Abstract
A series of berberine and tetrahydroberberine sulfonate derivatives were prepared and tested against the tyrosyl-DNA phosphodiesterase 1 (Tdp1) DNA-repair enzyme. The berberine derivatives inhibit the Tdp1 enzyme in the low micromolar range; this is the first reported berberine based Tdp1 inhibitor. A structure–activity relationship analysis revealed the importance of bromine substitution in the 12-position on the tetrahydroberberine scaffold. Furthermore, it was shown that the addition of a sulfonate group containing a polyfluoroaromatic moiety at position 9 leads to increased potency, while most of the derivatives containing an alkyl fragment at the same position were not active. According to the molecular modeling, the bromine atom in position 12 forms a hydrogen bond to histidine 493, a key catalytic residue. The cytotoxic effect of topotecan, a clinically important topoisomerase 1 inhibitor, was doubled in the cervical cancer HeLa cell line by derivatives 11g and 12g; both displayed low toxicity without topotecan. Derivatives 11g and 12g can therefore be used for further development to sensitize the action of clinically relevant Topo1 inhibitors.
Keywords:
berberine; tetrahydroberberine; Tdp1 inhibitor; cancer; molecular modeling; DNA repair enzyme; SAR 1. Introduction
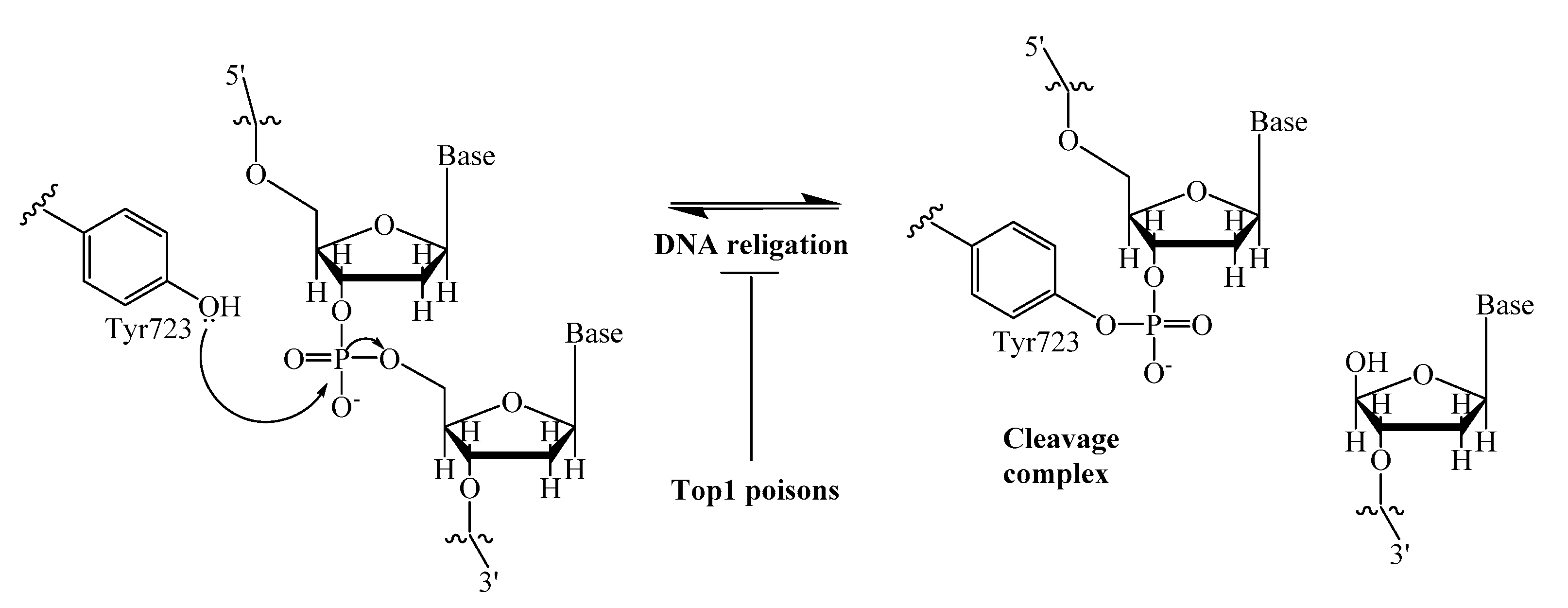
A promising strategy to enhance the efficacy of anticancer therapy is the inhibition of various DNA repair enzymes, which counteract the effect of many anticancer drugs [1,2]. This is particularly important where resistance to chemotherapy is observed. An interesting example is the poly (ADP-ribose) polymerase (PARP), which inhibitors were studied both in combination with chemotherapeutic agents and as individual drugs. Now olaparib, rucaparib and niraparib are in clinical use for the treatment of ovarian cancers [3]. Another DNA repair enzyme, tyrosyl-DNA phosphodiesterase 1 (Tdp1) has attracted considerable interest in the last few years mainly due to its ability to repair DNA lesions caused topoisomerase 1 (Top1) poisons, a well-established class of anticancer drugs [4]. The anticancer activity of Top1 poisons, camptothecin and its clinically important derivatives, topotecan and irinotecan [5,6], is based on their ability to bind to the covalent intermediate complex Top1/DNA and prevent the restoring of DNA integrity. This leads to the stabilization of the covalent bond between catalytic tyrosine (Y723) of Top1 and the 3′- end of DNA (Figure 1). The Tdp1 mechanism of action is the phosphotyrosyl bond hydrolysis [7], resulting in the resumption of DNA replication and cell division.
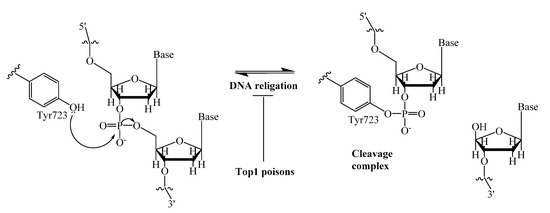
Figure 1.
DNA single-strand cleavage by nucleophilic attack of Tyr723 of Top1, and covalent cleavage complex formation.
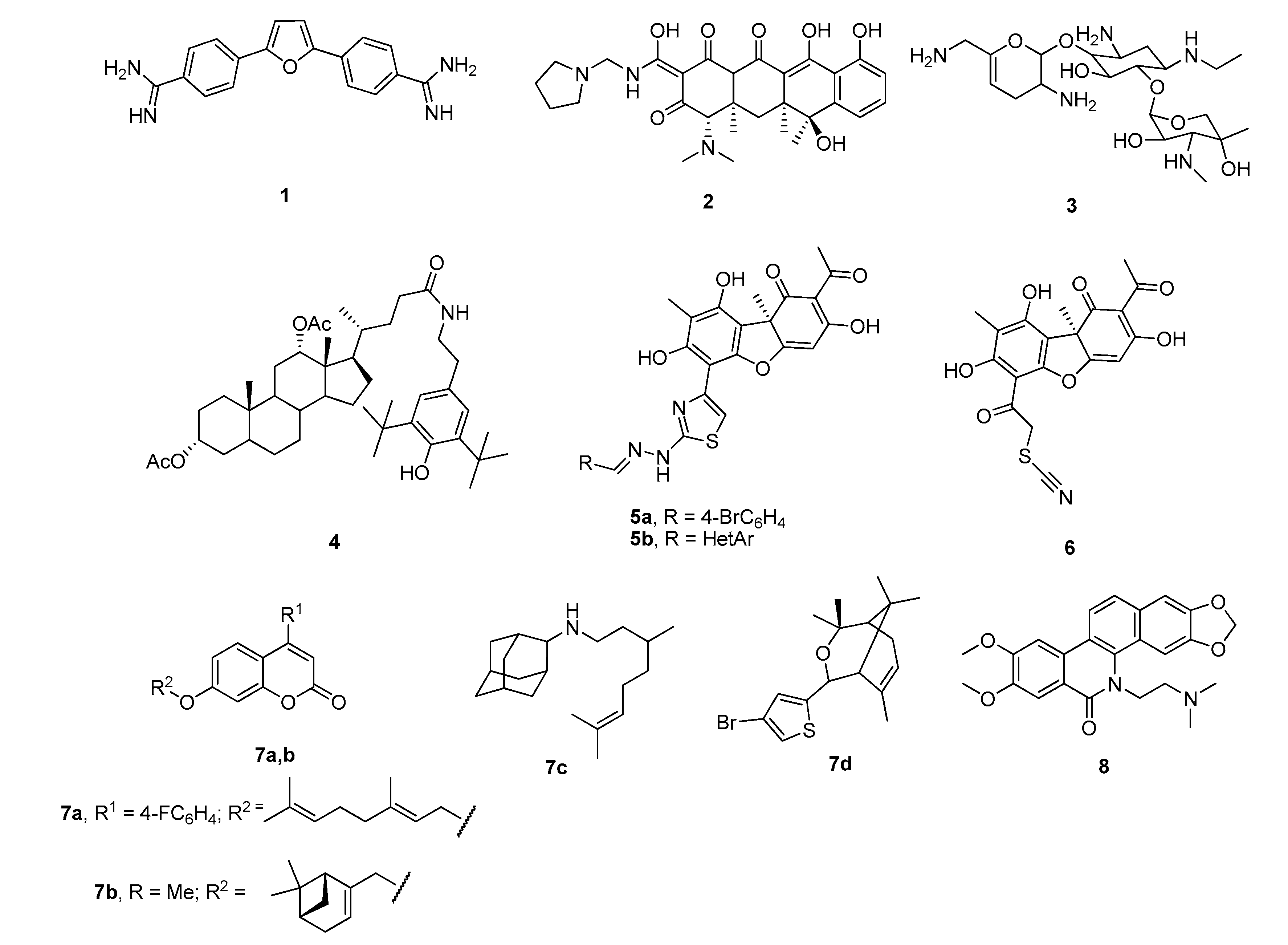
A few classes of Tdp1 inhibitors are known such as furamidines (compound 1, Figure 2), tetracyclines (compound 2), aminoglycosides (compound 3) [8,9]. Also, natural products of various types have been found to inhibit Tdp1, including derivatives of bile acids 4 [10,11], of lichen metabolite usnic acid 5a,b [12,13] and 6 [14], monoterpenoid derivatives 7 [15,16,17,18] and oxinitidine 8 [19] with inhibitory activity in the micro- or submicromolar range. Importantly, the hydrazinothiazole derivative of usnic acid 5a [13,20] and monoterpene-substituted 4-arylcoumarin 7a [21] significantly increased topotecan efficacy in vivo.
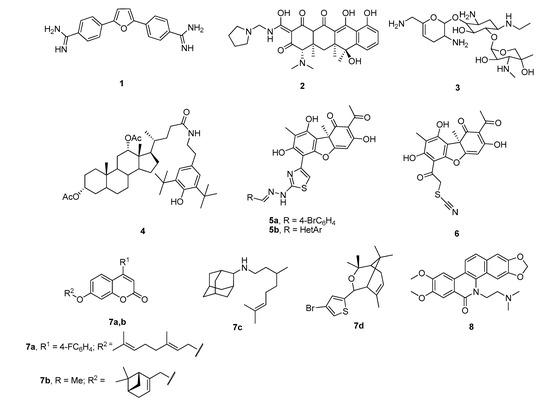
Figure 2.
Examples of established Tdp1 inhibitors 1–8.
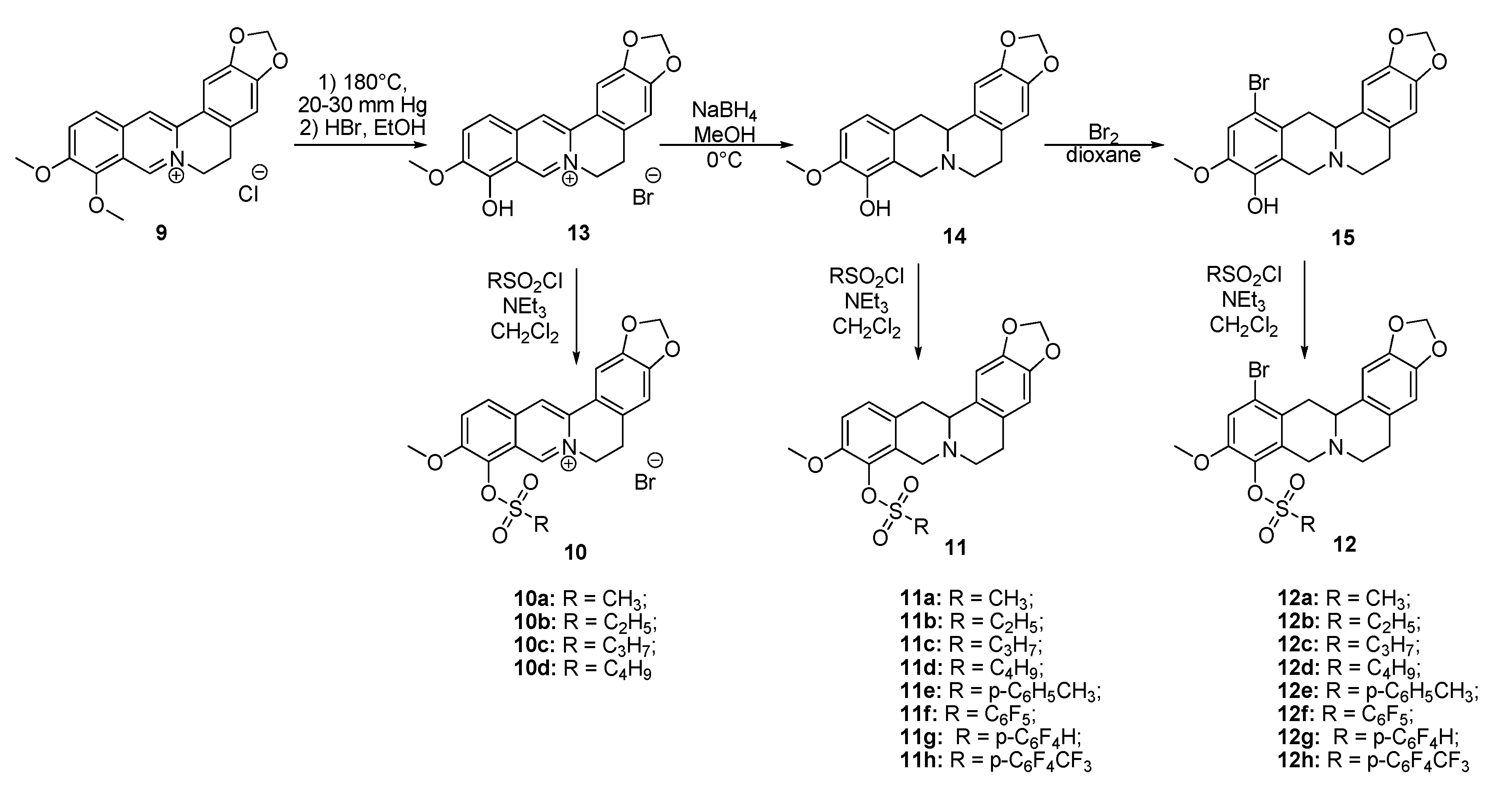
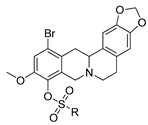
The aim of this study was to establish the potency of a novel structural class of natural products, the derivatives of berberine 9 (Scheme 1), which like their usnic acid counterparts are phenolic compounds. It is known that the isoquinoline plant alkaloid berberines have many beneficial physiological effects, e.g., they are hypocholesterolemic, antibacterial, hypoglycemic agents, antioxidants and, finally, they suppress tumor growth [22,23,24,25,26]. Interestingly, the sulfonate derivatives of berberines and tetrahydroberberines have been reported as promising hypocholesterolemic agents [27,28]. Although berberine derivatives never used as Tdp1 inhibitors, a preliminary molecular modeling study indicated that berberines with 9-sulfonate group would bind to Tdp1. In the present study, 9-sulfonate-berberine and tetrahydroberberine derivatives 10–12 with aliphatic and aromatic substitutes were synthesized and their potency against Tdp1 was tested. To the best of our knowledge, this has not been done previously. Using the HeLa cervical cancer cell line, derivatives 11g and 12g were found to be nontoxic and sensitized the cancer cells to topotecan.
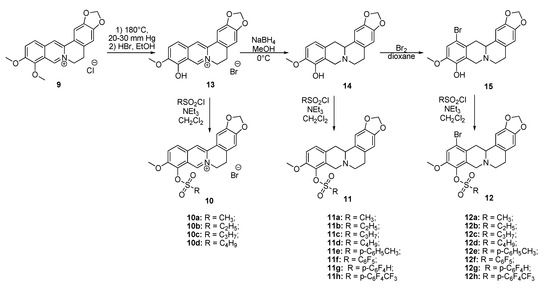
Scheme 1.
The synthetic pathways to sulfonates 10, 11 and 12.
2. Results and Discussion
2.1. Chemistry
Sulfonates 10–12 were synthesized in accordance with previously reported methods [27]. For this purpose, berberine 9 was selectively demethylated at 190 °C under vacuum as previously described [29]. After treatment with HBr, berberrubine hydrobromide 13 was isolated in 89% yield (Scheme 1). Compound 13 was reduced with sodium borohydride in methanol according to a procedure described previously [30] yielding tetrahydroberberrubine 14. The bromination of compound 14 with a bromine in dioxane solution afforded 12-bromotetrahydroberberrubine 15 in 52% yield. The reaction of tetrahydroderivatives 14 and 15 with polyfluoroaryl and alkyl sulfonylchlorides as well as with tosylchloride in dichloromethane in the presence of triethylamine produced tetrahydroberberrubine 9-O-sulfonates 11a–h (44–84% yields) and 12-bromotetrahydroberberrubine 9-O-sulfonates 12a–h (49–93% yields). New berberine type 10 derivatives were synthesized by the reaction of berberrubine hydrobromide 13 with different alkyl sulfochlorides. The reactions were carried out in dichloromethane in the presence of triethylamine for 5 h at room temperature. Sulfonates 10 were isolated by precipitation from the reaction mixtures.
The structures of the new compounds were confirmed by 1H-NMR,13C-NMR, IR and HRMS methods; the results are shown in the experimental section and the supplementary information.
2.2. Effects of the Berberine Sulfonates on Tdp1 Activity
An oligonucleotide real-time biosensor was used based on the ability of Tdp1 to remove fluorophore quenchers from the 3′-end of DNA, as previously described [31]. The hexadecameric oligonucleotide carried 5(6)-carboxyfluorescein (FAM) at the 5′-end and the fluorophore quencher BHQ1 (Black Hole Quencher-1) at the 3′-end. Tdp1 inhibitors prevent removal of fluorophore quenchers, thus reducing fluorescence intensity. The results of the Tdp1 assay for derivatives 10–12 and cytotoxic effects are shown in Table 1 and Table 2.

Table 1.
Tdp1 inhibiting activity (IC50—half maximal inhibitory concentration) of sulfonate berberine derivatives with aliphatic substituents and their cytotoxicity (CC50—half maximal cytotoxic concentration) in HeLa cells. Furamidine was used as a positive control (IC50 = 1.2 ± 0.3 µM).

Table 2.
Tdp 1 inhibiting activity (IC50) of sulfonate berberine derivatives with polyfluoroaromatic substituents and their cytotoxicity (CC50) in HeLa cells. Furamidine was used as a positive control (IC50 = 1.2 ± 0.3 µM).
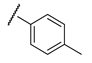
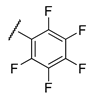
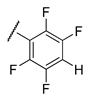
It is clear from the data in Table 1 that both alkyl sulfonates of berberine and tetrahydroberberine are not very active with the exception of the tetrahydroberberine derivatives containing both sufficiently long alkyl substituent and bromine in a para-position to the sulfonate substituent, 12c and 12d. The favorable substitution pattern of these derivatives is confirmed by the results of the tetrahydroberberine sulfonates with aromatic and polyfluoroaromatic substituents as shown in Table 2. Both sulfonates with para-toluenesulfonyl substituent (11e and 12e) are inactive, but all three fluorinated sulfonates (11f, 11g, 11h) show good inhibitory activity with IC50 values of ~1 µM. Bromine substitution is not important for 12e–h activity as comparison with their non-brominated analogues 11e–h shows.
Based on the data shown in Table 1 and Table 2 it can be stated that some berberine derivatives inhibit Tdp1. It was found that the structure of the substituent in the sulfonate affects the inhibitory activity. Polyfluorinated arylsulfonates 11f–h and 12f–h exhibited inhibitory activity in the low micromolar range, while their non-fluorinated analogs (11e, 12e) were inactive at these concentrations. In the series of alkylsulfonates, inhibitory activity was found only for compounds with a sufficiently long alkyl substituent (propyl-, butyl-) in the sulfonate group and in the presence of bromine substituent at position 12 (compounds 12c,d). To explain the observed effects, a molecular modeling study was carried out.
2.3. Molecular Modeling of the Berberine Derivatives
Twenty-three berberine derivatives were docked into the binding site of Tdp1 (PDB ID: 6DIE, resolution 1.78 Å) [32] with three water molecules (HOH814, 821 and 1078). It is established that keeping these crystalline water molecules improves the prediction quality of the docking scaffold (for further information see the Methodology section) [13]. The binding predictions of the scoring functions used are given in Table S1, all the ligands show reasonable scores.
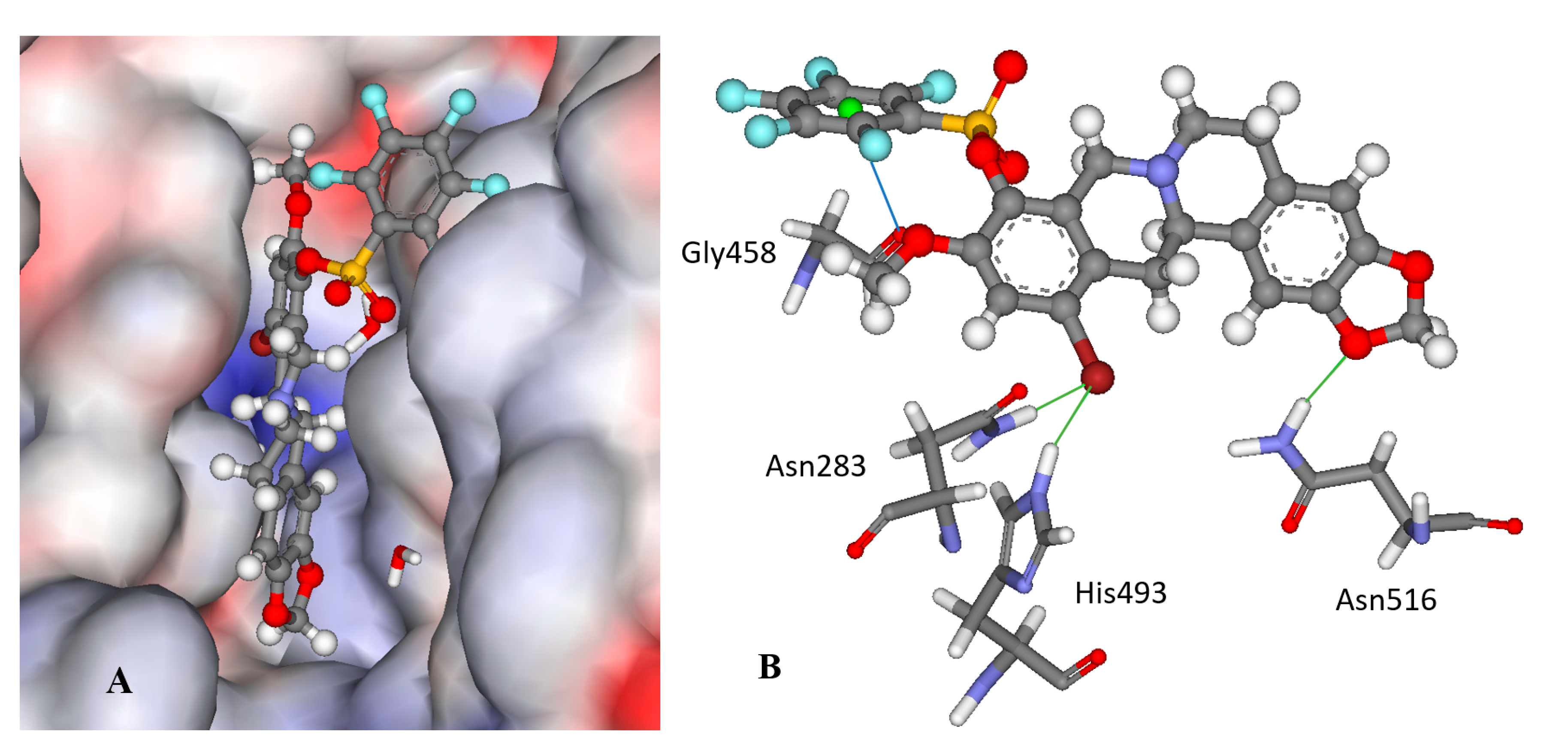
Compound 12f is the ligand with the best IC50 value and according to the docking; 12f fits neatly in the catalytic region as shown in Figure 3A. This region contains the catalytic histidine 263 and 493 amino acid residues thus the ligand blocs any activity of the enzyme. Indeed, 12f forms a weak H-bond with the His493 imidazole site group via the bromine substituent as shown in Figure 3B as well as with Asn283’s amide side chain. Finally, the amide group of Asn516 forms an H-bond with one of the oxygen atoms in the 1.3-benzodioxole moiety of the ligand. It is worth mentioning that the methoxy group on 12f can potentially form a weak H-bond with the thiol on Cys205, if the flexibility of the protein would be accounted for, and the same methoxy group has lipophilic contacts with Ile285 aliphatic side chain, stabilizing the binding mode. Interestingly, the modeling of the 11f ligand, which does not contain a bromine group, but also with a good IC50 value, did not give consistent results across the scoring functions used, i.e., different conformations were predicted. This strongly indicates that the bromine group with its weak H-bonds to Asn283 and His493 is essential for anchoring the ligand in the catalytic site.
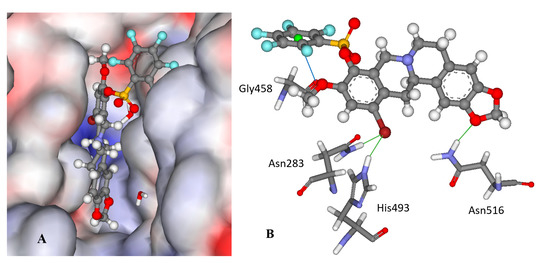
Figure 3.
The docked configuration of 12f in the binding site of Tdp1 as predicted by the ChemPLP scoring function. (A) The protein surface is rendered; blue depicts a hydrophilic region with a partial positive charge on the surface; red depicts hydrophobic region with a partial negative charge and grey shows neutral areas. The ligand occupies the catalytic pocket blocking access to it. (B) H-bonds are shown as green lines between 12f and the amino acids Asn283, His493 and Asn516 side chains. A potential lone pair-π stacking interaction is shown as a blue line between the carboxylic backbone group in Gly458 and the centroid (green ball) of the fluorinated phenyl group (3.5 Å).
Interestingly, derivatives 11e and 12e are essentially inactive (IC50 >15 μM); they are structural analogues of the 11f/12f pair with hydrogens on the phenyl ring as well as a para methyl substitution instead of fluorine groups. According to the modeling, the 11e/12e pair has a different binding mode from 11f/12f with the bromine moiety pointing into the aqueous phase for the former pair as shown in Figure S1 in the Supplementary Information. This indicates the importance of the fluorine substitution on the phenyl rings; the modeling suggests that the phenyl ring is leaning against the carboxyl moiety in the Gly458 backbone, which can form an interaction between the electron deficient fluoride substituted ring and the lone pairs of the oxygen atom in the carboxylic group as shown in Figure 3B.
To investigate the binding stability of the ligands molecular dynamic (MD) runs were conducted using the docked conformations of 11f, 12f and 12e for 10 ps at 1000 K. In general, the ligands are stable within the binding pocket and not ejected; the most stable intramolecular bond being between the fluorinated phenyl ring and the Gly458 carboxyl group for 12f. In contrast, the phenyl group in 12e is very mobile. The other H-bonding interactions predicted are often broken to be reestablished during the MD run.
2.4. Cytotoxicity
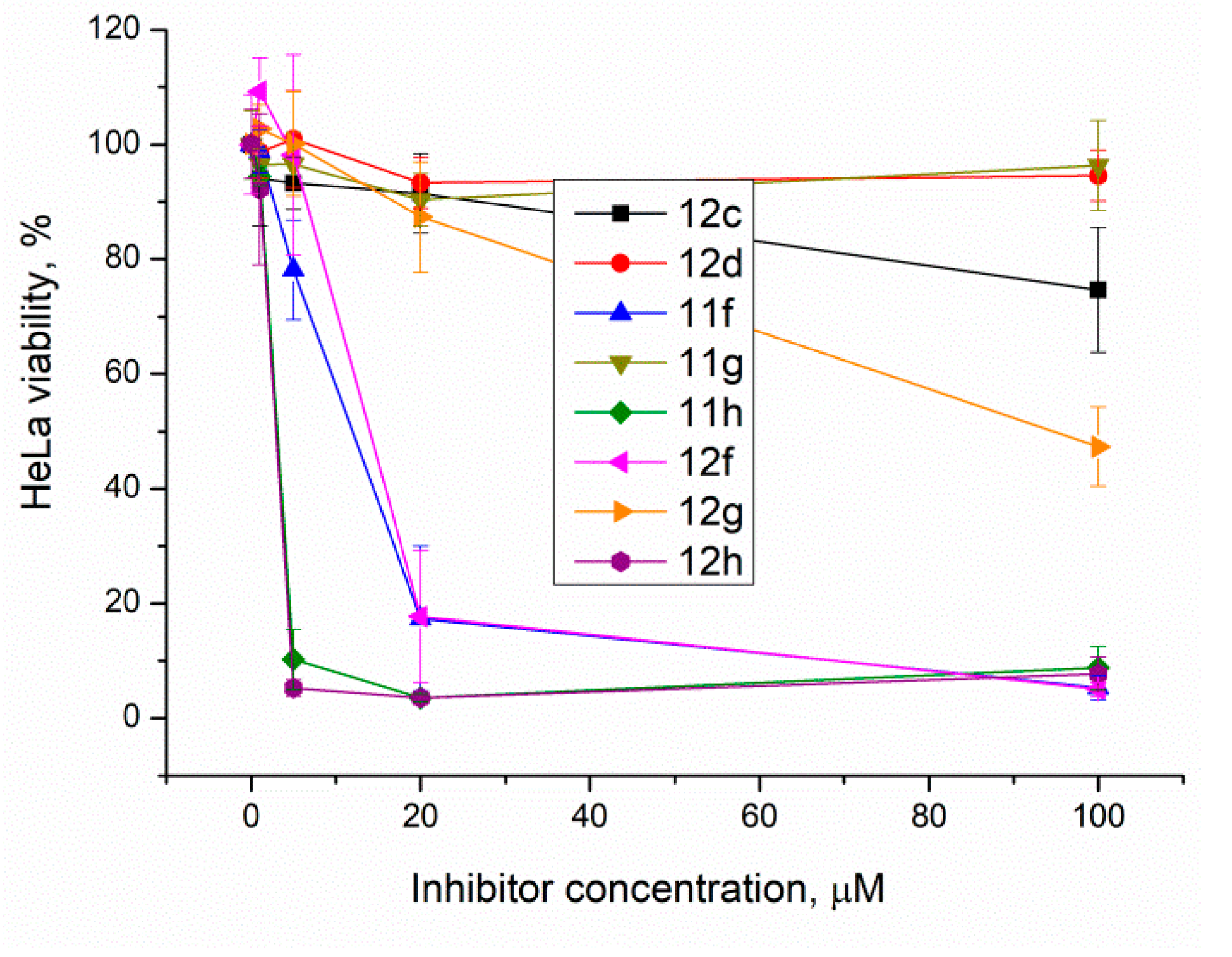
Top1 poisons are used as anticancer drugs for the treatment for various oncological diseases [33,34,35]. Since Tdp1 is involved in the removal of DNA damage caused by Top1 poisons, the activity of Tdp1 can lead to the development of drug resistance [36]. Thus, it is believed that Tdp1 inhibition can enhance the efficacy of Top1 poisons [37]. Tdp1 inhibitors should have the lowest possible intrinsic toxicity to minimize potential side effects. Therefore, we studied the intrinsic cytotoxicity of the compounds against HeLa cells (cervical carcinoma). EZ4U cell proliferation and cytotoxicity assay results are shown in Figure 4 for the ligands with Tdp1 inhibitory activity.
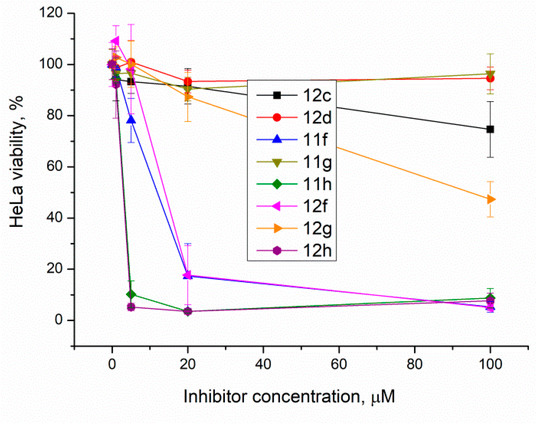
Figure 4.
The berberine derivatives’ cytotoxicity according to the EZ4U test. Error bars show standard deviations.
The results show that 9-O-sulfonates of 12-bromotetrahydoberberine with aliphatic substituents 12c and 12d, (Table 1 and Figure 4, black and red traces) are non-toxic up to 100 μM. The toxicity of 9-O-sulfonates strongly depends on the structure of the polyfluorinated fragment. Compounds with a CF3-group in the para-position were the most toxic; CC50 values of 2.6 μM and 2.2 μM for 11h and 12h, respectively (Table 2 and Figure 4, green and brown traces). The replacement of the trifluoromethyl group with a fluorine atom in the para-position reduced toxicity with CC50 values of 10 μM for 11f and 12f (Table 2 and Figure 4, blue and magenta traces, respectively). The compounds with a hydrogen atom in the para-position were non-toxic (11g, Table 2 and Figure 4, violet trace) or moderately toxic (12g, Table 2 and Figure 4, orange trace).
2.5. Sensitizing Effects
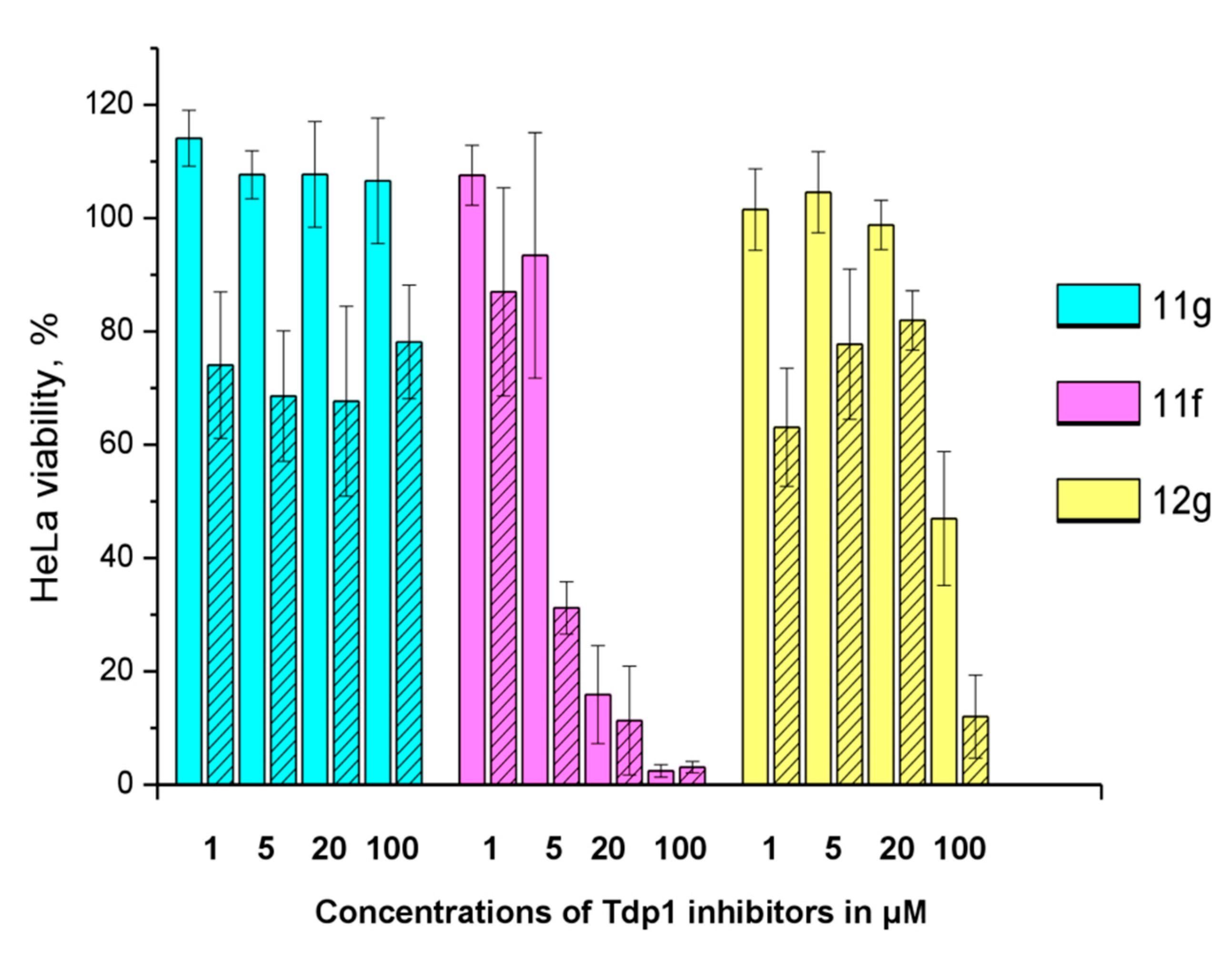
The sensitizing effect of the berberine inhibitors on topotecan’s cytotoxic potential was investigated. In order to determine the optimal concentration for the inhibitors to provide the maximum sensitizing effect, but remaining non-toxic, their concentrations were varied with topotecan concentration of 2 μM, its CC50 for HeLa cells. Topotecan significantly increased the cytotoxicity of compounds 11g, 12g, and 11f, the reliability was confirmed by the Mann–Whitney U-test, p = 0.05 and the results are shown in Figure 5. The original data are given in Table S2.
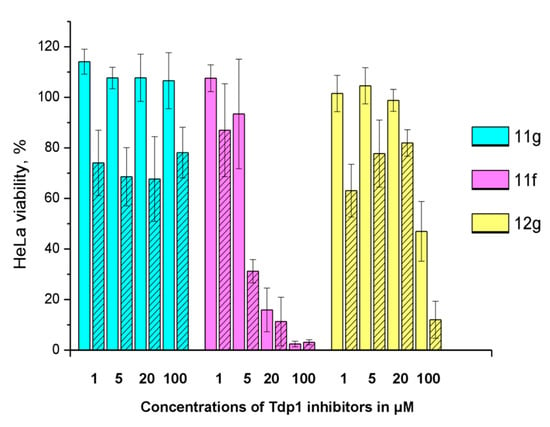
Figure 5.
The influence of 2 μM topotecan on Tdp1 inhibitors’ cytotoxicity. The unshaded histogram bars denote cell viability in the presence of a Tdp1 inhibitor. The hatched histogram bars indicate cell viability in the presence of a combination of a Tdp1 inhibitor with 2 μM of topotecan. The concentration of the Tdp1 inhibitor in μM is given under each pair of bars. Error bars show standard deviations.
11g is non-toxic in the concentration range used; 100% of living cells with a 100 μM maximum concentration. In the presence of topotecan, a significant decrease in cell survival is observed (~30%) at all concentrations. Compound 12g has a low toxicity potential of 95 μM (CC50). Topotecan weakly, but significantly reduces this value to 62 μM. Compound 11f is inherently toxic, and 90% of the cells die at 20 μM. At lower concentrations, the effect of topotecan is significant. CC50 value for 11f decreases three fold, from 11 to 3.7 μM, in the presence of topotecan. For other ligands (11h, 12c, d, f, h), the effect of topotecan was negligible or unreliable.
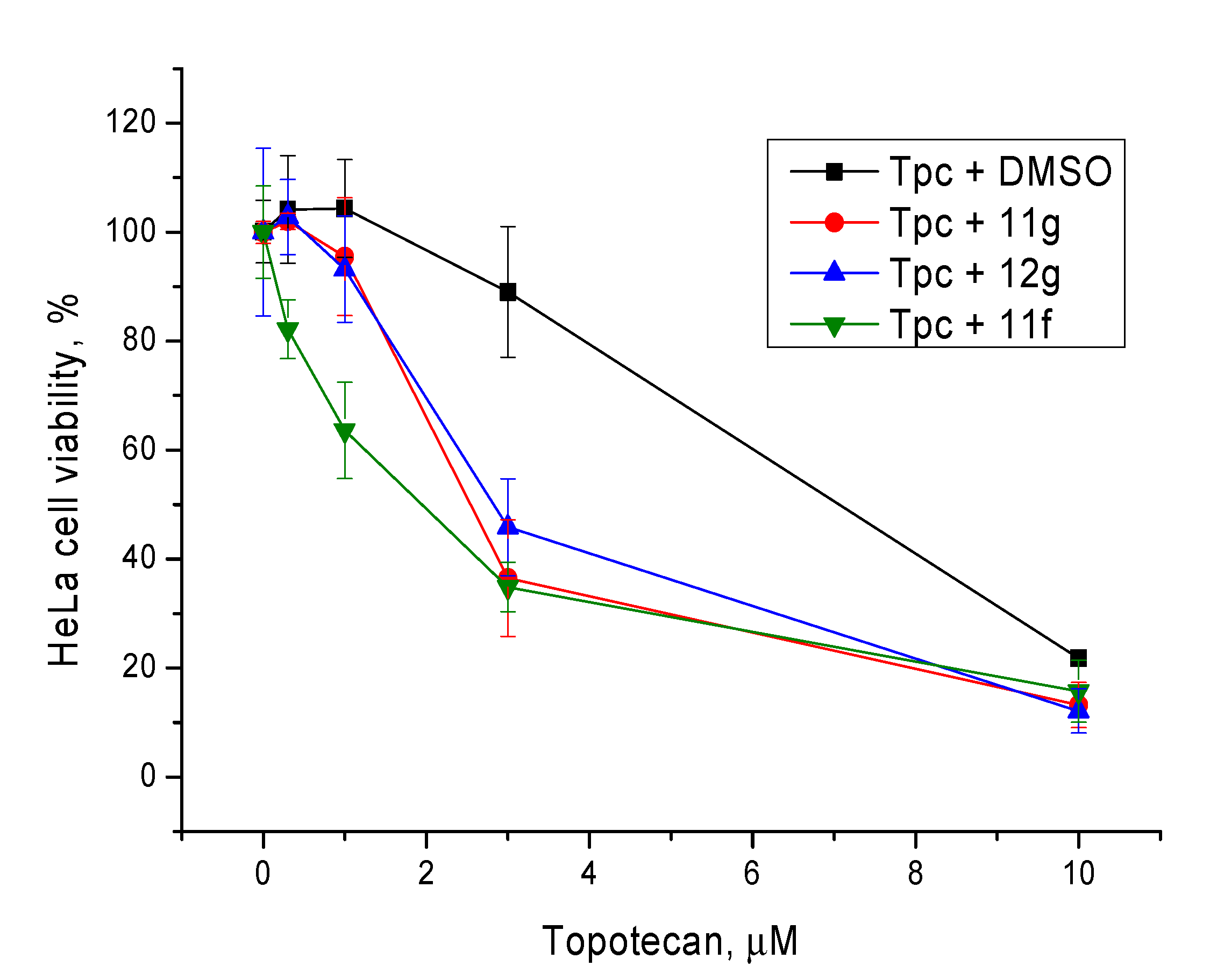
Non-toxic concentrations of the berberine derivatives (5 μM) were then tested at different concentrations of topotecan. The most toxic compound 11f caused 20% cell death at this concentration; the rest of the compounds were not toxic. In general, our Tdp1 inhibitors doubled the cytotoxic potential of topotecan as can be seen in Figure 6 and Table 3.
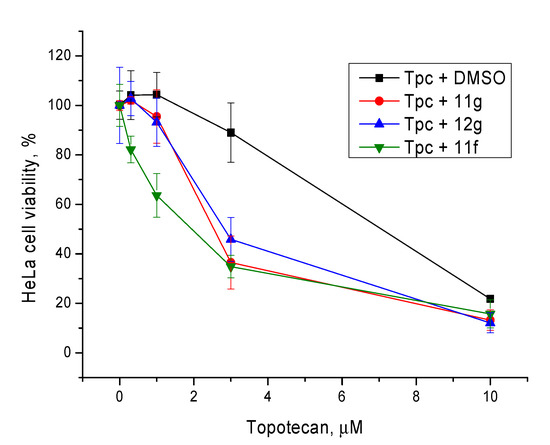
Figure 6.
The influence of the Tdp1 inhibitors at 5 µM on topotecan cytotoxicity. Error bars show standard deviations.

Table 3.
The influence of the Tdp1 inhibitors at 5 µM on the cytotoxic potential of topotecan (Tpc).
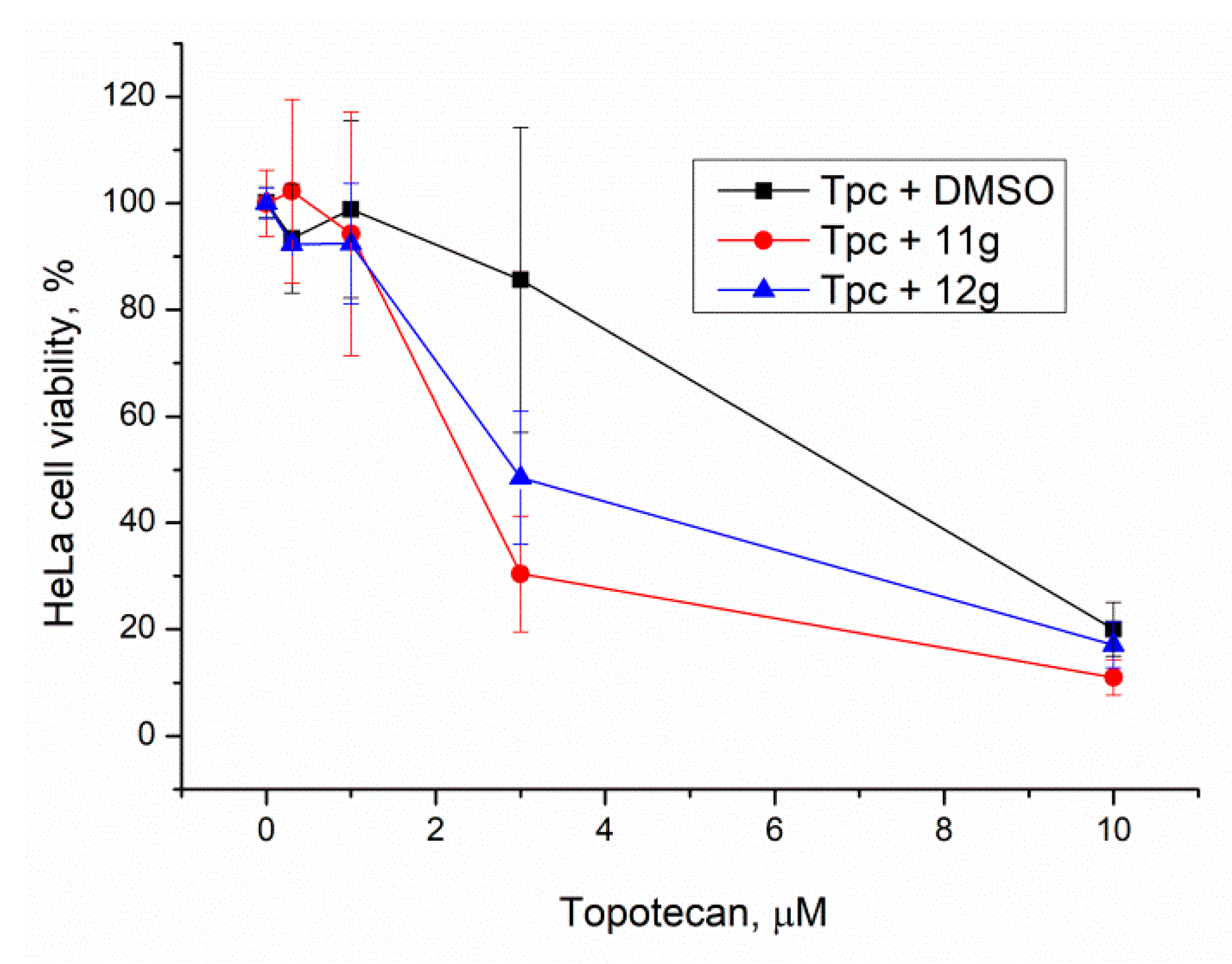
For comparison, the concentration of the inhibitors was increased to 20 μM. Compound 11f was not used due to its high toxicity. Again, compounds 12g and 11g had a significant effect, p < 0.05, confirmed by the Mann–Whitney U-test (Figure 7 and Table 3). It is interesting to note that the sensitizing effect of Tdp1 inhibitors was practically the same at 5 and 20 μM.
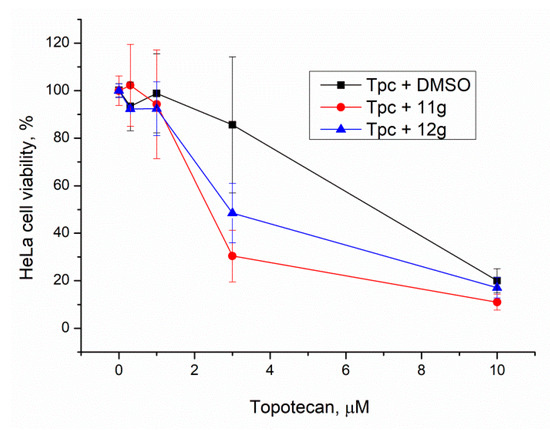
Figure 7.
Influence of Tdp1 inhibitors at 20 μM on topotecan cytotoxicity. Error bars show standard deviations.
Derivatives 11g and 12g can be considered to be the lead compounds since they are, unlike 11f, non-toxic (11g) or moderately toxic (12g) and have a pronounced sensitizing effect on topotecan.
2.6. Chemical Space
The calculated molecular descriptors MW (molecular weight), log P (water-octanol partition coefficient), HD (hydrogen bond donors), HA (hydrogen bond acceptors), PSA (polar surface area) and RB (rotatable bonds)) are given in Table S3. The MW of the ligands lies between 325.4 and 684.4 g mol−1 and falls into drug-like and Known Drug Space (KDS) regions. Log P spans from 1.6 to 5.4, i.e., over the three defined volumes in chemical space, with most in lead-like chemical space and only one ligand 12h in KDS. The HD, HA, RB and PSA values are within the lead- and drug-like definitions (for the definition of lead-like, drug-like and Known Drug Space regions see ref. [38] and Table S4).
The Known Drug Indexes (KDIs) for the ligands were calculated to gauge the balance of the molecular descriptors (MW, log P, HD, HA, PSA and RB). This method is based on the analysis of drugs in clinical use, i.e., the statistical distribution of each descriptor is fitted to a Gaussian function and normalized to 1 resulting in a weighted index. Both the summation of the indexes (KDI2a—Equation (1)) and multiplication (KDI2b—Equation (2)) methods were used [39]. The numerical results are given in Table S3 in the supplementary information.
KDI2a = IMW + Ilog P + IHD+ IHA + IRB + IPSA
KDI2b = IMW × Ilog P × IHD× IHA × IRB × IPSA
The KDI2a values range from 4.35 to 5.70 with a theoretical maximum of 6 and the average of 4.08 for known drugs. KDI2b range from 0.06 to 0.73, with a theoretical maximum of 1 and with KDS average of 0.18. The berberine ligands can be considered reasonably well balanced in terms of their molecular descriptors and therefore biocompatible. The most active compound 12f, has a KDI2a value of 4.58 and KDI2b of 0.12; the relatively low KDI2b value can be explained by its high MW, which is compensated by the favorable values for the other descriptors resulting in a good KDI2a value. The KDI2b index is sensitive to any outliers since multiplication of small numbers leads to small numbers.
3. Materials and Methods
3.1. Chemistry
Berberine chloride was purchased from Tokyo Chemical Industry Co., Ltd (Tokyo, Japan). Methanesulfochloride and ethanesulfochloride were purchased from Acros Organics (Belgium), 1-propanesulfochloride and 1-buthanesulfochloride were purchased from Alfa Aesar (Karlsruhe, Germany). 48% aqueous HBr solution was purchased from Acros Organics (the Netherlands). All solvents used in the reactions were purified and dried. Bromine, sodium borohydride and triethylamine were purchased from Sigma-Aldrich. Column chromatography was carried out on neutral alumina LL40/250. All reactions were monitored by TLC analysis using Merck Aluminium oxide 60 F254 plastic sheets (Darmstadt, Germany), eluent CH2Cl2-MeOH.
The 1H and 13C NMR spectra were recorded on a Bruker AM-400 spectrometer (400.13 and 100.61 MHz) for 5–10% solutions of the compounds in CDCl3 or DMSO-d6 using the signal of the solvent CDCl3 as the standard (δ 7.24 for 1H and δ 76.90 for 13C). The IR spectra were measured on a Vector 22 FTIR spectrometer in KBr pellets. High-resolution electrospray ionization (HRESI) mass spectra were carried out using a time-of-flight high-resolution mass spectrometer micrOTOF-Q (Bruker Daltonics, Germany) with an Agilent 1200 liquid chromatograph (Agilent Technologies, USA/Germany). Positive ion scanning in the range m/z = 100–3000. Drying gas (nitrogen) flow rate of 4 L/min; temperature—190 ℃; sprayer pressure—1.0 bar.
The spectroscopic and analytical measurements were carried out at the Multi-access Chemical Service Center of the Siberian Branch of the Russian Academy of Sciences.
3.1.1. General Synthesis of Compounds 11a–g and 12a–g
Desired compounds 11a–d, f, g and 12a–g were synthesized according to the general procedure described previously [27], compound 11e was also synthesized according to the procedure described previously [30].
3.1.2. General Procedure for the Synthesis of Sulfonates 10a–d
2.5 mmol of triethylamine was added to the suspension of 1 mmol of berberrubine hydrobromide in 25 mL of methylene chloride. 1.5 mmol of sulfochloride in 3 mL of methylene chloride was added under stirring at room temperature. After 5 h the precipitate was filtered, washed with methylene chloride and the pure product 10a–d was isolated.
3.1.3. 10-Methoxy-9-methanesulfonyloxy-2,3-methylenedioxyprotoberberine chloride (10a)
Yield: 7%
Spectra NMR 1H (400 MHz, DMSO-d6, δ, ppm, J/Hz): 3.22 (2H, t, J = 6.07 Hz, H-5), 3.73 (3H, s, SCH3), 4.12 (3H, s, OCH3), 5.00 (2H, t, J = 5.72 Hz, H-6), 6.19 (2H, s, OCH2O), 7.11 (1H, s, H-4), 7.82 (1H, s, H-1), 8.27 (1H, d, J = 9.06 Hz, H-12), 8.34 (1H, d, J = 9.06 Hz, H-11), 9.08 (1H, s, H-13), 9.72 (1H, s, H-8). Spectra NMR 13C (100 MHz, DMSO-d6, δ, ppm): 26.18 (C-5), 40.02 (SO2CH3), 55.70 (C-6), 57.44 (OCH3), 102.23 (OCH2O), 105.60 (C-1), 108.49 (C-4), 120.20 (C-13b), 120.86 (C-13), 121.79 (C-8a), 126.47 (C-12), 128.03 (C-11), 131.05 (C-4a), 131.44 (C-12a), 133.31 (C-13a), 138.59 (C-9), 144.04 (C-8), 147.77 (C-2), 150.17 (C-3), 151.89 (C-10). Spectra IR (cm−1): 804.21, 925.71, 1033.71, 1097.35, 1224.63, 1263.20, 1365.42, 1506.20, 1610.34, 1623.84, 3039.40, 3320.97, 3635.33. MS (ESI): m/z (M+) calcd for C20H18NO6S+, 400.085 found: 400.081.
3.1.4. 9-Ethanesulfonyloxy-10-methoxy-2,3-methylenedioxyprotoberberine chloride (10b)
Yield: 25%
Spectra NMR 1H (400 MHz, DMSO-d6, δ, ppm, J/Hz): 1.52 (3H, t, J = 7.32 Hz, CH2CH3), 3.22 (2H, t, J = 6.05 Hz, H-5), 3.89 (2H, q, J1 = 3.50 Hz, J2 = 10.96 Hz, CH2CH3), 4.12 (3H, s, OCH3), 5.01 (2H, t, J = 5.91 Hz, H-6), 6.18 (2H, s, OCH2O), 7.11 (1H, s, H-4), 7.82 (1H, s, H–1), 8.27 (1H, d, J = 9.48 Hz, H-12), 8.34 (1H, d, J = 9.48 Hz, H-11), 9.10 (1H, s, H-13), 9.64 (1H, s, H–8). Spectra NMR 13C (100 MHz, DMSO-d6, δ, ppm): 8.18 (CH2CH3), 26.19 (C-5), 47.00 (SCH2), 55.83 (C–6), 57.44 (OCH3), 102.20 (OCH2O), 105.61 (C-1), 108.45 (C–4), 120.18 (C-13b), 120.90 (C-13), 121.98 (C-8a), 126.42 (C-12), 128.02 (C-11), 131.02 (C-4a), 131.27 (C-12a), 133.36 (C-13a), 138.63 (C-9), 143.91 (C-8), 147.75 (C-2), 150.17 (C-3), 151.71 (C-10). Spectra IR (cm−1): 806.14, 923.78, 1033.71, 1097.35, 1164.85, 1222.70, 1278.2063, 1363.49, 1506.20, 1608.42, 1621.92, 3023.98, 3423.19, 3629.54. MS (ESI): m/z (M+) calcd for C21H20NO6S+, 414.101 found: 414.096.
3.1.5. 10-Methoxy-2,3-methylenedioxy-9-((propane-1-sulfonyl)oxy)protoberberine chloride (10c)
Yield: 34%
Spectra NMR 1H (400 MHz, DMSO-d6, δ, ppm, J/Hz): 1.11 (3H, t, J = 7.45 Hz, CH2CH3), 1.94–2.03 (2H, m, CH2CH3), 3.22 (2H, t, J = 5.72 Hz, H-5), 3.87 (2H, t, J = 7.58 Hz, SCH2CH2), 4.12 (3H, s, OCH3), 5.01 (2H, t, J = 6.02 Hz, H-6), 6.19 (2H, s, OCH2O), 7.11 (1H, s, H-4), 7.82 (1H, s, H-1), 8.27 (1H, d, J = 9.33 Hz, H-12), 8.34 (1H, d, J = 9.33 Hz, H-11), 9.10 (1H, s, H-13), 9.64 (1H, s, H-8). Spectra NMR 13C (100 MHz, DMSO-d6, δ, ppm): 12.49 (CH2CH3), 17.17 (CH2CH2CH3), 26.16 (C-5), 53.27 (SCH2), 55.80 (C-6), 57.46 (OCH3), 102.18 (OCH2O), 105.60 (C-1), 108.42 (C-4), 120.14 (C-13b), 120.86 (C-13), 121.93 (C-8a), 126.37 (C-12), 127.98 (C-11), 130.96 (C-4a), 131.23 (C-12a), 133.33 (C-13a), 138.55 (C-9), 143.91 (C-8), 147.70 (C-2), 150.12 (C-3), 151.69 (C-10). Spectra IR (cm−1): 817.71, 925.71, 1037.56, 1095.42, 1162.92, 1222.70, 1263.20, 1363.49, 1506.20, 1610.34, 1621.92, 3023.98, 3427.04. MS (ESI): m/z (M+) calcd for C22H22NO6S+, 428.116 found: 428.121.
3.1.6. 9-(Butane-1-sulfonyl)oxy-10-methoxy-2,3-methylenedioxy protoberberine chloride (10d)
Yield: 50%
Spectra NMR 1H (400 MHz, DMSO-d6, δ, ppm, J/Hz): 0.97 (3H, t, J = 7.34 Hz, CH2CH3), 1.48–1.58 (2H, m, CH2CH3), 1.89–1.97 (2H, m, SCH2CH2), 3.22 (2H, t, J = 5.96 Hz, H-5), 3.91 (2H, t, J = 7.68 Hz, SCH2), 4.11 (3H, s, OCH3), 5.02 (2H, t, J = 6.04 Hz), 6.18 (2H, c, OCH2O), 7.10 (1H, s, H-4), 7.82 (1H, s, H-1), 8.27 (1H, d, J = 9.21 Hz, H-12), 8.34 (1H, d, J = 9.21 Hz, H-11), 9.13 (1H, s, H-13), 9.67 (1H, s, H-8). Spectra NMR 13C (100 MHz, DMSO-d6, δ, ppm): 13.44 (CH3CH2), 20.74 (CH3CH2), 25.29 (SCH2CH2), 26.19 (C-5), 51.75 (SCH2), 55.80 (C-6), 57.46 (OCH3), 102.20 (OCH2O), 105.62 (C-1), 108.45 (C-4), 120.19 (C-13b), 120.91 (C-13), 121.98 (C-8a), 126.42 (C-12), 128.01 (C-11), 131.02 (C-4a), 131.30 (C-12a), 133.36 (C-13a), 138.62 (C-9), 143.97 (C-8), 147.75 (C-2), 150.16 (C-3), 151.72 (C-10). Spectra IR (cm−1): 811.92, 925.71, 1035.63, 1099.28, 1222.70, 1265.13, 1365.42, 1504.27, 1610.34, 1619.99, 2960.33, 3425.11. MS (ESI): m/z (M+) calcd for C23H24NO6S+, 442.132 found: 442.133.
3.2. Biology
3.2.1. Detection of Tdp1 Activity
The methodology has been reported in our previous work [30] and consists of fluorescence intensity measurement in a reaction of quencher removal from a fluorophore quencher-coupled DNA oligonucleotide catalyzed by Tdp1. The reaction was carried out at different concentrations of inhibitors (the control samples contained 1% of DMSO, Sigma, St. Louis, MO, USA). The reaction mixtures contained Tdp1 buffer (50 mM Tris-HCl pH 8.0, 50 mM NaCl, and 7 mM β-mercaptoethanol), 50 nM biosensor, and an inhibitor being tested. Purified Tdp1 (1.5 nM) triggered the reaction. The biosensor (5′-[FAM] AAC GTC AGGGTC TTC C [BHQ]-3′) was synthesized in the Laboratory of Biomedical Chemistry at the Institute of Chemical Biology and Fundamental Medicine (Novosibirsk, Russia).
The reactions were incubated on a POLARstar OPTIMA fluorimeter (BMG LABTECH, GmbH, Ortenberg, Germany) to measure fluorescence every 55 s (ex. 485/em. 520 nm) during the linear phase (here, data from minute 0 to minute 8). The values of IC50 were determined using a six-point concentration response curve in minimum three independent experiments and were calculated using MARS Data Analysis 2.0 (BMG LABTECH, GmbH, Ortenberg, Germany).
3.2.2. Cytotoxicity Assays
Cytotoxicity of the compounds to HeLa (human cervical cancer) cell line was examined using the EZ4U Cell Proliferation and Cytotoxicity Assay (Biomedica, Vienna, Austria), according to the manufacturer’s protocols. The cells were grown in Iscove’s modified Dulbecco’s medium (IMDM) with 40 μg/mL gentamicin, 50 IU/mL penicillin, 50 μg/mL streptomycin (MP Biomedicals, Santa Ana, CA, USA), and 10% of fetal bovine serum (Biolot, St. Petersburg, Russia) in a 5% CO2 atmosphere. After formation of a 30–50%-monolayer, the tested compounds were added to the medium. The volume of the added reagents was 1/100 of the total volume of the culture medium, and the amount of DMSO (Sigma, St. Louis, MO, USA) was 1% of the final volume. Control cells were grown in the presence of 1% DMSO. The cell culture was monitored for 3 days. To assess the influence of the inhibitors on the cytotoxic effect of topotecan (ACTAVIS GROUP PTC ehf., Bucharest, Romania), 50% cytotoxic concentrations of topotecan and of each inhibitor were determined to attain a defined single-agent effect. Then, minimum two independent tests were performed with each inhibitor in combination with topotecan. When using a combination of drugs, Tdp1 inhibitors were first added, then topotecan was added immediately (within 10–15 min).
3.3. Molecular Modeling
The compounds were docked against the crystal structure of TDP1 Tdp1 (PDB ID: 6DIE, resolution 1.78 Å) [32] which was obtained from the Protein Data Bank (PDB) [40,41]. The Scigress version FJ 2.6 program [42] was used to prepare the crystal structure for docking, i.e., the hydrogen atoms were added, the co-crystallized ligand benzene-1,2,4-tricarboxylic acid was removed as well as crystallographic water molecules except HOH 814, 821 and 1078. The waters were set on toggle—bound or displaced by the ligand during docking—and spin—automatic optimization of the orientation of the hydrogen atoms. The Scigress software suite was also used to build the inhibitors and the MM2 [43] force field was used to optimize the structures. Furthermore, Scigress was used for the 10 ps MD runs at 1000 K; the MM2 force filed was used, 5 Å radius was defined around the ligand and allowed to be flexible whereas the rest of the protein structure was held ridged (locked). First the binding pocket with the ligand was structurally optimized followed by the MD run. The docking center was defined as the position of a carbon on the ring of the co-crystallized benzene-1, 2, 4-tricarboxylic acid (x = −6.052, y = −14.428, z = 33.998) with 10 Å radius. Fifty docking runs were allowed for each ligand with default search efficiency (100%). The basic amino acids lysine and arginine were defined as protonated. Furthermore, aspartic and glutamic acids were assumed deprotonated. The GoldScore (GS) [44] and ChemScore (CS) [45,46] ChemPLP (Piecewise Linear Potential) [47] and ASP (Astex Statistical Potential) [48] scoring functions were implemented to predict the binding modes and relative energies of the ligands using the GOLD v5.4.1 software suite.
The QikProp 3.2 [49] software package was used to calculate the molecular descriptors of the molecules. The reliability of it QikProp established for the calculated descriptors. [50] The Known Drug Indexes (KDI) were calculated from the molecular descriptors as described by Eurtivong and Reynisson [40]. Five of the compounds (9 and 10a–d) carry a positive charge and QikProp does not compute charged molecules. The MW, Log P, HD and HA were derived using the Scigress version FJ 2.6 program [42] software, PSA and RB are not available in this software suite.
4. Conclusions
The berberine and tetrahydroberberine sulfonates and their brominated analogues were tested to evaluate their Tdp1 inhibitory activity. To our knowledge, this is the first report that documents berberine-based compounds as inhibitors of this enzyme. The IC50 values are in the 0.53 to 4 μM range. Of the alkyl sulfonates, only derivatives with bromine substitution on site 12 of tetrahydroberberrubine are active. While both tetrahydroberberrubine and 12-bromtetrahydroberberrubine derivatives containing polyfluoroaromatic substituents all have inhibitory activity with IC50 values of ~1 μM. According to the inhibitory activity, toxicity data and ability to sensitize topotecan against the HeLa cancer cell line the two most promising derivatives were identified—11g and 12g. These results indicate that sulfonates of tetrahydroberberine have potential to be developed as new agents for anticancer therapy due to their inhibitory activity and lack of toxicity.
Supplementary Materials
Supplementary Materials can be found at https://www.mdpi.com/1422-0067/21/19/7162/s1.
Author Contributions
Chemistry investigation, E.D.G., I.V.N., R.A.B. and O.A.L.; in vitro investigation, A.L.Z., A.A.C., E.S.I., N.S.D., E.M.M. and R.O.A.; modeling, J.R.; methodology, N.F.S. and O.I.L.; project administration, K.P.V.; supervision, K.P.V.; writing—original draft, E.D.G., A.L.Z. and O.A.L.; writing—review and editing, K.P.V., J.R., N.F.S. and O.I.L. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Russian Science Foundation grant № 19-13-00040.
Acknowledgments
Authors would like to acknowledge the Multi-Access Chemical Research Center SB RAS for spectral and analytical measurements.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Abbreviations
| Tdp1 | Tyrosyl-DNA phosphodiesterase 1 |
| PARP | Poly (ADP-ribose) polymerase |
References
- Zakharenko, A.L.; Lebedeva, N.A.; Lavrik, O.I. DNA Repair Enzymes as Promising Targets in Oncotherapy. Russ. J. Org. Chem. 2018, 44, 1–18. [Google Scholar] [CrossRef]
- Ferri, A.; Stagni, V.; Barilà, D. Targeting the DNA Damage Response to Overcome Cancer Drug Resistance in Glioblastoma. Int. J. Mol. Sci. 2020, 21, 4910. [Google Scholar] [CrossRef]
- Jiang, X.; Li, W.; Li, X.; Bai, H.; Zhang, Z. Current status and future prospects of PARP inhibitor clinical trials in ovarian cancer. Cancer Manag. Res. 2019, 1, 4371–4390. [Google Scholar] [CrossRef] [PubMed]
- Comeaux, E.Q.; Waardenburg, R.C. Tyrosyl-DNA phosphodiesterase I resolves both naturally and chemically induced DNA adducts and its potential as a therapeutic target. Drug Metab. Rev. 2014, 46, 494–507. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y.; Leo, E.; Zhang, H.; Marchand, C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010, 17, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Kciuk, M.; Marciniakand, B.; Kontek, R. Irinotecan—Still an Important Player in Cancer Chemotherapy: A Comprehensive Overview. Int. J. Mol. Sci. 2020, 21, 4919. [Google Scholar] [CrossRef]
- Pommier, Y.; Huang, S.-y.N.; Gao, R.; Das, B.B.; Murai, J.; Marchand, C. Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2). DNA Repair 2014, 19, 114–129. [Google Scholar] [CrossRef] [PubMed]
- Laev, S.S.; Salakhutdinov, N.F.; Lavrik, O.I. Tyrosyl-DNA phosphodiesterase inhibitors: Progress and potential. Bioorg. Med. Chem. 2016, 24, 5017–5027. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.N.; Pommier, Y.; Marchand, C. Tyrosyl-DNA Phosphodiesterase 1 (Tdp1) inhibitors. Expert Opin. Ther. Pat. 2011, 21, 1285–1292. [Google Scholar] [CrossRef] [PubMed]
- Salomatina, O.V.; Popadyuk, I.I.; Zakharenko, A.L.; Zakharova, O.D.; Fadeev, D.S.; Komarova, N.I.; Reynisson, J.; Arabshahi, H.I.; Chand, R.; Volcho, K.P.; et al. Novel Semisynthetic Derivatives of Bile Acids as Effective Tyrosyl-DNA Phosphodiesterase 1 Inhibitors. Molecules 2018, 23, 679. [Google Scholar] [CrossRef]
- Xiao, L.-G.; Zhang, Y.; Zhang, H.-L.; Li, D.; Gu, Q.; Tang, G.-H.; Yu, Q.; An, L.-K. Spiroconyone A, a new phytosterol with a spiro [5,6] ring system from Conyza japonica. Org. Biomol. Chem. 2020, 18, 5130–5136. [Google Scholar] [CrossRef] [PubMed]
- Zakharenko, A.; Luzina, O.; Koval, O.; Nilov, D.; Gushchina, I.; Dyrkheeva, N.; Švedas, V.; Salakhutdinov, N.; Lavrik, O. Tyrosyl-DNA Phosphodiesterase 1 Inhibitors: Usnic Acid Enamines Enhance the Cytotoxic Effect of Camptothecin. J. Nat. Prod. 2016, 79, 2961–2967. [Google Scholar] [CrossRef] [PubMed]
- Filimonov, A.S.; Chepanova, A.A.; Luzina, O.A.; Zakharenko, A.L.; Zakharova, O.D.; Ilina, E.S.; Dyrkheeva, N.S.; Kuprushkin, M.S.; Kolotaev, A.V.; Khachatryan, D.S.; et al. New Hydrazinothiazole Derivatives of Usnic Acid as Potent Tdp1 Inhibitors. Molecules 2019, 24, 3711. [Google Scholar] [CrossRef] [PubMed]
- Zakharenko, A.L.; Luzina, O.A.; Sokolov, D.N.; Zakharova, O.D.; Rakhmanova, M.E.; Chepanova, A.A.; Dyrkheeva, N.S.; Lavrik, O.I.; Salakhutdinov, N.F. Usnic acid derivatives are effective inhibitors of tyrosyl-DNA phosphodiesterase 1. Russ. J. Bioorg Chem. 2017, 43, 84–90. [Google Scholar] [CrossRef]
- Khomenko, T.; Zakharenko, A.; Odarchenko, T.; Arabshahi, H.J.; Sannikova, V.; Zakharova, O.; Korchagina, D.; Reynisson, J.; Volcho, K.; Salakhutdinov, N.; et al. New inhibitors of tyrosyl-DNA phosphodiesterase I (Tdp 1) combining 7-hydroxycoumarin and monoterpenoid moieties. Bioorg. Med. Chem. 2016, 24, 5573–5581. [Google Scholar] [CrossRef]
- Ponomarev, K.Y.; Suslov, E.V.; Zakharenko, A.L.; Zakharova, O.D.; Rogachev, A.D.; Korchagina, D.V.; Zafar, A.; Reynisson, J.; Nefedov, A.A.; Volcho, K.P.; et al. Aminoadamantanes containing monoterpene-derived fragments as potent tyrosyl-DNA phosphodiesterase 1 inhibitors. Bioorg. Chem. 2018, 76, 392–399. [Google Scholar] [CrossRef]
- Chepanova, A.A.; Li-Zhulanov, N.S.; Sukhikh, A.S.; Zafar, A.; Reynisson, J.; Zakharenko, A.L.; Zakharova, O.D.; Korchagina, D.V.; Volcho, K.P.; Salakhutdinov, N.F.; et al. Effective Inhibitors of Tyrosy l-DNA Phosphodiesterase 1 Based on Monoterpenoids as Potential Agents for Antitumor Therapy. Russ. J. Bioorg. Chem. 2019, 45, 647–655. [Google Scholar] [CrossRef]
- Il’ina, I.V.; Dyrkheeva, N.S.; Zakharenko, A.L.; Sidorenko, A.Y.; Li-Zhulanov, N.S.; Korchagina, D.V.; Chand, R.; Ayine-Tora, D.M.; Chepanova, A.A.; Zakharova, O.D.; et al. Design, Synthesis, and Biological Investigation of Novel Classes of 3-Carene-Derived Potent Inhibitors of TDP1. Molecules 2020, 25, 3496. [Google Scholar] [CrossRef]
- Zhang, X.-R.; Wang, H.-W.; Tang, W.-L.; Zhang, Y.; Yang, H.; Hu, D.-X.; Ravji, A.; Marchand, C.; Kiselev, E.; Ofori-Atta, K.; et al. Discovery, Synthesis, and Evaluation of Oxynitidine Derivatives as Dual Inhibitors of DNA Topoisomerase IB (TOP1) and Tyrosyl-DNA Phosphodiesterase 1 (TDP1), and Potential Antitumor Agents. J. Med. Chem. 2018, 61, 9908–9930. [Google Scholar] [CrossRef]
- Zakharenko, A.L.; Luzina, O.A.; Sokolov, D.N.; Kaledin, V.I.; Nikolin, V.P.; Popova, N.A.; Patel, J.; Zakharova, O.D.; Chepanova, A.A.; Zafar, A.; et al. Novel tyrosyl-DNA phosphodiesterase 1 inhibitors enhance the therapeutic impact of topotecan on in vivo tumor models. Eur. J. Med. Chem. 2019, 161, 581–593. [Google Scholar] [CrossRef]
- Khomenko, T.M.; Zakharenko, A.L.; Chepanova, A.A.; Ilina, E.S.; Zakharova, O.D.; Kaledin, V.I.; Nikolin, V.P.; Popova, N.A.; Korchagina, D.V.; Reynisson, J.; et al. Promising New Inhibitors of Tyrosyl-DNA Phosphodiesterase I (Tdp 1) Combining 4-Arylcoumarin and Monoterpenoid Moieties as Components of Complex Antitumor Therapy. Int. J. Mol. Sci. 2020, 21, 126. [Google Scholar] [CrossRef] [PubMed]
- Pang, B.; Zhao, L.-H.; Zhou, Q.; Zhao, T.-Y.; Wang, H.; Gu, C.-J.; Tong, X.-L. Application of Berberine on Treating Type 2 Diabetes Mellitus. Int. J. Endocrinol. 2015, 2015, 905749:1–905749:13. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.; Zhang, M.-B.; Liu, Y.-C.; Kang, J.-R.; Chu, Z.-Y.; Yin, K.-L.; Ding, L.-Y.; Ding, R.; Xiao, R.-X.; Yin, Y.-N.; et al. Role of Berberine in the Treatment of Methicillin-Resistant Staphylococcus aureus Infections. Sci. Rep. 2016, 6, 24748:1–24748:9. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.H.; Kim, K.J.; Cha, J.D.; Kim, H.K.; Lee, Y.E.; Choi, N.Y.; You, Y.O. Antimicrobial activity of berberine alone and in combination with ampicillin or oxacillin against methicillin-resistant Staphylococcus aureus. J. Med. Food. 2005, 8, 454–461. [Google Scholar] [CrossRef]
- Peng, L.; Kang, S.; Yin, Z.; Jia, R.; Song, X.; Li, L.; Li, Z.; Zou, Y.; Liang, X.; Li, L.; et al. Antibacterial activity and mechanism of berberine against Streptococcus agalactiae. Int. J. Clin. Exp. Pathol. 2015, 8, 5217–5223. [Google Scholar]
- Tan, W.; Li, Y.; Chen, M.; Wang, Y. Berberine hydrochloride: Anticancer activity and nanoparticulate delivery system. Int. J. Nanomed. 2011, 6, 1773–1777. [Google Scholar] [CrossRef]
- Nechepurenko, I.V.; Shirokova, E.D.; Khvostov, M.V.; Frolova, T.S.; Sinitsyna, O.I.; Maksimov, A.M.; Bredikhin, R.A.; Komarova, N.I.; Fadeev, D.S.; Luzina, O.A.; et al. Synthesis, hypolipidemic and antifungal activity of tetrahydroberberrubine sulfonates. Russ. Chem. Bull. Int. Ed. 2019, 68, 1052–1060. [Google Scholar] [CrossRef]
- Liu, H.; Wang, J.; Zhang, R.; Cairns, N.; Liu, J. Compounds, Compositions and Methods for Reducing Lipid Levels. International Patent Application No. PCT/US2008/067762, 20 June 2008. [Google Scholar]
- Iwasa, K.; Kamigauchi, M. 13-hydroxylation of tetrahydroberberine in cell suspension cultures of some Corydalis species. Phytochemistry 1996, 41, 1511–1515. [Google Scholar] [CrossRef]
- Nechepurenko, I.V.; Boyarskikh, U.A.; Khvostov, M.V.; Baev, D.S.; Komarova, N.I.; Filipenko, M.L.; Tolstikova, T.G.; Salakhutdinov, N.F. Hypolipidemic Berberine Derivatives with a Reduced Aromatic Ring C. Chem. Nat. Comp. 2015, 51, 916–922. [Google Scholar] [CrossRef]
- Zakharenko, A.L.; Khomenko, T.M.; Zhukova, S.V.; Koval, O.A.; Zakharova, O.D.; Anarbaev, R.O.; Lebedeva, N.A.; Korchagina, D.V.; Komarova, N.I.; Vasiliev, V.G.; et al. Synthesis and biological evaluation of novel tyrosyl-DNA phosphodiesterase 1 inhibitors with a benzopentathiepine moiety. Bioorg. Med. Chem. 2015, 23, 2044–2052. [Google Scholar] [CrossRef]
- Lountos, G.T.; Zhao, X.Z.; Kiselev, E.; Tropea, J.E.; Needle, D.; Pommier, Y.; Burke, T.R.; Waugh, D.S. Identification of a Ligand Binding Hot Spot and Structural Motifs Replicating Aspects of Tyrosyl-DNA Phosphodiesterase I (TDP1) Phosphoryl Recognition by Crystallographic Fragment Cocktail Screening. Nucleic Acids Res. 2019, 47, 10134–10150. [Google Scholar] [CrossRef] [PubMed]
- Martino, E.; Della Volpe, S.; Terribile, E.; Benetti, E.; Sakaj, M.; Centamore, A.; Sala, A.; Collina, S. The long story of camptothecin: From traditional medicine to drugs. Bioorg. Med. Chem. Lett. 2017, 27, 701–707. [Google Scholar] [CrossRef]
- Bjornsti, M.A.; Kaufmann, S.H. Topoisomerases and cancer chemotherapy: Recent advances and unanswered questions. F1000Research 2019, 8, 1704. [Google Scholar] [CrossRef] [PubMed]
- Bailly, C. Irinotecan: 25 years of cancer treatment. Pharmacol. Res. 2019, 148, 104398:1–104398:11. [Google Scholar] [CrossRef] [PubMed]
- Perego, P.; Cossa, G.; Tinelli, S.; Corna, E.; Carenini, N.; Gatti, L.; De Cesare, M.; Ciusani, E.; Zunino, F.; Luison, E.; et al. Role of tyrosyl-DNA phosphodiesterase 1 and inter-players in regulation of tumor cell sensitivity to topoisomerase I inhibition. Biochem. Pharmacol. 2012, 83, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Brettrager, E.J.; van Waardenburg, R.C.A.M. Targeting Tyrosyl-DNA phosphodiesterase I to enhance toxicity of phosphodiester linked DNA-adducts. Cancer Drug Resist. 2019, 2, 1153–1163. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Logan, G.; Reynisson, J. Wine Compounds as a Source for HTS Screening Collections. A Feasibility Study. Mol. Inf. 2012, 31, 847–855. [Google Scholar] [CrossRef]
- Eurtivong, C.; Reynisson, J. The Development of a Weighted Index to Optimise Compound Libraries for High Throughput Screening. Mol. Inf. 2018, 37, 1800068:1–1800068:10. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Berman, H.; Henrick, K.; Nakamura, H. Announcing the Worldwide Protein Data Bank. Nat. Struct. Biol. 2003, 10, 980. [Google Scholar] [CrossRef]
- Scigress Ultra V. F.J 2.6. (EU 3.1.7) Fujitsu Limited 2008–2016.
- Allinger, N.L. Conformational Analysis. 130. MM2. A Hydrocarbon Force Field Utilizing V1 and V2 Torsional Terms. J. Am. Chem. Soc. 1977, 99, 8127–8134. [Google Scholar] [CrossRef]
- Jones, G.; Willet, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and Validation of a Genetic Algorithm for Flexible Docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [PubMed]
- Eldridge, M.D.; Murray, C.; Auton, T.R.; Paolini, G.V.; Mee, P.M. Empirical Scoring Functions: I. The Development of a Fast Empirical Scoring Function to Estimate the Binding Affinity of Ligands in Receptor Complexes. J. Comput. Aided Mol. Des. 1997, 11, 425–445. [Google Scholar] [CrossRef]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved Protein-Ligand Docking using GOLD. Proteins 2003, 52, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Korb, O.; Stutzle, T.; Exner, T.E. Empirical Scoring Functions for Advanced Protein−Ligand Docking with PLANTS. J. Chem. Inf. Model. 2009, 49, 84–96. [Google Scholar] [CrossRef]
- Mooij, W.T.M.; Verdonk, M.L. General and Targeted Statistical Potentials for Protein–ligand Interactions. Proteins 2005, 61, 272–287. [Google Scholar] [CrossRef]
- QikProp; Version 3.2; Schrödinger: New York, NY, USA, 2009.
- Ioakimidis, L.; Thoukydidis, L.; Naeem, S.; Mirza, A.; Reynisson, J. Benchmarking the Reliability of QikProp. Correlation between Experimental and Predicted Values. QSAR Comb. Sci. 2008, 27, 445–456. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).