Systematic Analysis of Gibberellin Pathway Components in Medicago truncatula Reveals the Potential Application of Gibberellin in Biomass Improvement
Abstract
1. Introduction
2. Results
2.1. Genome-Wide Identification of GA20ox, GA3ox, and GA2ox Genes in M. truncatula
2.2. Phylogenetic Analysis and Chromosomal Locations of MtGAox Genes
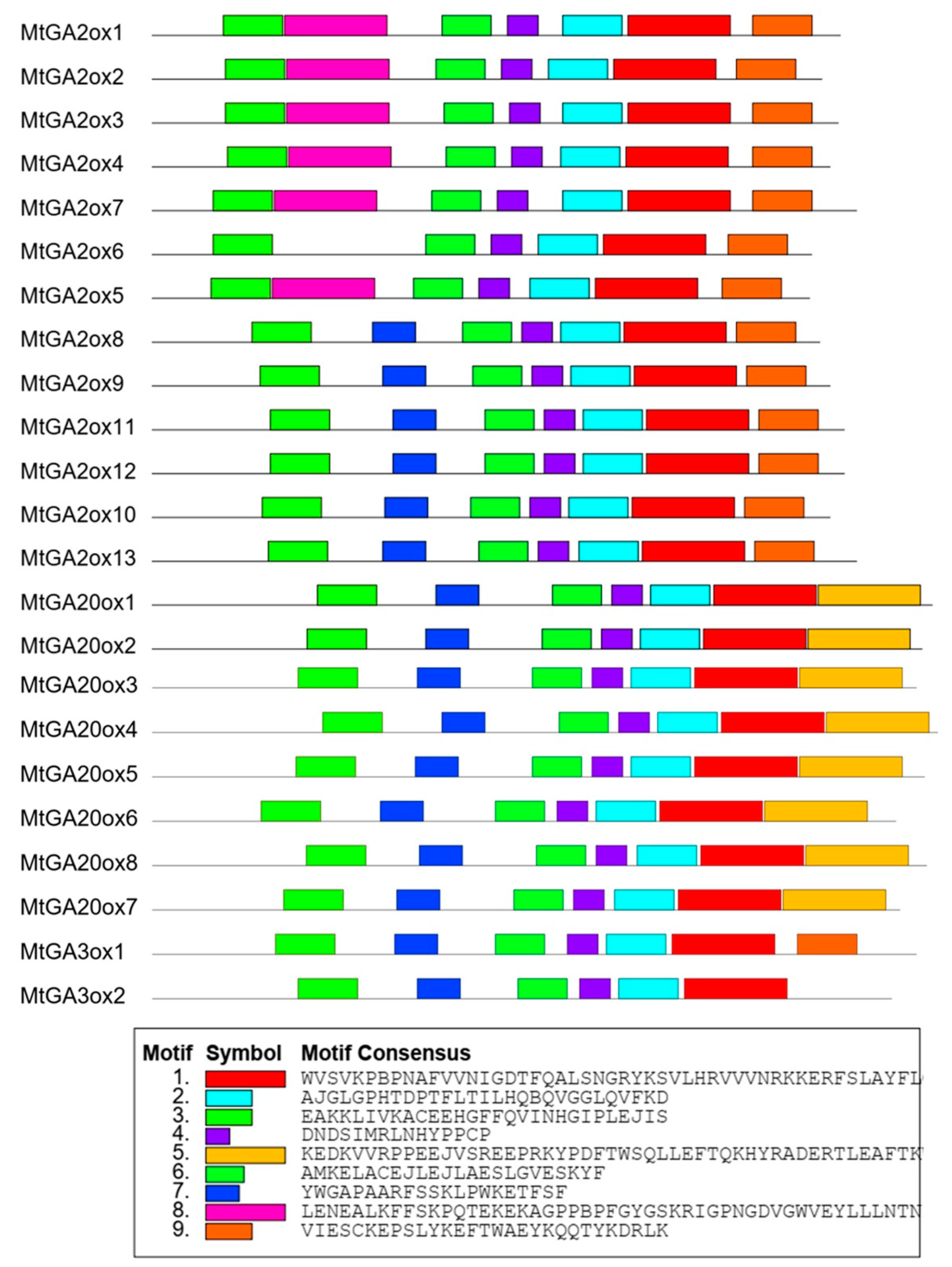
2.3. Gene Structures and Conserved Motifs of MtGAox in M. truncatula
2.4. Expression Patterns of MtGAox Genes
2.5. Identification of GA Receptor MtGID1 and MtDELLA Genes in M. truncatula
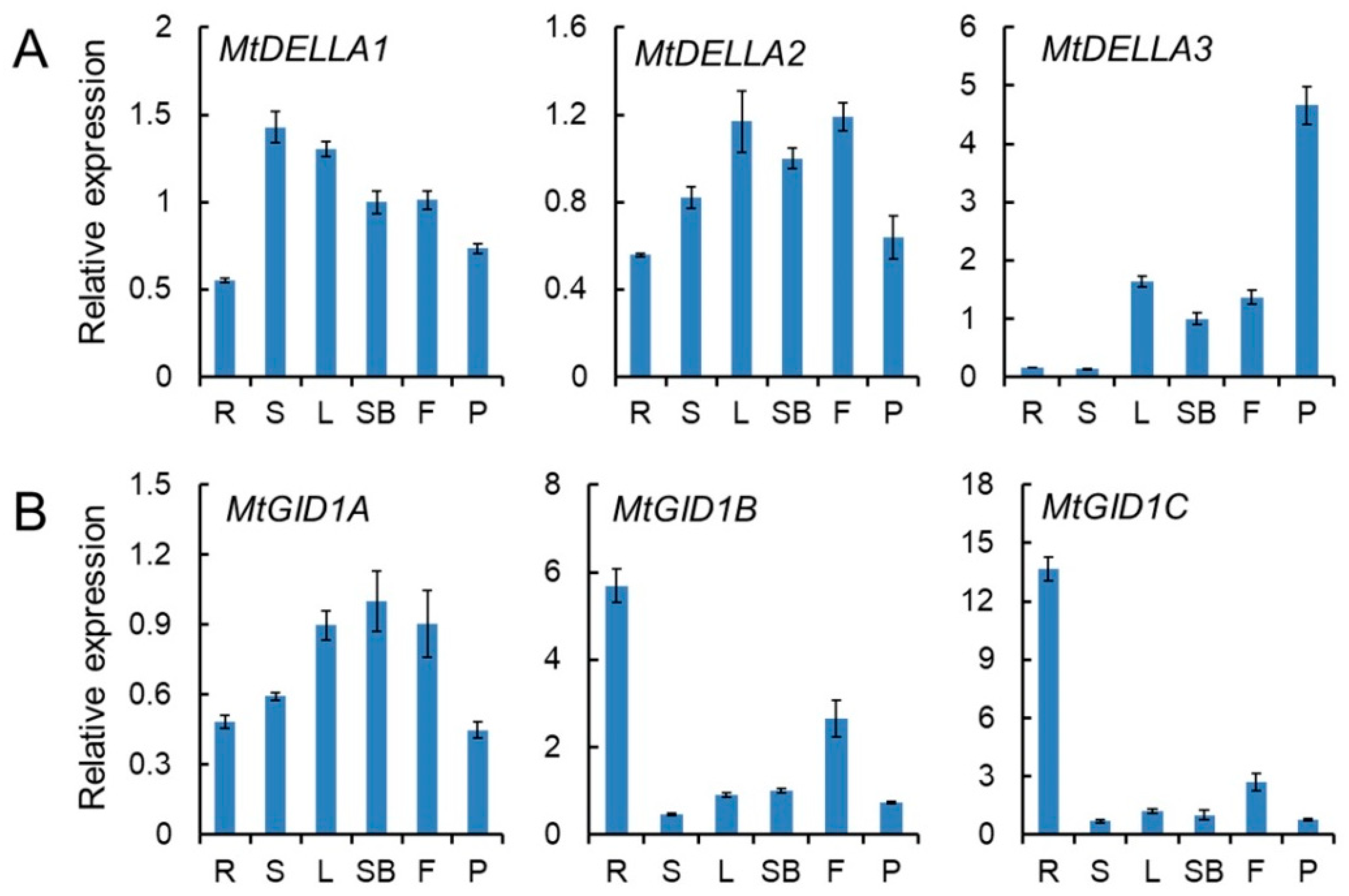
2.6. Expression Patterns of MtGID1 and MtDELLA Genes
2.7. The GA Receptors, MtGID1s, Were Able to Interact with MtDELLA Proteins
2.8. The Transcript Level of MtGAoxs Is Regulated by a Feedback Mechanism
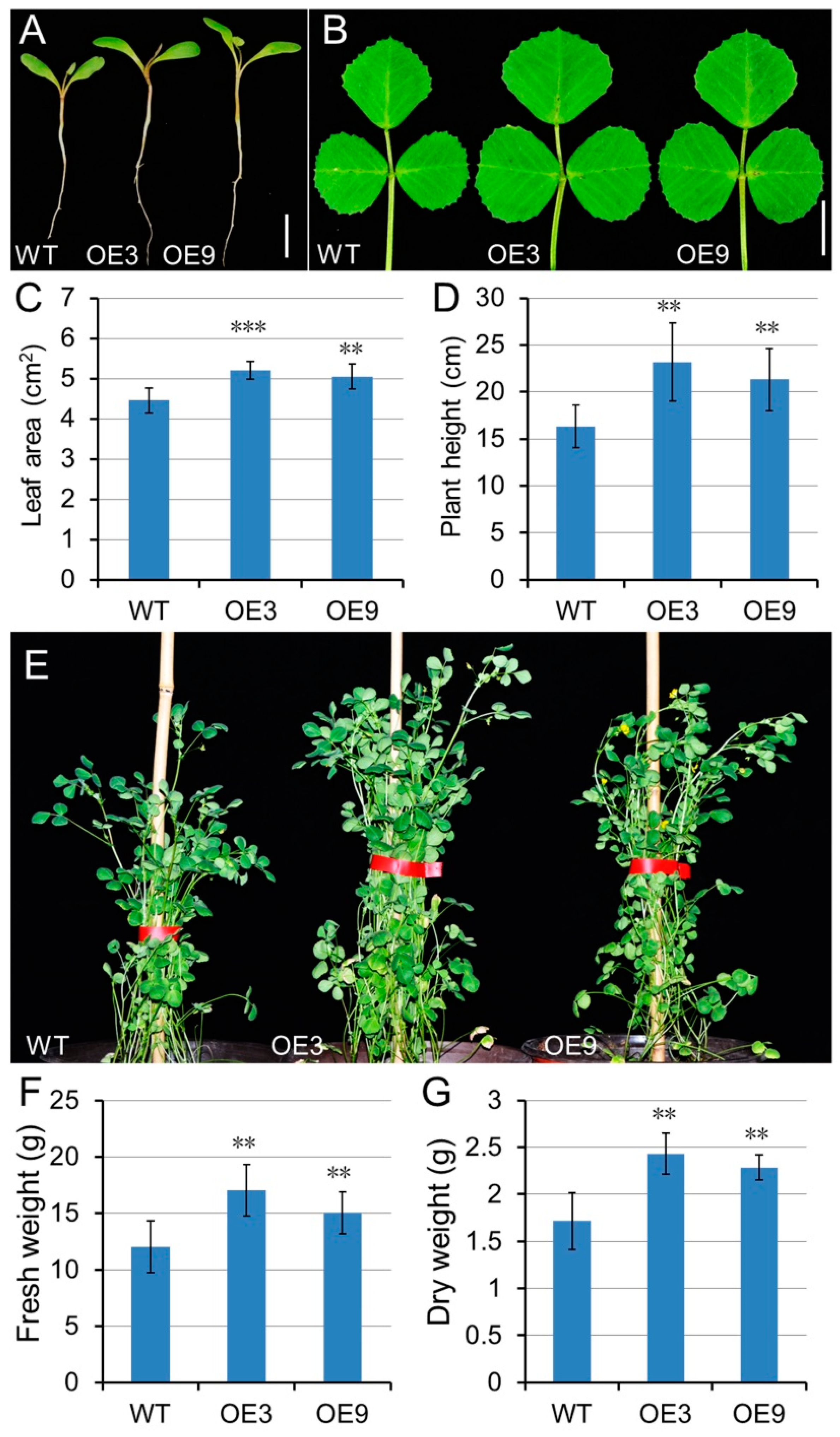
2.9. GA Has a Positive Effect on Growth and Biomass of M. truncatula and Alfalfa
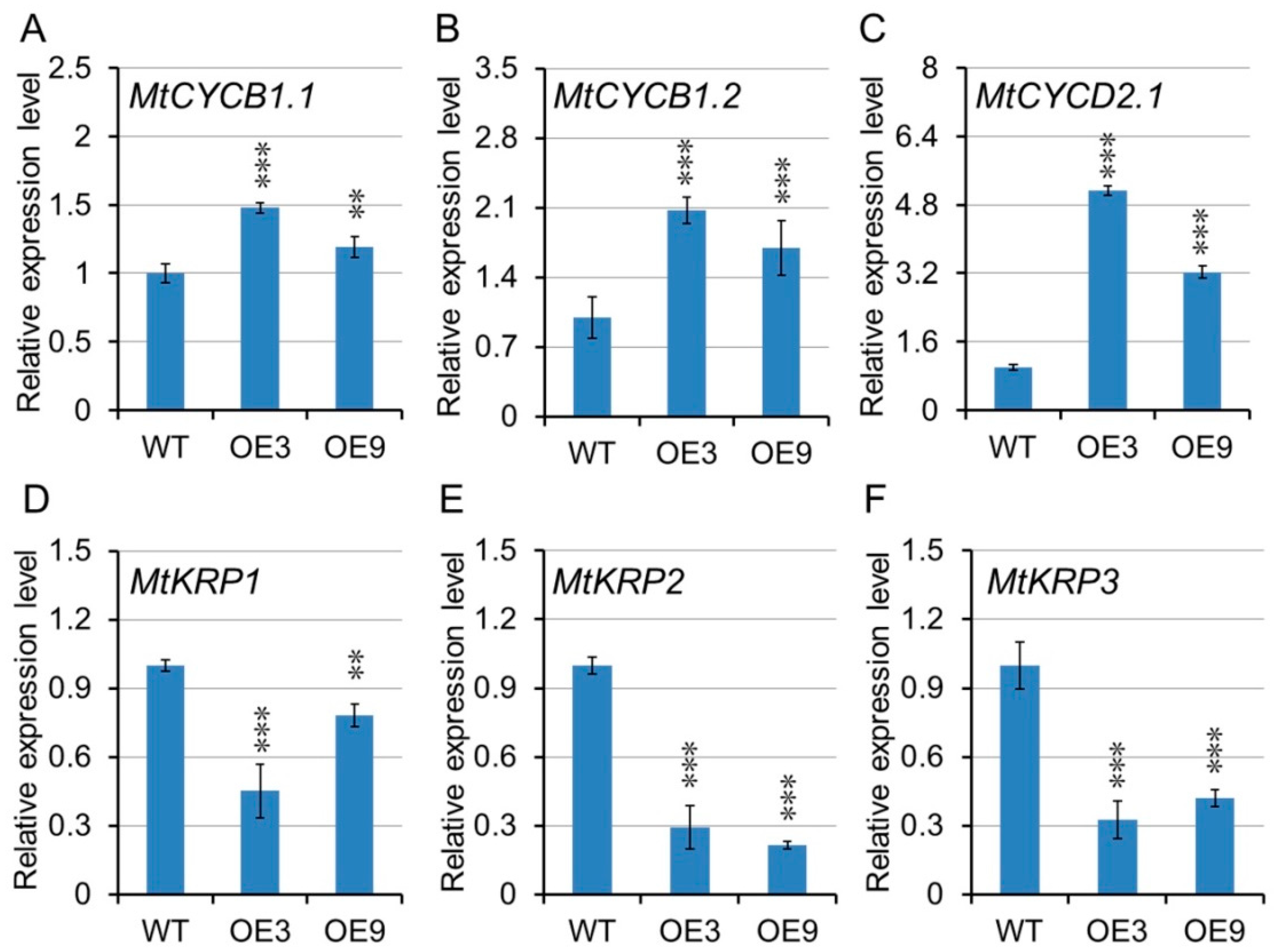
2.10. Transcription Analysis of Cell Development Related Genes in Transgenic Lines
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Identification and Phylogenetic Analysis of GA20ox, GA3ox, GA2ox, GID, and DELLA Genes in M. truncatula
4.3. Chromosome Location, Gene Structure, and Motif Detection
4.4. RNA Extraction and qRT-PCR
4.5. Vector Construction and Plant Transformation
4.6. Yeast Two-Hybrid
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yamaguchi, S. Gibberellin Metabolism and its Regulation. Annu. Rev. Plant Biol. 2008, 59, 225–251. [Google Scholar] [CrossRef]
- Sun, T.-P.; Gubler, F. MOLECULAR MECHANISM OF GIBBERELLIN SIGNALING IN PLANTS. Annu. Rev. Plant. Biol. 2004, 55, 197–223. [Google Scholar] [CrossRef] [PubMed]
- Olszewski, N.; Sun, T.P.; Gubler, F. Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant. Cell 2002, 14 (Suppl. 1), S61–S80. [Google Scholar] [CrossRef] [PubMed]
- McAdam, E.L.; Reid, J.B.; Foo, E. Gibberellins promote nodule organogenesis but inhibit the infection stages of nodulation. J. Exp. Bot. 2018, 69, 2117–2130. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, S.; Gresshoff, P.M.; Ferguson, B.J. Mechanistic action of gibberellins in legume nodulation. J. Integr. Plant Biol. 2014, 56, 971–978. [Google Scholar] [CrossRef] [PubMed]
- Fonouni-Farde, C.; Kisiala, A.; Brault, M.; Emery, R.J.N.; Diet, A.; Frugier, F. DELLA1-Mediated Gibberellin Signaling Regulates Cytokinin-Dependent Symbiotic Nodulation. Plant Physiol. 2017, 175, 1795–1806. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.J.; Foo, E.; Ross, J.J.; Reid, J.B. Relationship between gibberellin, ethylene and nodulation in Pisum sativum. New Phytol. 2011, 189, 829–842. [Google Scholar] [CrossRef]
- Maekawa, T.; Maekawa-Yoshikawa, M.; Takeda, N.; Imaizumi-Anraku, H.; Murooka, Y.; Hayashi, M. Gibberellin controls the nodulation signaling pathway in Lotus japonicus. Plant J. 2009, 58, 183–194. [Google Scholar] [CrossRef]
- Sun, T.P. Gibberellin metabolism, perception and signaling pathways in Arabidopsis. Arab. Book 2008, 6, e0103. [Google Scholar] [CrossRef]
- Salazar-Cerezo, S.; Martinez-Montiel, N.; Garcia-Sanchez, J.; Perez, Y.T.R.; Martinez-Contreras, R.D. Gibberellin biosynthesis and metabolism: A convergent route for plants, fungi and bacteria. Microbiol. Res. 2018, 208, 85–98. [Google Scholar] [CrossRef]
- Ueguchi-Tanaka, M.; Nakajima, M.; Motoyuki, A.; Matsuoka, M. Gibberellin receptor and its role in gibberellin signaling in plants. Annu. Rev. Plant Biol. 2007, 58, 183–198. [Google Scholar] [CrossRef] [PubMed]
- Vandenbussche, F.; Fierro, A.C.; Wiedemann, G.; Reski, R.; Van Der Straeten, D. Evolutionary conservation of plant gibberellin signalling pathway components. BMC Plant Biol. 2007, 7, 65. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, A.; Ashikari, M.; Ueguchi-Tanaka, M.; Itoh, H.; Nishimura, A.; Swapan, D.; Ishiyama, K.; Saito, T.; Kobayashi, M.; Khush, G.S.; et al. A mutant gibberellin-synthesis gene in rice. Nature 2002, 416, 701–702. [Google Scholar] [CrossRef] [PubMed]
- Rieu, I.; Ruiz-Rivero, O.; Fernandez-Garcia, N.; Griffiths, J.; Powers, S.J.; Gong, F.; Linhartova, T.; Eriksson, S.; Nilsson, O.; Thomas, S.G.; et al. The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 2008, 53, 488–504. [Google Scholar] [CrossRef] [PubMed]
- Mitchum, M.G.; Yamaguchi, S.; Hanada, A.; Kuwahara, A.; Yoshioka, Y.; Kato, T.; Tabata, S.; Kamiya, Y.; Sun, T.P. Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J. 2006, 45, 804–818. [Google Scholar] [CrossRef]
- Hu, Y.X.; Tao, Y.B.; Xu, Z.F. Overexpression of Jatropha Gibberellin 2-oxidase 6 (JcGA2ox6) Induces Dwarfism and Smaller Leaves, Flowers and Fruits in Arabidopsis and Jatropha. Front. Plant Sci. 2017, 8, 2103. [Google Scholar] [CrossRef]
- Yan, J.; Liao, X.; He, R.; Zhong, M.; Feng, P.; Li, X.; Tang, D.; Liu, X.; Zhao, X. Ectopic expression of GA 2-oxidase 6 from rapeseed (Brassica napus L.) causes dwarfism, late flowering and enhanced chlorophyll accumulation in Arabidopsis thaliana. Plant Physiol. Biochem. 2017, 111, 10–19. [Google Scholar] [CrossRef]
- Eriksson, M.E.; Israelsson, M.; Olsson, O.; Moritz, T. Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat. Biotechnol. 2000, 18, 784–788. [Google Scholar] [CrossRef]
- Hedden, P.; Kamiya, Y. GIBBERELLIN BIOSYNTHESIS: Enzymes, Genes and Their Regulation. Annu Rev. Plant Phys. 1997, 48, 431–460. [Google Scholar] [CrossRef]
- Penfield, S.; Li, Y.; Gilday, A.D.; Graham, S.; Graham, I.A. Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 2006, 18, 1887–1899. [Google Scholar] [CrossRef]
- Oh, E.; Yamaguchi, S.; Hu, J.; Yusuke, J.; Jung, B.; Paik, I.; Lee, H.S.; Sun, T.P.; Kamiya, Y.; Choi, G. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 2007, 19, 1192–1208. [Google Scholar] [CrossRef] [PubMed]
- Ueguchi-Tanaka, M.; Ashikari, M.; Nakajima, M.; Itoh, H.; Katoh, E.; Kobayashi, M.; Chow, T.Y.; Hsing, Y.I.; Kitano, H.; Yamaguchi, I.; et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 2005, 437, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Livne, S.; Lor, V.S.; Nir, I.; Eliaz, N.; Aharoni, A.; Olszewski, N.E.; Eshed, Y.; Weiss, D. Uncovering DELLA-Independent Gibberellin Responses by Characterizing New Tomato procera Mutants. Plant Cell 2015, 27, 1579–1594. [Google Scholar] [CrossRef]
- Itoh, H.; Ueguchi-Tanaka, M.; Sato, Y.; Ashikari, M.; Matsuoka, M. The Gibberellin Signaling Pathway Is Regulated by the Appearance and Disappearance of SLENDER RICE1 in Nuclei. Plant Cell 2002, 14, 57–70. [Google Scholar] [CrossRef]
- Harberd, N.P.; Belfield, E.; Yasumura, Y. The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: How an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell 2009, 21, 1328–1339. [Google Scholar] [CrossRef] [PubMed]
- Hauvermale, A.L.; Ariizumi, T.; Steber, C.M. Gibberellin signaling: A theme and variations on DELLA repression. Plant Physiol. 2012, 160, 83–92. [Google Scholar] [CrossRef]
- Eckardt, N.A. GA perception and signal transduction: Molecular interactions of the GA receptor GID1 with GA and the DELLA protein SLR1 in rice. Plant Cell 2007, 19, 2095–2097. [Google Scholar] [CrossRef][Green Version]
- Jiang, C.; Fu, X. GA action: Turning on de-DELLA repressing signaling. Curr. Opin. Plant Biol. 2007, 10, 461–465. [Google Scholar] [CrossRef]
- Sun, T.P. Gibberellin-GID1-DELLA: A pivotal regulatory module for plant growth and development. Plant Physiol. 2010, 154, 567–570. [Google Scholar] [CrossRef]
- Ueguchi-Tanaka, M.; Nakajima, M.; Katoh, E.; Ohmiya, H.; Asano, K.; Saji, S.; Hongyu, X.; Ashikari, M.; Kitano, H.; Yamaguchi, I.; et al. Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 2007, 19, 2140–2155. [Google Scholar] [CrossRef]
- Shimada, A.; Ueguchi-Tanaka, M.; Nakatsu, T.; Nakajima, M.; Naoe, Y.; Ohmiya, H.; Kato, H.; Matsuoka, M. Structural basis for gibberellin recognition by its receptor GID1. Nature 2008, 456, 520–523. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, J.; Murase, K.; Rieu, I.; Zentella, R.; Zhang, Z.L.; Powers, S.J.; Gong, F.; Phillips, A.L.; Hedden, P.; Sun, T.P.; et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 2006, 18, 3399–3414. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, M.; Shimada, A.; Takashi, Y.; Kim, Y.C.; Park, S.H.; Ueguchi-Tanaka, M.; Suzuki, H.; Katoh, E.; Iuchi, S.; Kobayashi, M.; et al. Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 2006, 46, 880–889. [Google Scholar] [CrossRef] [PubMed]
- Iuchi, S.; Suzuki, H.; Kim, Y.C.; Iuchi, A.; Kuromori, T.; Ueguchi-Tanaka, M.; Asami, T.; Yamaguchi, I.; Matsuoka, M.; Kobayashi, M.; et al. Multiple loss-of-function of Arabidopsis gibberellin receptor AtGID1s completely shuts down a gibberellin signal. Plant J. 2007, 50, 958–966. [Google Scholar] [CrossRef]
- Illouz-Eliaz, N.; Ramon, U.; Shohat, H.; Blum, S.; Livne, S.; Mendelson, D.; Weiss, D. Multiple Gibberellin Receptors Contribute to Phenotypic Stability under Changing Environments. Plant Cell 2019, 31, 1506–1519. [Google Scholar] [CrossRef]
- Sun, T.P. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol. 2011, 21, R338–R345. [Google Scholar] [CrossRef]
- McGinnis, K.M.; Thomas, S.G.; Soule, J.D.; Strader, L.C.; Zale, J.M.; Sun, T.P.; Steber, C.M. The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 2003, 15, 1120–1130. [Google Scholar] [CrossRef]
- Sasaki, A.; Itoh, H.; Gomi, K.; Ueguchi-Tanaka, M.; Ishiyama, K.; Kobayashi, M.; Jeong, D.-H.; An, G.; Kitano, H.; Ashikari, M.; et al. Accumulation of Phosphorylated Repressor for Gibberellin Signaling in an F-box Mutant. Science 2003, 299, 1896–1898. [Google Scholar] [CrossRef]
- Dill, A.; Thomas, S.G.; Hu, J.; Steber, C.M.; Sun, T.P. The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 2004, 16, 1392–1405. [Google Scholar] [CrossRef]
- Gomi, K.; Sasaki, A.; Itoh, H.; Ueguchi-Tanaka, M.; Ashikari, M.; Kitano, H.; Matsuoka, M. GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J. 2004, 37, 626–634. [Google Scholar] [CrossRef]
- Marti, C.; Orzaez, D.; Ellul, P.; Moreno, V.; Carbonell, J.; Granell, A. Silencing of DELLA induces facultative parthenocarpy in tomato fruits. Plant J. 2007, 52, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Willige, B.C.; Ghosh, S.; Nill, C.; Zourelidou, M.; Dohmann, E.M.; Maier, A.; Schwechheimer, C. The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 2007, 19, 1209–1220. [Google Scholar] [CrossRef]
- Bassel, G.W.; Mullen, R.T.; Bewley, J.D. Procera is a putative DELLA mutant in tomato (Solanum lycopersicum): Effects on the seed and vegetative plant. J. Exp. Bot. 2008, 59, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Jasinski, S.; Tattersall, A.; Piazza, P.; Hay, A.; Martinez-Garcia, J.F.; Schmitz, G.; Theres, K.; McCormick, S.; Tsiantis, M. PROCERA encodes a DELLA protein that mediates control of dissected leaf form in tomato. Plant J. 2008, 56, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Carol, P.; Richards, D.E.; King, K.E.; Cowling, R.J.; Murphy, G.P.; Harberd, N.P. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Gene Dev. 1997, 11, 3194–3205. [Google Scholar] [CrossRef]
- Asano, K.; Hirano, K.; Ueguchi-Tanaka, M.; Angeles-Shim, R.B.; Komura, T.; Satoh, H.; Kitano, H.; Matsuoka, M.; Ashikari, M. Isolation and characterization of dominant dwarf mutants, Slr1-d, in rice. Mol. Genet. Genom. 2009, 281, 223–231. [Google Scholar] [CrossRef]
- Fleishon, S.; Shani, E.; Ori, N.; Weiss, D. Negative reciprocal interactions between gibberellin and cytokinin in tomato. New Phytol. 2011, 190, 609–617. [Google Scholar] [CrossRef]
- Dill, A.; Sun, T.-P. Synergistic Derepression of Gibberellin Signaling by Removing RGA and GAI Function in Arabidopsis thaliana. Genetics 2001, 159, 777. [Google Scholar]
- Dill, A.; Jung, H.-S.; Sun, T.-P. The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc. Natl. Acad. Sci. USA 2001, 98, 14162–14167. [Google Scholar] [CrossRef]
- Pearce, S.; Saville, R.; Vaughan, S.P.; Chandler, P.M.; Wilhelm, E.P.; Sparks, C.A.; Al-Kaff, N.; Korolev, A.; Boulton, M.I.; Phillips, A.L.; et al. Molecular characterization of Rht-1 dwarfing genes in hexaploid wheat. Plant Physiol. 2011, 157, 1820–1831. [Google Scholar] [CrossRef]
- Peng, J.; Richards, D.E.; Hartley, N.M.; Murphy, G.P.; Devos, K.M.; Flintham, J.E.; Beales, J.; Fish, L.J.; Worland, A.J.; Pelica, F.; et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 1999, 400, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Biemelt, S.; Tschiersch, H.; Sonnewald, U. Impact of altered gibberellin metabolism on biomass accumulation, lignin biosynthesis, and photosynthesis in transgenic tobacco plants. Plant Physiol. 2004, 135, 254–265. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, J.; Ban, L.; Wu, Y.; Wu, X.; Wang, Y.; Wen, H.; Chapurin, V.; Dzyubenko, N.; Li, Z.; et al. Functional characterization of a gibberellin receptor and its application in alfalfa biomass improvement. Sci. Rep. 2017, 7, 41296. [Google Scholar] [CrossRef]
- Do, P.T.; De Tar, J.R.; Lee, H.; Folta, M.K.; Zhang, Z.J. Expression of ZmGA20ox cDNA alters plant morphology and increases biomass production of switchgrass (Panicum virgatum L.). Plant Biotechnol. J. 2016, 14, 1532–1540. [Google Scholar] [CrossRef] [PubMed]
- Daviere, J.M.; Achard, P. A Pivotal Role of DELLAs in Regulating Multiple Hormone Signals. Mol. Plant 2016, 9, 10–20. [Google Scholar] [CrossRef]
- Locascio, A.; Blázquez, M.A.; Alabadí, D. Genomic Analysis of DELLA Protein Activity. Plant Cell Physiol. 2013, 54, 1229–1237. [Google Scholar] [CrossRef]
- Gazara, R.K.; Moharana, K.C.; Bellieny-Rabelo, D.; Venancio, T.M. Expansion and diversification of the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1 (GID1) family in land plants. Plant Mol. Biol. 2018, 97, 435–449. [Google Scholar] [CrossRef]
- Conti, L.; Nelis, S.; Zhang, C.; Woodcock, A.; Swarup, R.; Galbiati, M.; Tonelli, C.; Napier, R.; Hedden, P.; Bennett, M.; et al. Small Ubiquitin-like Modifier protein SUMO enables plants to control growth independently of the phytohormone gibberellin. Dev. Cell 2014, 28, 102–110. [Google Scholar] [CrossRef]
- Hedden, P.; Phillips, A.L. Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 2000, 5, 523–530. [Google Scholar] [CrossRef]
- Plackett, A.R.; Powers, S.J.; Fernandez-Garcia, N.; Urbanova, T.; Takebayashi, Y.; Seo, M.; Jikumaru, Y.; Benlloch, R.; Nilsson, O.; Ruiz-Rivero, O.; et al. Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell 2012, 24, 941–960. [Google Scholar] [CrossRef]
- Richards, D.E.; King, K.E.; Ait-ali, T.; Harberd, N.P. HOW GIBBERELLIN REGULATES PLANT GROWTH AND DEVELOPMENT: A Molecular Genetic Analysis of Gibberellin Signaling. Annu Rev. Plant Phys. 2001, 52, 67–88. [Google Scholar] [CrossRef] [PubMed]
- Shwartz, I.; Levy, M.; Ori, N.; Bar, M. Hormones in tomato leaf development. Dev. Biol. 2016, 419, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Hedden, P.; Sponsel, V. A Century of Gibberellin Research. J. Plant Growth Regul. 2015, 34, 740–760. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.L.; Ward, D.A.; Uknes, S.; Appleford, N.; Lange, T.; Huttly, A.K.; Gaskin, P.; Graebe, J.E.; Hedden, P. Isolation and Expression of Three Gibberellin 20-Oxidase cDNA Clones from Arabidopsis. Plant Physiol. 1995, 108, 1049–1057. [Google Scholar] [CrossRef]
- Thomas, S.G.; Phillips, A.L.; Hedden, P. Molecular cloning and functional expression of gibberellin 2- oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA 1999, 96, 4698–4703. [Google Scholar] [CrossRef]
- Sun, H.; Pang, B.; Yan, J.; Wang, T.; Wang, L.; Chen, C.; Li, Q.; Ren, Z. Comprehensive Analysis of Cucumber Gibberellin Oxidase Family Genes and Functional Characterization of CsGA20ox1 in Root Development in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 3135. [Google Scholar] [CrossRef]
- Boudet, N.; Aubourg, S.; Toffano-Nioche, C.; Kreis, M.; Lecharny, A. Evolution of intron/exon structure of DEAD helicase family genes in Arabidopsis, Caenorhabditis, and Drosophila. Genome Res. 2001, 11, 2101–2114. [Google Scholar] [CrossRef]
- Babenko, V.N.; Rogozin, I.B.; Mekhedov, S.L.; Koonin, E.V. Prevalence of intron gain over intron loss in the evolution of paralogous gene families. Nucleic Acids Res. 2004, 32, 3724–3733. [Google Scholar] [CrossRef]
- He, H.; Liang, G.; Lu, S.; Wang, P.; Liu, T.; Ma, Z.; Zuo, C.; Sun, X.; Chen, B.; Mao, J. Genome-Wide Identification and Expression Analysis of GA2ox, GA3ox, and GA20ox Are Related to Gibberellin Oxidase Genes in Grape (Vitis Vinifera L.). Genes 2019, 10, 680. [Google Scholar] [CrossRef]
- Pan, C.; Tian, K.; Ban, Q.; Wang, L.; Sun, Q.; He, Y.; Yang, Y.; Pan, Y.; Li, Y.; Jiang, J.; et al. Genome-Wide Analysis of the Biosynthesis and Deactivation of Gibberellin-Dioxygenases Gene Family in Camellia sinensis (L.) O. Kuntze. Genes 2017, 8, 235. [Google Scholar] [CrossRef]
- Murase, K.; Hirano, Y.; Sun, T.P.; Hakoshima, T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 2008, 456, 459–463. [Google Scholar] [CrossRef]
- Fonouni-Farde, C.; Tan, S.; Baudin, M.; Brault, M.; Wen, J.; Mysore, K.S.; Niebel, A.; Frugier, F.; Diet, A. DELLA-mediated gibberellin signalling regulates Nod factor signalling and rhizobial infection. Nat. Commun. 2016, 7, 12636. [Google Scholar] [CrossRef]
- Jin, Y.; Liu, H.; Luo, D.; Yu, N.; Dong, W.; Wang, C.; Zhang, X.; Dai, H.; Yang, J.; Wang, E. DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways. Nat. Commun. 2016, 7, 12433. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Liu, W.; Nandety, R.S.; Crook, A.D.; Mysore, K.S.; Pislariu, C.I.; Frugoli, J.A.; Dickstein, R.; Udvardi, M.K. Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 2019. [Google Scholar] [CrossRef] [PubMed]
- Hay, A.; Kaur, H.; Phillips, A.; Hedden, P.; Hake, S.; Tsiantis, M. The Gibberellin Pathway Mediates KNOTTED1-Type Homeobox Function in Plants with Different Body Plans. Curr. Biol. 2002, 12, 1557–1565. [Google Scholar] [CrossRef]
- Han, Y.; Teng, K.; Nawaz, G.; Feng, X.; Usman, B.; Wang, X.; Luo, L.; Zhao, N.; Liu, Y.; Li, R. Generation of semi-dwarf rice (Oryza sativa L.) lines by CRISPR/Cas9-directed mutagenesis of OsGA20ox2 and proteomic analysis of unveiled changes caused by mutations. 3 Biotech. 2019, 9, 387. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, Y.; Yin, Y.; Liu, Q.; Li, N.; Liu, X.; Li, X.; Guo, C.; Hao, D. Development of dwarfish and yield-effective GM maize through passivation of bioactive gibberellin. Transgenic Res. 2019, 28, 589–599. [Google Scholar] [CrossRef]
- Chen, X.; Tian, X.; Xue, L.; Zhang, X.; Yang, S.; Traw, M.B.; Huang, J. CRISPR-Based Assessment of Gene Specialization in the Gibberellin Metabolic Pathway in Rice. Plant Physiol. 2019, 180, 2091–2105. [Google Scholar] [CrossRef]
- Garcia-Hurtado, N.; Carrera, E.; Ruiz-Rivero, O.; Lopez-Gresa, M.P.; Hedden, P.; Gong, F.; Garcia-Martinez, J.L. The characterization of transgenic tomato overexpressing gibberellin 20-oxidase reveals induction of parthenocarpic fruit growth, higher yield, and alteration of the gibberellin biosynthetic pathway. J. Exp. Bot. 2012, 63, 5803–5813. [Google Scholar] [CrossRef]
- Mauriat, M.; Moritz, T. Analyses of GA20ox- and GID1-over-expressing aspen suggest that gibberellins play two distinct roles in wood formation. Plant J. 2009, 58, 989–1003. [Google Scholar] [CrossRef]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Jiang, H.; Xu, Y.; Wang, Y.; Zhu, L.; Yu, X.; Kong, F.; Zhou, C.; Han, L. Systematic Analysis of Gibberellin Pathway Components in Medicago truncatula Reveals the Potential Application of Gibberellin in Biomass Improvement. Int. J. Mol. Sci. 2020, 21, 7180. https://doi.org/10.3390/ijms21197180
Wang H, Jiang H, Xu Y, Wang Y, Zhu L, Yu X, Kong F, Zhou C, Han L. Systematic Analysis of Gibberellin Pathway Components in Medicago truncatula Reveals the Potential Application of Gibberellin in Biomass Improvement. International Journal of Molecular Sciences. 2020; 21(19):7180. https://doi.org/10.3390/ijms21197180
Chicago/Turabian StyleWang, Hongfeng, Hongjiao Jiang, Yiteng Xu, Yan Wang, Lin Zhu, Xiaolin Yu, Fanjiang Kong, Chuanen Zhou, and Lu Han. 2020. "Systematic Analysis of Gibberellin Pathway Components in Medicago truncatula Reveals the Potential Application of Gibberellin in Biomass Improvement" International Journal of Molecular Sciences 21, no. 19: 7180. https://doi.org/10.3390/ijms21197180
APA StyleWang, H., Jiang, H., Xu, Y., Wang, Y., Zhu, L., Yu, X., Kong, F., Zhou, C., & Han, L. (2020). Systematic Analysis of Gibberellin Pathway Components in Medicago truncatula Reveals the Potential Application of Gibberellin in Biomass Improvement. International Journal of Molecular Sciences, 21(19), 7180. https://doi.org/10.3390/ijms21197180




