocheteau, P.; Gayraud-Morel, B.; Siegl-Cachedenier, I.; Blasco, M.A.; Tajbakhsh, S. A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell 2012, 148, 112–125. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, D.; Scimè, A. Mitochondrial Function in Muscle Stem Cell Fates. Front. Cell Dev. Biol. 2020, 8, 480. [Google Scholar] [CrossRef]
- Baldelli, S.; Aquilano, K.; Ciriolo, M. PGC-1α buffers ROS-mediated removal of mitochondria during myogenesis. Cell Death Dis. 2014, 5, e1515. [Google Scholar] [CrossRef] [Green Version]
- Le Moal, E.; Pialoux, V.; Juban, G.; Groussard, C.; Zouhal, H.; Chazaud, B.; Mounier, R. Redox control of skeletal muscle regeneration. Antioxid. Redox Signal. 2017, 27, 276–310. [Google Scholar] [CrossRef] [PubMed]
- Naghdi, S.; Hajnóczky, G. VDAC2-specific cellular functions and the underlying structure. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2016, 1863, 2503–2514. [Google Scholar] [CrossRef] [PubMed]
- Ivankovic, D.; Chau, K.Y.; Schapira, A.H.; Gegg, M.E. Mitochondrial and lysosomal biogenesis are activated following PINK 1/parkin-mediated mitophagy. J. Neurochem. 2016, 136, 388–402. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.W.; Erlich, A.T.; Crilly, M.J.; Hood, D.A. Parkin is required for exercise-induced mitophagy in muscle: Impact of aging. Am. J. Physiol.-Endocrinol. Metab. 2018, 315, E404–E415. [Google Scholar] [CrossRef]
- Rana, A.; Rera, M.; Walker, D.W. Parkin overexpression during aging reduces proteotoxicity, alters mitochondrial dynamics, and extends lifespan. Proc. Natl. Acad. Sci. USA 2013, 110, 8638–8643. [Google Scholar] [CrossRef] [Green Version]
- Goldberg, M.S.; Fleming, S.M.; Palacino, J.J.; Cepeda, C.; Lam, H.A.; Bhatnagar, A.; Meloni, E.G.; Wu, N.; Ackerson, L.C.; Klapstein, G.J. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J. Biol. Chem. 2003, 278, 43628–43635. [Google Scholar] [CrossRef] [Green Version]
- Dunn, K.W.; Kamocka, M.M.; McDonald, J.H. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 2011, 300, C723–C742. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esteca, M.V.; Severino, M.B.; Silvestre, J.G.; Palmeira dos Santos, G.; Tamborlin, L.; Luchessi, A.D.; Moriscot, A.S.; Gustafsson, Å.B.; Baptista, I.L. Loss of Parkin Results in Altered Muscle Stem Cell Differentiation during Regeneration. Int. J. Mol. Sci. 2020, 21, 8007. https://doi.org/10.3390/ijms21218007
Esteca MV, Severino MB, Silvestre JG, Palmeira dos Santos G, Tamborlin L, Luchessi AD, Moriscot AS, Gustafsson ÅB, Baptista IL. Loss of Parkin Results in Altered Muscle Stem Cell Differentiation during Regeneration. International Journal of Molecular Sciences. 2020; 21(21):8007. https://doi.org/10.3390/ijms21218007
Chicago/Turabian StyleEsteca, Marcos V., Matheus B. Severino, João G. Silvestre, Gustavo Palmeira dos Santos, Letícia Tamborlin, Augusto D. Luchessi, Anselmo S. Moriscot, Åsa B. Gustafsson, and Igor L. Baptista. 2020. "Loss of Parkin Results in Altered Muscle Stem Cell Differentiation during Regeneration" International Journal of Molecular Sciences 21, no. 21: 8007. https://doi.org/10.3390/ijms21218007





