Sustained Incompatibility between MAPK Signaling and Pathogen Effectors
Abstract
:1. Introduction
2. MAPKs as Targets of Virulent and Avirulent Effectors
3. MAPK Activities during ETI: General or Peculiar?
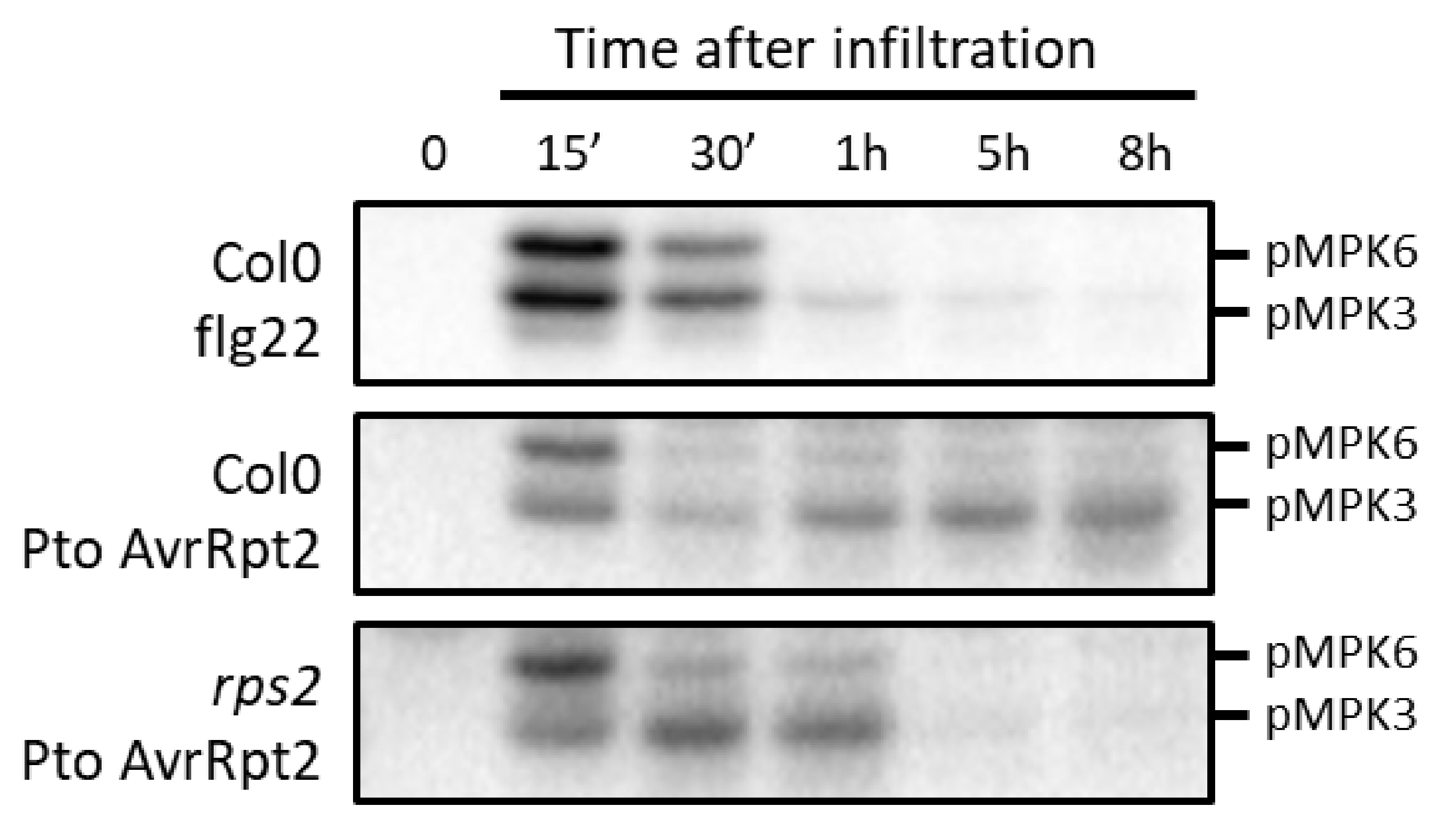
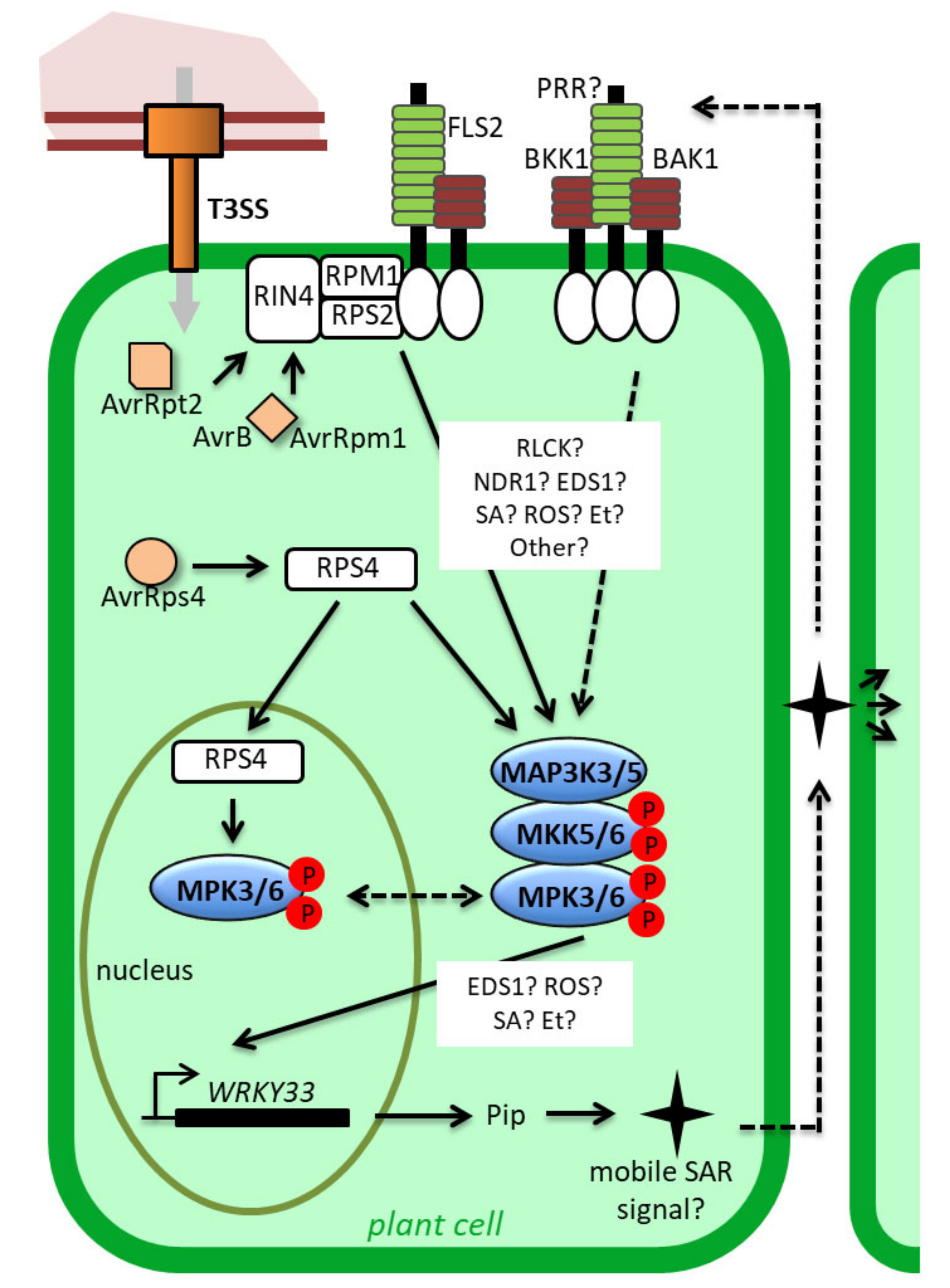
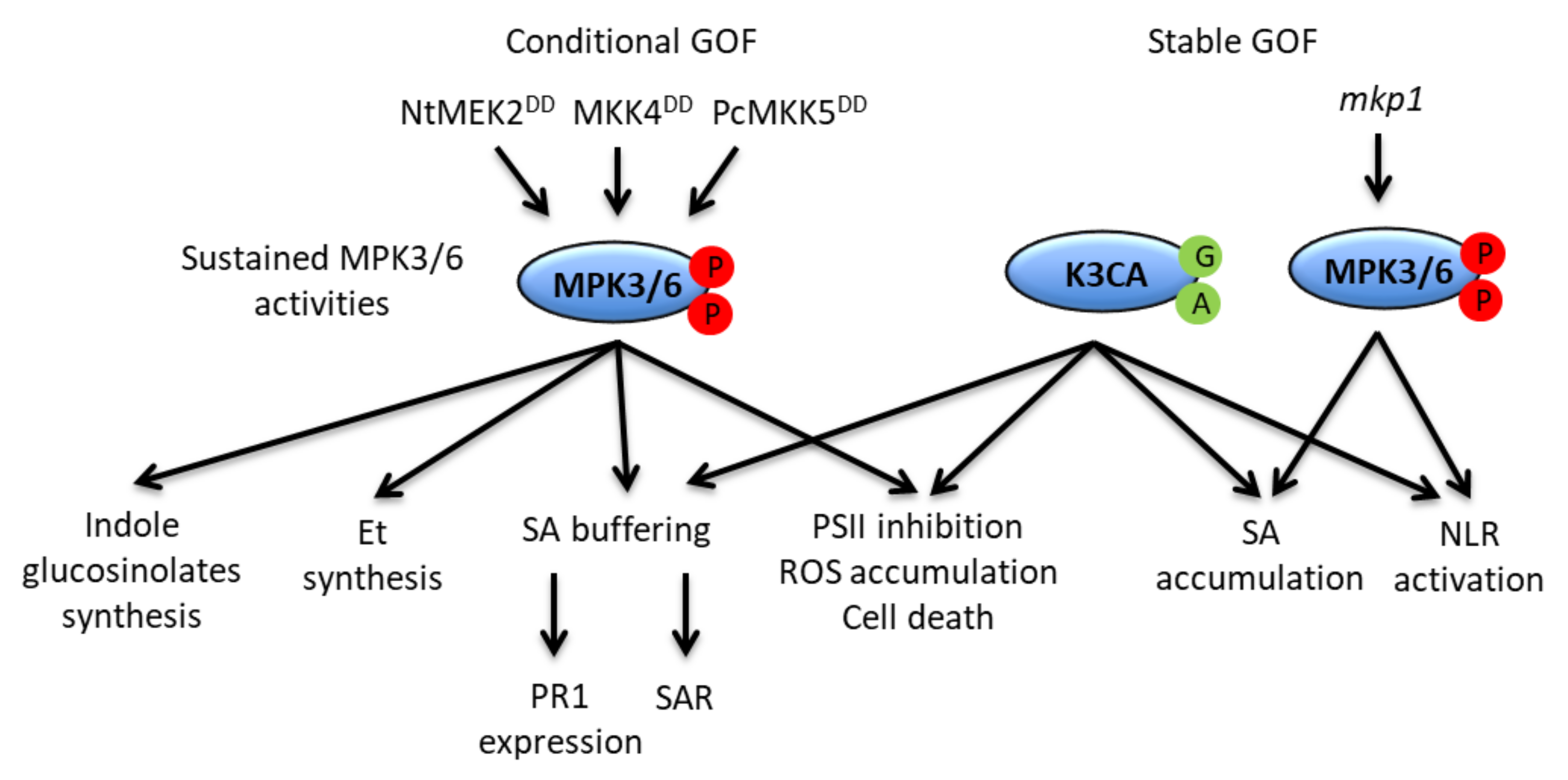
4. Functions of MAPKs during ETI: Robustness and Cell Death
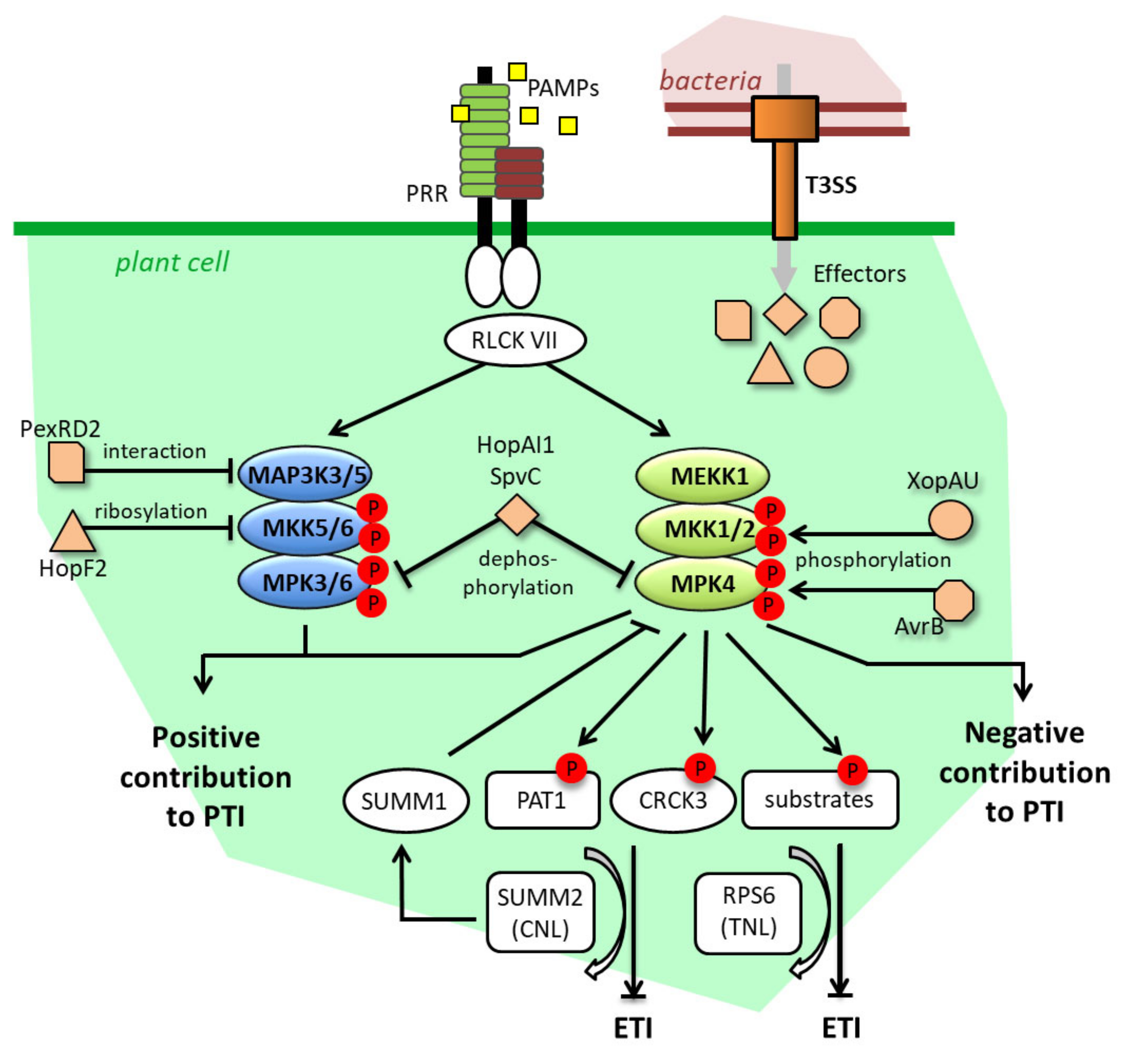
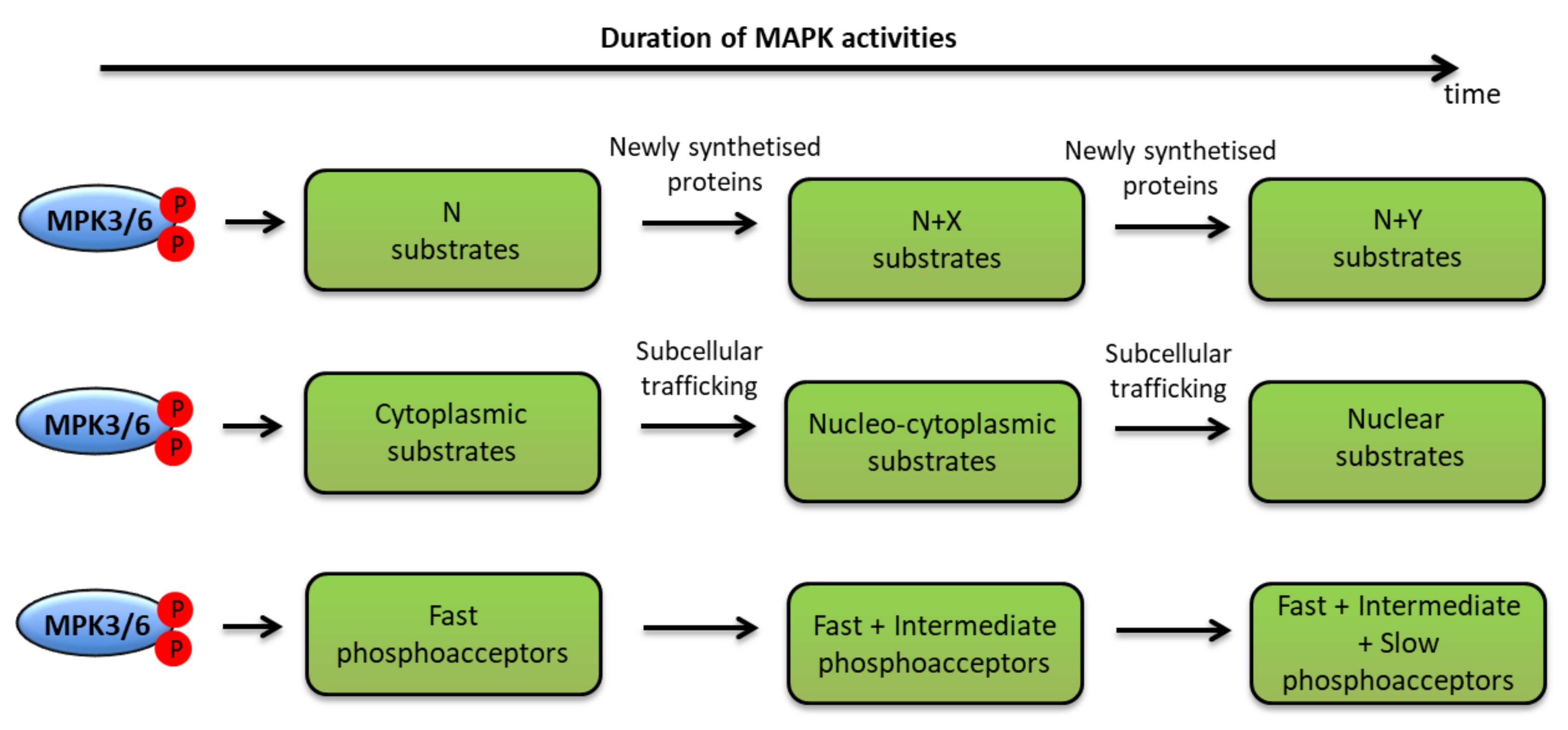
5. MAPK Substrates during ETI: Is the Temporal Phosphocode Relevant?
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CNL | Coiled-Coil NLR |
| Et | Ethylene |
| ETI | Effector-Triggered Immunity |
| HR | Hypersensitive Response |
| IGS | Indole Glucosinolate |
| MAPK | Mitogen-Activated Protein Kinases |
| NLR | Nucleotide-Binding Leucine-Rich Repeat Receptors |
| PAMP | Pathogen-Associated Molecular Patterns |
| PRR | Pattern Recognition Receptor |
| PSII | PhotoSystem II |
| PTI | PAMP-Triggered Immunity |
| R | Resistance |
| RLCK | Receptor-Like Cytoplasmic Kinase |
| ROS | Reactive Oxygen Species |
| SA | Salicylic Acid |
| SAR | Systemic Acquired Resistance |
| SIPK | SA-induced Protein Kinase, homologue of MPK6 |
| SUMM | Suppressor of mkk1mkk2 |
| TMV | Tobacco Mosaic Virus |
| TNL | Toll Interleukin-1 Receptor NLR |
| WIPK | Wounding-Induced Protein Kinase, homologue of MPK3 |
| WT | Wild Type |
References
- Jones, J.D.G.; Dangl, J.L. The Plant Immune System. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Flor, H.H. Current Status of the Gene-For-Gene Concept. 1971. Available online: https://doi.org/10.1146/annurev.py.09.090171.001423 (accessed on 20 October 2020).
- Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 2014, 35, 345–351. [Google Scholar] [CrossRef]
- Bigeard, J.; Colcombet, J.; Hirt, H. Signaling Mechanisms in Pattern-Triggered Immunity (PTI). Mol. Plant 2015, 8, 521–539. [Google Scholar] [CrossRef] [Green Version]
- Kourelis, J.; van der Hoorn, R.A.L. Defended to the Nines: 25 Years of Resistance Gene Cloning Identifies Nine Mechanisms for R Protein Function. Plant Cell 2018, 30, 285–299. [Google Scholar] [CrossRef] [Green Version]
- Su, J.; Spears, B.J.; Kim, S.H.; Gassmann, W. Constant vigilance: Plant functions guarded by resistance proteins. Plant J. 2018, 93, 637–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, J.D.G.; Vance, R.E.; Dangl, J.L. Intracellular innate immune surveillance devices in plants and animals. Science 2016, 354. [Google Scholar] [CrossRef] [Green Version]
- Aarts, N.; Metz, M.; Holub, E.; Staskawicz, B.J.; Daniels, M.J.; Parker, J.E. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 1998, 95, 10306–10311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adachi, H.; Derevnina, L.; Kamoun, S. NLR singletons, pairs, and networks: Evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr. Opin. Plant Biol. 2019, 50, 121–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venugopal, S.C.; Jeong, R.D.; Mandal, M.K.; Zhu, S.; Chandra-Shekara, A.C.; Xia, Y.; Hersh, M.; Stromberg, A.J.; Navarre, D.R.; Kachroo, A.; et al. Enhanced disease susceptibility 1 and salicylic acid act redundantly to regulate resistance gene-mediated signaling. PLoS Genet. 2009, 5, 22–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhandari, D.D.; Lapin, D.; Kracher, B.; von Born, P.; Bautor, J.; Niefind, K.; Parker, J.E. An EDS1 heterodimer signalling surface enforces timely reprogramming of immunity genes in Arabidopsis. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Kapos, P.; Devendrakumar, K.T.; Li, X. Plant NLRs: From discovery to application. Plant Sci. 2019, 279, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, K.; Katagiri, F. Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 2010, 13, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Van Wersch, R.; Zhang, Y. Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Mol. Plant Microbe Interact. 2018, 31, 403–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-Triggered Immunity: From Pathogen Perception to Robust Defense. Annu. Rev. Plant Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef] [PubMed]
- Mapk Group. Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 2002, 7, 301–308. [Google Scholar] [CrossRef]
- Colcombet, J.; Hirt, H. Arabidopsis MAPKs: A complex signalling network involved in multiple biological processes. Biochem. J. 2008, 413, 217–226. [Google Scholar] [CrossRef]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.-L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002, 415, 977–983. [Google Scholar] [CrossRef]
- Sun, T.; Nitta, Y.; Zhang, Q.; Wu, D.; Tian, H.; Lee, J.S.; Zhang, Y. Antagonistic interactions between two MAP kinase cascades in plant development and immune signaling. EMBO Rep. 2018, 19, e45324. [Google Scholar] [CrossRef]
- Gao, M.; Liu, J.; Bi, D.; Zhang, Z.; Cheng, F.; Chen, S.; Zhang, Y. MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 2008, 18, 1190–1198. [Google Scholar] [CrossRef] [Green Version]
- Frei dit Frey, N.; Garcia, A.V.; Bigeard, J.; Zaag, R.; Bueso, E.; Garmier, M.; Pateyron, S.; de Tauzia-Moreau, M.-L.; Brunaud, V.; Balzergue, S.; et al. Functional analysis of Arabidopsisimmune-related MAPKs uncovers a role for MPK3 as negative regulator of inducible defences. Genome Biol. 2014, 15, R87. [Google Scholar] [CrossRef] [Green Version]
- Toruño, T.Y.; Stergiopoulos, I.; Coaker, G. Plant-Pathogen Effectors: Cellular Probes Interfering with Plant Defenses in Spatial and Temporal Manners. Annu. Rev. Phytopathol. 2016, 54, 419–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laflamme, B.; Dillon, M.M.; Martel, A.; Almeida, R.N.D.; Desveaux, D.; Guttman, D.S. The pan-genome effector-triggered immunity landscape of a host-pathogen interaction. Science 2020, 367, 763–768. [Google Scholar] [CrossRef] [PubMed]
- WHITE, F.F.; POTNIS, N.; JONES, J.B.; KOEBNIK, R. The type III effectors of Xanthomonas. Mol. Plant Pathol. 2009, 10, 749–766. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shao, F.; Li, Y.; Cui, H.; Chen, L.; Li, H.; Zou, Y.; Long, C.; Lan, L.; Chai, J.; et al. A Pseudomonas syringae Effector Inactivates MAPKs to Suppress PAMP-Induced Immunity in Plants. Cell Host Microbe 2007, 1, 175–185. [Google Scholar] [CrossRef] [Green Version]
- Neumann, C.; Fraiture, M.; Hernàndez-Reyes, C.; Akum, F.N.; Virlogeux-Payant, I.; Chen, Y.; Pateyron, S.; Colcombet, J.; Kogel, K.-H.; Hirt, H.; et al. The Salmonella effector protein SpvC, a phosphothreonine lyase is functional in plant cells. Front. Microbiol. 2014, 5, 548. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, J.; Hou, S.; Wang, X.; Li, Y.; Ren, D.; Chen, S.; Tang, X.; Zhou, J.M. A pseudomonas syringae ADP-ribosyltransferase inhibits arabidopsis mitogen-activated protein kinase kinases. Plant Cell 2010, 22, 2033–2044. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Wu, S.; Chen, X.; Liu, C.; Sheen, J.; Shan, L.; He, P. The Pseudomonas syringae effector HopF2 suppresses Arabidopsis immunity by targeting BAK1. Plant J. 2014, 77, 235–245. [Google Scholar] [CrossRef] [Green Version]
- Xiang, T.; Zong, N.; Zou, Y.; Wu, Y.; Zhang, J.; Xing, W.; Li, Y.; Tang, X.; Zhu, L.; Chai, J.; et al. Pseudomonas syringae Effector AvrPto Blocks Innate Immunity by Targeting Receptor Kinases. Curr. Biol. 2008, 18, 74–80. [Google Scholar] [CrossRef] [Green Version]
- Gimenez-Ibanez, S.; Hann, D.R.; Ntoukakis, V.; Petutschnig, E.; Lipka, V.; Rathjen, J.P. AvrPtoB Targets the LysM Receptor Kinase CERK1 to Promote Bacterial Virulence on Plants. Curr. Biol. 2009, 19, 423–429. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Li, W.; Xiang, T.; Liu, Z.; Laluk, K.; Ding, X.; Zou, Y.; Gao, M.; Zhang, X.; Chen, S.; et al. Receptor-like Cytoplasmic Kinases Integrate Signaling from Multiple Plant Immune Receptors and Are Targeted by a Pseudomonas syringae Effector. Cell Host Microbe 2010, 7, 290–301. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Wu, Y.; Gao, M.; Zhang, J.; Kong, Q.; Liu, Y.; Ba, H.; Zhou, J.; Zhang, Y. Disruption of PAMP-induced MAP kinase cascade by a pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 2012, 11, 253–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, Q.; Qu, N.; Gao, M.; Zhang, Z.; Ding, X.; Yang, F.; Li, Y.; Dong, O.X.; Chen, S.; Li, X.; et al. The MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in Arabidopsis. Plant Cell 2012, 24, 2225–2236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, S.H.; Bush, S.M.; Zaman, N.; Stecker, K.; Sussman, M.R.; Krysan, P. Deletion of a tandem gene family in Arabidopsis: Increased MEKK2 abundance triggers autoimmunity when the MEKK1-MKK1/2-MPK4 signaling cascade is disrupted. Plant Cell 2013, 25, 1895–1910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nitta, Y.; Qiu, Y.; Yaghmaiean, H.; Zhang, Q.; Huang, J.; Adams, K.; Zhang, Y. MEKK2 inhibits activation of MAP kinases in Arabidopsis. Plant J. 2020, 103, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, Y.; Huang, H.; Gao, M.; Wu, D.; Kong, Q.; Zhang, Y. The NLR protein SUMM 2 senses the disruption of an immune signaling MAP kinase cascade via CRCK 3. EMBO Rep. 2017, 18, 292–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roux, M.E.; Rasmussen, M.W.; Palma, K.; Lolle, S.; Regué, À.M.; Bethke, G.; Glazebrook, J.; Zhang, W.; Sieburth, L.; Larsen, M.R.; et al. The mRNA decay factor PAT 1 functions in a pathway including MAP kinase 4 and immune receptor SUMM 2. EMBO J. 2015, 34, 593–608. [Google Scholar] [CrossRef] [Green Version]
- Takagi, M.; Hamano, K.; Takagi, H.; Morimoto, T.; Akimitsu, K.; Terauchi, R.; Shirasu, K.; Ichimura, K. Disruption of the MAMP-Induced MEKK1-MKK1/MKK2-MPK4 Pathway Activates the TNL Immune Receptor SMN1/RPS6. Plant Cell Physiol. 2019, 60, 778–787. [Google Scholar] [CrossRef]
- Brodersen, P.; Petersen, M.; Bjørn Nielsen, H.; Zhu, S.; Newman, M.-A.; Shokat, K.M.; Rietz, S.; Parker, J.; Mundy, J. Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J. 2006, 47, 532–546. [Google Scholar] [CrossRef] [Green Version]
- Cui, H.; Wang, Y.; Xue, L.; Chu, J.; Yan, C.; Fu, J.; Chen, M.; Innes, R.W.; Zhou, J. A pseudomonass syringae protein perturbs Arabidopsis hormone signalling by activating MAP KINASE 4. Cell Host Microbe 2012, 7, 164–175. [Google Scholar] [CrossRef] [Green Version]
- Teper, D.; Girija, A.M.; Bosis, E.; Popov, G.; Savidor, A.; Sessa, G. The Xanthomonas euvesicatoria type III effector XopAU is an active protein kinase that manipulates plant MAP kinase signaling. PLoS Pathog. 2018, 14, e1006880. [Google Scholar] [CrossRef] [Green Version]
- Berriri, S.; Garcia, A.V.; dit Frey, N.F.; Rozhon, W.; Pateyron, S.; Leonhardt, N.; Montillet, J.L.; Leung, J.; Hirt, H.; Colcombet, J. Constitutively active mitogen-activated protein kinase versions reveal functions of Arabidopsis MPK4 in pathogen defense signaling. Plant Cell 2012, 24, 4281–4293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Jiang, S.; Yu, X.; Cheng, C.; Chen, S.; Cheng, Y.; Yuan, J.S.; Jiang, D.; He, P.; Shan, L. Phosphorylation of Trihelix Transcriptional Repressor ASR3 by MAP KINASE4 Negatively Regulates Arabidopsis Immunity. Plant Cell 2015, 27, 839–856. [Google Scholar] [CrossRef] [Green Version]
- Du, Y.; Chen, X.; Guo, Y.; Zhang, X.; Zhang, H.; Li, F.; Huang, G.; Meng, Y.; Shan, W. Phytophthora infestans RXLR effector PITG20303 targets a potato MKK1 protein to suppress plant immunity. New Phytol. 2020. [Google Scholar] [CrossRef] [PubMed]
- King, S.R.F.; McLellan, H.; Boevink, P.C.; Armstrong, M.R.; Bukharov, T.; Sukarta, O.; Win, J.; Kamoun, S.; Birch, P.R.J.; Banfield, M.J. Phytophthora infestans RXLR effector PexRD2 interacts with host MAPKKKε to suppress plant immune signaling. Plant Cell 2014, 26, 1345–1359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melech-Bonfil, S.; Sessa, G. Tomato MAPKKKε is a positive regulator of cell-death signaling networks associated with plant immunity. Plant J. 2010, 64, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Klessig, D.F. Resistance gene N-mediated de novo synthesis and activation of a tobacco mitogen-activated protein kinase by tobacco mosaic virus infection. Proc. Natl. Acad. Sci. USA 1998, 95, 7433–7438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Liu, Y.; Klessig, D.F. Multiple levels of tobacco WIPK activation during the induction of cell death by fungal elicitins. Plant J. 2000, 23, 339–347. [Google Scholar] [CrossRef] [Green Version]
- Tsuda, K.; Mine, A.; Bethke, G.; Igarashi, D.; Botanga, C.J.; Tsuda, Y.; Glazebrook, J.; Sato, M.; Katagiri, F. Dual Regulation of Gene Expression Mediated by Extended MAPK Activation and Salicylic Acid Contributes to Robust Innate Immunity in Arabidopsis thaliana. PLoS Genet. 2013, 9. [Google Scholar] [CrossRef]
- Ren, D.; Liu, Y.; Yang, K.-Y.; Han, L.; Mao, G.; Glazebrook, J.; Zhang, S. A Fungal-Responsive MAPK Cascade Regulates Phytoalexin Biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2008, 105, 5638–5643. [Google Scholar] [CrossRef] [Green Version]
- Han, L.; Li, G.-J.; Yang, K.-Y.; Mao, G.; Wang, R.; Liu, Y.; Zhang, S. Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J. 2010, 64, 114–127. [Google Scholar] [CrossRef]
- Genenncher, B.; Wirthmueller, L.; Roth, C.; Klenke, M.; Ma, L.; Sharon, A.; Wiermer, M. Nucleoporin-regulated MAP kinase signaling in immunity to a necrotrophic fungal pathogen. Plant Physiol. 2016, 172, 1293–1305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Underwood, W.; Zhang, S.; He, S.Y. The Pseudomonas syringae type III effector tyrosine phosphatase HopAO1 suppresses innate immunity in Arabidopsis thaliana. Plant J. 2007, 52, 658–672. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Menke, F.L.H.; Yoshioka, K.; Moder, W.; Shirano, Y.; Klessig, D.F. High humidity suppresses ssi4-mediated cell death and disease resistance upstream of MAP kinase activation, H2O2 production and defense gene expression. Plant J. 2004, 39, 920–932. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Yang, L.; Zhu, Q.; Wu, H.; He, Y.; Liu, Y.; Xu, J.; Jiang, D.; Zhang, S. Active photosynthetic inhibition mediated by MPK3/MPK6 is critical to effector-triggered immunity. PLoS Biol. 2018, 16, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Schuck, S.; Wu, J.; Yang, P.; Döring, A.C.; Zeier, J.; Tsuda, K. A mpk3/6-wrky33-ald1-pipecolic acid regulatory loop contributes to systemic acquired resistance. Plant Cell 2018, 30, 2480–2494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ngou, B.P.M.; Ahn, H.K.; Ding, P.; Redkar, A.; Brown, H.; Ma, Y.; Youles, M.; Tomlinson, L.; Jones, J.D.G. Estradiol-inducible AvrRps4 expression reveals distinct properties of TIR-NLR-mediated effector-triggered immunity. J. Exp. Bot. 2020, 71, 2186–2197. [Google Scholar] [CrossRef]
- Cui, H.; Gobbato, E.; Kracher, B.; Qiu, J.; Bautor, J.; Parker, J.E. A core function of EDS1 with PAD4 is to protect the salicylic acid defense sector in Arabidopsis immunity. New Phytol. 2017, 213, 1802–1817. [Google Scholar] [CrossRef] [Green Version]
- Mine, A.; Seyfferth, C.; Kracher, B.; Berens, M.L.; Becker, D.; Tsuda, K. The defense phytohormone signaling network enables rapid, high-amplitude transcriptional reprogramming during effector-triggered immunity. Plant Cell 2018, 30, 1199–1219. [Google Scholar] [CrossRef] [Green Version]
- Yoo, H.; Greene, G.H.; Yuan, M.; Xu, G.; Burton, D.; Liu, L.; Marqués, J.; Dong, X. Translational Regulation of Metabolic Dynamics during Effector-Triggered Immunity. Mol. Plant 2019, 88–98. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Zhang, Y.; Guan, R.; Li, S.; Xu, X.; Zhang, S.; Xu, J. Co-regulation of indole glucosinolates and camalexin biosynthesis by CPK5/CPK6 and MPK3/MPK6 signaling pathways. J. Integr. Plant Biol. 2020. [Google Scholar] [CrossRef]
- Lassowskat, I.; Böttcher, C.; Eschen-Lippold, L.; Scheel, D.; Lee, J. Sustained mitogen-activated protein kinase activation reprograms defense metabolism and phosphoprotein profile in Arabidopsis thaliana. Front. Plant Sci. 2014, 5, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, W.; Li, B.; Li, S.; Liang, Y.; Wu, X.; Ma, M.; Wang, J.; Gao, J.; Cai, Y.; Zhang, Y.; et al. Mitogen-Activated Protein Kinase Cascade MKK7-MPK6 Plays Important Roles in Plant Development and Regulates Shoot Branching by Phosphorylating PIN1 in Arabidopsis. PLoS Biol. 2016, 14, 1–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, L.; Li, Y.; Wang, Q.; Xu, J.; Chen, Y.; Yang, H.; Ren, D. Activation of MKK9-MPK3/MPK6 enhances phosphate acquisition in Arabidopsis thaliana. New Phytol. 2014, 203, 1146–1160. [Google Scholar] [CrossRef] [PubMed]
- Eschen-Lippold, L.; Jiang, X.; Elmore, J.M.; Mackey, D.; Shan, L.; Coaker, G.; Scheel, D.; Lee, J. Bacterial AvrRpt2-like cysteine proteases block activation of the arabidopsis mitogen-activated protein kinases, MPK4 and MPK11. Plant Physiol. 2016, 171, 2223–2238. [Google Scholar] [CrossRef] [Green Version]
- Qi, Y.; Tsuda, K.; Glazebrook, J.; Katagiri, F. Physical association of pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) immune receptors in Arabidopsis. Mol. Plant Pathol. 2011, 12, 702–708. [Google Scholar] [CrossRef]
- Roberts, R.; Hind, S.R.; Pedley, K.F.; Diner, B.A.; Szarzanowicz, M.J.; Luciano-Rosario, D.; Majhi, B.B.; Popov, G.; Sessa, G.; Oh, C.-S.; et al. Mai1 Protein Acts Between Host Recognition of Pathogen Effectors and Mitogen-Activated Protein Kinase Signaling. Mol. Plant-Microbe Interact. 2019, 32, 1496–1507. [Google Scholar] [CrossRef]
- Bi, G.; Zhou, Z.; Wang, W.; Li, L.; Rao, S.; Wu, Y.; Zhang, X.; Menke, F.L.H.; Chen, S.; Zhou, J.-M. Receptor-Like Cytoplasmic Kinases Directly Link Diverse Pattern Recognition Receptors to the Activation of Mitogen-Activated Protein Kinase Cascades in Arabidopsis. Plant Cell 2018, 30, 1543–1561. [Google Scholar] [CrossRef] [Green Version]
- Wirthmueller, L.; Zhang, Y.; Jones, J.D.G.; Parker, J.E. Nuclear Accumulation of the Arabidopsis Immune Receptor RPS4 Is Necessary for Triggering EDS1-Dependent Defense. Curr. Biol. 2007, 17, 2023–2029. [Google Scholar] [CrossRef] [Green Version]
- Hoser, R.; Zurczak, M.; Lichocka, M.; Zuzga, S.; Dadlez, M.; Samuel, M.A.; Ellis, B.E.; Stuttmann, J.; Parker, J.E.; Hennig, J.; et al. Nucleocytoplasmic partitioning of tobacco N receptor is modulated by SGT1. New Phytol. 2013, 200, 158–171. [Google Scholar] [CrossRef]
- Day, B.; Dahlbeck, D.; Staskawicz, B.J. NDR1 Interaction with RIN4 Mediates the Differential Activation of Multiple Disease Resistance Pathways in Arabidopsis. Plant Cell 2006, 18, 2782–2791. [Google Scholar] [CrossRef] [Green Version]
- Knepper, C.; Savory, E.A.; Day, B. Arabidopsis NDR1 Is an Integrin-Like Protein with a Role in Fluid Loss and Plasma Membrane-Cell Wall Adhesion. Plant Physiol. 2011, 156, 286–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robatzek, S.; Chinchilla, D.; Boller, T. Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 2006, 20, 537–542. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Lu, D.; Shan, L. Ubiquitination of pattern recognition receptors in plant innate immunity. Mol. Plant Pathol. 2014, 15, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.D.; Sacks, D.B. Protein scaffolds in MAP kinase signalling. Cell. Signal. 2009, 21, 462–469. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Z.; Li, J.-F.; Niu, Y.; Zhang, X.-C.; Woody, O.Z.; Xiong, Y.; Djonović, S.; Millet, Y.; Bush, J.; McConkey, B.J.; et al. Pathogen-secreted proteases activate a novel plant immune pathway. Nature 2015, 521, 213–216. [Google Scholar] [CrossRef]
- El-Shetehy, M.; Wang, C.; Shine, M.B.; Yu, K.; Kachroo, A.; Kachroo, P. Nitric oxide and reactive oxygen species are required for systemic acquired resistance in plants. Plant Signal. Behav. 2015, 10, e998544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wittek, F.; Hoffmann, T.; Kanawati, B.; Bichlmeier, M.; Knappe, C.; Wenig, M.; Schmitt-Kopplin, P.; Parker, J.E.; Schwab, W.; Vlot, A.C. Arabidopsis ENHANCED DISEASE SUSCEPTIBILITY1 promotes systemic acquired resistance via azelaic acid and its precursor 9-oxo nonanoic acid. J. Exp. Bot. 2014, 65, 5919–5931. [Google Scholar] [CrossRef] [Green Version]
- Verberne, M.C.; Hoekstra, J.; Bol, J.F.; Linthorst, H.J.M. Signaling of systemic acquired resistance in tobacco depends on ethylene perception. Plant J. 2003, 35, 27–32. [Google Scholar] [CrossRef]
- Efroni, I.; Birnbaum, K.D. The potential of single-cell profiling in plants. Genome Biol. 2016, 17, 65. [Google Scholar] [CrossRef] [Green Version]
- Krysan, P.J.; Colcombet, J. Cellular Complexity in MAPK Signaling in Plants: Questions and Emerging Tools to Answer Them. Front. Plant Sci. 2018, 9, 1674. [Google Scholar] [CrossRef]
- Zaman, N.; Seitz, K.; Kabir, M.; George-Schreder, L.S.; Shepstone, I.; Liu, Y.; Zhang, S.; Krysan, P.J. A Förster resonance energy transfer sensor for live-cell imaging of mitogen-activated protein kinase activity in Arabidopsis. Plant J. 2019, 97, 970–983. [Google Scholar] [CrossRef] [PubMed]
- Mao, G.; Meng, X.; Liu, Y.; Zheng, Z.; Chen, Z.; Zhang, S. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 2011, 23, 1639–1653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mine, A.; Berens, M.L.; Nobori, T.; Anver, S.; Fukumoto, K.; Winkelmüller, T.M.; Takeda, A.; Becker, D.; Tsuda, K. Pathogen exploitation of an abscisic acid- and jasmonate-inducible MAPK phosphatase and its interception by Arabidopsis immunity. Proc. Natl. Acad. Sci. USA 2017, 114, 7456–7461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eckardt, N.A. Phosphatase AP2C1 Is a Key Component of MAPK Signaling in Arabidopsis. Plant Cell 2007, 19, 2098. [Google Scholar] [CrossRef] [Green Version]
- Brock, A.K.; Willmann, R.; Kolb, D.; Grefen, L.; Lajunen, H.M.; Bethke, G.; Lee, J.; Nürnberger, T.; Gust, A.A. The Arabidopsis Mitogen-Activated Protein Kinase Phosphatase PP2C5 Affects Seed Germination, Stomatal Aperture, and Abscisic Acid-Inducible Gene Expression. Plant Physiol. 2010, 153, 1098–1111. [Google Scholar] [CrossRef] [Green Version]
- Bartels, S.; Anderson, J.C.; González Besteiro, M.A.; Carreri, A.; Hirt, H.; Buchala, A.; Métraux, J.P.; Peck, S.C.; Ulm, R. Map kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1 -mediated responses in Arabidopsis. Plant Cell 2009, 21, 2884–2897. [Google Scholar] [CrossRef] [Green Version]
- Lumbreras, V.; Vilela, B.; Irar, S.; Solé, M.; Capellades, M.; Valls, M.; Coca, M.; Pagès, M. MAPK phosphatase MKP2 mediates disease responses in Arabidopsis and functionally interacts with MPK3 and MPK6. Plant J. 2010, 63, 1017–1030. [Google Scholar] [CrossRef]
- Furlan, G.; Nakagami, H.; Eschen-Lippold, L.; Jiang, X.; Majovsky, P.; Kowarschik, K.; Hoehenwarter, W.; Lee, J.; Trujillo, M. Changes in PUB22 Ubiquitination Modes Triggered by MITOGEN-ACTIVATED PROTEIN KINASE3 Dampen the Immune Response. Plant Cell 2017, 29, 726–745. [Google Scholar] [CrossRef] [Green Version]
- Kadota, Y.; Liebrand, T.W.H.; Goto, Y.; Sklenar, J.; Derbyshire, P.; Menke, F.L.H.; Torres, M.A.; Molina, A.; Zipfel, C.; Coaker, G.; et al. Quantitative phosphoproteomic analysis reveals common regulatory mechanisms between effector- and PAMP-triggered immunity in plants. New Phytol. 2019, 221, 2160–2175. [Google Scholar] [CrossRef]
- McConnell, E.W.; Berg, P.; Westlake, T.J.; Wilson, K.M.; Popescu, G.V.; Hicks, L.M.; Popescu, S.C. Proteome-Wide Analysis of Cysteine Reactivity during Effector-Triggered Immunity. Plant Physiol. 2019, 179, 1248–1264. [Google Scholar] [CrossRef] [Green Version]
- Yang, K.Y.; Liu, Y.; Zhang, S. Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc. Natl. Acad. Sci. USA 2001, 98, 741–746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Jin, H.; Yang, K.Y.; Kim, C.Y.; Baker, B.; Zhang, S. Interaction between two mitogen-activated protein kinases during tobacco defense signaling. Plant J. 2003, 34, 149–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, H.; Liu, Y.; Yang, K.Y.; Kim, C.Y.; Baker, B.; Zhang, S. Function of a mitogen-activated protein kinase pathway in N gene-mediated resistance in tobacco. Plant J. 2003, 33, 719–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, D.; Yang, K.Y.; Li, G.J.; Liu, Y.; Zhang, S. Activation of Ntf4, a tobacco mitogen-activated protein kinase, during plant defense response and its involvement in hypersensitive response-like cell death. Plant Physiol. 2006, 141, 1482–1493. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Ren, D.; Pike, S.; Pallardy, S.; Gassmann, W.; Zhang, S. Chloroplast-generated reactive oxygen species are involved in hypersensitive response-like cell death mediated by a mitogen-activated protein kinase cascade. Plant J. 2007, 51, 941–954. [Google Scholar] [CrossRef]
- Del Pozo, O.; Pedley, K.F.; Martin, G.B. MAPKKKα is a positive regulator of cell death associated with both plant immunity and disease. EMBO J. 2004, 23, 3072–3082. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.Y.; Liu, Y.; Thorne, E.T.; Yang, H.; Fukushige, H.; Gassmann, W.; Hildebrand, D.; Sharp, R.E.; Zhang, S. Activation of a Stress-Responsive Mitogen-Activated Protein Kinase Cascade Induces the Biosynthesis of Ethylene in Plants. Plant Cell 2003, 15, 2707–2718. [Google Scholar] [CrossRef] [Green Version]
- Ren, D.; Yang, H.; Zhang, S. Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J. Biol. Chem. 2002, 277, 559–565. [Google Scholar] [CrossRef] [Green Version]
- Genot, B.; Lang, J.; Berriri, S.; Garmier, M.; Gilard, F.; Pateyron, S.; Haustraete, K.; Van Der Streaten, D.; Hirt, H.; Colcombet, J. Constitutively active arabidopsis MAP kinase 3 triggers defense responses involving salicylic acid and SUMM2 resistance protein. Plant Physiol. 2017, 174, 1238–1249. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Meng, J.; Meng, X.; Zhao, Y.; Liu, J.; Sun, T.; Liu, Y.; Wang, Q.; Zhang, S. Pathogen-Responsive MPK3 and MPK6 Reprogram the Biosynthesis of Indole Glucosinolates and Their Derivatives in Arabidopsis Immunity. Plant Cell 2016, 28, 1144–1162. [Google Scholar] [CrossRef] [Green Version]
- Gravino, M.; Savatin, D.V.; Macone, A.; De Lorenzo, G. Ethylene production in Botrytis cinerea- and oligogalacturonide-induced immunity requires calcium-dependent protein kinases. Plant J. 2015, 84, 1073–1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Meng, X.; Wang, R.; Mao, G.; Han, L.; Liu, Y.; Zhang, S. Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in arabidopsis. PLoS Genet. 2012, 8, e1002767. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, S. Phosphorylation of 1-Aminocyclopropane-1-Carboxylic Acid Synthase by MPK6, a Stress-Responsive Mitogen-Activated Protein Kinase, Induces Ethylene Biosynthesis in Arabidopsis. Plant Cell 2004, 16, 3386–3399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lang, J.; Genot, B.; Hirt, H.; Colcombet, J. Constitutive activity of the Arabidopsis MAP Kinase 3 confers resistance to Pseudomonas syringae and drives robust immune responses. Plant Signal. Behav. 2017, 12, e1356533. [Google Scholar] [CrossRef]
- Xu, J.; Xie, J.; Yan, C.; Zou, X.; Ren, D.; Zhang, S. A chemical genetic approach demonstrates that MPK3/MPK6 activation and NADPH oxidase-mediated oxidative burst are two independent signaling events in plant immunity. Plant J. 2014, 77, 222–234. [Google Scholar] [CrossRef]
- Li, S.; Han, X.; Yang, L.; Deng, X.; Wu, H.; Zhang, M.; Liu, Y.; Zhang, S.; Xu, J. Mitogen-activated protein kinases and calcium-dependent protein kinases are involved in wounding-induced ethylene biosynthesis in Arabidopsis thaliana. Plant. Cell Environ. 2018, 41, 134–147. [Google Scholar] [CrossRef]
- Zhang, J.; Gao, J.; Zhu, Z.; Song, Y.; Wang, X.; Wang, X.; Zhou, X. MKK4/MKK5-MPK1/MPK2 cascade mediates SA-activated leaf senescence via phosphorylation of NPR1 in Arabidopsis. Plant Mol. Biol. 2020, 102, 463–475. [Google Scholar] [CrossRef]
- Dóczi, R.; Bögre, L. The Quest for MAP Kinase Substrates: Gaining Momentum. Trends Plant Sci. 2018, 23, 918–932. [Google Scholar] [CrossRef]
- Pitzschke, A. Modes of MAPK substrate recognition and control. Trends Plant Sci. 2015, 20, 49–55. [Google Scholar] [CrossRef]
- Menzel, W.; Stenzel, I.; Helbig, L.M.; Krishnamoorthy, P.; Neumann, S.; Eschen-Lippold, L.; Heilmann, M.; Lee, J.; Heilmann, I. A PAMP-triggered MAPK cascade inhibits phosphatidylinositol 4,5-bisphosphate production by PIP5K6 in Arabidopsis thaliana. New Phytol. 2019, 224, 833–847. [Google Scholar] [CrossRef]
- Joo, S.; Liu, Y.; Lueth, A.; Zhang, S. MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J. 2008, 54, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Xu, J.; He, Y.; Yang, K.-Y.; Mordorski, B.; Liu, Y.; Zhang, S. Phosphorylation of an ERF Transcription Factor by Arabidopsis MPK3/MPK6 Regulates Plant Defense Gene Induction and Fungal Resistance. Plant Cell 2013, 25, 1126–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Shang, Y.; Fan, B.; Yu, J.Q.; Chen, Z. Arabidopsis LIP5, a Positive Regulator of Multivesicular Body Biogenesis, Is a Critical Target of Pathogen-Responsive MAPK Cascade in Plant Basal Defense. PLoS Pathog. 2014, 10, e1004243. [Google Scholar] [CrossRef]
- Perraki, A.; DeFalco, T.A.; Derbyshire, P.; Avila, J.; Séré, D.; Sklenar, J.; Qi, X.; Stransfeld, L.; Schwessinger, B.; Kadota, Y.; et al. Phosphocode-dependent functional dichotomy of a common co-receptor in plant signalling. Nature 2018, 561, 248–252. [Google Scholar] [CrossRef]
- Brunet, A.; Roux, D.; Lenormand, P.; Dowd, S.; Keyse, S.; Pouysségur, J. Nuclear translocation of p42/p44 mitogen-activated protein kinase is required for growth factor-induced gene expression and cell cycle entry. EMBO J. 1999, 18, 664–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Rudd, J.J.; Macioszek, V.K.; Scheel, D. Dynamic Changes in the Localization of MAPK Cascade Components Controlling Pathogenesis-related (PR) Gene Expression during Innate Immunity in Parsley. J. Biol. Chem. 2004, 279, 22440–22448. [Google Scholar] [CrossRef] [Green Version]
- Marshall, C.J. Specificity of receptor tyrosine kinase signaling: Transient versus sustained extracellular signal-regulated kinase activation. Cell 1995, 80, 179–185. [Google Scholar] [CrossRef] [Green Version]
- Mylona, A.; Theillet, F.X.; Foster, C.; Cheng, T.M.; Miralles, F.; Bates, P.A.; Selenko, P.; Treisman, R. Opposing effects of Elk-1 multisite phosphorylation shape its response to ERK activation. Science 2016, 354, 233–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, S.-D.; Cho, Y.-H.; Tena, G.; Xiong, Y.; Sheen, J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 2008, 451, 789–795. [Google Scholar] [CrossRef]
- Hoehenwarter, W.; Thomas, M.; Nukarinen, E.; Egelhofer, V.; Rohrig, H.; Weckwerth, W.; Conrath, U.; Beckers, G.J.M. Identification of novel in vivo MAP kinase substrates in arabidopsis thaliana through use of tandem metal oxide affinity chromatography. Mol. Cell. Proteomics 2013, 12, 369–380. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Eschen-Lippold, L.; Lassowskat, I.; Böttcher, C.; Scheel, D. Cellular reprogramming through mitogen-activated protein kinases. Front. Plant Sci. 2015, 6, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pecher, P.; Eschen-Lippold, L.; Herklotz, S.; Kuhle, K.; Naumann, K.; Bethke, G.; Uhrig, J.; Weyhe, M.; Scheel, D.; Lee, J. The Arabidopsis thaliana mitogen-activated protein kinases MPK3 and MPK6 target a subclass of ‘VQ-motif’-containing proteins to regulate immune responses. New Phytol. 2014, 203, 592–606. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, X.; He, Y.; Sang, T.; Wang, P.; Dai, S.; Zhang, S.; Meng, X. Differential Phosphorylation of the Transcription Factor WRKY33 by the Protein Kinases CPK5/CPK6 and MPK3/MPK6 Cooperatively Regulates Camalexin Biosynthesis in Arabidopsis. Plant Cell 2020, 32, 2621–2638. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lang, J.; Colcombet, J. Sustained Incompatibility between MAPK Signaling and Pathogen Effectors. Int. J. Mol. Sci. 2020, 21, 7954. https://doi.org/10.3390/ijms21217954
Lang J, Colcombet J. Sustained Incompatibility between MAPK Signaling and Pathogen Effectors. International Journal of Molecular Sciences. 2020; 21(21):7954. https://doi.org/10.3390/ijms21217954
Chicago/Turabian StyleLang, Julien, and Jean Colcombet. 2020. "Sustained Incompatibility between MAPK Signaling and Pathogen Effectors" International Journal of Molecular Sciences 21, no. 21: 7954. https://doi.org/10.3390/ijms21217954




