The Role of miR-342 in Vascular Health. Study in Subclinical Cardiovascular Disease in Mononuclear Cells, Plasma, Inflammatory Cytokines and PANX2
Abstract
1. Introduction
2. Results
2.1. Patient Phenotypes
2.2. MicroRNA-342 Isoform Expression
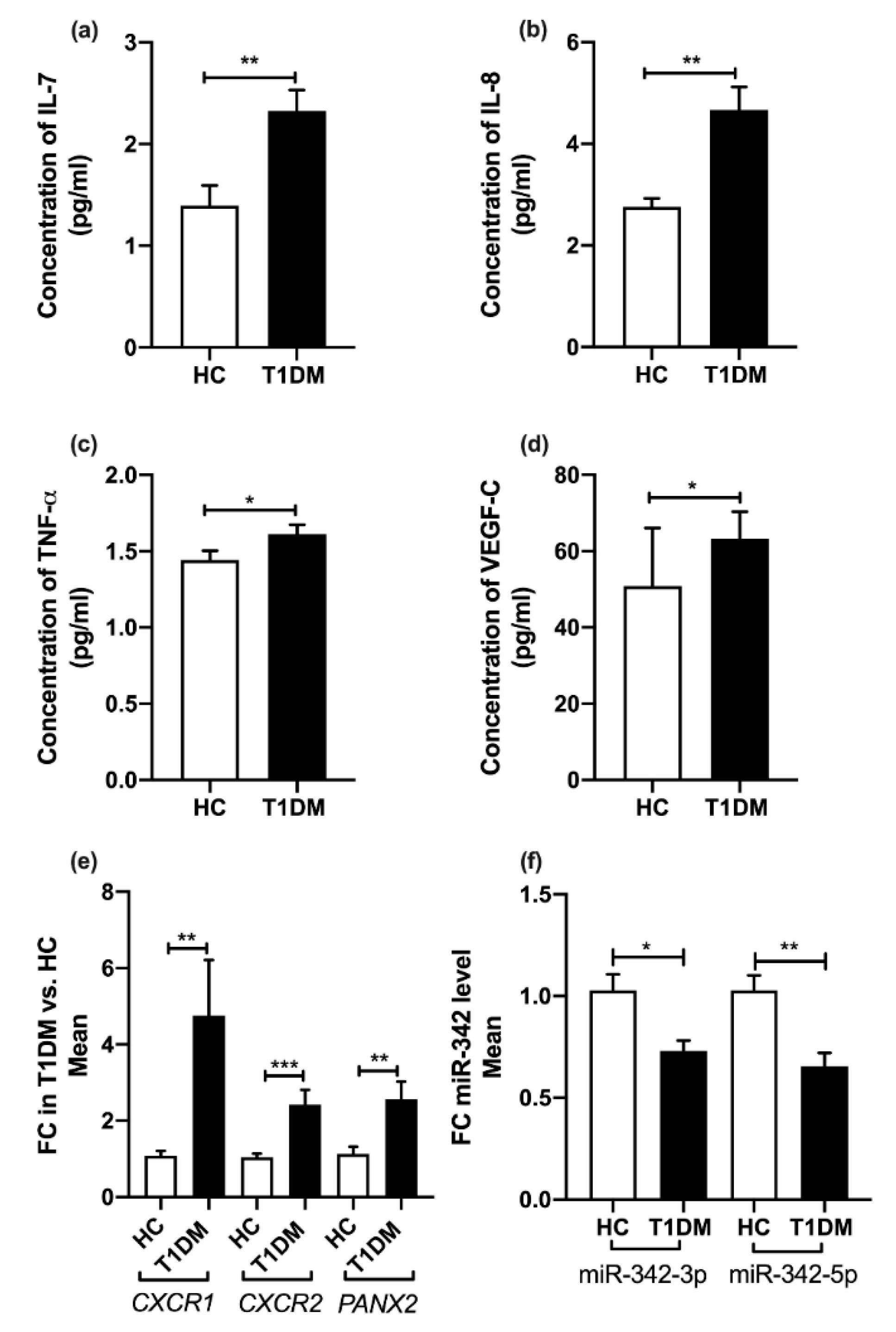
2.3. Cytokine Profiles
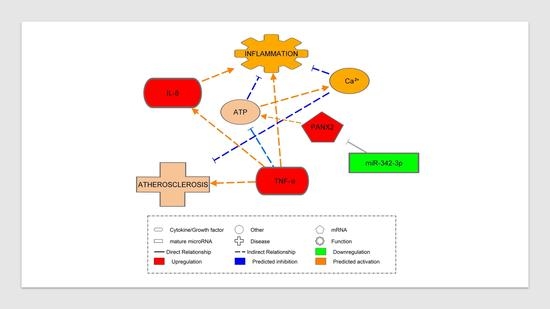
2.4. Pannexin-2 (PANX2) (Target of miR-342-3p) mRNA Expression
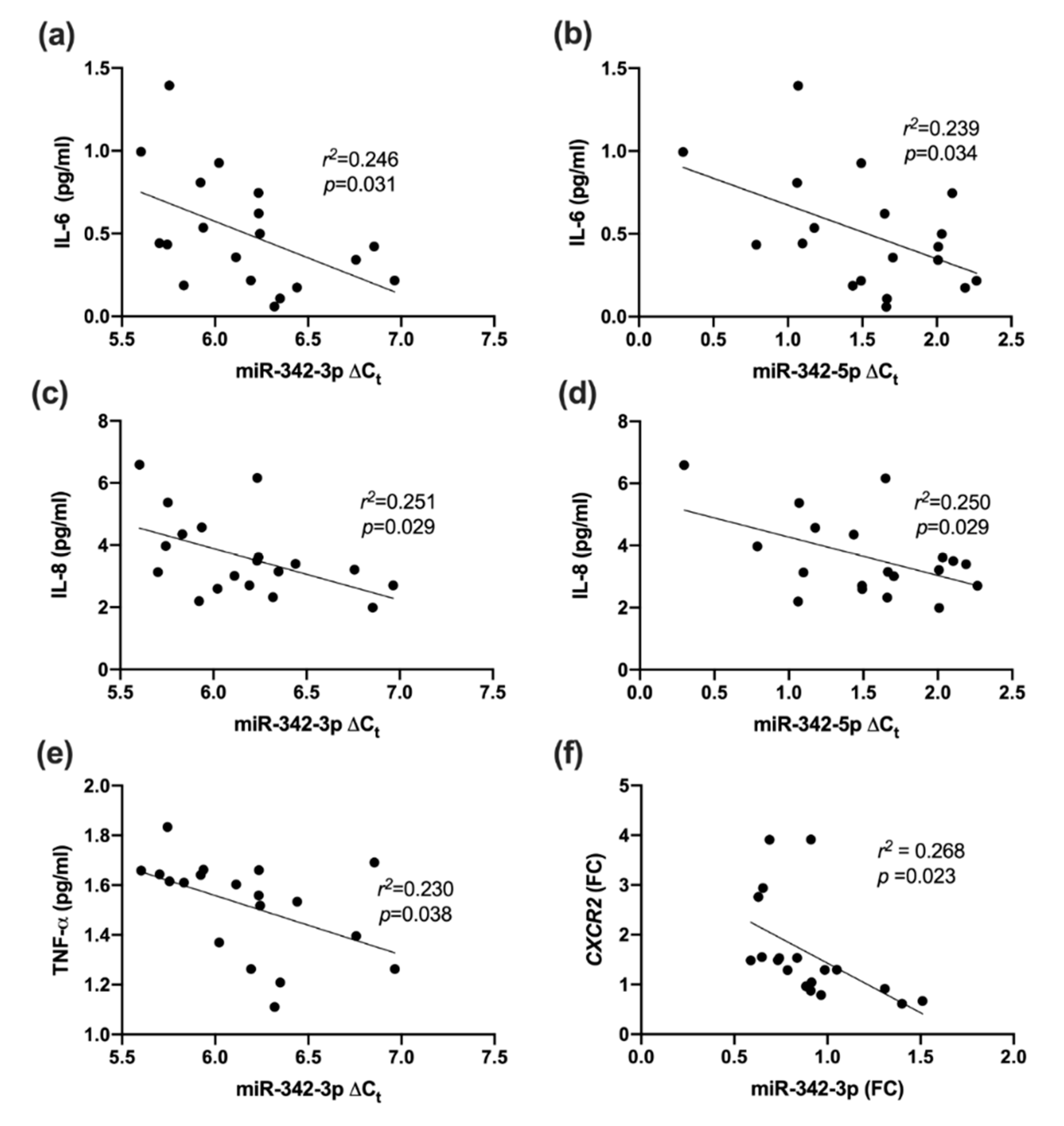
2.5. Association between miR-342 Isoforms and Inflammatory Markers
2.6. Association between miR-342 Isoforms and Inflammatory Receptors
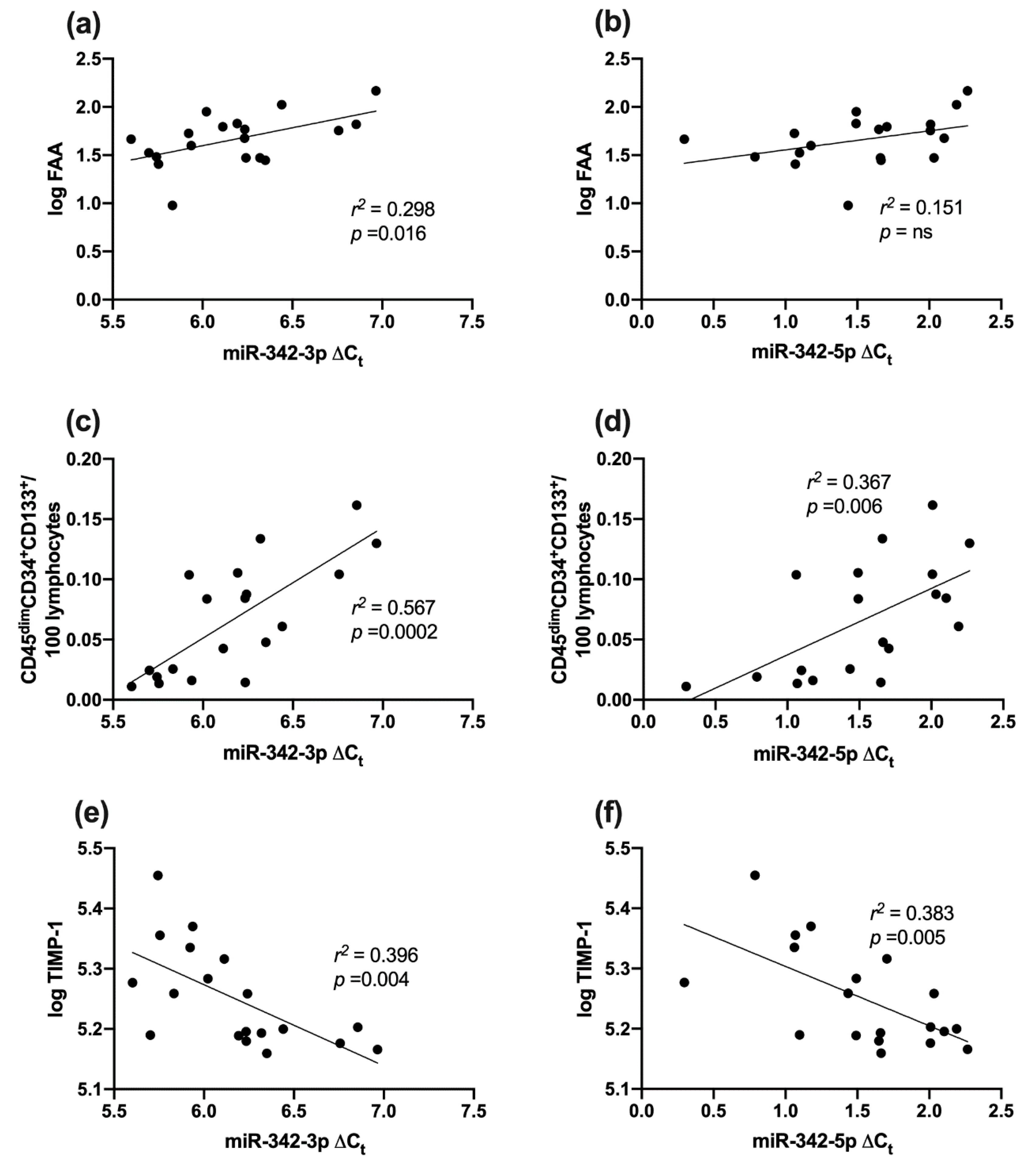
2.7. Association between miR-342 Isoforms and Vascular Health (Fibronectin Adhesion Assay, CD45dimCD34+133+ Cells and TIMP-1)
2.8. Correlation between miR-342 Isoforms and HbA1c
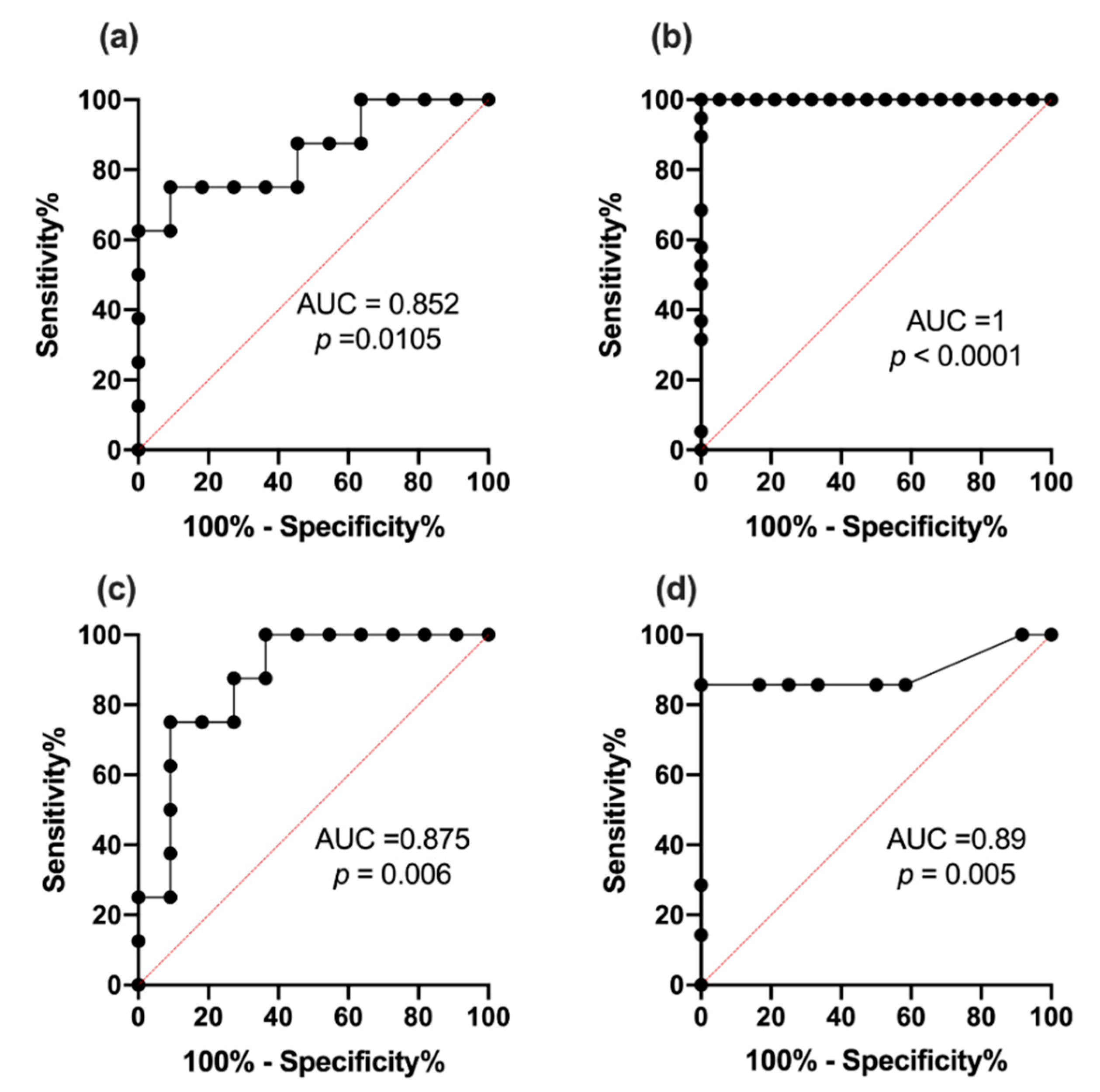
2.9. Receiver Operating Characteristic Curve Analysis for miR342-3p and 342-5p
3. Discussion
3.1. Inflammatory Markers in T1DM
3.2. MiR-342-3p Expression in PBMC in T1DM
3.3. MiR-342-5p Expression in PBMCs in T1DM
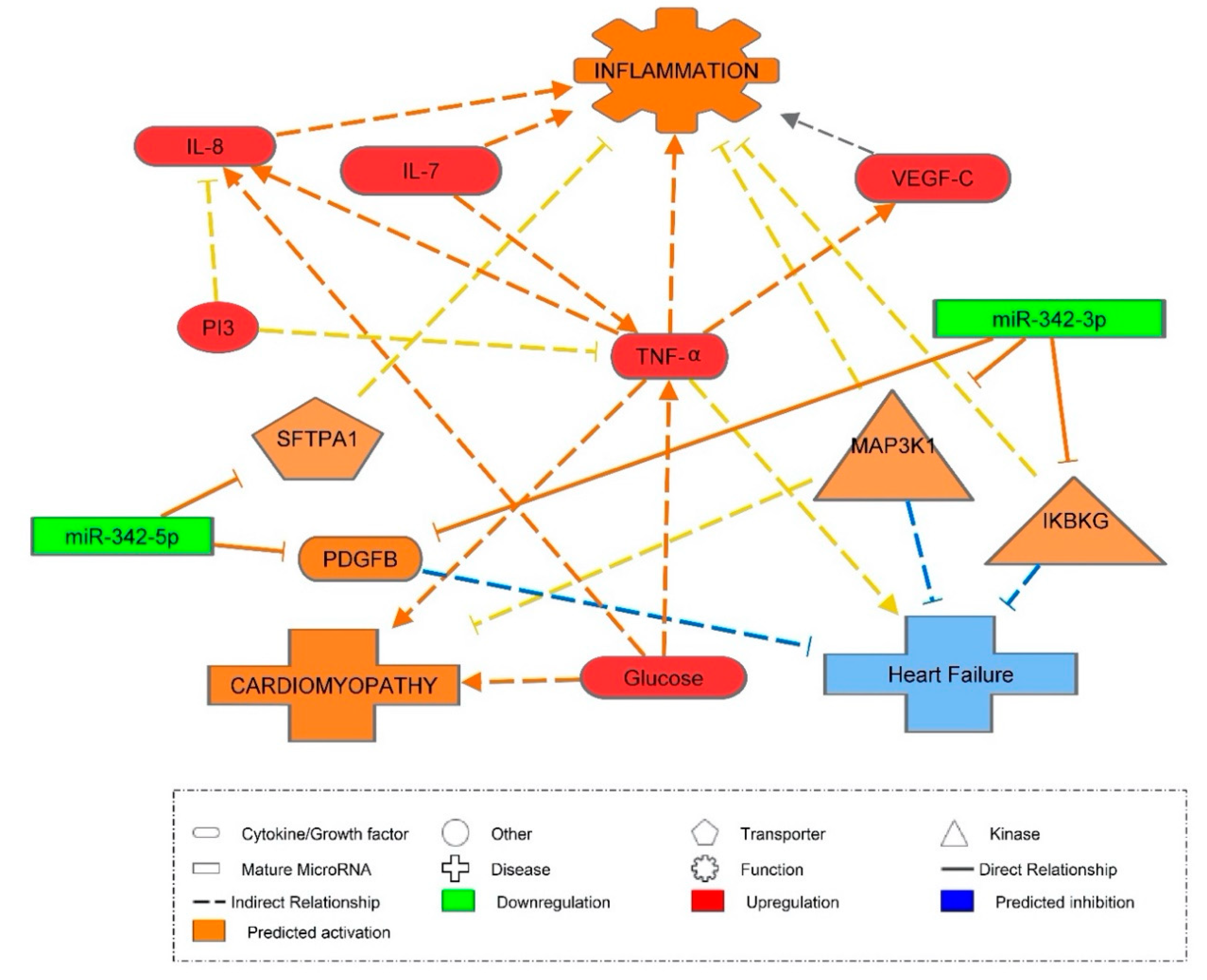
3.4. Inflammatory Marker Associations with miR-342
3.5. Vascular Health and miR-342
3.6. miR-342 Affecting Hyperglycemia
3.7. ROC Analysis
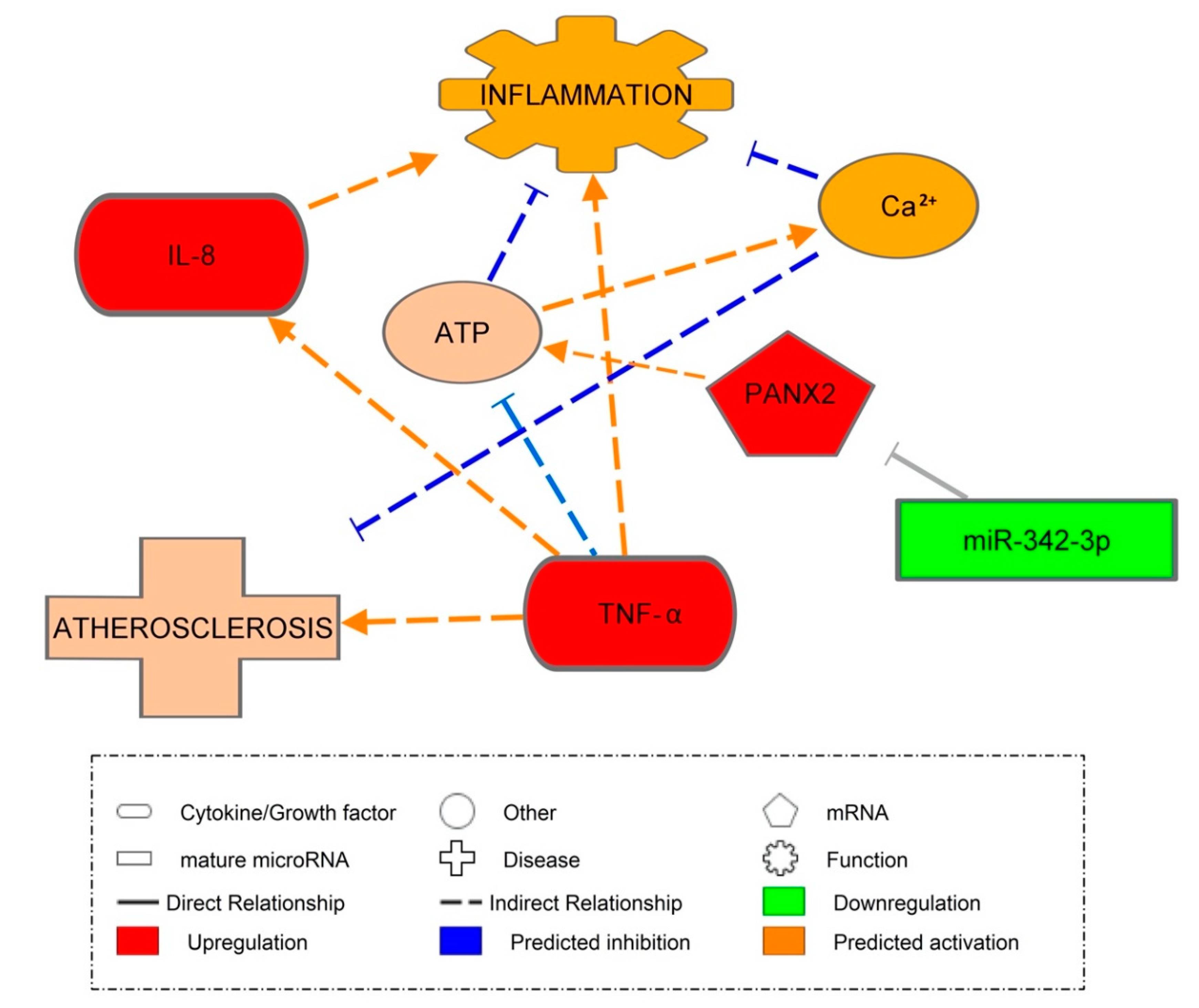
3.8. Pannexin-2 mRNA Target of miR-342-3p
4. Materials and Methods
4.1. Meso Scale Discovery (MSD) Assay
4.2. Flow Cytometric Evaluation of Circulating Endothelial Progenitor Cells
4.3. Fibronectin Adhesion Assay (FAA)
4.4. Extraction of MicroRNAs from Plasma
4.5. Extraction of MicroRNA and mRNA from PBMCs
4.6. Assay of miRNA and mRNA Using Real-Time Quantitative PCR
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| cEPC | Circulating endothelial cell |
| CFU-Hill | Colony forming Unit-Hills colony |
| CVD | Cardiovascular disease |
| CXCR1/2 | Chemokine receptor |
| FAA | Fibronectin Adhesion Assay |
| IKBKG | Inhibitor of nuclear factor kappa B kinase regulatory subunit Gamma |
| IPA | Ingenuity Pathway Analysis |
| MAP3K1 | Mitogen-activated protein 3 kinase 1 |
| PANX2 | Pannexin-2 |
| PBMCs | Peripheral blood mononuclear cells |
| SFTPA1 | Surfactant protein A1 |
| T1DM | Type 1 diabetes mellitus |
| TIMP-1 | Tissue inhibitor of metalloproteases |
References
- Schram, M.T.; Chaturvedi, N.; Schalkwijk, C.G.; Fuller, J.H.; Stehouwer, C.D.; Group, E.P.C.S. Markers of inflammation are cross-sectionally associated with microvascular complications and cardiovascular disease in type 1 diabetes--the EURODIAB Prospective Complications Study. Diabetologia 2005, 48, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Livingstone, S.J.; Levin, D.; Looker, H.C.; Lindsay, R.S.; Wild, S.H.; Joss, N.; Leese, G.; Leslie, P.; McCrimmon, R.J.; Metcalfe, W.; et al. Estimated life expectancy in a Scottish cohort with type 1 diabetes, 2008-2010. JAMA 2015, 313, 37–44. [Google Scholar] [CrossRef]
- Sibal, L.; Aldibbiat, A.; Agarwal, S.C.; Mitchell, G.; Oates, C.; Razvi, S.; Weaver, J.U.; Shaw, J.A.; Home, P.D. Circulating endothelial progenitor cells, endothelial function, carotid intima-media thickness and circulating markers of endothelial dysfunction in people with type 1 diabetes without macrovascular disease or microalbuminuria. Diabetologia 2009, 52, 1464–1473. [Google Scholar] [CrossRef]
- Ahmed, F.W.; Rider, R.; Glanville, M.; Narayanan, K.; Razvi, S.; Weaver, J.U. Metformin improves circulating endothelial cells and endothelial progenitor cells in type 1 diabetes: MERIT study. Cardiovasc. Diabetol. 2016, 15, 116. [Google Scholar] [CrossRef] [PubMed]
- West, D.J.; Campbell, M.D.; Gonzalez, J.T.; Walker, M.; Stevenson, E.J.; Ahmed, F.W.; Wijaya, S.; Shaw, J.A.; Weaver, J.U. The inflammation, vascular repair and injury responses to exercise in fit males with and without Type 1 diabetes: An observational study. Cardiovasc. Diabetol. 2015, 14, 71. [Google Scholar] [CrossRef] [PubMed]
- Werner, N.; Kosiol, S.; Schiegl, T.; Ahlers, P.; Walenta, K.; Link, A.; Bohm, M.; Nickenig, G. Circulating endothelial progenitor cells and cardiovascular outcomes. N. Engl. J. Med. 2005, 353, 999–1007. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.M.; Zalos, G.; Halcox, J.P.; Schenke, W.H.; Waclawiw, M.A.; Quyyumi, A.A.; Finkel, T. Circulating endothelial progenitor cells, vascular function, and cardiovascular risk. N. Engl. J. Med. 2003, 348, 593–600. [Google Scholar] [CrossRef]
- Clark, M.; Kroger, C.J.; Tisch, R.M. Type 1 Diabetes: A Chronic Anti-Self-Inflammatory Response. Front. Immunol. 2017, 8, 1898. [Google Scholar] [CrossRef]
- Hulsmans, M.; Holvoet, P. MicroRNA-containing microvesicles regulating inflammation in association with atherosclerotic disease. Cardiovasc. Res. 2013, 100, 7–18. [Google Scholar] [CrossRef]
- Schulte, C.; Barwari, T.; Joshi, A.; Theofilatos, K.; Zampetaki, A.; Barallobre-Barreiro, J.; Singh, B.; Sorensen, N.A.; Neumann, J.T.; Zeller, T.; et al. Comparative Analysis of Circulating Noncoding RNAs Versus Protein Biomarkers in the Detection of Myocardial Injury. Circ. Res. 2019, 125, 328–340. [Google Scholar] [CrossRef]
- Kantharidis, P.; Wang, B.; Carew, R.M.; Lan, H.Y. Diabetes complications: The microRNA perspective. Diabetes 2011, 60, 1832–1837. [Google Scholar] [CrossRef]
- Assmann, T.S.; Recamonde-Mendoza, M.; De Souza, B.M.; Crispim, D. MicroRNA expression profiles and type 1 diabetes mellitus: Systematic review and bioinformatic analysis. Endocr. Connect. 2017, 6, 773–790. [Google Scholar] [CrossRef] [PubMed]
- Butz, H.; Kinga, N.; Racz, K.; Patocs, A. Circulating miRNAs as biomarkers for endocrine disorders. J. Endocrinol. Investig. 2016, 39, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Simonson, B.; Das, S. MicroRNA Therapeutics: The Next Magic Bullet? Mini Rev. Med. Chem. 2015, 15, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.Y.; Kim, K.C.; Park, M.H.; Seo, Y.; Park, H.; Park, M.H.; Chang, J.; Hwang, D.Y.; Han, S.B.; Kim, S.; et al. Atherosclerosis is exacerbated by chitinase-3-like-1 in amyloid precursor protein transgenic mice. Theranostics 2018, 8, 749–766. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xia, J.W.; Ke, Z.P.; Zhang, B.H. Blockade of NEAT1 represses inflammation response and lipid uptake via modulating miR-342-3p in human macrophages THP-1 cells. J. Cell. Physiol. 2019, 234, 5319–5326. [Google Scholar] [CrossRef] [PubMed]
- Lozano, C.; Estibals, V.; Duroux-Richard, I.; Apparailly, F. miR-342-3p promotes cell survival and motility of osteoclast precursors. Ann. Rheum. Dis. 2019, 78, 27–28. [Google Scholar] [CrossRef]
- Takahashi, P.; Xavier, D.J.; Evangelista, A.F.; Manoel-Caetano, F.S.; Macedo, C.; Collares, C.V.; Foss-Freitas, M.C.; Foss, M.C.; Rassi, D.M.; Donadi, E.A.; et al. MicroRNA expression profiling and functional annotation analysis of their targets in patients with type 1 diabetes mellitus. Gene 2014, 539, 213–223. [Google Scholar] [CrossRef]
- Hezova, R.; Slaby, O.; Faltejskova, P.; Mikulkova, Z.; Buresova, I.; Raja, K.R.; Hodek, J.; Ovesna, J.; Michalek, J. microRNA-342, microRNA-191 and microRNA-510 are differentially expressed in T regulatory cells of type 1 diabetic patients. Cell. Immunol. 2010, 260, 70–74. [Google Scholar] [CrossRef]
- Chartoumpekis, D.V.; Zaravinos, A.; Ziros, P.G.; Iskrenova, R.P.; Psyrogiannis, A.I.; Kyriazopoulou, V.E.; Habeos, I.G. Differential expression of microRNAs in adipose tissue after long-term high-fat diet-induced obesity in mice. PLoS ONE 2012, 7, e34872. [Google Scholar] [CrossRef]
- Versari, D.; Daghini, E.; Virdis, A.; Ghiadoni, L.; Taddei, S. Endothelial dysfunction as a target for prevention of cardiovascular disease. Diabetes Care 2009, 32 (Suppl. 2), S314–S321. [Google Scholar] [CrossRef]
- Calzascia, T.; Pellegrini, M.; Lin, A.; Garza, K.M.; Elford, A.R.; Shahinian, A.; Ohashi, P.S.; Mak, T.W. CD4 T cells, lymphopenia, and IL-7 in a multistep pathway to autoimmunity. Proc. Natl. Acad. Sci. USA 2008, 105, 2999–3004. [Google Scholar] [CrossRef] [PubMed]
- Monti, P.; Bonifacio, E. Interleukin-7 and type 1 diabetes. Curr. Diab. Rep. 2014, 14, 518. [Google Scholar] [CrossRef] [PubMed]
- Cieri, N.; Camisa, B.; Cocchiarella, F.; Forcato, M.; Oliveira, G.; Provasi, E.; Bondanza, A.; Bordignon, C.; Peccatori, J.; Ciceri, F.; et al. IL-7 and IL-15 instruct the generation of human memory stem T cells from naive precursors. Blood 2013, 121, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Sudo, T.; Nishikawa, S.; Ohno, N.; Akiyama, N.; Tamakoshi, M.; Yoshida, H.; Nishikawa, S. Expression and function of the interleukin 7 receptor in murine lymphocytes. Proc. Natl. Acad. Sci. USA 1993, 90, 9125–9129. [Google Scholar] [CrossRef] [PubMed]
- Lo, H.C.; Lin, S.C.; Wang, Y.M. The relationship among serum cytokines, chemokine, nitric oxide, and leptin in children with type 1 diabetes mellitus. Clin. Biochem. 2004, 37, 666–672. [Google Scholar] [CrossRef] [PubMed]
- Ha, H.; Debnath, B.; Neamati, N. Role of the CXCL8-CXCR1/2 Axis in Cancer and Inflammatory Diseases. Theranostics 2017, 7, 1543–1588. [Google Scholar] [CrossRef]
- Williams, R.O.; Feldmann, M.; Maini, R.N. Anti-tumor necrosis factor ameliorates joint disease in murine collagen-induced arthritis. Proc. Natl. Acad. Sci. USA 1992, 89, 9784–9788. [Google Scholar] [CrossRef]
- Ahmadi, R.; Heidarian, E.; Fadaei, R.; Moradi, N.; Malek, M.; Fallah, S. miR-342-5p Expression Levels in Coronary Artery Disease Patients and its Association with Inflammatory Cytokines. Clin. Lab. 2018, 64, 603–609. [Google Scholar] [CrossRef]
- Wei, Y.; Nazari-Jahantigh, M.; Chan, L.; Zhu, M.; Heyll, K.; Corbalan-Campos, J.; Hartmann, P.; Thiemann, A.; Weber, C.; Schober, A. The microRNA-342-5p fosters inflammatory macrophage activation through an Akt1- and microRNA-155-dependent pathway during atherosclerosis. Circulation 2013, 127, 1609–1619. [Google Scholar] [CrossRef]
- Goon, P.K.; Lip, G.Y.; Boos, C.J.; Stonelake, P.S.; Blann, A.D. Circulating endothelial cells, endothelial progenitor cells, and endothelial microparticles in cancer. Neoplasia 2006, 8, 79–88. [Google Scholar] [CrossRef]
- Avogaro, A.; Albiero, M.; Menegazzo, L.; de Kreutzenberg, S.; Fadini, G.P. Endothelial dysfunction in diabetes: The role of reparatory mechanisms. Diabetes Care 2011, 34 (Suppl. 2), S285–S290. [Google Scholar] [CrossRef]
- McClung, J.A.; Naseer, N.; Saleem, M.; Rossi, G.P.; Weiss, M.B.; Abraham, N.G.; Kappas, A. Circulating endothelial cells are elevated in patients with type 2 diabetes mellitus independently of HbA(1)c. Diabetologia 2005, 48, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Werner, N.; Nickenig, G. Influence of cardiovascular risk factors on endothelial progenitor cells: Limitations for therapy? Arterioscler. Thromb. Vasc. Biol. 2006, 26, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, P.R.; Timms, P.M.; Chandran, S.; Gordon, D. Peripheral blood level alterations of TIMP-1, MMP-2 and MMP-9 in patients with type 1 diabetes. Diabet. Med. 2001, 18, 777–780. [Google Scholar] [CrossRef] [PubMed]
- Ries, C. Cytokine functions of TIMP-1. Cell. Mol. Life Sci. 2014, 71, 659–672. [Google Scholar] [CrossRef]
- Cheng, S.; Cui, Y.; Fan, L.; Mu, X.; Hua, Y. T2DM inhibition of endothelial miR-342-3p facilitates angiogenic dysfunction via repression of FGF11 signaling. Biochem. Biophys. Res. Commun. 2018, 503, 71–78. [Google Scholar] [CrossRef]
- Colagiuri, S.; Lee, C.M.; Wong, T.Y.; Balkau, B.; Shaw, J.E.; Borch-Johnsen, K.; Group, D.-C.W. Glycemic thresholds for diabetes-specific retinopathy: Implications for diagnostic criteria for diabetes. Diabetes Care 2011, 34, 145–150. [Google Scholar] [CrossRef]
- Expert Committee on Drug Dependence; Classification of Diabetes Mellitus. Report of the expert committee on the diagnosis and classification of diabetes mellitus. Diabetes Care 2003, 26 (Suppl. 1), S5–S20. [Google Scholar] [CrossRef]
- D’Hondt, C.; Ponsaerts, R.; De Smedt, H.; Vinken, M.; De Vuyst, E.; De Bock, M.; Wang, N.; Rogiers, V.; Leybaert, L.; Himpens, B.; et al. Pannexin channels in ATP release and beyond: An unexpected rendezvous at the endoplasmic reticulum. Cell. Signal. 2011, 23, 305–316. [Google Scholar] [CrossRef]
- Makarenkova, H.P.; Shestopalov, V.I. The role of pannexin hemichannels in inflammation and regeneration. Front. Physiol. 2014, 5, 63. [Google Scholar] [CrossRef] [PubMed]
- Berchtold, L.A.; Miani, M.; Diep, T.A.; Madsen, A.N.; Cigliola, V.; Colli, M.; Krivokapic, J.M.; Pociot, F.; Eizirik, D.L.; Meda, P.; et al. Pannexin-2-deficiency sensitizes pancreatic beta-cells to cytokine-induced apoptosis in vitro and impairs glucose tolerance in vivo. Mol. Cell. Endocrinol. 2017, 448, 108–121. [Google Scholar] [CrossRef] [PubMed]
- Bond, S.R.; Naus, C.C. The pannexins: Past and present. Front. Physiol. 2014, 5, 58. [Google Scholar] [CrossRef] [PubMed]
- Kanter, J.E.; Bornfeldt, K.E. Inflammation and diabetes-accelerated atherosclerosis: Myeloid cell mediators. Trends Endocrinol. Metab. 2013, 24, 137–144. [Google Scholar] [CrossRef] [PubMed]

| Variable | Healthy Controls (n = 20) | Type 1 Diabetes Mellitus (n = 29) | p-Value |
|---|---|---|---|
| Age (years) | 46.5 ± 11.7 | 47.2 ± 12.7 | 0.8 |
| BMI (kg/m2) | 26.0 ± 4.5 | 28.4 ± 6.7 | 0.3 |
| Gender (M/F) n | 9/11 | 14/15 | - |
| HbA1c (mmol/mol) | 35.1 ± 2.8 | 57.3 ± 7.6 | <0.001 *** |
| HbA1c (%) | 5.4 ± 0.3 | 7.4 ± 0.7 | <0.001 *** |
| Glucose (mmol/L) | 4.6 ± 1.0 | 9.7 ± 3.7 | <0.001 *** |
| Hemoglobin (g/dL) | 14.3 ± 1.2 | 14.5 ±1.2 | 1.0 |
| Triglycerides (mmol/L) | 1.4 ± 0.7 | 0.9 ± 0.4 | 0.008 ** |
| Systolic Blood Pressure (mmHg) | 117.5 ± 14.0 | 127.5 ± 9.4 | 0.005 ** |
| CD45dimCD34+ CD133+ per 100 lymphocytes | 0.009 ± 0.03 | 0.02 ± 0.01 | <0.001 *** |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ray, S.L.; Coulson, D.J.; Yeoh, M.L.Y.; Tamara, A.; Latief, J.S.; Bakhashab, S.; Weaver, J.U. The Role of miR-342 in Vascular Health. Study in Subclinical Cardiovascular Disease in Mononuclear Cells, Plasma, Inflammatory Cytokines and PANX2. Int. J. Mol. Sci. 2020, 21, 7217. https://doi.org/10.3390/ijms21197217
Ray SL, Coulson DJ, Yeoh MLY, Tamara A, Latief JS, Bakhashab S, Weaver JU. The Role of miR-342 in Vascular Health. Study in Subclinical Cardiovascular Disease in Mononuclear Cells, Plasma, Inflammatory Cytokines and PANX2. International Journal of Molecular Sciences. 2020; 21(19):7217. https://doi.org/10.3390/ijms21197217
Chicago/Turabian StyleRay, Sabina L., David J. Coulson, Megan Li Yuen Yeoh, Alice Tamara, Jevi Septyani Latief, Sherin Bakhashab, and Jolanta U. Weaver. 2020. "The Role of miR-342 in Vascular Health. Study in Subclinical Cardiovascular Disease in Mononuclear Cells, Plasma, Inflammatory Cytokines and PANX2" International Journal of Molecular Sciences 21, no. 19: 7217. https://doi.org/10.3390/ijms21197217
APA StyleRay, S. L., Coulson, D. J., Yeoh, M. L. Y., Tamara, A., Latief, J. S., Bakhashab, S., & Weaver, J. U. (2020). The Role of miR-342 in Vascular Health. Study in Subclinical Cardiovascular Disease in Mononuclear Cells, Plasma, Inflammatory Cytokines and PANX2. International Journal of Molecular Sciences, 21(19), 7217. https://doi.org/10.3390/ijms21197217