The Status of Soil Microbiome as Affected by the Application of Phosphorus Biofertilizer: Fertilizer Enriched with Beneficial Bacterial Strains
Abstract
1. Introduction
2. Results
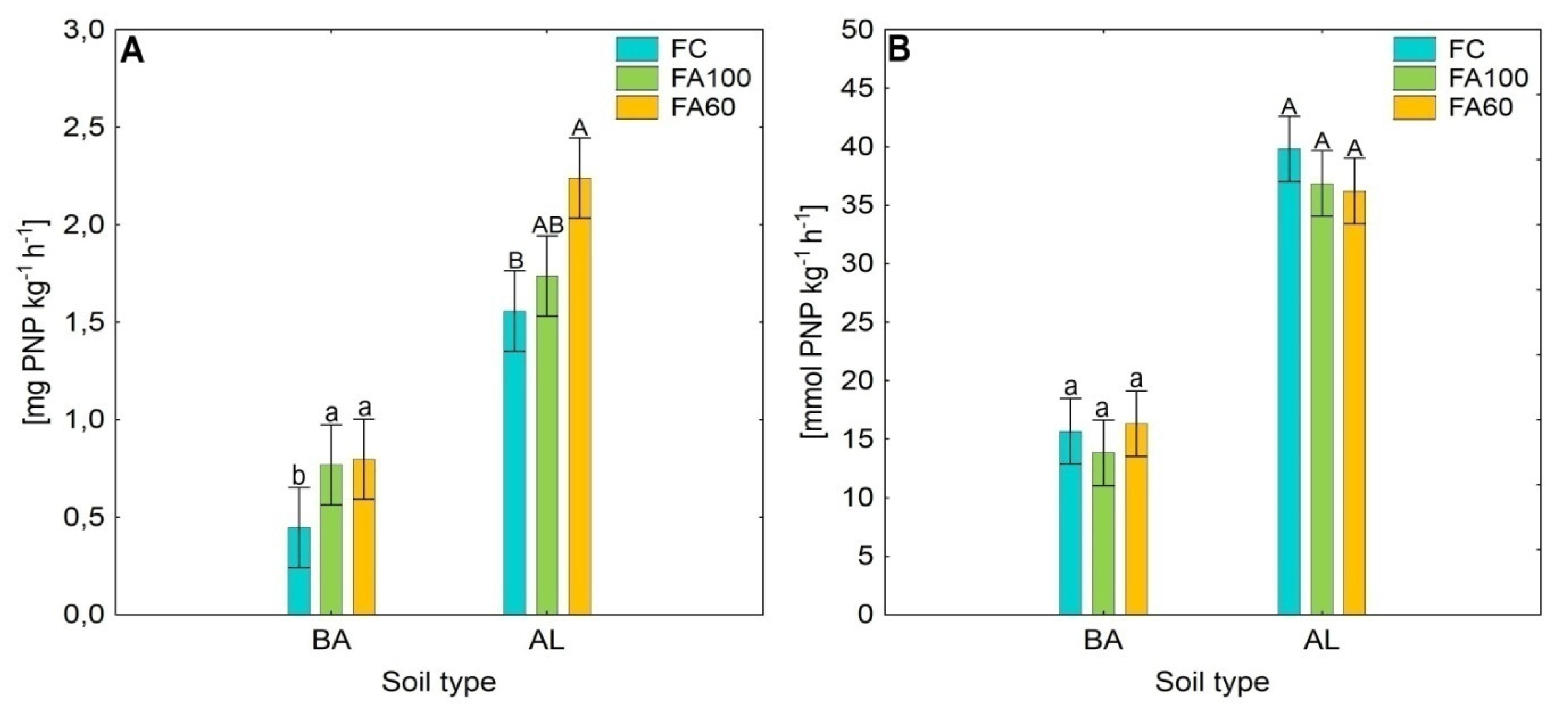
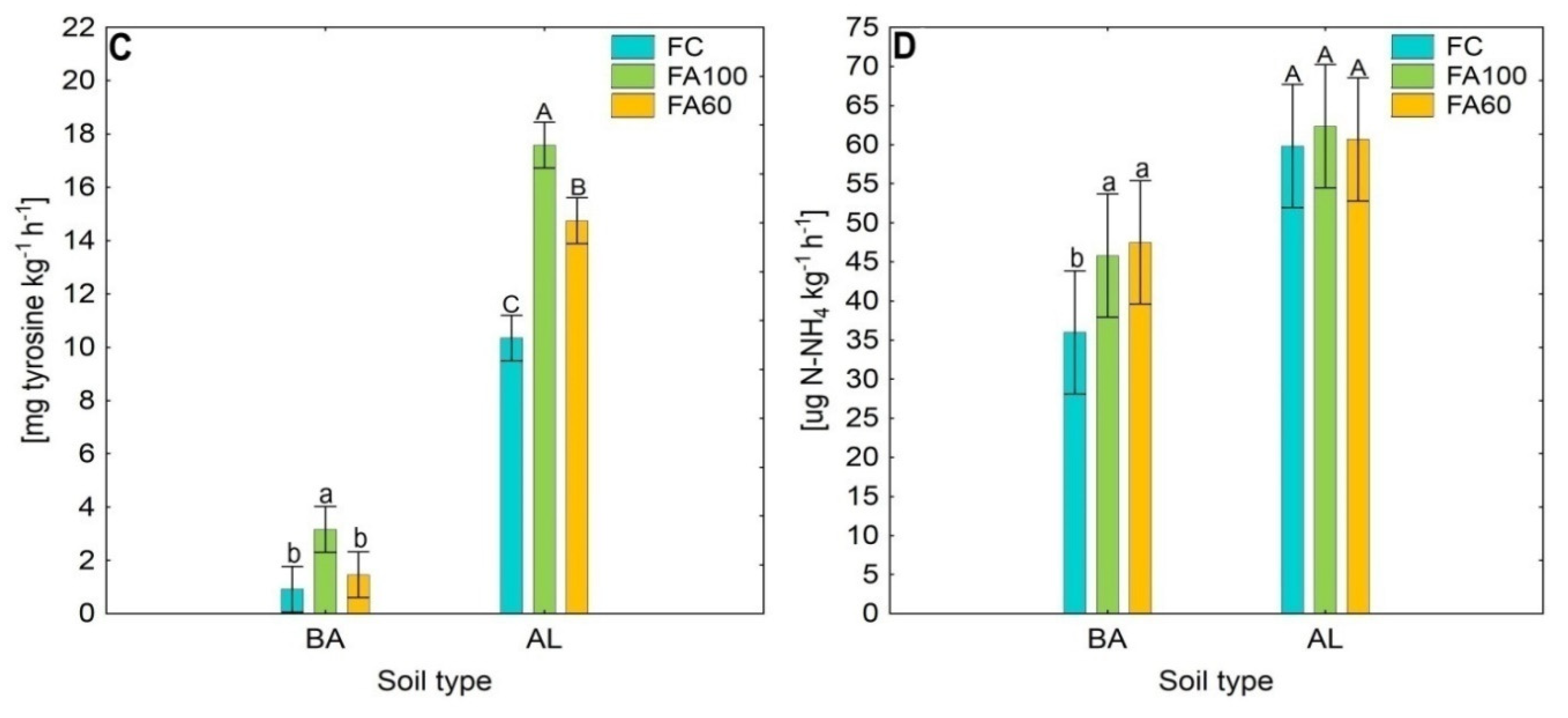
2.1. Enzymatic Activity
2.2. Community Level Physiological Profiles (CLPP)
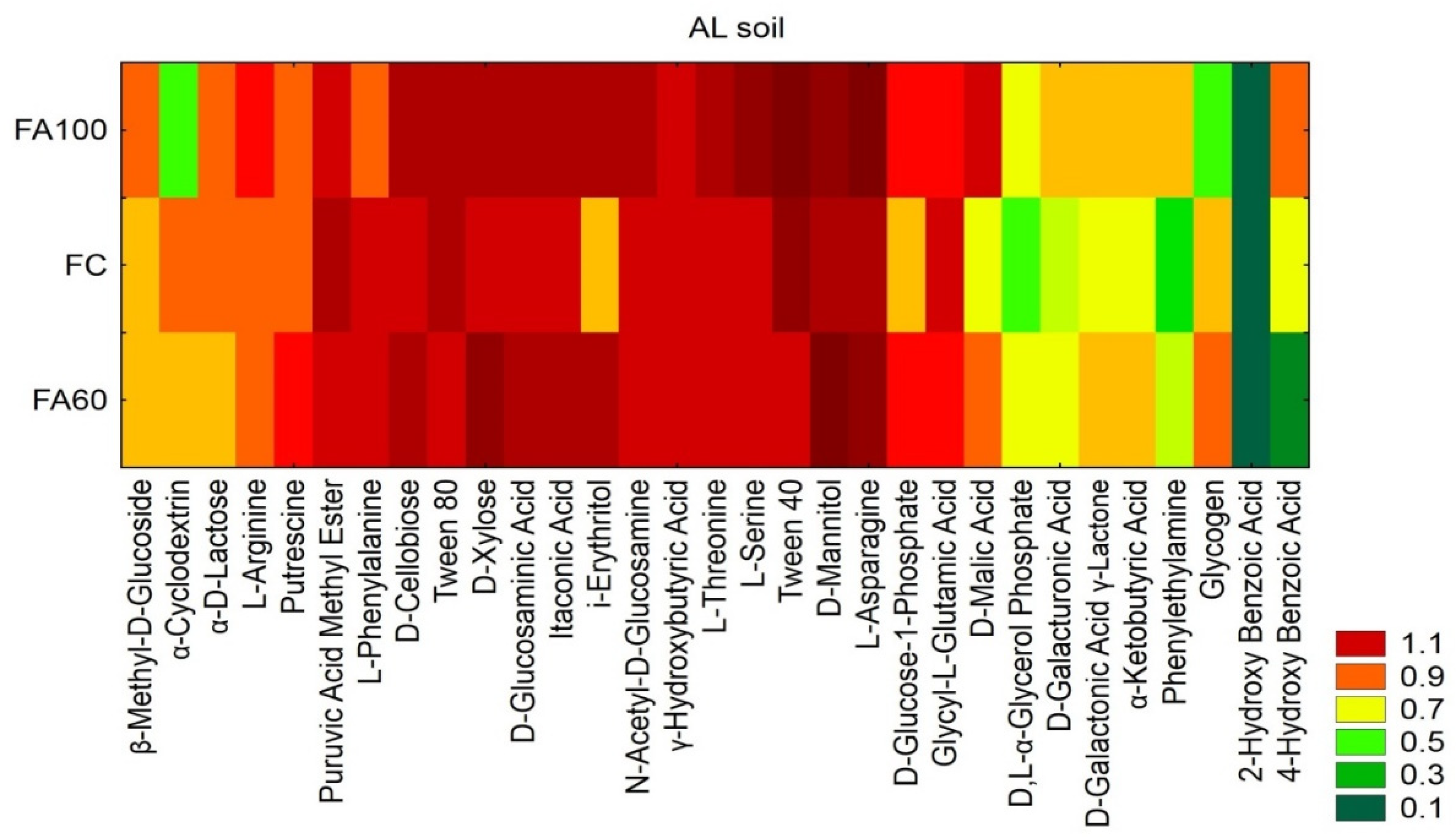
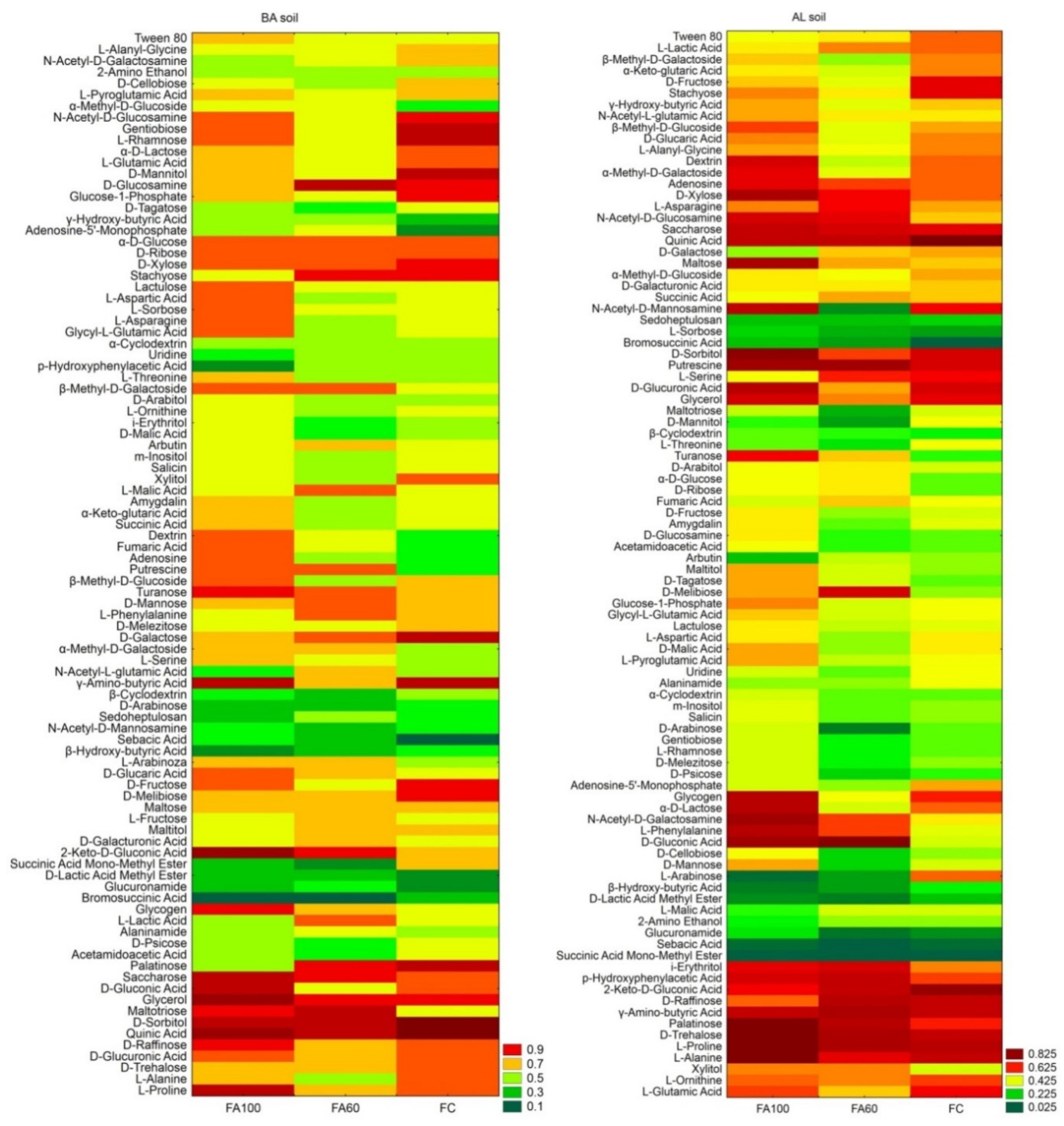
2.2.1. Metabolic Potential of the Soil Bacterial Community
2.2.2. Growth Pattern of Soil Fungal Community on Different Carbon Sources
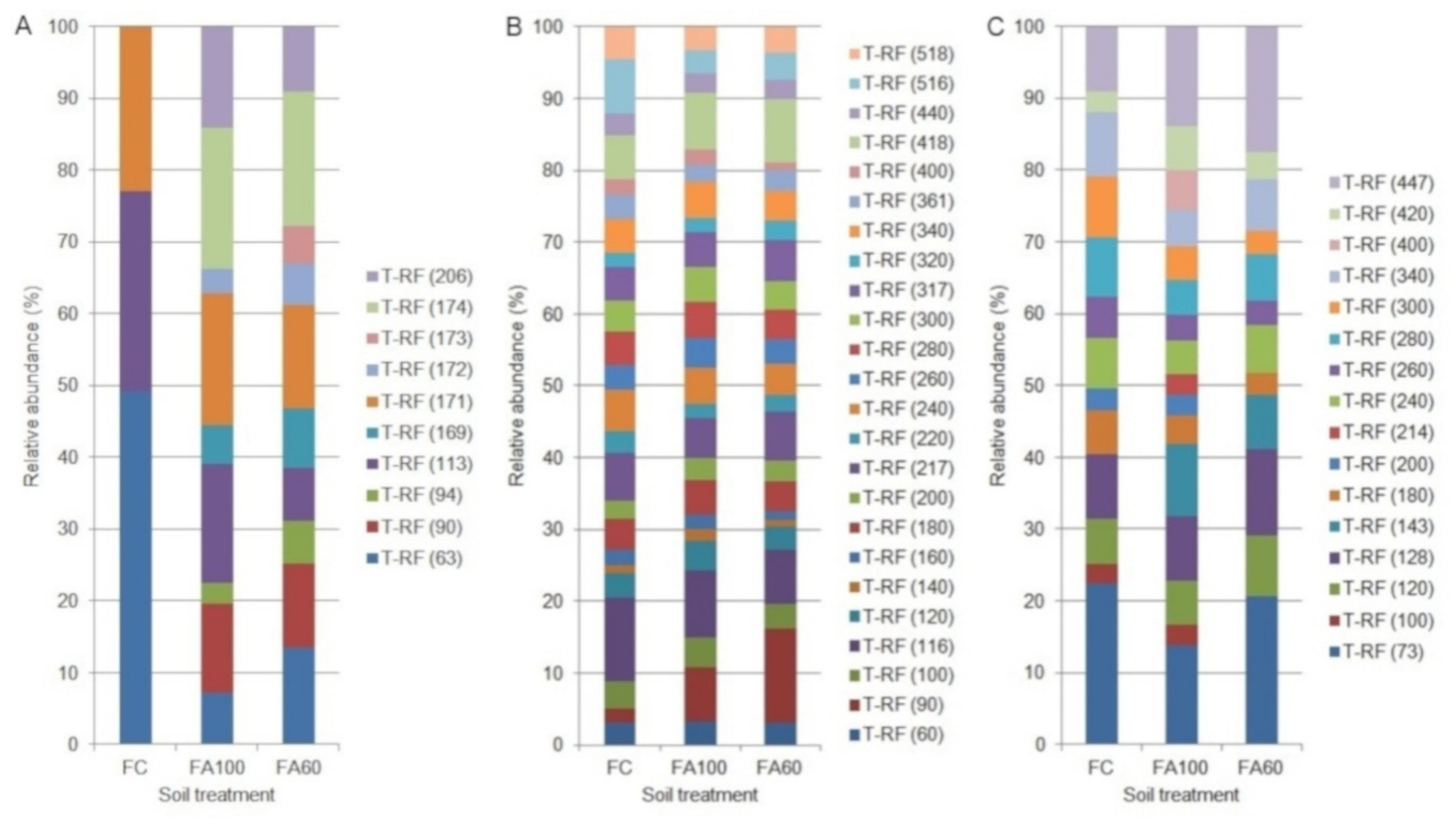
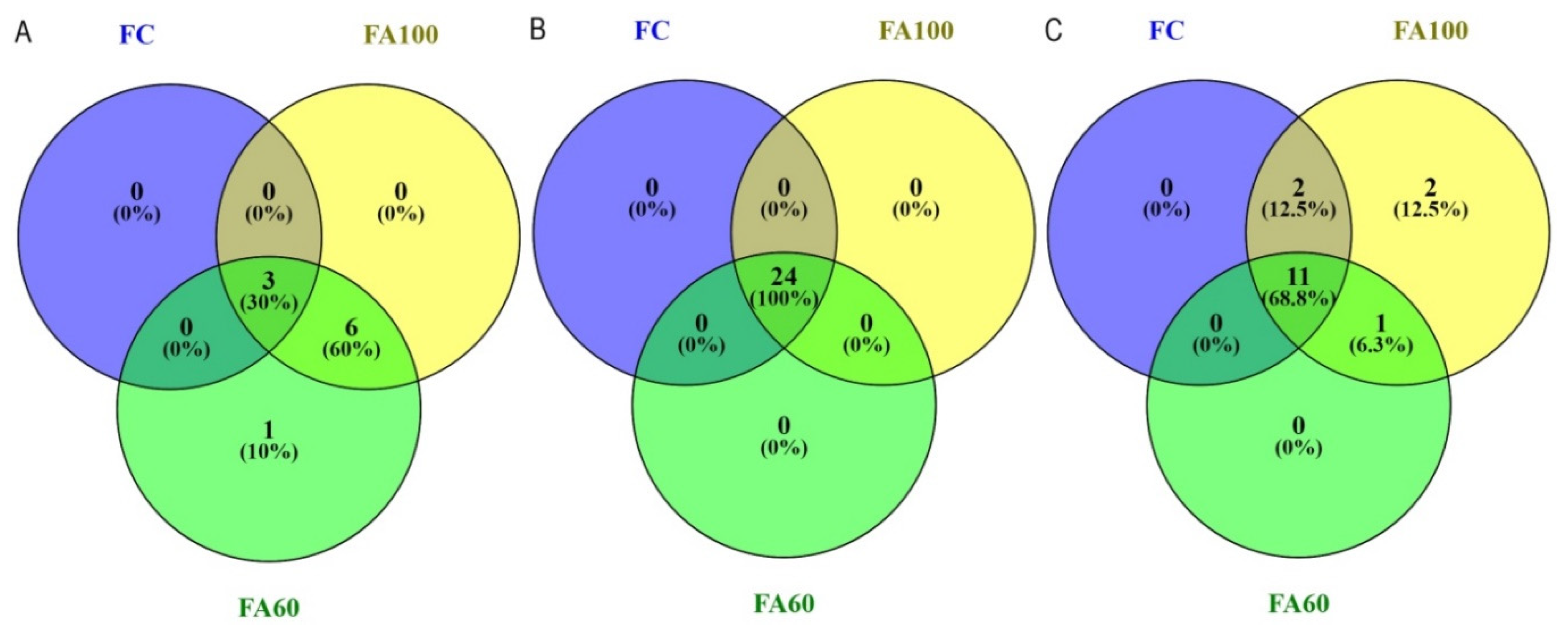
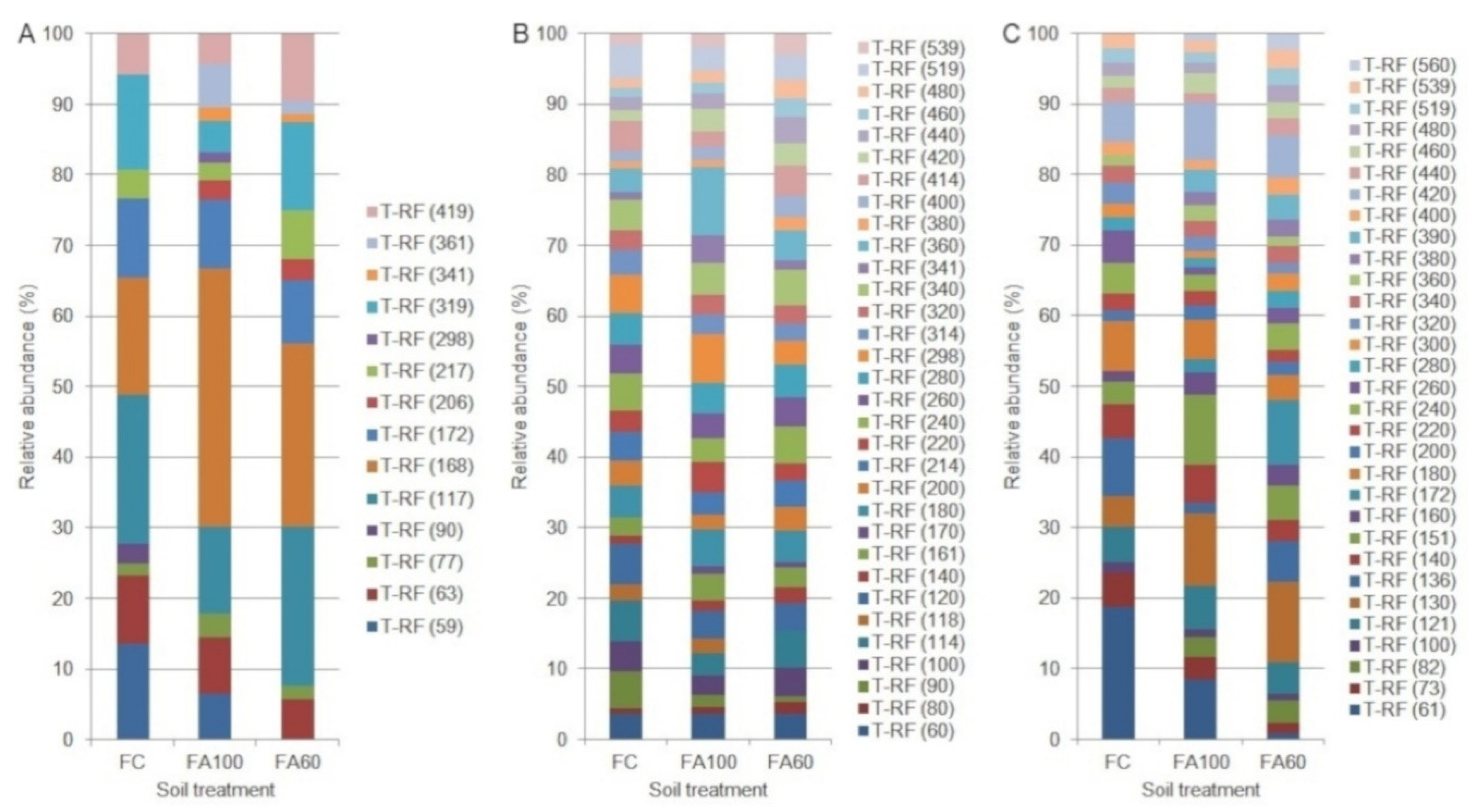
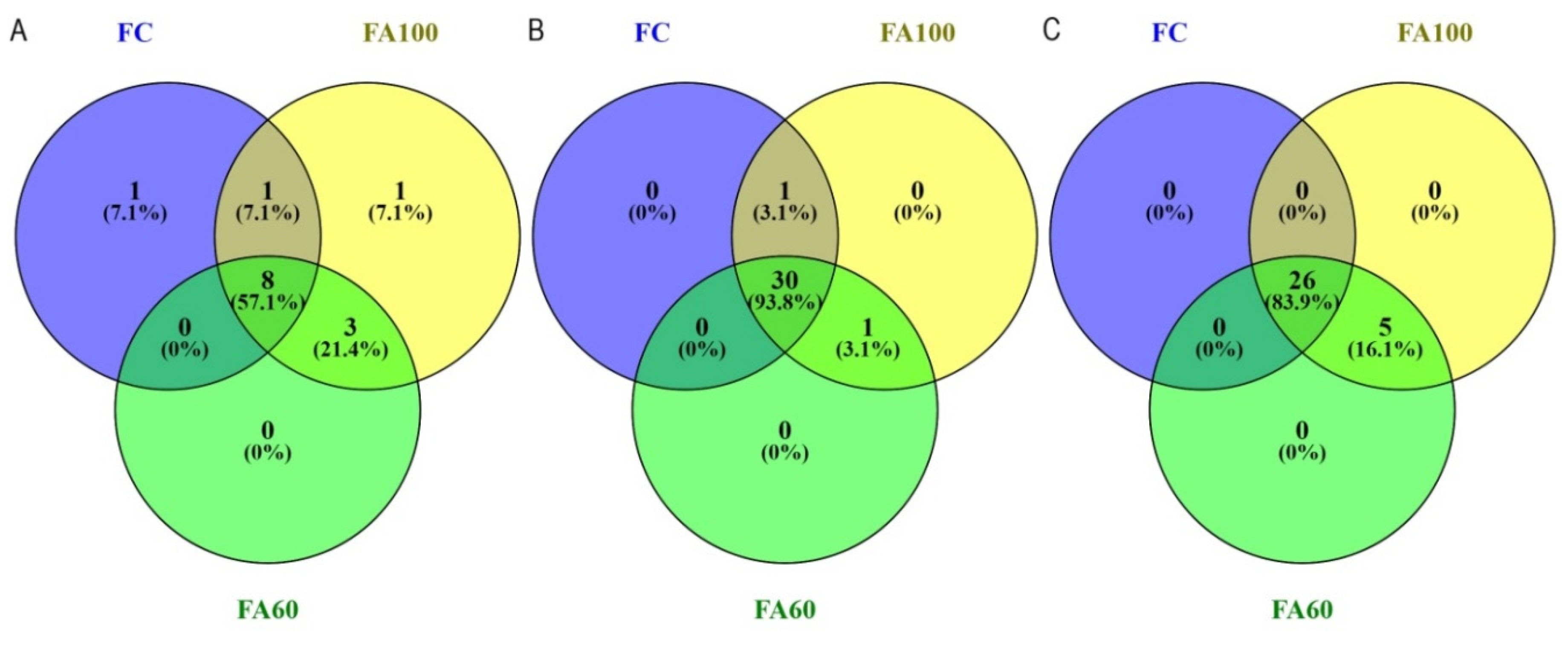
2.3. Terminal Restriction Fragment Length Polymorphism
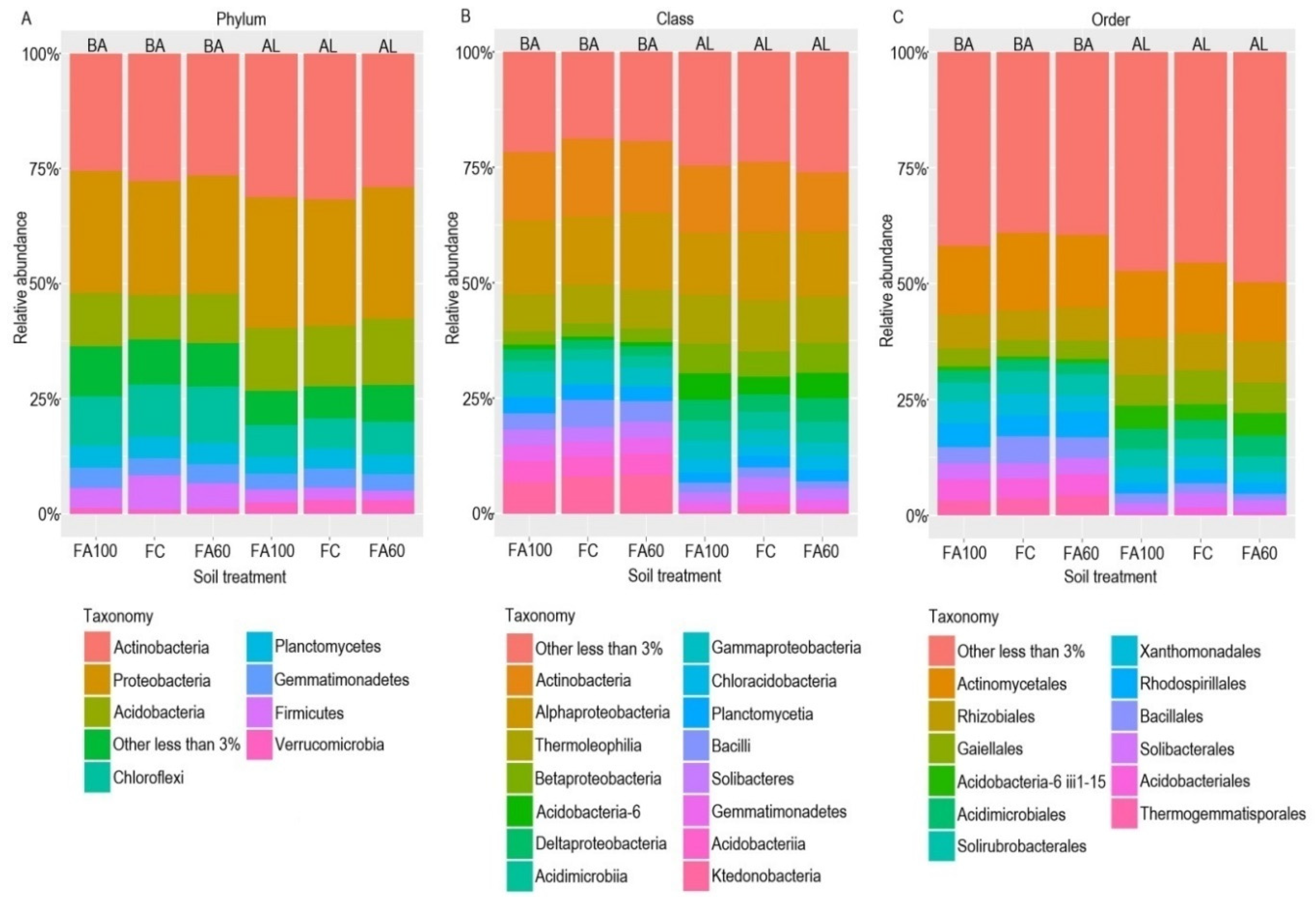
2.4. Next Generation Sequencing
2.4.1. Alpha Diversity
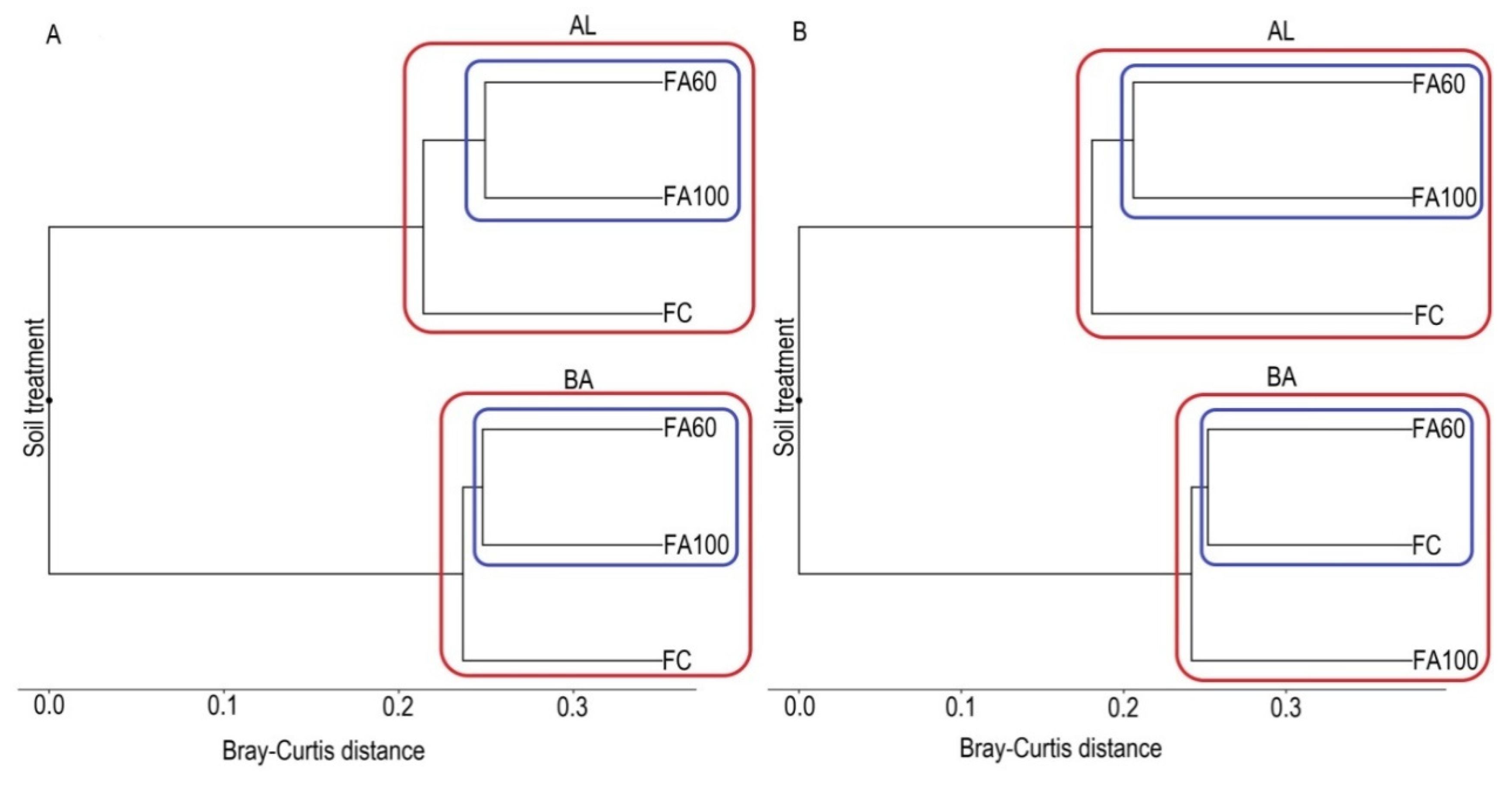
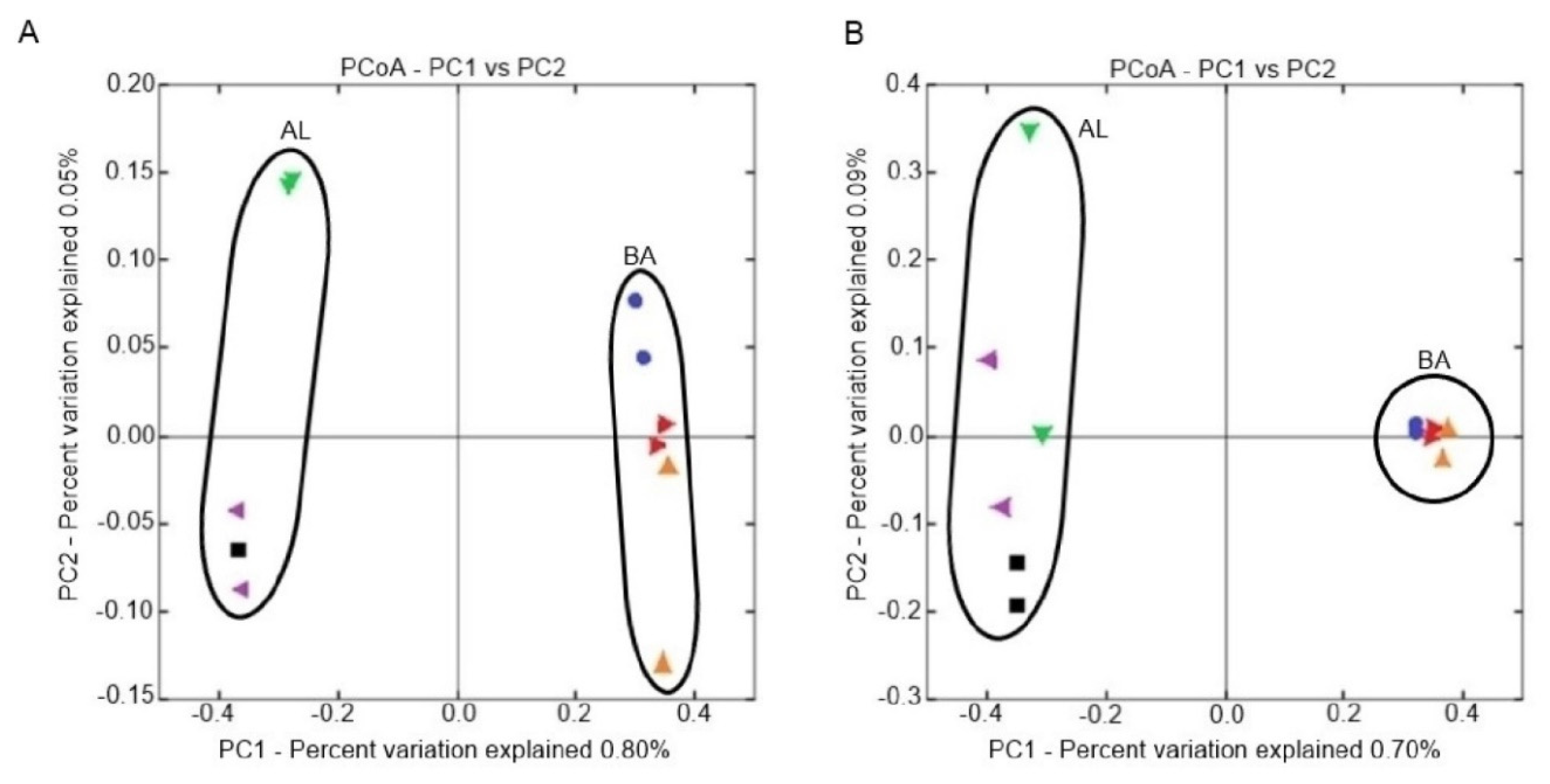
2.4.2. Beta Diversity
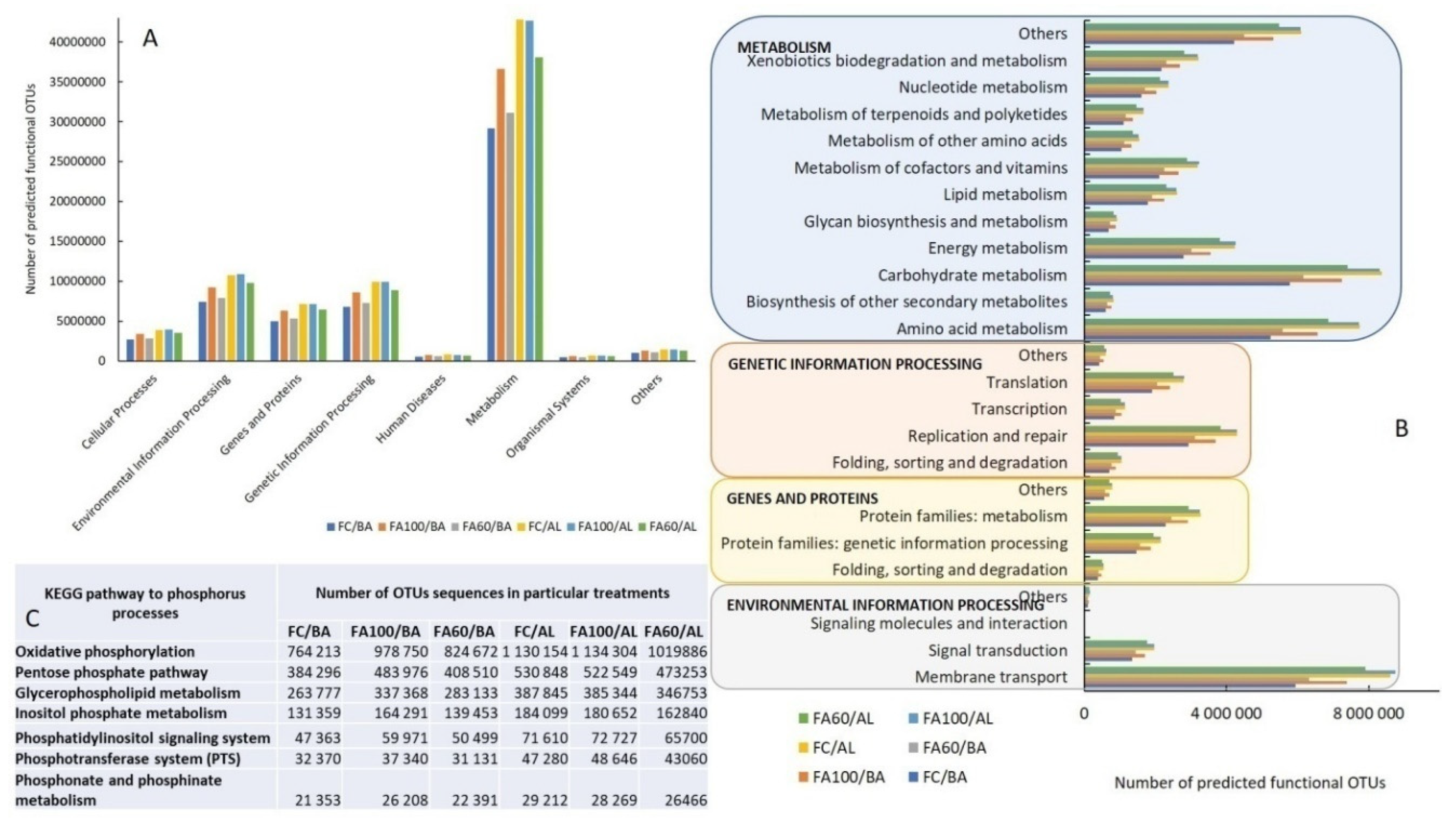
2.4.3. Functional Prediction of the Bacterial Community
2.4.4. Functional Guilds Prediction of the Fungal Community
3. Discussion
4. Materials and Methods
4.1. Study Site and Soil Sampling
4.2. Enzymatic Activities
4.3. Community Level Physiological Profiling (CLPP)
4.4. DNA Extraction
4.5. Multiplex Terminal Restriction Fragment Length Polymorphism (M-tRFLP)
4.6. Next Generation Sequencing (NGS)
4.7. Statistical and Bioinformatics Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Maron, P.-A.; Sarr, A.; Kaisermann, A.; Lévêque, J.; Mathieu, O.; Guigue, J.; Karimi, B.; Bernard, L.; Dequiedt, S.; Terrat, S.; et al. High microbial diversity promotes soil ecosystem functioning. Appl. Environ. Microbiol. 2018, 84, e02738. [Google Scholar] [CrossRef]
- Harrison, P.A.; Berry, P.M.; Simpson, G.; Haslett, J.R.; Blicharska, M.; Bucur, M.; Dunford, R.; Egoh, B.; Garcia-Llorente, M.; Geamănă, N.; et al. Linkages between biodiversity attributes and ecosystem services: A systematic review. Ecosyst. Serv. 2014, 9, 191–203. [Google Scholar] [CrossRef]
- Bindraban, P.S.; Dimkpa, C.; Nagarajan, L.; Roy, A.; Rabbinge, R. Revisiting fertilisers and fertilisation strategies for improved nutrient uptake by plants. Biol. Fertil. Soils 2015, 51, 897–911. [Google Scholar] [CrossRef]
- Chourasiya, D.; Sharma, M.P.; Maheshwari, H.S.; Ramesh, A.; Sharma, S.K.; Adhya, T.K. Microbial diversity and soil health in tropical agroecosystems. In Advances in Soil Microbiology: Recent Trends and Future Prospects Volume 2: Soil-Microbe-Plant Interaction Microorganisms for Sustainability 4; Adhya, T.K., Mishra, B.B., Annapurna, K., Verma, D.K., Kumar, U., Eds.; Springer: Singapore, 2017; pp. 19–35. ISBN 9789811073809. [Google Scholar]
- Frąc, M.; Hannula, S.E.; Belka, M.; Jȩdryczka, M. Fungal biodiversity and their role in soil health. Front. Microbiol. 2018, 9, 707. [Google Scholar] [CrossRef]
- Hendgen, M.; Hoppe, B.; Döring, J.; Friedel, M.; Kauer, R.; Frisch, M.; Dahl, A.; Kellner, H. Effects of different management regimes on microbial biodiversity in vineyard soils. Sci. Rep. 2018, 8, 9393. [Google Scholar] [CrossRef]
- White, P.J.; Brown, P.H. Plant nutrition for sustainable development and global health. Ann. Bot. 2010, 105, 1073–1080. [Google Scholar] [CrossRef]
- Chun, J.H.; Kim, S.; Arasu, M.V.; Al-Dhabi, N.A.; Chung, D.Y.; Kim, S.J. Combined effect of Nitrogen, Phosphorus and Potassium fertilizers on the contents of glucosinolates in rocket salad (Eruca sativa Mill.). Saudi J. Biol. Sci. 2017, 24, 436–443. [Google Scholar] [CrossRef]
- Peram, N.; Dayal, A.; Thomas, N.; Ramteke, P.W. Effect of different sources and levels of sulphur on growth and yield of maize (Zea mays L.) varieties. Int. J. Curr. Microbiol. Appl. Sci. 2018, 6, 1593–1600. [Google Scholar]
- Saif, S.; Khan, M.S.; Zaidi, A.; Ahmad, E. Role of Phosphate-Solubilizing Actinomycetes in plant growth promotion: Current perspective. In Phosphate Solubilizing Microorganisms: Principles and Application of Microphos Technology; Khan, M.S., Zaidi, A., Musarrat, J., Eds.; Springer: Cham, Germany, 2014; pp. 137–156. ISBN 9783319082165. [Google Scholar]
- Taliman, N.A.; Dong, Q.; Echigo, K.; Raboy, V.; Saneoka, H. Effect of Phosphorus Fertilization on the growth, photosynthesis, nitrogen fixation, mineral accumulation, seed yield, and seed quality of a soybean low-phytate line. Plants 2019, 8, 119. [Google Scholar] [CrossRef]
- Malhotra, H.; Vandana; Sharma, S.; Pandey, R. Phosphorus nutrition: Plant growth in response to deficiency and excess. In Plant Nutrients and Abiotic Stress Tolerance; Hasanuzzaman, M., Fujita, M., Oku, H., Nahar, K., Hawrylak-Nowak, B., Eds.; Springer: Singapore, 2018; pp. 171–190. ISBN 9789811090448. [Google Scholar]
- Geng, Y.; Cao, G.; Wang, L.; Wang, S. Effects of equal chemical fertilizer substitutions with organic manure on yield, dry matter, and nitrogen uptake of spring maize and soil nitrogen distribution. PLoS ONE 2019, 14, e02195. [Google Scholar] [CrossRef]
- Yousaf, M.; Li, J.; Lu, J.; Ren, T.; Cong, R.; Fahad, S.; Li, X. Effects of fertilization on crop production and nutrient-supplying capacity under rice-oilseed rape rotation system. Sci. Rep. 2017, 7, 1270. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Tian, H. Global nitrogen and phosphorus fertilizer use for agriculture production in the past half century: Shifted hot spots and nutrient imbalance. Earth Syst. Sci. Data 2017, 9, 181–192. [Google Scholar] [CrossRef]
- European Commission. Communication from the commission to the European Parliament, the council, the European economic and social committee and the committee of the regions. In A Farm to Fork Strategy for a Fair, Healthy and Environmentally-Friendly Food System; European Commission: Brussels, Belgium, 2020; ISBN 9789896540821. [Google Scholar]
- Bhardwaj, D.; Ansari, M.W.; Sahoo, R.K.; Tuteja, N. Biofertilizers function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop productivity. Microb. Cell Fact. 2014, 13, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Itelima, J.U.; Bang, W.J.; Onyimba, I.A.; Sila, M.D.; Egbere, O.J. A review: Biofertilizers–A key player in enhancing soil fertility and crop productivity. J. Microbiol. Biotechnol. Rep. 2018, 2, 22–28. [Google Scholar]
- García-Fraile, P.; Menéndez, E.; Rivas, R. Role of bacterial biofertilizers in agriculture and forestry. AIMS Bioeng. 2015, 2, 183–205. [Google Scholar] [CrossRef]
- Radhakrishnan, R.; Hashem, A.; Abd Allah, E.F. Bacillus: A biological tool for crop improvement through bio-molecular changes in adverse environments. Front. Physiol. 2017, 8, 667. [Google Scholar] [CrossRef]
- Liu, X.; Li, Q.; Li, Y.; Guan, G.; Chen, S. Paenibacillus strains with nitrogen fixation and multiple beneficial properties for promoting plant growth. PeerJ 2019, 7, e7445. [Google Scholar] [CrossRef]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.C. Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Fact. 2016, 15, 203. [Google Scholar] [CrossRef] [PubMed]
- Daud, N.S.; Mohd Din, A.R.J.; Rosli, M.A.; Azam, Z.M.; Othman, N.Z.; Sarmidi, M.R. Paenibacillus polymyxa bioactive compounds for agricultural and biotechnological applications. Biocatal. Agric. Biotechnol. 2019, 18, 101092. [Google Scholar] [CrossRef]
- Vinci, G.; Cozzolino, V.; Mazzei, P.; Monda, H.; Savy, D.; Drosos, M.; Piccolo, A. Effects of Bacillus amyloliquefaciens and different phosphorus sources on Maize plants as revealed by NMR and GC-MS based metabolomics. Plant Soil 2018, 429, 437–450. [Google Scholar] [CrossRef]
- Chen, L.; Liu, Y.; Wu, G.; Veronican Njeri, K.; Shen, Q.; Zhang, N.; Zhang, R. Induced maize salt tolerance by rhizosphere inoculation of Bacillus amyloliquefaciens SQR9. Physiol. Plant. 2016, 158, 34–44. [Google Scholar] [CrossRef]
- Saeid, A.; Prochownik, E.; Dobrowolska-Iwanek, J. Phosphorus solubilization by Bacillus species. Molecules 2018, 23, 2897. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Martínez-Hidalgo, P.; Ice, T.A.; Maymon, M.; Humm, E.A.; Nejat, N.; Sanders, E.R.; Kaplan, D.; Hirsch, A.M. Antifungal activity of Bacillus species against fusarium and analysis of the potential mechanisms used in biocontrol. Front. Microbiol. 2018, 9, 2363. [Google Scholar] [CrossRef]
- Gupta, N.; Debnath, S.; Sharma, S.; Sharma, P.; Purohit, J. Role of nutrients in controlling the plant diseases in sustainable agriculture. In Agriculturally Important Microbes for Sustainable Agriculture Volume 2: Applications in Crop Production and Protection; Meena, V.S., Mishra, P.K., Bisht, J.K., Pattanayak, A., Eds.; Springer: Singapore, 2017; pp. 217–262. ISBN 9789811053436. [Google Scholar]
- Mahdi, S.S.; Hassan, G.I.; Samoon, S.A.; Rather, H.A.; Dar, S.A.; Zehra, B. Bio-fertilizers in organic agriculture. J. Phytol. 2010, 2, 42–54. [Google Scholar]
- Schmidt, J.E.; Gaudin, A.C.M. What is the agronomic potential of biofertilizers for maize? A meta-analysis. FEMS Microbiol. Ecol. 2018, 94, 1–10. [Google Scholar] [CrossRef]
- Mondal, T.; Datta, J.K.; Mondal, N.K. Chemical fertilizer in conjunction with biofertilizer and vermicompost induced changes in morpho-physiological and bio-chemical traits of mustard crop. J. Saudi Soc. Agric. Sci. 2017, 16, 135–144. [Google Scholar] [CrossRef]
- Junier, P.; Junier, T.; Witzel, K.P. TRiFLe, a Program for in silico terminal restriction fragment length polymorphism analysis with user-defined sequence sets. Appl. Environ. Microbiol. 2008, 74, 6452–6456. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov (accessed on 28 July 2020).
- Conn, V.M.; Franco, C.M.M. Analysis of the Endophytic actinobacterial population in the roots of wheat (Triticum aestivum L.) by terminal restriction fragment length polymorphism and sequencing of 16S rRNA clones. Appl. Environ. Microbiol. 2004, 70, 1787–1794. [Google Scholar] [CrossRef]
- Gryta, A.; Frąc, M.; Oszust, K. Genetic and metabolic diversity of soil microbiome in response to exogenous organic matter amendments. Agronomy 2020, 10, 546. [Google Scholar] [CrossRef]
- Gryta, A.; Frąc, M.; Oszust, K. Community shift in structure and functions across soil profile in response to organic waste and mineral fertilization strategies. Appl. Soil Ecol. 2019, 143, 55–60. [Google Scholar] [CrossRef]
- Holík, L.; Hlisnikovský, L.; Honzík, R.; Trögl, J.; Burdová, H.; Popelka, J. Soil microbial communities and enzyme activities after long-term application of inorganic and organic fertilizers at different depths of the soil profile. Sustainability 2019, 11, 3251. [Google Scholar] [CrossRef]
- Mącik, M.; Gryta, A.; Frąc, M. Biofertilizers in agriculture: An overview on concepts, strategies and effects on soil microorganisms. In Advances in Agronomy; Sparks, D.L., Ed.; Academic Press: Cambridge, MA, USA; San Diego, CA, USA; Oxford, UK; London, UK, 2020; Volume 162, pp. 31–87. [Google Scholar]
- Dong, W.-Y.; Zhang, X.-Y.; Dai, X.-Q.; Fu, X.-L.; Yang, F.-T.; Liu, X.-Y.; Sun, X.-M.; Wen, X.-F.; Schaeffer, S. Changes in soil microbial community composition in response to fertilization of paddy soils in subtropical China. Appl. Soil Ecol. 2014, 84, 140–147. [Google Scholar] [CrossRef]
- Sharma, S.K.; Johri, B.N.; Ramesh, A.; Joshi, O.P.; Sai Prasad, S.V. Selection of plant growth-promoting Pseudomonas spp. That enhanced productivity of soybean-wheat cropping system in Central India. J. Microbiol. Biotechnol. 2011, 21, 1127–1142. [Google Scholar] [CrossRef]
- Walkiewicz, A.; Brzezińska, M.; Bieganowski, A.; Sas-Paszt, L.; Frąc, M. Early response of soil microbial biomass and activity to biofertilizer application in degraded brunic arenosol and abruptic luvisol of contrasting textures. Agronomy 2020, 10, 1347. [Google Scholar] [CrossRef]
- Santra, S.C.; Mallick, A.; Samal, A.C. Biofertilizer for bioremediation. In Recent Trends in Biofertitilizers; Pati, B.R., Mandal, S.M., Eds.; I K International Publishing House: New Delhi, India, 2016; pp. 205–234. [Google Scholar]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 2017, 8, 971. [Google Scholar] [CrossRef] [PubMed]
- Oszust, K.; Frąc, M.; Gryta, A.; Bilińska, N. The influence of ecological and conventional plant production systems on soil microbial quality under Hops (Humulus lupulus). Int. J. Mol. Sci. 2014, 15, 9907–9923. [Google Scholar] [CrossRef]
- Srinivasa Rao, C.; Grover, M.; Kundu, S.; Desai, S. Soil Enzymes. In Encyclopedia of Soil Science, 3rd ed.; Lal, R., Ed.; CRC Press: Boca Raton, FL, USA, 2017; pp. 2100–2107. ISBN 3120052906. [Google Scholar]
- Dotaniya, M.L.; Aparna, K.; Dotaniya, C.K.; Singh, M.; Regar, K.L. Role of Soil Enzymes in Sustainable Crop Production. In Enzymes in Food Biotechnology Production, Applications, and Future Prospects; Kuddus, M., Ed.; Academic Press: London, UK; San Diego, CA, USA; Cambridge, MA, USA; Oxford, UK, 2019; pp. 569–589. ISBN 9780128132807. [Google Scholar]
- Błońska, E.; Lasota, J.; Gruba, P. Enzymatic activity and stabilization of organic matter in soil with different detritus inputs. Soil Sci. Plant Nutr. 2017, 63, 242–247. [Google Scholar]
- Lin, S.; Wang, S.; Si, Y.; Yang, W.; Zhu, S.; Ni, W. Variations in eco-enzymatic stoichiometric and microbial characteristics in paddy soil as affected by long-term integrated organic-inorganic fertilization. PLoS ONE 2017, 12, e0189908. [Google Scholar] [CrossRef]
- Dal Cortivo, C.; Ferrari, M.; Visioli, G.; Lauro, M.; Fornasier, F.; Barion, G.; Panozzo, A.; Vamerali, T. Effects of seed-applied biofertilizers on rhizosphere biodiversity and growth of common wheat (Triticum aestivum L.) in the field. Front. Plant Sci. 2020, 11, 72. [Google Scholar] [CrossRef]
- Mukhtar, S.; Shahid, I.; Mehnaz, S.; Malik, K.A. Assessment of two carrier materials for phosphate solubilizing biofertilizers and their effect on growth of wheat (Triticum aestivum L.). Microbiol. Res. 2017, 205, 107–117. [Google Scholar] [CrossRef]
- Subhashini, D.V. Effect of NPK Fertilizers and Co-inoculation with phosphate-solubilizing arbuscular mycorrhizal fungus and potassium-mobilizing bacteria on growth, yield, nutrient acquisition, and quality of tobacco (Nicotiana tabacum L.). Commun. Soil Sci. Plant Anal. 2016, 47, 328–337. [Google Scholar] [CrossRef]
- Mengual, C.; Schoebitz, M.; Azcón, R.; Roldán, A. Microbial inoculants and organic amendment improves plant establishment and soil rehabilitation under semiarid conditions. J. Environ. Manag. 2014, 134, 1–7. [Google Scholar] [CrossRef]
- Devaraj, K.; Aathika, S.; Periyasamy, K.; Manickam Periyaraman, P.; Palaniyandi, S.; Subramanian, S. Production of thermostable multiple enzymes from Bacillus amyloliquefaciens KUB29. Nat. Prod. Res. 2018, 33, 1674–1677. [Google Scholar] [CrossRef] [PubMed]
- Raza, W.; Yang, W.; Shen, Q.R. Paenibacillus polymyxa: Antibiotics, hydrolytic enzymes and hazard assessment. J. Plant Pathol. 2008, 90, 419–430. [Google Scholar]
- Bradáčová, K.; Kandeler, E.; Berger, N.; Ludewig, U.; Neumann, G. Microbial consortia inoculants stimulate early growth of maize depending on nitrogen and phosphorus supply. Plant Soil Environ. 2020, 66, 105–112. [Google Scholar] [CrossRef]
- George, T.S.; Fransson, A.-M.; Hammond, J.P.; White, P.J. Phosphorus nutrition: Rhizosphere processes, plant response and adaptations. In Phosphorus in Action, Soil Biology 26; Bünemann, E., Oberson, A., Frossard, E., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 245–271. ISBN 978-3-642-15270-2. [Google Scholar]
- Vranova, V.; Rejsek, K.; Formanek, P. Proteolytic activity in soil: A review. Appl. Soil Ecol. 2013, 70, 23–32. [Google Scholar] [CrossRef]
- Escalas, A.; Hale, L.; Voordeckers, J.W.; Yang, Y.; Firestone, M.K.; Alvarez-Cohen, L.; Zhou, J. Microbial functional diversity: From concepts to applications. Ecol. Evol. 2019, 9, 12000–12016. [Google Scholar] [CrossRef]
- Gałązka, A.; Gawryjołek, K.; Grządziel, J.; Frąc, M.; Księżak, J. Microbial community diversity and the interaction of soil under maize growth in different cultivation techniques. Plant.Soil Environ. 2017, 63, 264–270. [Google Scholar]
- Gryta, A.; Frąc, M.; Oszust, K. The Application of the Biolog EcoPlate approach in ecotoxicological evaluation of dairy sewage sludge. Appl. Biochem. Biotechnol. 2014, 174, 1434–1443. [Google Scholar] [CrossRef]
- Muńiz, S.; Lacarta, J.; Pata, M.P.; Jiménez, J.J.; Navarro, E. Analysis of the diversity of substrate utilisation of soil bacteria exposed to Cd and earthworm activity using generalised additive models. PLoS ONE 2014, 9, e85057. [Google Scholar] [CrossRef]
- Floch, C.; Chevremont, A.C.; Joanico, K.; Capowiez, Y.; Criquet, S. Indicators of pesticide contamination: Soil enzyme compared to functional diversity of bacterial communities via Biolog® Ecoplates. Eur. J. Soil Biol. 2011, 47, 256–263. [Google Scholar] [CrossRef]
- Frąc, M.; Oszust, K.; Lipiec, J. Community level physiological profiles (CLPP), characterization and microbial activity of soil amended with dairy sewage sludge. Sensors 2012, 12, 3253–3268. [Google Scholar] [CrossRef] [PubMed]
- Stefanowicz, A. The biolog plates technique as a tool in ecological studies of microbial communities. Pol. J. Environ. Stud. 2006, 15, 669–676. [Google Scholar]
- Yang, M.; Yang, D.; Yu, X. Soil microbial communities and enzyme activities in sea-buckthorn (Hippophae rhamnoides) plantation at different ages. PLoS ONE 2018, 13, e0190959. [Google Scholar] [CrossRef]
- Siczek, A.; Frąc, M.; Gryta, A.; Kalembasa, S.; Kalembasa, D. Variation in soil microbial population and enzyme activities under faba bean as affected by pentachlorophenol. Appl. Soil Ecol. 2020, 150, 103466. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, X.; Jiang, Q.; Shen, G.; Ding, W. Legacy effects of continuous chloropicrin-fumigation for 3-years on soil microbial community composition and metabolic activity. AMB Express 2017, 7, 178. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Kumar, D.; Sharma, P. Effect of organics, biofertilizers and crop residue application on soil microbial activity in rice-wheat and rice-wheat mungbean cropping systems in the Indo-Gangetic plains. Cogent Geosci. 2015, 1, 1085296. [Google Scholar] [CrossRef]
- Chaudhry, V.; Rehman, A.; Mishra, A.; Chauhan, P.S.; Nautiyal, C.S. Changes in bacterial community structure of agricultural land due to long-term organic and chemical amendments. Microb. Ecol. 2012, 64, 450–460. [Google Scholar] [CrossRef]
- Cao, X.; Ma, Q.; Zhong, C.; Yang, X.; Zhu, L.; Zhang, J.; Jin, Q.; Wu, L. Elevational variation in soil amino acid and inorganic nitrogen concentrations in Taibai Mountain, China. PLoS ONE 2016, 11, e0157979. [Google Scholar] [CrossRef]
- Vranova, V.; Zahradnickova, H.; Janous, D.; Skene, K.R.; Matharu, A.S.; Rejsek, K.; Formanek, P. The significance of D-amino acids in soil, fate and utilization by microbes and plants: Review and identification of knowledge gaps. Plant Soil 2012, 354, 21–39. [Google Scholar] [CrossRef]
- Cheng, M.; Zeng, G.; Huang, D.; Yang, C.; Lai, C.; Zhang, C.; Liu, Y. Tween 80 surfactant-enhanced bioremediation: Toward a solution to the soil contamination by hydrophobic organic compounds. Crit. Rev. Biotechnol. 2017, 38, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Pei, X.H.; Zhan, X.H.; Wang, S.M.; Lin, Y.S.; Zhou, L.X. Effects of a Biosurfactant and a synthetic surfactant on Phenanthrene degradation by a Sphingomonas strain. Pedosphere 2010, 20, 771–779. [Google Scholar] [CrossRef]
- Da Silva Lima, L.; Olivares, F.L.; Rodrigues de Oliveira, R.; Vega, M.R.G.; Aguiar, N.O.; Canellas, L.P. Root exudate profiling of maize seedlings inoculated with Herbaspirillum seropedicaeand humic acids. Chem. Biol. Technol. Agric. 2014, 1, 23. [Google Scholar] [CrossRef]
- Gunina, A.; Kuzyakov, Y. Sugars in soil and sweets for microorganisms: Review of origin, content, composition and fate. Soil Biol. Biochem. 2015, 90, 87–100. [Google Scholar] [CrossRef]
- Lipiec, J.; Frąc, M.; Brzezińska, M.; Turski, M.; Oszust, K. Linking microbial enzymatic activities and functional diversity of soil around earthworm burrows and casts. Front. Microbiol. 2016, 7, 1361. [Google Scholar] [CrossRef]
- Pimentel, T.; Marcelino, J.; Ricardo, F.; Soares, A.M.V.M.; Calado, R. Bacterial communities 16S rDNA fingerprinting as a potential tracing tool for cultured seabass Dicentrarchus labrax. Sci. Rep. 2017, 7, 11862. [Google Scholar] [CrossRef]
- Paul, D.; Kumar, S.; Mishra, M.; Parab, S.; Banskar, S.; Shouche, Y.S. Molecular genomic techniques for identification of soil microbial community structure and dynamics. In Advances in Soil Microbiology: Recent Trends and Future Prospects, Microorganisms for Sustainability 3; Adhya, T.K., Ed.; Springer Nature: Singapore, 2018; pp. 9–33. ISBN 9789811061783. [Google Scholar]
- Gryta, A.; Frąc, M. Methodological aspects of multiplex terminal restriction fragment length polymorphism-technique to describe the genetic diversity of soil bacteria, archaea and fungi. Sensors 2020, 20, 3292. [Google Scholar] [CrossRef]
- Trabelsi, D.; Ben Ammar, H.; Mengoni, A.; Mhamdi, R. Appraisal of the crop-rotation effect of rhizobial inoculation on potato cropping systems in relation to soil bacterial communities. Soil Biol. Biochem. 2012, 54, 1–6. [Google Scholar] [CrossRef]
- Soliman, T.; Yang, S.Y.; Yamazaki, T.; Jenke-Kodama, H. Profiling soil microbial communities with next-generation sequencing: The influence of DNA kit selection and technician technical expertise. PeerJ 2017, 5, e4178. [Google Scholar] [CrossRef]
- Feng, G.; Xie, T.; Wang, X.; Bai, J.; Tang, L.; Zhao, H.; Wei, W.; Wang, M.; Zhao, Y. Metagenomic analysis of microbial community and function involved in cd-contaminated soil. BMC Microbiol. 2018, 18, 11. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Y.; Zhao, L.; Zhang, W.; Liu, L. Change of soil microbial community under long-term fertilization in a reclaimed sandy agricultural ecosystem. PeerJ 2019, 7, e6497. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Zhou, J.; Ongena, M.; Liu, W.; Wei, D.; Zhao, B.; Guan, D.; Jiang, X.; Li, J. Effect of long-term fertilization strategies on bacterial community composition in a 35-year field experiment of Chinese Mollisols. AMB Express 2018, 8, 20. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Wang, J.; Wang, C.; Wang, Q. The 19-years inorganic fertilization increased bacterial diversity and altered bacterial community composition and potential functions in a paddy soil. Appl. Soil Ecol. 2019, 144, 60–67. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, C.; Yu, W.W.; Turak, A.; Chen, D.; Huang, Y.; Ao, J.; Jiang, Y.; Huang, Z. Effects of nitrogen and phosphorus inputs on soil bacterial abundance, diversity, and community composition in chinese fir plantations. Front. Microbiol. 2018, 9, 1543. [Google Scholar] [CrossRef] [PubMed]
- Itävaara, M.; Salavirta, H.; Marjamaa, K.; Ruskeeniemi, T. Geomicrobiology and metagenomics of terrestrial deep subsurface microbiomes. In Advances in Applied Microbiology; Academic Press: Cambridge, MA, USA; San Diego, CA, USA; Oxford, UK; London, UK, 2016; Volume 94, pp. 1–77. [Google Scholar]
- Deng, J.; Yin, Y.; Zhu, W.; Zhou, Y. Variations in soil bacterial community diversity and structures among different revegetation types in the Baishilazi nature reserve. Front. Microbiol. 2018, 9, 2874. [Google Scholar] [CrossRef]
- Normand, P.; Benson, D.R.; Berry, A.M.; Tisa, L.S. The Family Frankiaceae. In The Prokaryotes: Actinobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 339–356. ISBN 9783642301384. [Google Scholar]
- De Souza, J.A.M.; Alves, L.M.C.; de Mello Varani, A.; de Macedo Lemos, E.G. The Family Bradyrhizobiaceae. In The Prokaryotes: Alphaproteobacteria and Betaproteobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 135–154. ISBN 9783642301971. [Google Scholar]
- Zhou, S.; Han, L.; Wang, Y.; Yang, G.; Li, Z.; Hu, P. Azospirillum humicireducens sp. nov., a nitrogen-fixing bacterium isolated from a microbial fuel cell. Int. J. Syst. Evol. Microbiol. 2013, 63, 2618–2624. [Google Scholar] [CrossRef] [PubMed]
- Kerou, M.; Offre, P.; Valledor, L.; Abby, S.S.; Melcher, M.; Nagler, M.; Weckwerth, W.; Schleper, C. Proteomics and comparative genomics of Nitrososphaera viennensis reveal the core genome and adaptations of archaeal ammonia oxidizers. Proc. Natl. Acad. Sci. USA 2016, 113, E7937–E7946. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Liang, Y.; Huang, D. Organic farming improves soil microbial abundance and diversity under greenhouse condition: A case study in Shanghai (Eastern China). Sustainability 2018, 10, 3825. [Google Scholar] [CrossRef]
- Oren, A.; Xu, X.-W. The Family Hyphomicrobiaceae. In The Prokaryotes: Alphaproteobacteria and Betaproteobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 247–281. ISBN 9783642301971. [Google Scholar]
- Kuever, J. The Family Syntrophobacteraceae. In The Prokaryotes: Deltaproteobacteria and Epsilonproteobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 289–299. ISBN 9783642390449. [Google Scholar]
- Taylor, T.N.; Krings, M.; Taylor, E.L. Ascomycota. In Fossil Fungi; Taylor, T.N., Krings, M., Taylor, E.L., Eds.; Academic Press: London, UK; San Diego, CA, USA; Waltham, MA, USA; Oxford, UK, 2015; pp. 129–171. ISBN 9780123877314. [Google Scholar]
- Ellegaard-Jensen, L.; Aamand, J.; Kragelund, B.B.; Johnsen, A.H.; Rosendahl, S. Strains of the soil fungus Mortierella show different degradation potentials for the phenylurea herbicide diuron. Biodegradation 2013, 24, 765–774. [Google Scholar] [CrossRef]
- Nakagawa, A.; Osawa, S.; Hirata, T.; Yamagishi, Y.; Hosoda, J.; Horikoshi, T. 2,4-Dichlorophenol degradation by the soil fungus Mortierella sp. Biosci. Biotechnol. Biochem. 2006, 70, 525–527. [Google Scholar] [CrossRef]
- Tamayo-Velez, A.; Osorio, N.W. Co-inoculation with an arbuscular mycorrhizal fungus and a phosphate-solubilizing fungus promotes the plant growth and phosphate uptake of avocado plantlets in a nursery. Botany 2017, 95, 539–545. [Google Scholar] [CrossRef]
- Li, F.; Chen, L.; Redmile-Gordon, M.; Zhang, J.; Zhang, C.; Ning, Q.; Li, W. Mortierella elongata’s roles in organic agriculture and crop growth promotion in a mineral soil. Land Degrad. Dev. 2018, 29, 1642–1651. [Google Scholar] [CrossRef]
- Osorio, N.W. Effectiveness of Phosphate Solubilizing Microorganism in Increasing Plant Phosphate Uptake and Growth in Tropical Soils. In Bacteria in Agrobiology: Plant Nutrient Management; Maheshwari, D.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 65–80. ISBN 9783642210617. [Google Scholar]
- Osorio, N.W.; Habte, M. Soil phosphate desorption induced by a phosphate-solubilizing fungus. Commun. Soil Sci. Plant Anal. 2014, 45, 451–460. [Google Scholar] [CrossRef]
- Nguyen, T.T.T.; Park, S.W.; Pangging, M.; Lee, H.B. Molecular and morphological confirmation of three undescribed species of mortierella from Korea. Mycobiology 2019, 47, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; Sun, X.R.; Wu, X.Q.; Li, G.E.; Wang, Z.; Li, D.W. The phosphate-solubilizing ability of Penicillium guanacastense and its effects on the growth of Pinus massoniana in phosphate-limiting conditions. Biol. Open 2019, 8, bio046797. [Google Scholar] [CrossRef]
- Radhakrishnan, R.; Kang, S.M.; Baek, I.Y.; Lee, I.J. Characterization of plant growth-promoting traits of Penicillium species against the effects of high soil salinity and root disease. J. Plant Interact. 2014, 9, 754–762. [Google Scholar] [CrossRef]
- Sangale, M.K.; Shahnawaz, M.; Ade, A.B. Potential of fungi isolated from the dumping sites mangrove rhizosphere soil to degrade polythene. Sci. Rep. 2019, 9, 5390. [Google Scholar] [CrossRef]
- Elsharkawy, M.M.; Shimizu, M.; Takahashi, H.; Hyakumachi, M. Induction of systemic resistance against Cucumber mosaic virus by Penicillium simplicissimum GP17-2 in Arabidopsis and tobacco. Plant Pathol. 2012, 61, 964–976. [Google Scholar] [CrossRef]
- Kotowicz, N.K.; Frąc, M.; Lipiec, J. The importance of Fusarium Fungi in wheat cultivation–pathogenicity and mycotoxins production: A review. J. Anim. Plant Sci. 2014, 21, 3326–3343. [Google Scholar]
- Rana, A.; Sahgal, M.; Johri, B.N. Fusarium oxysporum: Genomics, Diversity and Plant–Host Interaction. In Developments in Fungal Biology and Applied Mycology; Satyanarayana, T., Deshmukh, S.K., Johri, B.N., Eds.; Springer: Singapore, 2017; pp. 159–199. ISBN 9789811047688. [Google Scholar]
- Qiu, M.; Zhang, R.; Xue, C.; Zhang, S.; Li, S.; Zhang, N.; Shen, Q. Application of bio-organic fertilizer can control Fusarium wilt of cucumber plants by regulating microbial community of rhizosphere soil. Biol. Fertil. Soils 2012, 48, 807–816. [Google Scholar] [CrossRef]
- Zhang, S.; Raza, W.; Yang, X.; Hu, J.; Huang, Q.; Xu, Y.; Liu, X.; Ran, W.; Shen, Q. Control of Fusarium wilt disease of cucumber plants with the application of a bioorganic fertilizer. Biol. Fertil. Soils 2008, 44, 1073–1080. [Google Scholar] [CrossRef]
- Weng, Q.; Zhang, X.; Chen, W.; Hu, Q. Secondary Metabolites and the risks of Isaria fumosorosea and Isaria farinosa. Molecules 2019, 24, 664. [Google Scholar] [CrossRef]
- Kuźniar, T.; Ropek, D.; Krysa, A. The use of entomopathogenic fungus Isaria fumosorosea to reduce harmfulness of European corn borer (Ostrinia nubilalis Hbn.) in cultivation of sweet maize. Progess Plant Prot. 2012, 52, 354–359. [Google Scholar]
- Liao, X.; Fang, W.; Lin, L.; Lu, H.L.; Leger, R.J.S. Metarhizium robertsii produces an extracellular invertase (MrINV) that plays a pivotal role in rhizospheric interactions and root colonization. PLoS ONE 2013, 8, e78118. [Google Scholar] [CrossRef]
- Sasan, R.K.; Bidochka, M.J. The insect-pathogenic fungus Metarhizium robertsii (Clavicipitaceae) is also an endophyte that stimulates plant root development. Am. J. Bot. 2012, 99, 101–107. [Google Scholar] [CrossRef]
- Badali, H.; Gueidan, C.; Najafzadeh, M.J.; Bonifaz, A.; Gerrits van den Ende, A.H.G.; de Hoog, G.S. Biodiversity of the genus Cladophialophora. Stud. Mycol. 2008, 61, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Kantarcioǧlu, A.S.; De Hoog, G.S. Infections of the central nervous system by melanized fungi: A review of cases presented between 1999 and 2004. Mycoses 2004, 47, 4–13. [Google Scholar] [PubMed]
- Romero, H.; Viscaya, L.; Ferrara, G. Bacillus subtilis inhibits growth of Cladophialophora carrionii in vitro. J. Mycol. Med. 2006, 16, 92–94. [Google Scholar] [CrossRef]
- Stosiek, N.; Terebieniec, A.; Ząbek, A.; Młynarz, P.; Cieśliński, H.; Klimek-Ochab, M. N-phosphonomethylglycine utilization by the psychrotolerant yeast Solicoccozyma terricola M 3.1.4. Bioorg. Chem. 2019, 93, 102866. [Google Scholar] [CrossRef] [PubMed]
- Devi, K.; Brahma, J.; Shrivastava, K. Documentation of four hitherto unreported wild edible macro fungi from Chirang district of Assam, North-East India. Int. J. Conserv. Sci. 2016, 7, 709–718. [Google Scholar]
- Schindler, F.; Gube, M.; Kothe, E. Bioremediation and heavy metal uptake: Microbial approaches at field scale. In Bio-Geo Interactions in Metal-Contaminated Soils; Kothe, E., Varma, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 365–383. ISBN 9783642233272. [Google Scholar]
- Steffen, K.T.; Hatakka, A.; Hofrichter, M. Degradation of Benzo[a]pyrene by the litter-decomposing Basidiomycete Stropharia coronilla: Role of Manganese Peroxidase. Appl. Environ. Microbiol. 2003, 69, 3957–3964. [Google Scholar] [CrossRef] [PubMed]
- Borowik, K.; Rusek, P.; Schab, S.; Rusek, Ł. Biofertilizer and Method of its Production (Bionawóz i Sposób Jego Wytwarzania). Patent Application No. 431350, 10 March 2019. [Google Scholar]
- Conijn, J.G.; Bindraban, P.S.; Schröder, J.J.; Jongschaap, R.E.E. Can our global food system meet food demand within planetary boundaries? Agric. Ecosyst. Environ. 2018, 251, 244–256. [Google Scholar] [CrossRef]
- Zantua, M.I.; Bremner, J.M. Stability of urease in soils. Soil Biol. Biochem. 1977, 9, 135–140. [Google Scholar] [CrossRef]
- Ladd, J.N.; Butler, J.H.A. Short-term assays of soil proteolytic enzyme activities using proteins and dipeptide derivatives as substrates. Soil Biol. Biochem. 1972, 4, 19–30. [Google Scholar] [CrossRef]
- Alef, K.; Nannipieri, P. Enzyme activities. In Methods in Applied Soil Microbiology and Biochemistry; Alef, K., Nannipieri, P., Eds.; Academic Press: London, UK; Cambridge, MA, USA; San Diego, CA, USA; Oxford, UK, 1995; pp. 311–373. ISBN 0125138407. [Google Scholar]
- Eivazi, F.; Tabatabai, M.A. Glucosidases and galactosidases in soils. Soil Biol. Biochem. 1988, 20, 601–606. [Google Scholar] [CrossRef]
- Tabatabai, M.A.; Bremner, J.M. Use of p-nitrophenyl phosphate for assay of soil phosphatase activity. Soil Biol. Biochem. 1969, 1, 301–307. [Google Scholar] [CrossRef]
- Li, L.; Xu, M.; Ali, M.E.; Zhang, W.; Duan, Y.; Li, D. Factors affecting soil microbial biomass and functional diversity with the application of organic amendments in three contrasting cropland soils during a field experiment. PLoS ONE 2018, 13, e02038. [Google Scholar] [CrossRef]
- Sofo, A.; Ricciuti, P. A standardized method for estimating the functional diversity of soil bacterial community by Biolog® EcoPlatesTM assay-The case study of a sustainable olive orchard. Appl. Sci. 2019, 9, 4035. [Google Scholar] [CrossRef]
- Pertile, G.; Panek, J.; Oszust, K.; Siczek, A.; Frąc, M. Intraspecific functional and genetic diversity of Petriella setifera. PeerJ 2018, 6, e4420. [Google Scholar] [CrossRef]
- Insam, H. A new set of substrates proposed for community characterization in environmental samples. In Microbial Communities: Functional Versus Structural Approaches; Insam, H., Rangger, A., Eds.; Springer: Berlin/Heidelberg, Germany, 1997; pp. 259–260. [Google Scholar]
- Oszust, K.; Gryta, A.; Ziemiński, K.; Bilińska-Wielgus, N.; Gałązka, R.; Frąc, M. Characterization of microbial functional and genetic diversity as a novel strategy of biowaste ecotoxicological evaluation. Int. J. Environ. Sci. Technol. 2019, 16, 4261–4274. [Google Scholar] [CrossRef]
- Wolinska, A.; Frąc, M.; Oszust, K.; Szafranek-Nakonieczna, A.; Zielenkiewicz, U.; Stępniewska, Z. Microbial biodiversity of meadows under different modes of land use: Catabolic and genetic fingerprinting. World J. Microbiol. Biotechnol. 2017, 33, 154. [Google Scholar]
- Singh, B.K.; Nazaries, L.; Munro, S.; Anderson, I.C.; Campbell, C.D. Use of multiplex terminal restriction fragment length polymorphism for rapid and simultaneous analysis of different components of the soil microbial community. Appl. Environ. Microbiol. 2006, 72, 7278–7285. [Google Scholar] [PubMed]
- Marchesi, J.R.; Sato, T.; Weightman, A.J.; Martin, T.A.; Fry, J.C.; Hiom, S.J.; Wade, W.G. Design and evaluation of useful bacterium-specific PCR Primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 1998, 64, 795–799. [Google Scholar] [PubMed]
- Hauben, L.; Vauterin, L.; Swings, J.; Moore, E.R.B. Comparison of 16S Ribosomal DNA Sequences of All Xanthomonas Species. Int. J. Syst. Bacteriol. 1997, 47, 328–335. [Google Scholar]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [PubMed]
- White, T.J.; Bruns, T.D.; Lee, S.; Taylor, J. Analysis of phylogenetic relationships by amplification and direct sequencing of ribosomal genes. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfond, D.H., Sainsky, J.J., White, T.J., Eds.; Academic Press: London, UK, 1990; pp. 315–322. [Google Scholar]
- Jurgens, G.; Saano, A. Diversity of soil Archaea in boreal forest before, and after clear-cutting and prescribed burning. FEMS Microbiol. Ecol. 1999, 29, 205–213. [Google Scholar]
- Jurgens, G.; Lindström, K.; Saano, A. Novel group within the Kingdom Crenarchaeota from Boreal forest soil. Appl. Environ. Microbiol. 1997, 63, 803–805. [Google Scholar]
- Fish, J.A.; Chai, B.; Wang, Q.; Sun, Y.; Brown, C.T.; Tiedje, J.M.; Cole, J.R. FunGene: The functional gene pipeline and repository. Front. Microbiol. 2013, 4, 291. [Google Scholar] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar]
- Schmidt, P.A.; Bálint, M.; Greshake, B.; Bandow, C.; Römbke, J.; Schmitt, I. Illumina metabarcoding of a soil fungal community. Soil Biol. Biochem. 2013, 65, 128–132. [Google Scholar]
- Vilgalys Mycology Lab Conserved Primer Sequences for PCR Amplification and Sequencing from Nuclear Ribosomal RNA. Available online: https://sites.duke.edu/vilgalyslab/rdna_primers_for_fungi/ (accessed on 18 March 2020).
- Garland, J.L. Analytical approaches to the characterization of samples of microbial communities using patterns of potential C source utilization. Soil Biol. Biochem. 1996, 28, 213–221. [Google Scholar]
- Oszust, K.; Panek, J.; Pertile, G.; Siczek, A.; Oleszek, M.; Frąc, M. Metabolic and genetic properties of Petriella setifera precultured on waste. Front. Microbiol. 2018, 9, 115. [Google Scholar]
- Garland, J.L.; Mills, A.L. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl. Environ. Microbiol. 1991, 57, 2351–2359. [Google Scholar]
- Oliveros, J.C. 2007–2015 Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. Available online: https://bioinfogp.cnb.csic.es/tools/venny/ (accessed on 18 March 2020).
- Fredriksson, N.J.; Hermansson, M.; Wilén, B.M. Tools for T-RFLP data analysis using Excel. BMC Bioinform. 2014, 15, 361. [Google Scholar]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a Chimera-Checked 16S rRNA Gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D1, D259–D264. [Google Scholar]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [PubMed]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar]



| Soil Type/Treatment | |||||||
|---|---|---|---|---|---|---|---|
| BA | AL | ||||||
| FC | FA100 | FA60 | FC | FA100 | FA60 | ||
| ECO Plates | AWCD * | 0.81 ± 0.08 a | 0.70 ± 0.16 b | 0.80 ± 0.03 ab | 0.81 ± 0.32 a | 0.91 ± 0.06 a | 0.92 ± 0.13 a |
| R * | 20.00 ± 1.00 a | 19.00 ± 3.00 a | 19.00 ± 2.00 a | 21.00 ± 3.00 a | 21.00 ± 1.00 a | 22.00 ± 0.00 a | |
| H * | 2.84 ± 0.07 a | 2.87 ± 0.07 a | 2.74 ± 0.12 a | 3.05 ± 0.11 a | 2.92 ± 0.29 a | 3.04 ± 0.02 a | |
| FF Plates | AWDD * | 0.55 ± 0.25 a | 0.60 ± 0.09 a | 0.51 ± 0.11 a | 0.40 ± 0.17 a | 0.47 ± 0.08 a | 0.38 ± 0.06 a |
| R * | 52.00 ± 13.00 a | 58.00 ± 9.00 a | 56.00 ± 10.00 a | 52.00 ± 13.00 a | 58.00 ± 4.00 a | 51.00 ± 6.00 a | |
| H * | 3.82 ± 0.41 a | 3.92 ± 0.16 a | 3.88 ± 0.13 a | 3.88 ± 0.14 a | 3.94 ± 0.10 a | 3.83 ± 0.15 a | |
| NGS 16S | OUT ** | 2549 min 2507; max 2590 a | 2744 min 2736; max 2750 a | 2578 min 2565; max 2590 a | 3037 min 2998; max 3976 a | 2928 min 2925; max 2930 a | 3038 min 3035; max 3040 a |
| H * | 8.78 ± 0.01 a | 8.94 ± 0.01 a | 8.79 ± 0.01 a | 9.14 ± 0.02 a | 9.12 ± 0.01 a | 9.19 ± 0.01 a | |
| NGS ITS1 | OUT ** | 1234 min 1215; max 1251 a | 1342 min 1325; max 1358 a | 1338 min 1329; max 1345 a | 1385 min 1296; max 1471 a | 1411 min 1351; max 1470 a | 1441 min 1424; max 1458 a |
| H ** | 6.47 min 6.38; max 6.56 b | 6.83 min 6.83; max 6.84 a | 6.75 min 6.74; max 6.75 ab | 7.23 min 7.16; max 7.30 a | 7.51 min 7.45; max 7.57 a | 7.61 min 7.50; max 7.71 a | |
| t-RFLP bacteria | R ** | 3.00 min 2.96; max 3.00 b | 7.00 min 4.98; max 8.96 ab | 10.00 min 9.98; max 10.00 a | 8.00 min 6.99; max 8.00 a | 11.00 min 10.99; max 11.00 a | 9.00 min 6.65; max 10.98 a |
| H ** | 1.04 min 1.03; max 1.04 b | 1.86 min 1.59; max 2.13 ab | 2.21 min 2.20; max 2.22 a | 1.86 min 1.84; max 1.87 a | 2.02 min 2.00; max 2.04 a | 1.91 min 1.72; max 2.09 a | |
| t-RFLP archaea | R * | 24.00 ± 1.00 a | 24.00 ± 0.00 a | 23.00 ± 2.00 a | 26.00 ± 4.00 a | 25.00 ± 3.00 a | 30.00 ± 3.00 a |
| H * | 3.01 ± 0.03 ab | 3.04 ± 0.00 a | 2.93 ± 0.08 b | 3.16 ± 0.17 a | 3.10 ± 0.19 a | 3.33 ± 0.04 a | |
| t-RFLP fungi | R ** | 11.00 min 9.97; max 11.99 ab | 16.00 min 15.98; max 16.00 a | 10.00 min 7.99; max 11.01 b | 19.00 min 11.96; max 26.00 a | 25.00 min 19.90; max 28.93 a | 29.00 min 26.98; max 29.92 a |
| H ** | 2.26 min 2.20; max 2.32 a | 2.62 min 2.58; max 2.65 a | 2.11 min 1.88; max 2.32 a | 2.69 min 2.16; max 3.20 a | 2.93 min 2.71; max 3.15 a | 3.15 min 3.11; max 3.17 a | |
| Soil Type | ||||||
|---|---|---|---|---|---|---|
| BrunicArenosol (BA) | Abruptic Luvisol (AL) | |||||
| Treatment | Bacteria | Archaea | Fungi | Bacteria | Archaea | Fungi |
| FC-FA100 | 0.33 | 1 | 0.81 | 0.64 | 0.97 | 0.84 |
| FC-FA60 | 0.30 | 1 | 0.61 | 0.62 | 0.94 | 0.84 |
| FA100-FA60 | 0.90 | 1 | 0.75 | 0.85 | 0.97 | 1 |
| Treatment | Brunic Arenosol (BA) | Abruptic Luvisol (AL) |
|---|---|---|
| Optimal dose (FC) | 125 kg phosphate mineral fertilizer; 365 kg urea; 290 potassium salt | 150 kg phosphate mineral fertilizer; 360 kg urea; 284 kg potassium salt |
| Optimal dose enriched with microorganisms (FA100) | 156.25 kg phosphate mineral fertilizer enriched with beneficial bacterial strains; 365 kg urea; 290 potassium salt | 187.5 kg phosphate mineral fertilizer enriched with beneficial bacterial strains; 360 kg urea; 284 kg potassium salt |
| Dose reduced by 40% enriched with microorganisms (FA60) | 93.75 kg phosphate mineral fertilizer enriched with beneficial strains; 365 kg urea; 290 potassium salt | 112.5 kg phosphate mineral fertilizer enriched with beneficial bacterial strains; 360 kg urea; 284 kg potassium salt |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mącik, M.; Gryta, A.; Sas-Paszt, L.; Frąc, M. The Status of Soil Microbiome as Affected by the Application of Phosphorus Biofertilizer: Fertilizer Enriched with Beneficial Bacterial Strains. Int. J. Mol. Sci. 2020, 21, 8003. https://doi.org/10.3390/ijms21218003
Mącik M, Gryta A, Sas-Paszt L, Frąc M. The Status of Soil Microbiome as Affected by the Application of Phosphorus Biofertilizer: Fertilizer Enriched with Beneficial Bacterial Strains. International Journal of Molecular Sciences. 2020; 21(21):8003. https://doi.org/10.3390/ijms21218003
Chicago/Turabian StyleMącik, Mateusz, Agata Gryta, Lidia Sas-Paszt, and Magdalena Frąc. 2020. "The Status of Soil Microbiome as Affected by the Application of Phosphorus Biofertilizer: Fertilizer Enriched with Beneficial Bacterial Strains" International Journal of Molecular Sciences 21, no. 21: 8003. https://doi.org/10.3390/ijms21218003
APA StyleMącik, M., Gryta, A., Sas-Paszt, L., & Frąc, M. (2020). The Status of Soil Microbiome as Affected by the Application of Phosphorus Biofertilizer: Fertilizer Enriched with Beneficial Bacterial Strains. International Journal of Molecular Sciences, 21(21), 8003. https://doi.org/10.3390/ijms21218003







