Repurposing Benzbromarone for Familial Amyloid Polyneuropathy: A New Transthyretin Tetramer Stabilizer
Abstract
1. Introduction
2. Results and Discussion
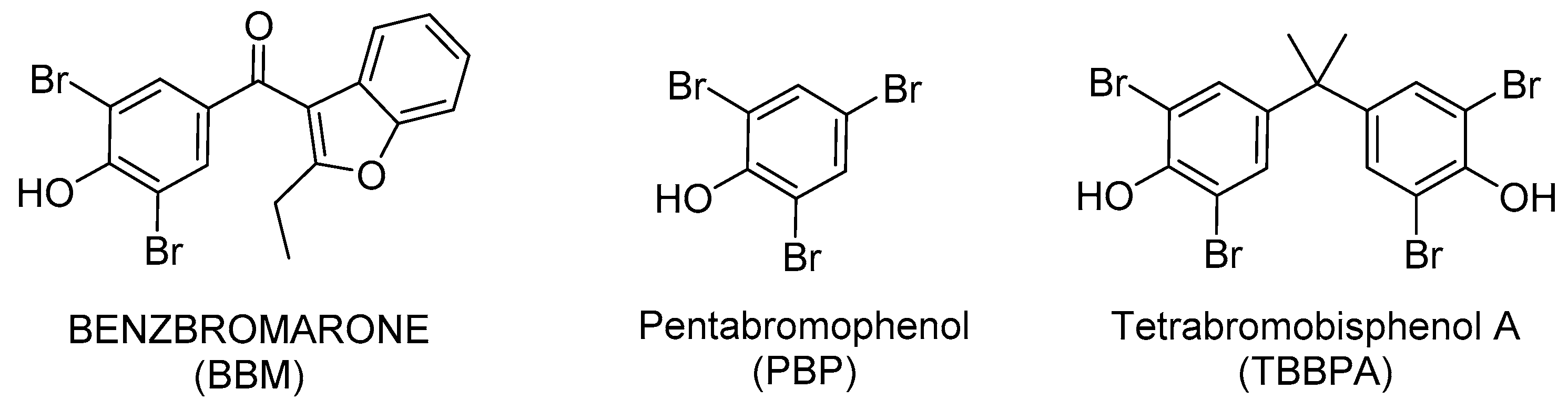
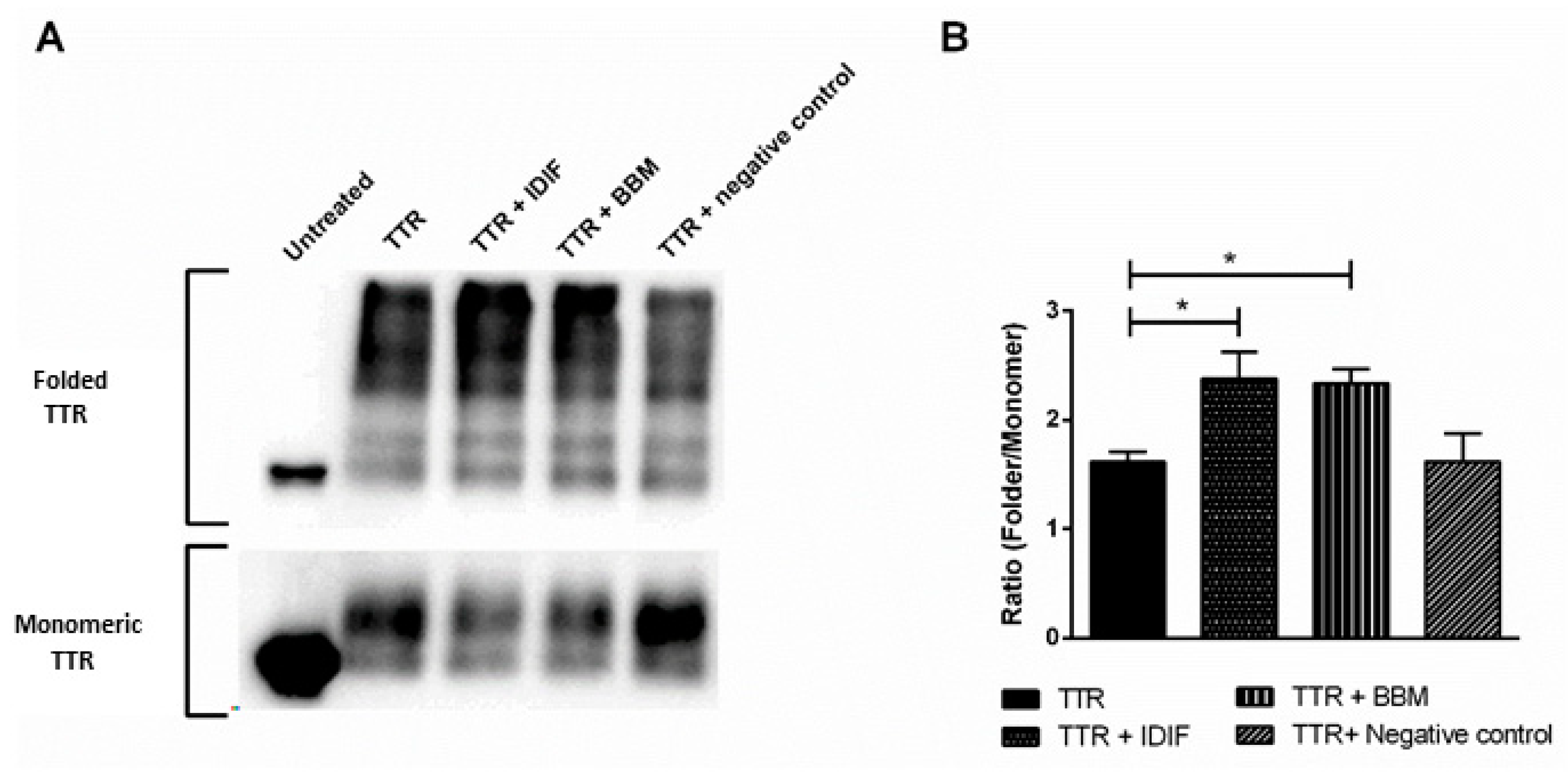
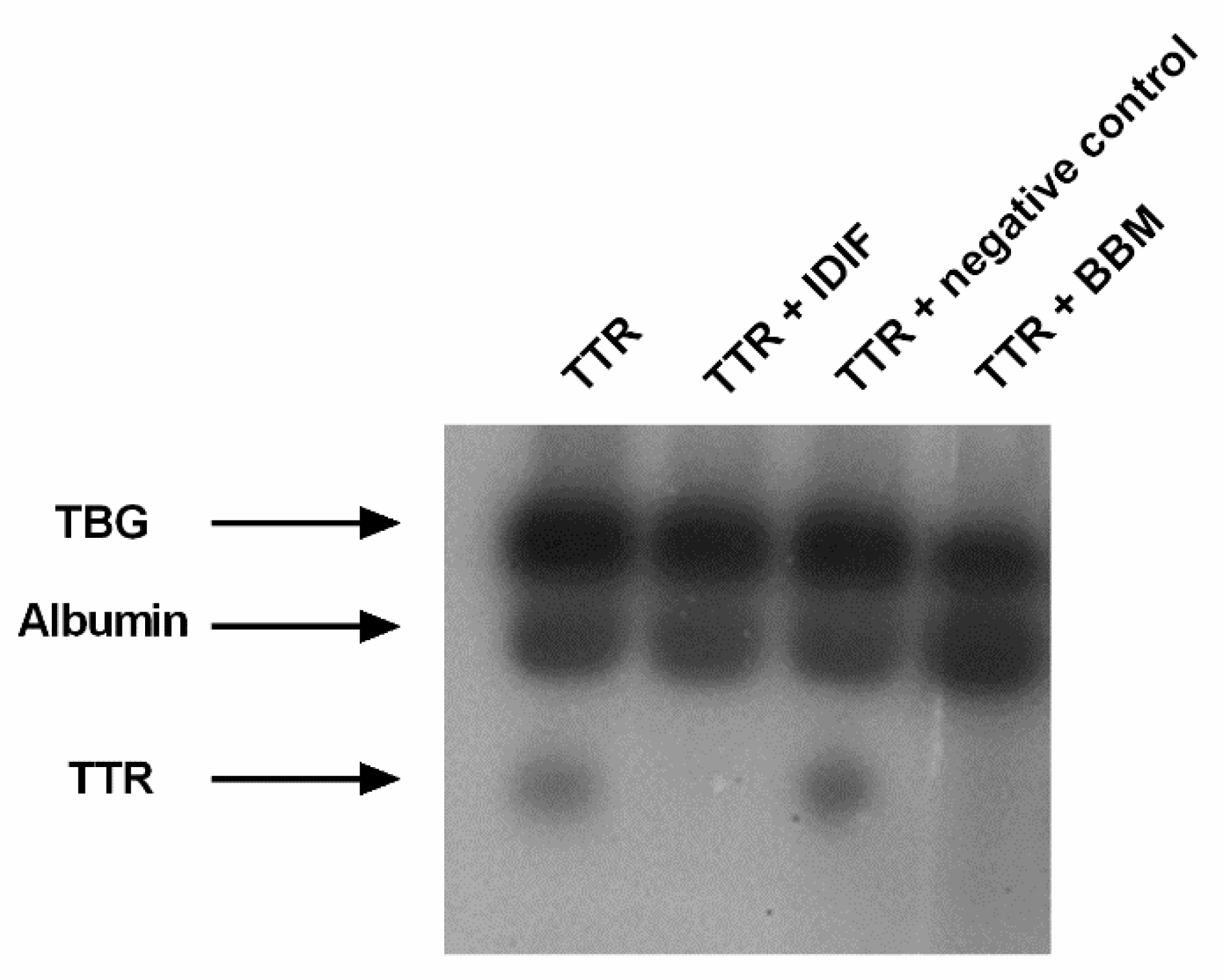
2.1. Benzbromarone (BBM) Stabilizes the TTR Tetrameric Fold
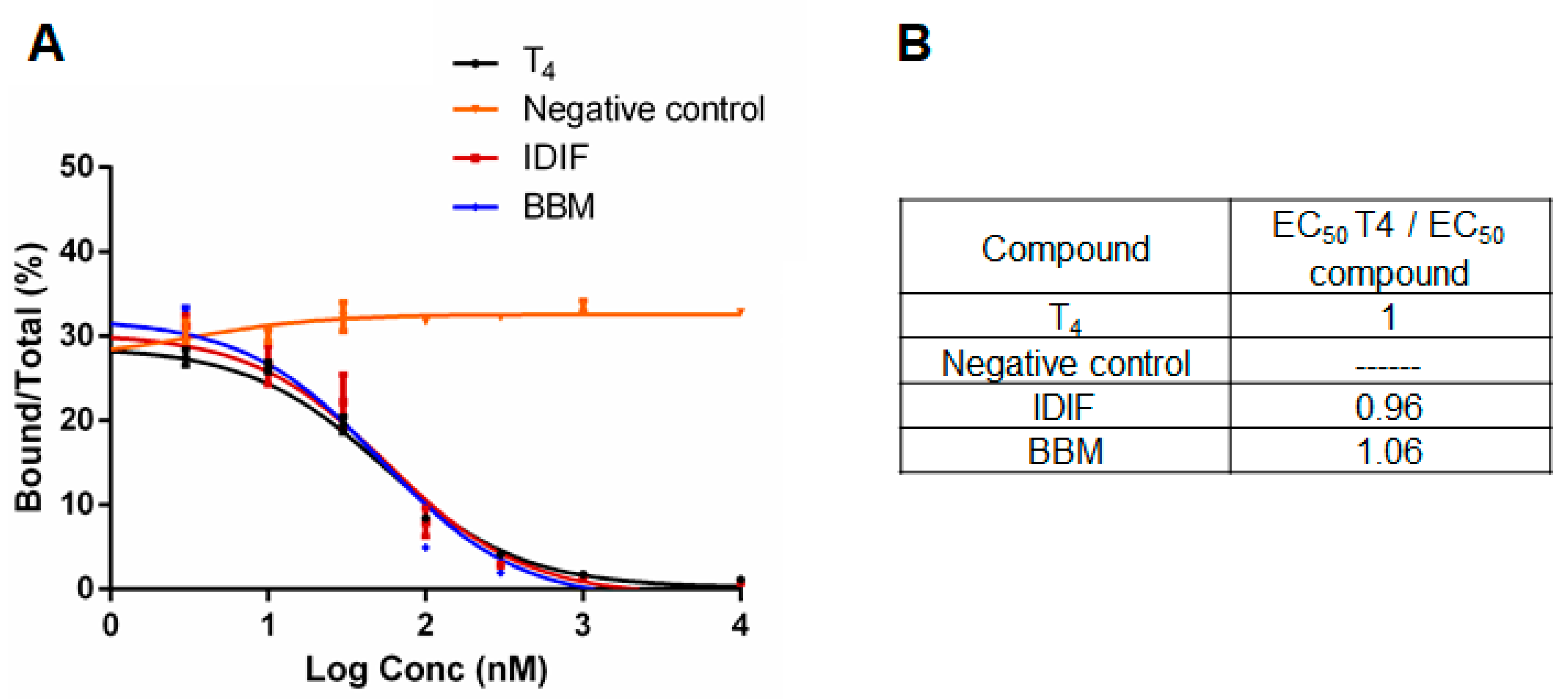
2.2. Benzbromarone (BBM) Binds TTR in the T4 Central Binding Pocket
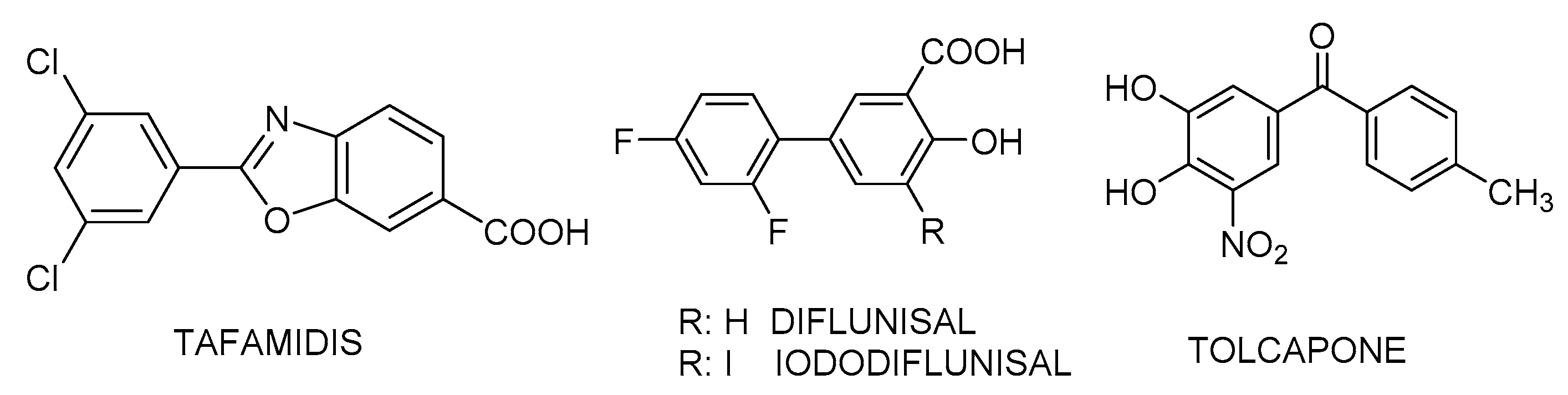
2.3. Isothermal Titration Calorimetry (ITC) Studies of the Binary Interaction between TTR and the Small-Molecule Drug Benzbromarone (BBM). Comparison with other TTR Binary Interactions with Iododiflunisal (IDIF), Tolcapone and Tafamidis
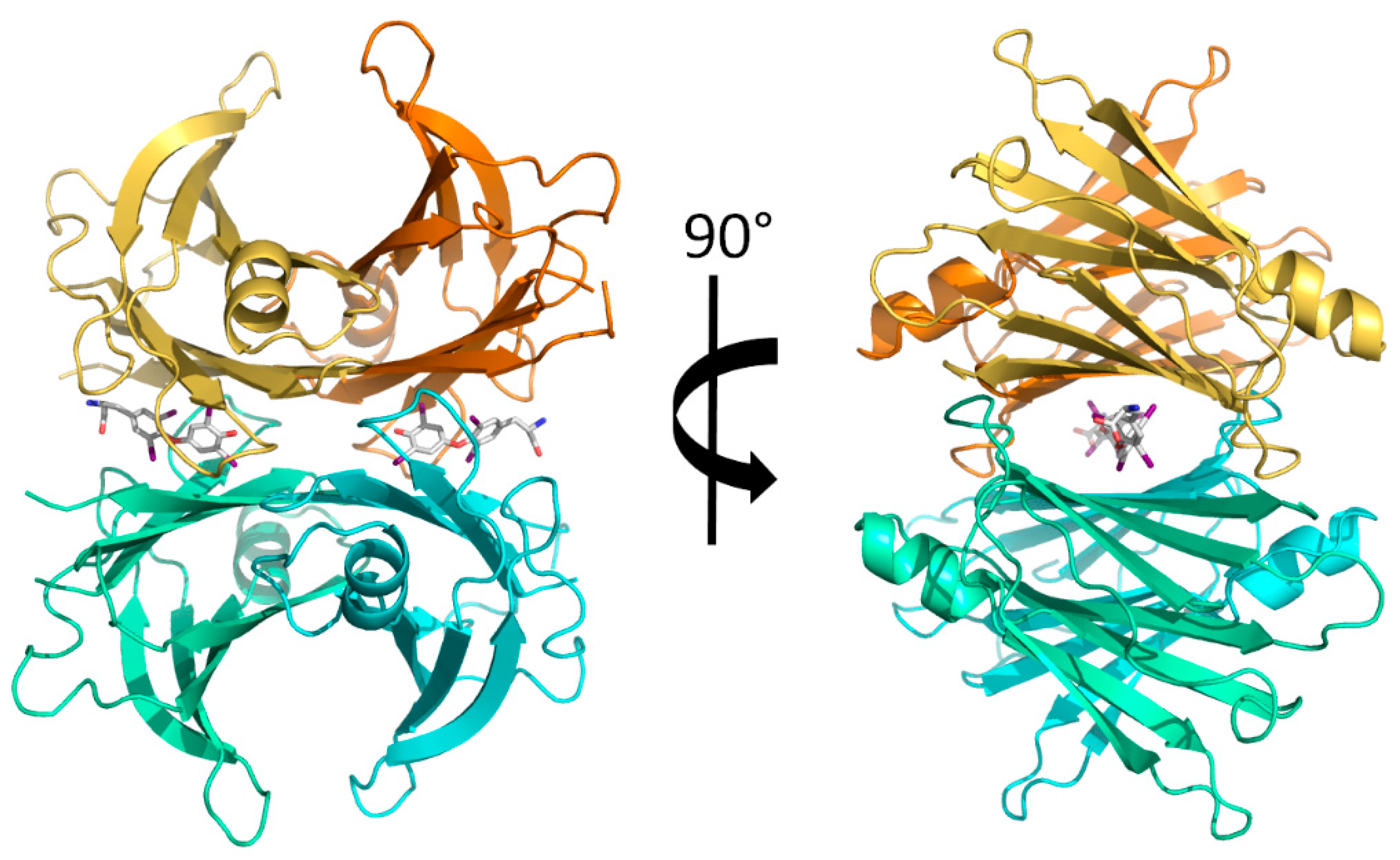
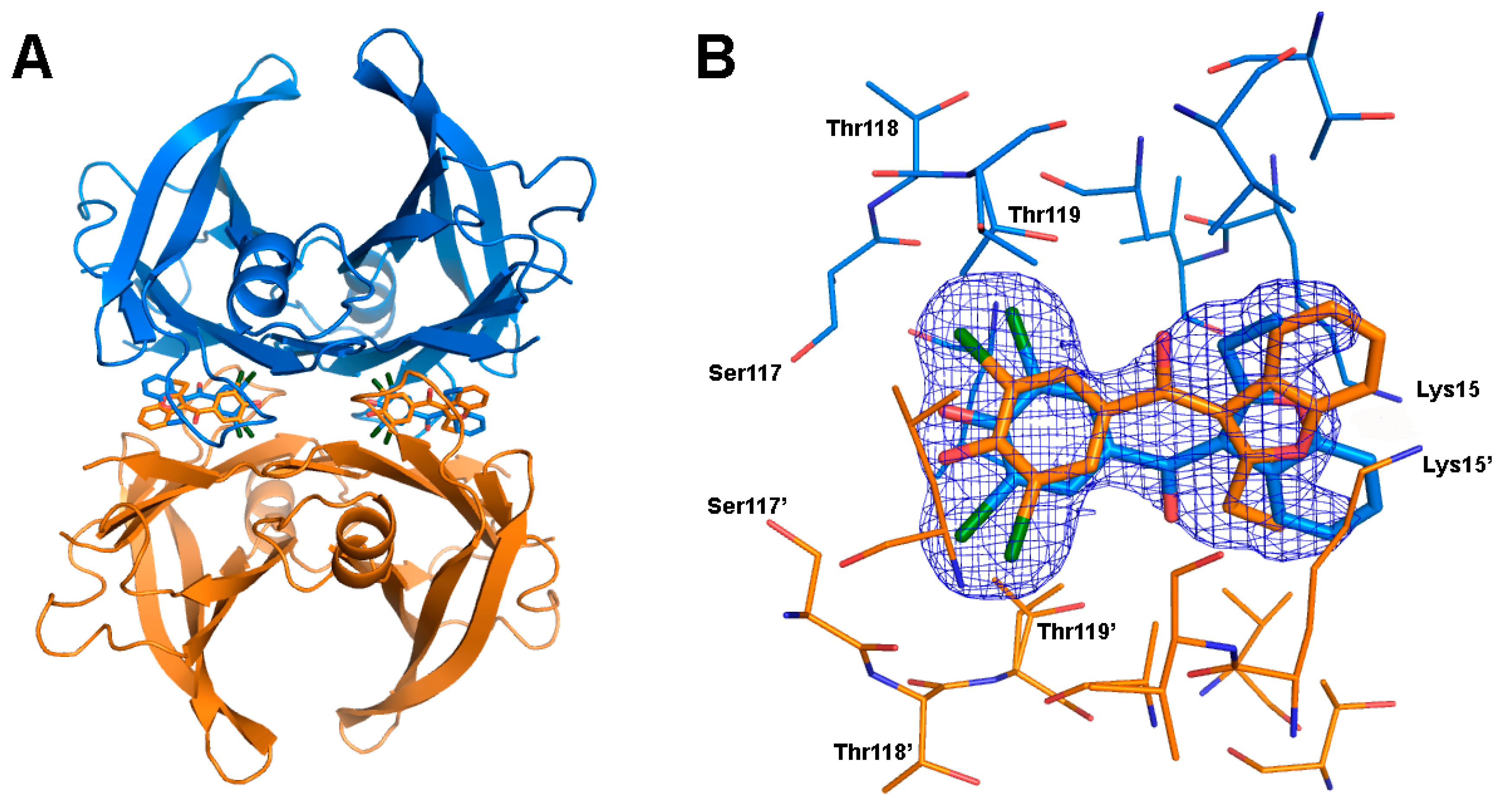
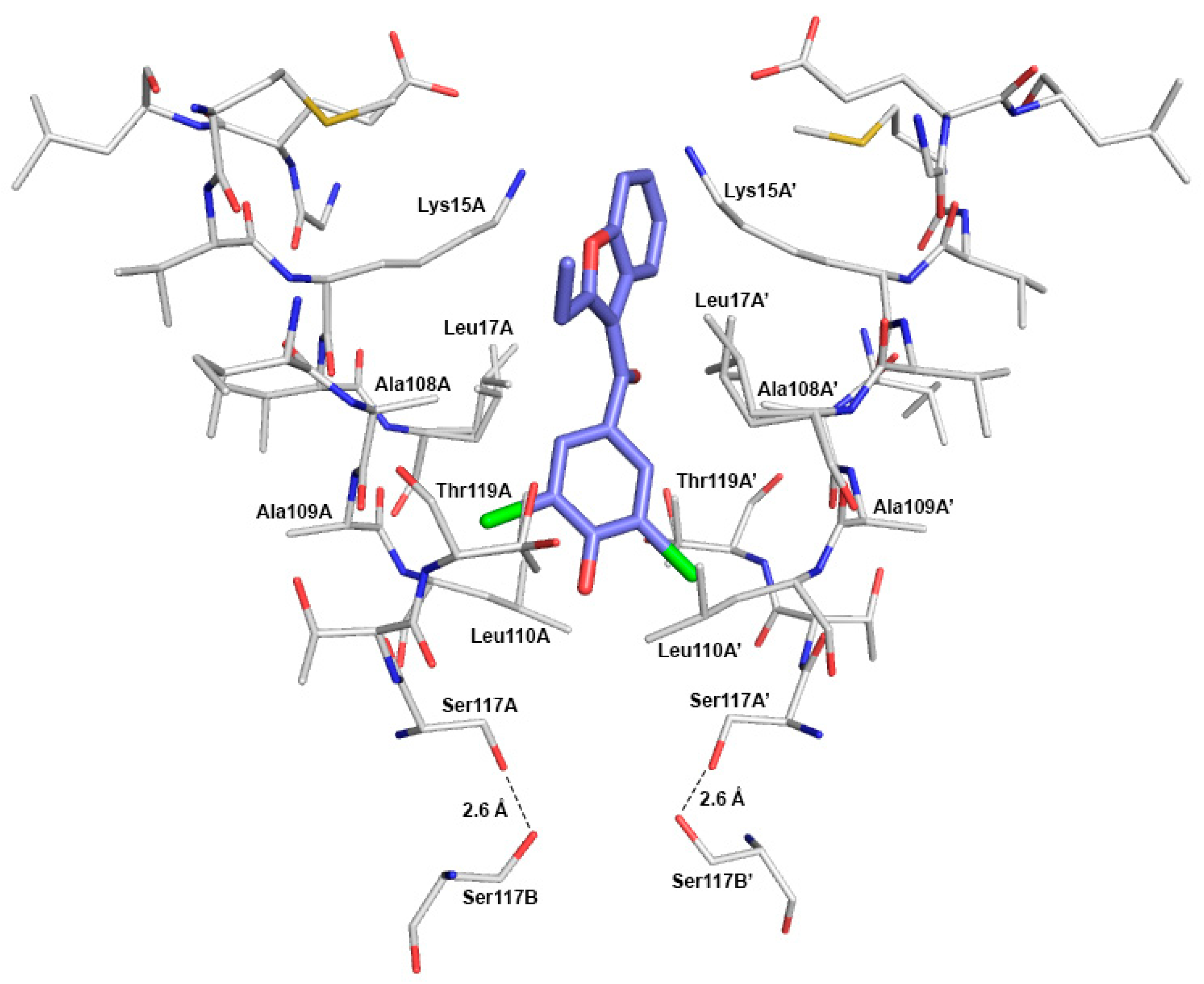
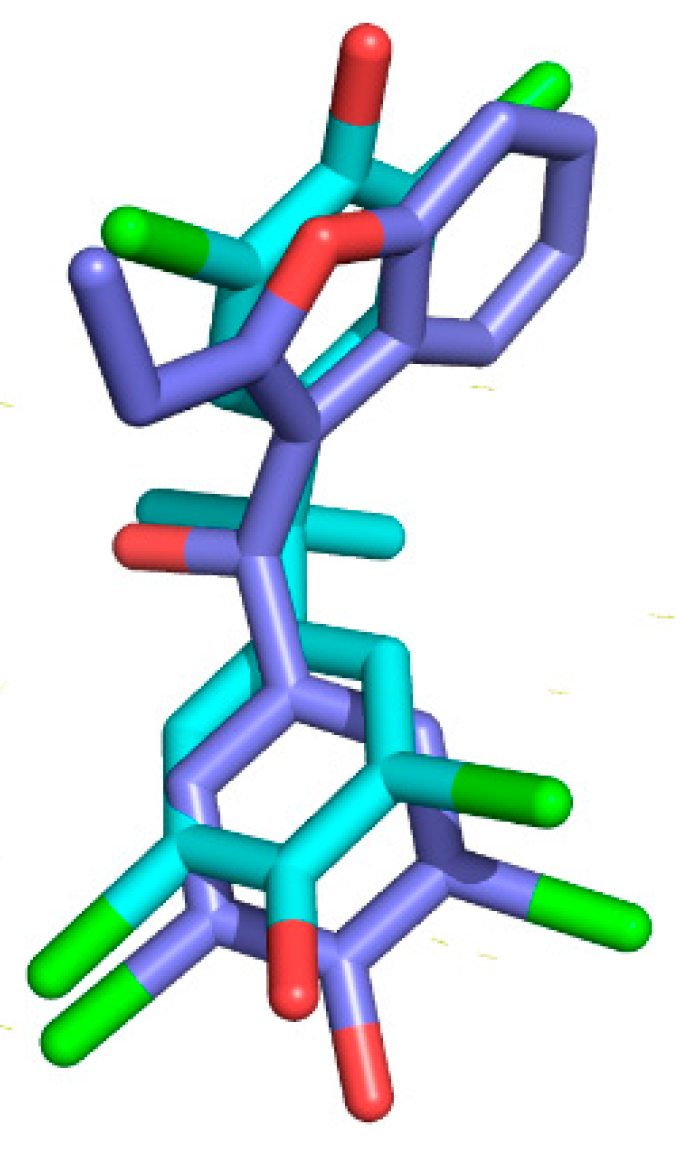
2.4. Crystal Structure of TTR in Complex with BBM (TTR:BBM Complex)
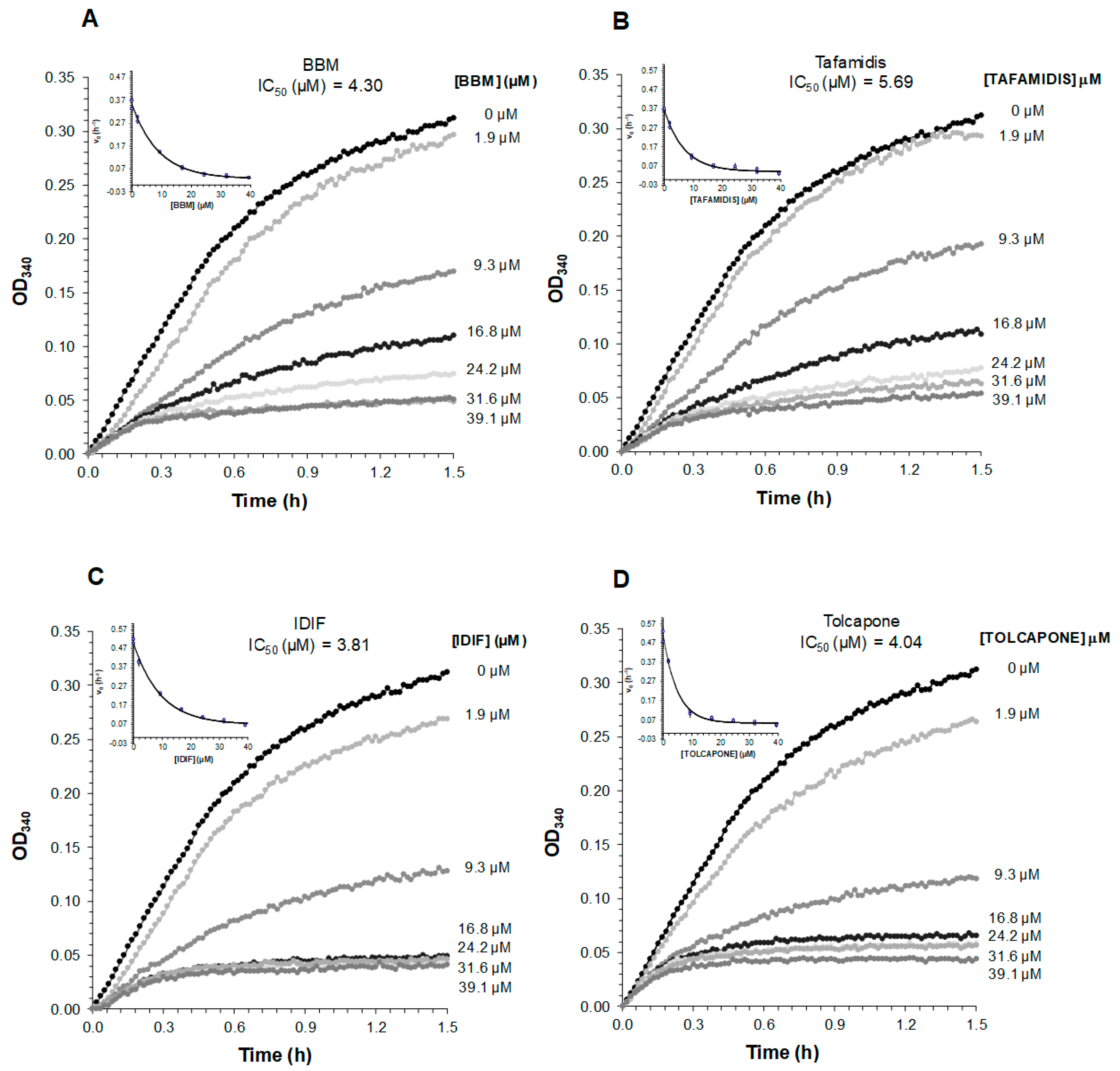
2.5. Comparative Analysis of the Ability of BBM to Inhibit TTR Fibrillogenesis at Acidic pH
3. Materials and Methods
3.1. Compounds
3.2. TTR Production and Purification
3.3. TTR Stability Assay
3.4. Thyroxine Binding Assays
3.5. Isothermal Titration Calorimetry (ITC) Studies
3.6. Co-Crystallization
3.7. X-ray Diffraction Data Collection, Processing and Structure Refinement
3.8. Kinetic Turbidity Assay
3.9. Statistics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s disease |
| BBM | Benzbromarone |
| BSA | Bovine serum albumin |
| COMT | Catechol-O-methyltransferase |
| CSF | Cerebrospinal fluid |
| DIF | Diflunisal |
| DM | Dried milk |
| DMSO | Dimethyl sulfoxide |
| ECL | Enhanced chemiluminescence |
| EDTA | Ethylenediaminetetraacetic acid |
| FAP | Familial amyloid polyneuropathy |
| HBP | Halogen-binding pocket |
| HEPES | N-(2-Hydroxyethyl) piperazine-N′-(2-ethanesulfonic acid) |
| HPLC | High-performance liquid chromatography |
| IDIF | Iododiflunisal |
| ITC | Isothermal titration calorimetry |
| NMR | Nuclear magnetic resonance |
| NSAID | Non-steroidal anti-inflammatory drug |
| PBP | Pentabromophenol |
| PBS | Phosphate-buffered saline |
| PDB | Protein Data Bank |
| rpm | revolutions per minute |
| RT | Room temperature |
| SDS | Sodium dodecyl sulphate |
| TBBPA | Tetrabromobisphenol A |
| T4 | Thyroxine |
| TBG | Thyroxine-binding globulin |
| TTR | Transthyretin |
| UPLC-TOF-MS | Ultra-high performance liquid chromatography time of flight mass spectrometry |
References
- Blake, C.C.; Geisow, M.J.; Oatley, S.J.; Rerat, B.; Rerat, C. Structure of prealbumin: Secondary, tertiary and quaternary interactions determined by Fourier refinement at 1.8 Å. J. Mol. Biol. 1978, 121, 339–356. [Google Scholar] [CrossRef]
- Hamilton, J.A.; Benson, M.D. Transthyretin: A review from a structural perspective. Cell. Mol. Life Sci. 2001, 58, 1491–1521. [Google Scholar] [CrossRef] [PubMed]
- Herbert, J.; Wilcox, J.N.; Pham, K.T.; Fremeau, R.T., Jr.; Zeviani, M.; Dwork, A.; Soprano, D.R.; Makover, A.; Goodman, D.S.; Zimmerman, E.A. Transthyretin: A choroid plexus-specific transport protein in human brain. The 1986 S. Weir Mitchell award. Neurology 1986, 36, 900–911. [Google Scholar] [CrossRef] [PubMed]
- Monaco, H.L.; Rizzi, M.; Coda, A. Structure of a complex of two plasma proteins: Transthyretin and retinol binding protein. Science 1995, 268, 1039–1041. [Google Scholar] [CrossRef] [PubMed]
- Zanotti, G.; Berni, R. Plasma retinol-binding protein: Structure and interactions with retinol, retinoids, and transthyretin. Vitam. Horm. 2004, 69, 271–295. [Google Scholar]
- Hagen, G.A.; Elliott, W.J. Transport of thyroid hormones in serum and cerebrospinal fluid. J. Clin. Endocrinol. Metab. 1973, 7, 415–422. [Google Scholar] [CrossRef]
- Gião, T.; Saavedra, J.; Cotrina, E.; Quintana, J.; Llop, J.; Arsequell, G.; Cardoso, I. Undiscovered Roles for Transthyretin: From a Transporter Protein to a New Therapeutic Target for Alzheimer’s Disease. Int. J. Mol. Sci. 2020, 21, 2075. [Google Scholar] [CrossRef]
- Schwarzman, A.L.; Gregori, L.; Vitek, M.P.; Lyubski, S.; Strittmatter, W.J.; Enghilde, J.J.; Bhasin, R.; Silverman, J.; Weisgraber, K.H.; Coyle, P.K.; et al. Transthyretin sequesters amyloid beta protein and prevents amyloid formation. Proc. Nat. Acad. Sci. USA 1994, 91, 8368–8372. [Google Scholar] [CrossRef]
- Garai, K.; Posey, A.E.; Li, X.; Buxbaum, J.N.; Pappu, R.V. Inhibition of amyloid beta fibril formation by monomeric human transthyretin. Protein. Sci. 2018, 27, 1252–1261. [Google Scholar] [CrossRef]
- Alemi, M.; Silva, S.C.; Santana, I.; Cardoso, I. Transthyretin stability is critical in assisting beta amyloid clearance—Relevance of transthyretin stabilization in Alzheimer’s disease. CNS Neurosci. Ther. 2017, 23, 605–619. [Google Scholar] [CrossRef]
- Adams, D.; Koike, H.; Slama, M.; Coelho, T. Hereditary transthyretin amyloidosis: A model of medical progress for a fatal disease. Nat. Rev. Neurol. 2019, 15, 387–404. [Google Scholar] [CrossRef] [PubMed]
- Plante-Bordeneuve, V. Transthyretin familial amyloid polyneuropathy: An update. J. Neurol. 2018, 265, 976–983. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, O.K.; Ruberg, F.L. Cardiac amyloidosis: An update on pathophysiology, diagnosis, and treatment. Trends Cardiovasc. Med. 2018, 28, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Westermark, P.; Sletten, K.; Johansson, B.; Cornwell, G.G., 3rd. Fibril in senile systemic amyloidosis is derived from normal transthyretin. Proc. Natl. Acad. Sci. USA 1990, 87, 2843–2845. [Google Scholar] [CrossRef]
- Ziskin, J.L.; Greicius, M.D.; Zhu, W.; Okumu, A.N.; Adams, C.M.; Plowey, E.D. Neuropathologic analysis of Tyr69His TTR variant meningovascular amyloidosis with dementia. Acta Neuropathol. Commun. 2015, 3, 43. [Google Scholar] [CrossRef]
- Connelly, S.; Choi, S.; Johnson, S.M.; Kelly, J.W.; Wilson, I.A. Structure-based design of kinetic stabilizers that ameliorate the transthyretin amyloidoses. Curr. Opin. Struct. Biol. 2010, 20, 54–62. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Palaninathan, S.K. Nearly 200 X-Ray Crystal Structures of Transthyretin: What Do They Tell Us about This Protein and the Design of Drugs for TTR Amyloidoses? Curr. Med. Chem. 2012, 19, 2324–2342. [Google Scholar] [CrossRef]
- Nencetti, S.; Orlandini, E. TTR fibril formation inhibitors: Is there a SAR? Curr. Med. Chem. 2012, 19, 2356–2379. [Google Scholar]
- Guo, X.; Liu, Z.; Zheng, Y.; Li, Y.; Li, L.; Liu, H.; Chen, Z.; Wu, L. Review on the Structures and Activities of Transthyretin Amyloidogenesis Inhibitors. Drug Des. Devel. Ther. 2020, 14, 1057–1081. [Google Scholar] [CrossRef]
- Wojtczak, A.; Cody, V.; Luft, J.; Pangborn, W. Structures of human transthyretin complexed with thyroxine at 2.0 Å resolution and 3’, 5’-dinitro-N-acetyl-L-thyronine at 2.2 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 1996, 52, 758–765. [Google Scholar] [CrossRef] [PubMed]
- Waddington Cruz, M.; Amass, L.; Keohane, D.; Schwartz, J.; Li, H.; Gundapaneni, B. Early intervention with tafamidis provides long-term (5.5-year) delay of neurologic progression in transthyretin hereditary amyloid polyneuropathy. Amyloid 2016, 23, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Berk, J.L.; Suhr, O.B.; Obici, L.; Sekijima, Y.; Zeldenrust, S.R.; Yamashita, T.; Heneghan, M.A.; Gorevic, P.D.; Litchy, W.J.; Wiesman, J.F.; et al. Diflunisal Trial Consortium. Repurposing diflunisal for familial amyloid polyneuropathy: A randomized clinical trial. JAMA 2013, 310, 2658–2667. [Google Scholar] [CrossRef] [PubMed]
- Sant’Anna, R.; Gallego, P.; Robinson, L.Z.; Pereira-Henriques, A.; Ferreira, N.; Pinheiro, F.; Esperante, S.; Pallares, I.; Huertas, O.; Almeida, M.R.; et al. Repositioning tolcapone as a potent inhibitor of transthyretin amyloidogenesis and associated cellular toxicity. Nat. Commun. 2016, 7, 10787. [Google Scholar] [CrossRef]
- Fox, J.C.; Hellawell, J.L.; Rao, S.; O’Reilly, T.; Lumpkin, R.; Jernelius, J.; Gretler, D.; Sinha, U. First-in-Human Study of AG10, a Novel, Oral, Specific, Selective, and Potent Transthyretin Stabilizer for the Treatment of Transthyretin Amyloidosis: A Phase 1 Safety, Tolerability, Pharmacokinetic, and Pharmacodynamic Study in Healthy Adult Volunteers. Clin. Pharmacol. Drug Dev. 2020, 9, 115–129. [Google Scholar] [CrossRef]
- Almeida, M.R.; Macedo, B.; Cardoso, I.; Alves, I.; Valencia, G.; Arsequell, G.; Planas, A.; Saraiva, M.J. Selective binding to transthyretin and tetramer stabilization in serum from patients with familial amyloidotic polyneuropathy by an iodinated diflunisal derivative. Biochem. J. 2004, 381, 351–356. [Google Scholar] [CrossRef]
- Gales, L.; Macedo-Ribeiro, S.; Arsequell, G.; Valencia, G.; Saraiva, M.J.; Damas, A.M. Human transthyretin in complex with iododiflunisal: Structural features associated with a potent amyloid inhibitor. Biochem. J. 2005, 388, 615–621. [Google Scholar] [CrossRef]
- Mairal, T.; Nieto, J.; Pinto, M.; Almeida, M.R.; Gales, L.; Ballesteros, A.; Barluenga, J.; Pérez, J.J.; Vázquez, J.T.; Centeno, N.B.; et al. Iodine Atoms: A New Molecular Feature for the Design of Potent Transthyretin Fibrillogenesis Inhibitors. PLoS ONE 2009, 4, e4124. [Google Scholar] [CrossRef]
- Ghosh, M.; Meerts, I.A.; Cook, A.; Bergman, A.; Brouwer, A.; Johnson, L.N. Structure of human transthyretin complexed with bromophenols: A new mode of binding. Acta Crystallogr. Sect. D Biol. Crystallogr. 2000, 56, 1085–1095. [Google Scholar] [CrossRef]
- Iakovleva, I.; Begum, A.; Brännström, K.; Wijsekera, A.; Nilsson, L.; Zhang, J.; Andersson, P.L.; Sauer-Eriksson, A.E.; Olofsson, A. Tetrabromobisphenol A Is an Efficient Stabilizer of the Transthyretin Tetramer. PLoS ONE 2016, 11, e0153529. [Google Scholar] [CrossRef]
- Ciccone, L.; Nencetti, S.; Rossello, A.; Stura, E.A.; Orlandini, E. Synthesis and structural analysis of halogen substituted fibril formation inhibitors of Human Transthyretin (TTR). J. Enzym. Inhib. Med. Chem. 2016, 31, 40–51. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nilsson, L.; Larsson, A.; Begum, A.; Iakovleva, I.; Carlsson, M.; Brännström, K.; Sauer-Eriksson, A.E.; Olofsson, A. Modifications of the 7-Hydroxyl Group of the Transthyretin Ligand Luteolin Provide Mechanistic Insights into Its Binding Properties and High Plasma Specificity. PLoS ONE 2016, 11, e0153112. [Google Scholar] [CrossRef] [PubMed]
- Cotrina, E.Y.; Pinto, M.; Bosch, L.; Vilà, M.; Blasi, D.; Quintana, J.; Centeno, N.B.; Arsequell, G.; Planas, A.; Valencia, G. Modulation of the fibrillogenesis inhibition properties of two transthyretin ligands by halogenation. J. Med. Chem. 2013, 56, 9110–9121. [Google Scholar] [CrossRef]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Masseoud, D.; Rott, K.; Liu-Bryan, R.; Agudelo, C. Overview of hyperuricaemia and gout. Curr. Pharm. Des. 2005, 11, 4117–4124. [Google Scholar] [CrossRef]
- Strilchuk, L.; Fogacci, F.; Cicero, A.F. Safety and tolerability of available urate-lowering drugs: A critical review. Expert Opin. Drug Saf. 2019, 18, 261–271. [Google Scholar] [CrossRef]
- López, L.C.; Varea, O.; Navarro, S.; Carrodeguas, J.A.; Sanchez de Groot, N.; Ventura, S.; Sancho, J. Benzbromarone, Quercetin, and Folic Acid Inhibit Amylin Aggregation. Int. J. Mol. Sci. 2016, 17, 964. [Google Scholar] [CrossRef]
- Petrukhin, K. Transthyretin Ligands Capable of Inhibiting Retinol-Dependent RBP4-TTR Interaction for Treatment of Age-Related Macular Degeneration, Stargardt Disease, and Other Retinal Disease Characterized by Excessive Lipofuscin Accumulation. US 20150057320 A1, 26 February 2015. [Google Scholar]
- Santos, L.; Rodrigues, D.; Alemi, M.; Silva, S.; Ribeiro, C.; Cardoso, I. Resveratrol administration increases Transthyretin protein levels ameliorating AD features-importance of transthyretin tetrameric stability. Mol. Med. 2016, 22, 597–607. [Google Scholar] [CrossRef]
- Doyle, M.L. Characterization of binding interactions by isothermal titration calorimetry. Curr. Opin. Biotechnol. 1997, 8, 31–35. [Google Scholar] [CrossRef]
- Miller, M.; Pal, A.; Albusairi, W.; Joo, H.; Pappas, B.; Haque Tuhin, M.T.; Liang, D.; Jampala, R.; Liu, F.; Khan, J.; et al. Enthalpy-Driven Stabilization of Transthyretin by AG10 Mimics a Naturally Occurring Genetic Variant That Protects from Transthyretin Amyloidosis. J. Med. Chem. 2018, 61, 7862–7876. [Google Scholar] [CrossRef]
- Bulawa, C.E.; Connelly, S.; Devit, M.; Wang, L.; Weigel, C.; Fleming, J.A.; Packman, J.; Powers, E.T.; Wiseman, R.L.; Foss, T.R.; et al. Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade. Proc. Natl. Acad. Sci. USA 2012, 109, 9629–9634. [Google Scholar] [CrossRef] [PubMed]
- Dolado, I.; Nieto, J.; Saraiva, M.J.M.; Arsequell, G.; Valencia, G.; Planas, A. Kinetic assay for high-throughput screening of in vitro transthyretin amyloid fibrillogenesis inhibitors. J. Comb. Chem. 2005, 7, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Razavi, H.; Palaninathan, S.K.; Powers, E.T.; Wiseman, R.L.; Purkey, H.E.; Mohamedmohaideen, N.N.; Deechongkit, S.; Chiang, K.P.; Dendle, M.T.A.; Sacchettini, J.C.; et al. Benzoxazoles as Transthyretin Amyloid Fibril Inhibitors: Synthesis, Evaluation, and Mechanism of Action. Angew. Chem. 2003, 115, 2864–2867. [Google Scholar] [CrossRef]
- Furuya, H.; Saraiva, M.J.; Gawinowicz, M.A.; Alves, I.L.; Costa, P.P.; Sasaki, H.; Goto, I.; Sakaki, Y. Production of recombinant human transthyretin with biological activities toward the understanding of the molecular basis of familial amyloidotic polyneuropathy (FAP). Biochemistry 1991, 30, 2415–2421. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.R.; Damas, A.M.; Lans, M.C.; Brouwer, A.; Saraiva, M.J. Thyroxine binding to transthyretin Met 119. Comparative studies of different heterozygotic carriers and structural analysis. Endocrine 1997, 6, 309–315. [Google Scholar] [CrossRef]
- Almeida, M.R.; Alves, I.L.; Terazaki, H.; Ando, Y.; Saraiva, M.J. Comparative Studies of Two Transthyretin Variants with Protective Effects on Familial Amyloidotic Polyneuropathy: TTR R104H and TTR T119M. Biochem. Biophys. Res. Commun. 2000, 270, 1024–1028. [Google Scholar] [CrossRef]
- Battye, T.G.; Kontogiannis, L.; Johnson, O.; Powell, H.R.; Leslie, A.G. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Bailey, S. The Ccp4 Suite-Programs for Protein Crystallography. Acta Crystallogr. D Biol Crystallogr. 1994, 50, 760–763. [Google Scholar]
- Brunger, A.T. Free R-Value-a Novel Statistical Quantity for Assessing the Accuracy of Crystal-Structures. Nature 1992, 355, 472–475. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger, L. The PyMOL Molecular Graphics System Version 1.3r1. 2010. Available online: https://pymol.org/2/ (accessed on 1 September 2020).
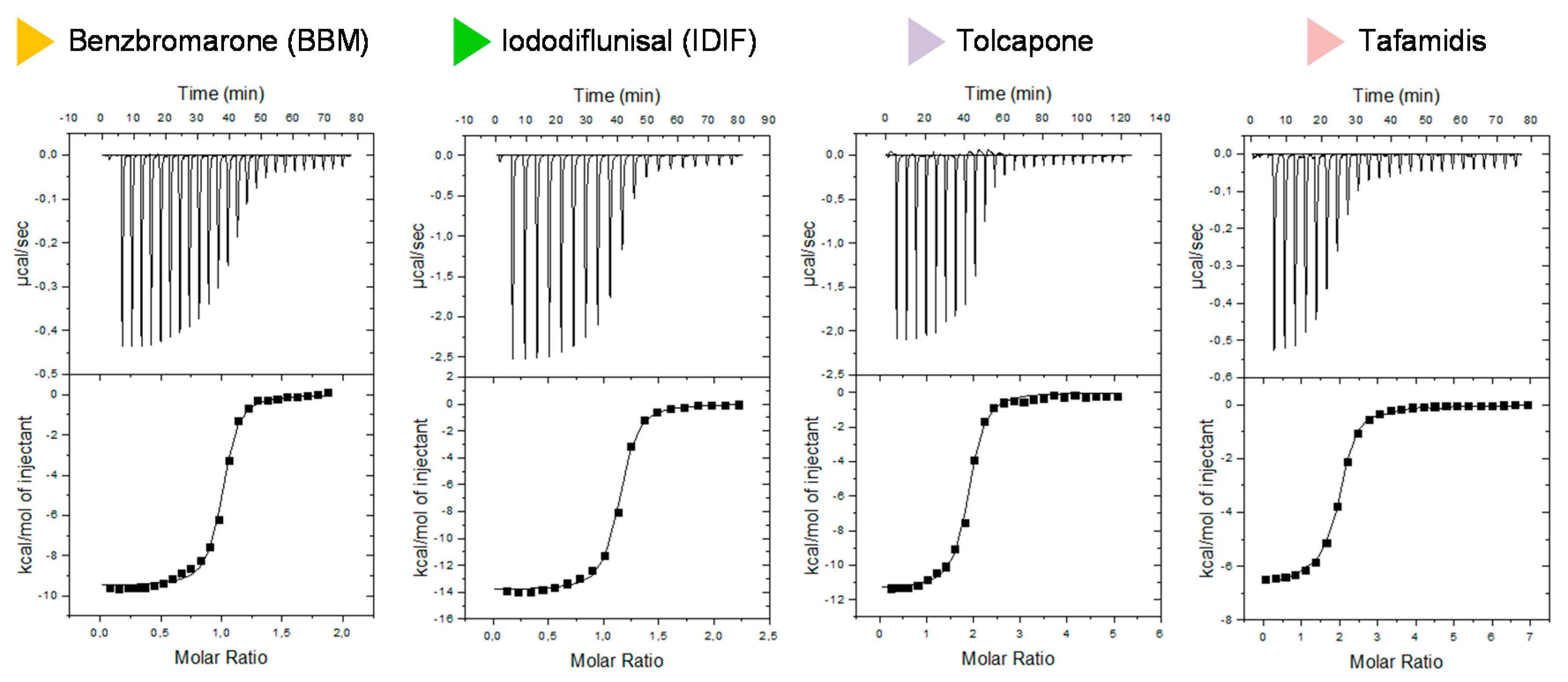
| Assay | n | Kd (nM) | ΔG (Kcal/mol) | ΔH (Kcal/mol) | TΔS (Kcal/mol) |
|---|---|---|---|---|---|
| wtTTR + BBM | 1 | 60 | −9.93 | −9.94 | −0.02 |
| wtTTR + IDIF | 1 | 120 | −9.49 | −13.10 | −3.61 |
| wtTTR + tafamidis | 2 | 200 | −9.16 | −6.50 | 2.66 |
| wtTTR + tolcapone | 2 | 270 | −8.98 | −12.30 | −3.36 |
| wtTTR + diflunisal | 1 | 900 | −8.25 | −11.96 | −3.70 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cotrina, E.Y.; Oliveira, Â.; Leite, J.P.; Llop, J.; Gales, L.; Quintana, J.; Cardoso, I.; Arsequell, G. Repurposing Benzbromarone for Familial Amyloid Polyneuropathy: A New Transthyretin Tetramer Stabilizer. Int. J. Mol. Sci. 2020, 21, 7166. https://doi.org/10.3390/ijms21197166
Cotrina EY, Oliveira Â, Leite JP, Llop J, Gales L, Quintana J, Cardoso I, Arsequell G. Repurposing Benzbromarone for Familial Amyloid Polyneuropathy: A New Transthyretin Tetramer Stabilizer. International Journal of Molecular Sciences. 2020; 21(19):7166. https://doi.org/10.3390/ijms21197166
Chicago/Turabian StyleCotrina, Ellen Y., Ângela Oliveira, José Pedro Leite, Jordi Llop, Luis Gales, Jordi Quintana, Isabel Cardoso, and Gemma Arsequell. 2020. "Repurposing Benzbromarone for Familial Amyloid Polyneuropathy: A New Transthyretin Tetramer Stabilizer" International Journal of Molecular Sciences 21, no. 19: 7166. https://doi.org/10.3390/ijms21197166
APA StyleCotrina, E. Y., Oliveira, Â., Leite, J. P., Llop, J., Gales, L., Quintana, J., Cardoso, I., & Arsequell, G. (2020). Repurposing Benzbromarone for Familial Amyloid Polyneuropathy: A New Transthyretin Tetramer Stabilizer. International Journal of Molecular Sciences, 21(19), 7166. https://doi.org/10.3390/ijms21197166





