Non-Coding RNAs in Brain Tumors, the Contribution of lncRNAs, circRNAs, and snoRNAs to Cancer Development—Their Diagnostic and Therapeutic Potential
Abstract
1. Introduction
2. Characterization of Selected Brain Tumors
2.1. Adult Gliomas
2.2. Glioblastoma
2.3. Pediatric Gliomas
2.4. Medulloblastoma
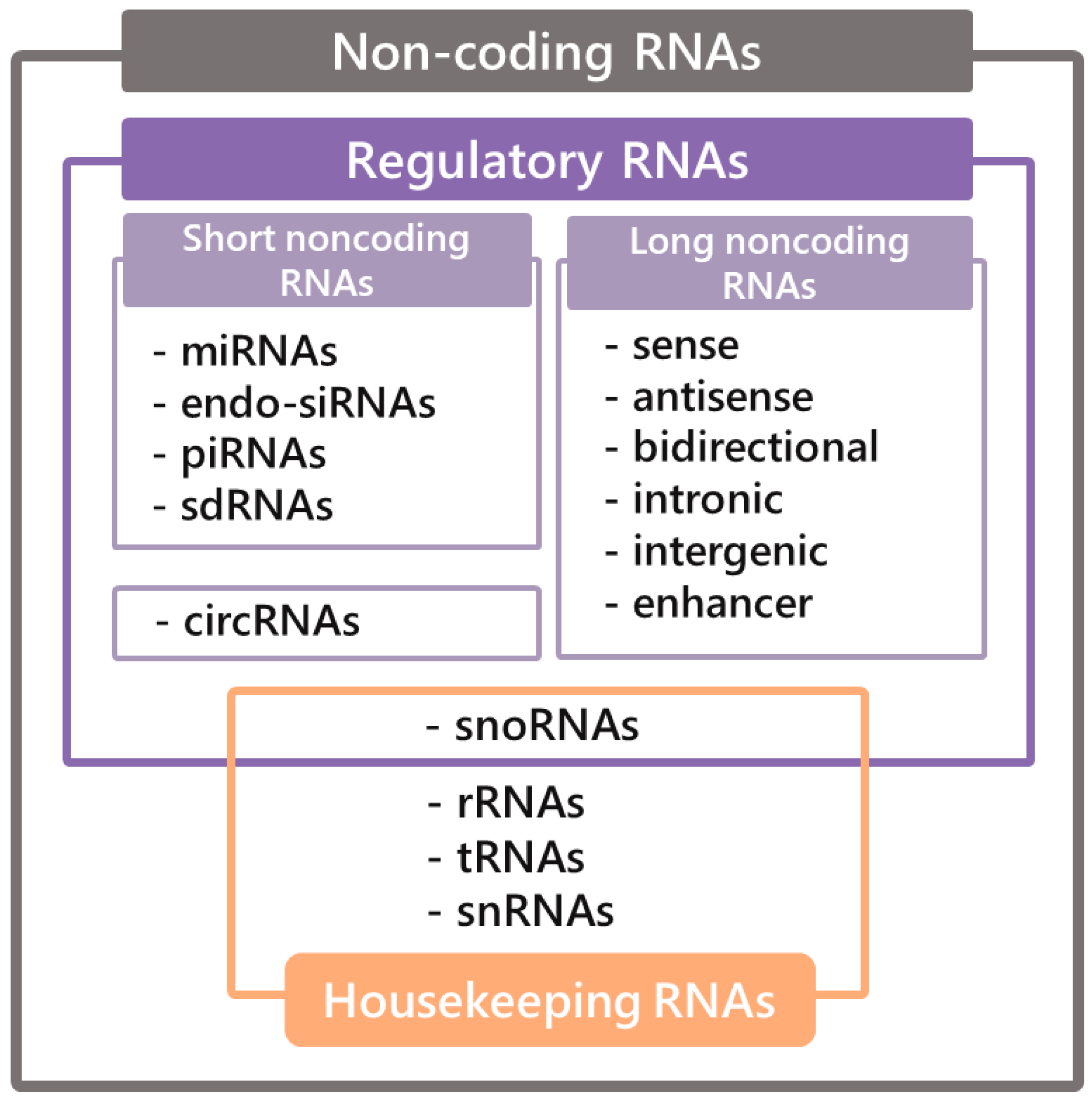
3. Brief Insight into the World of Non-Coding RNAs
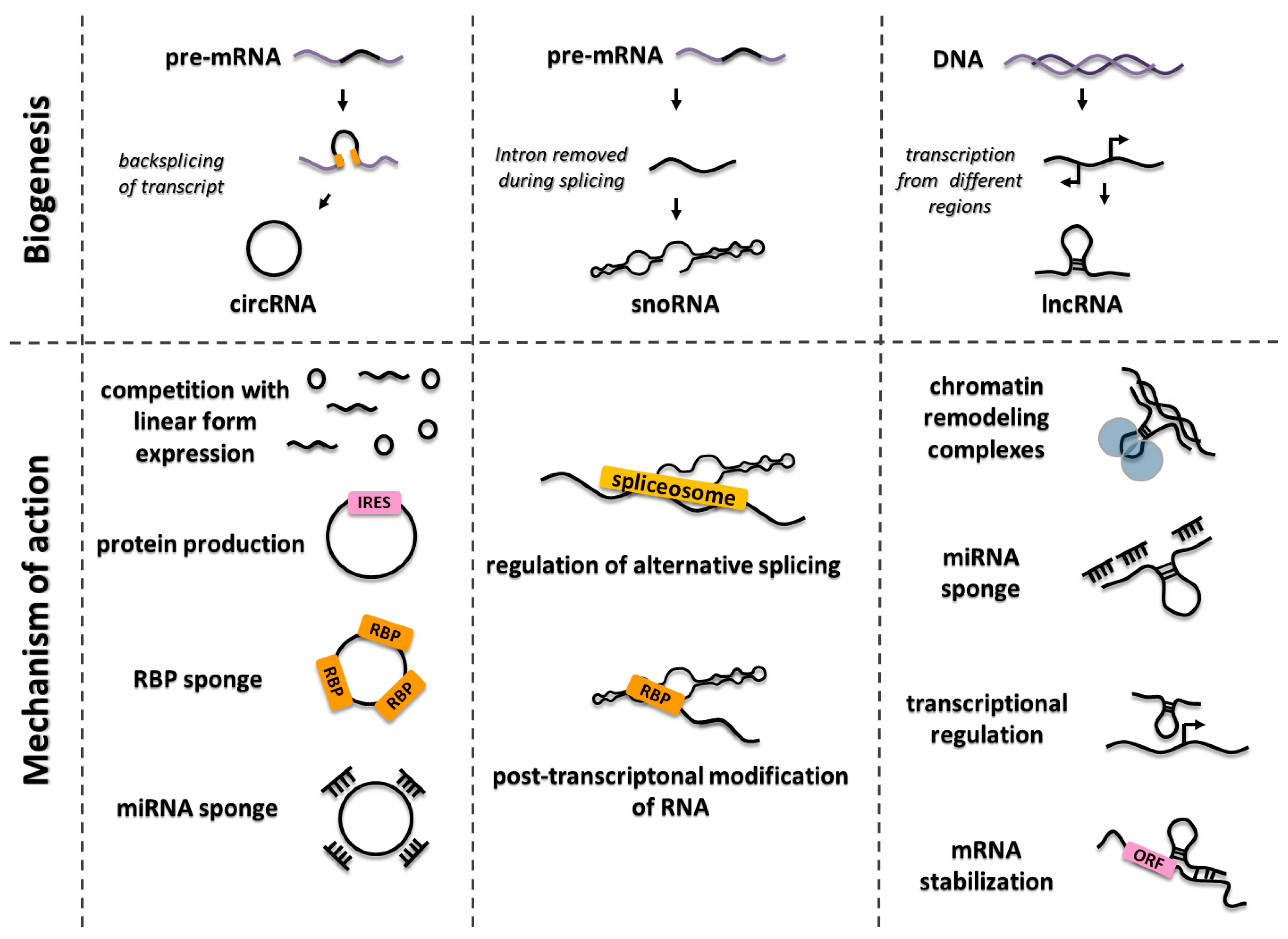
3.1. LncRNAs
3.2. CircRNAs
3.3. SnoRNAs
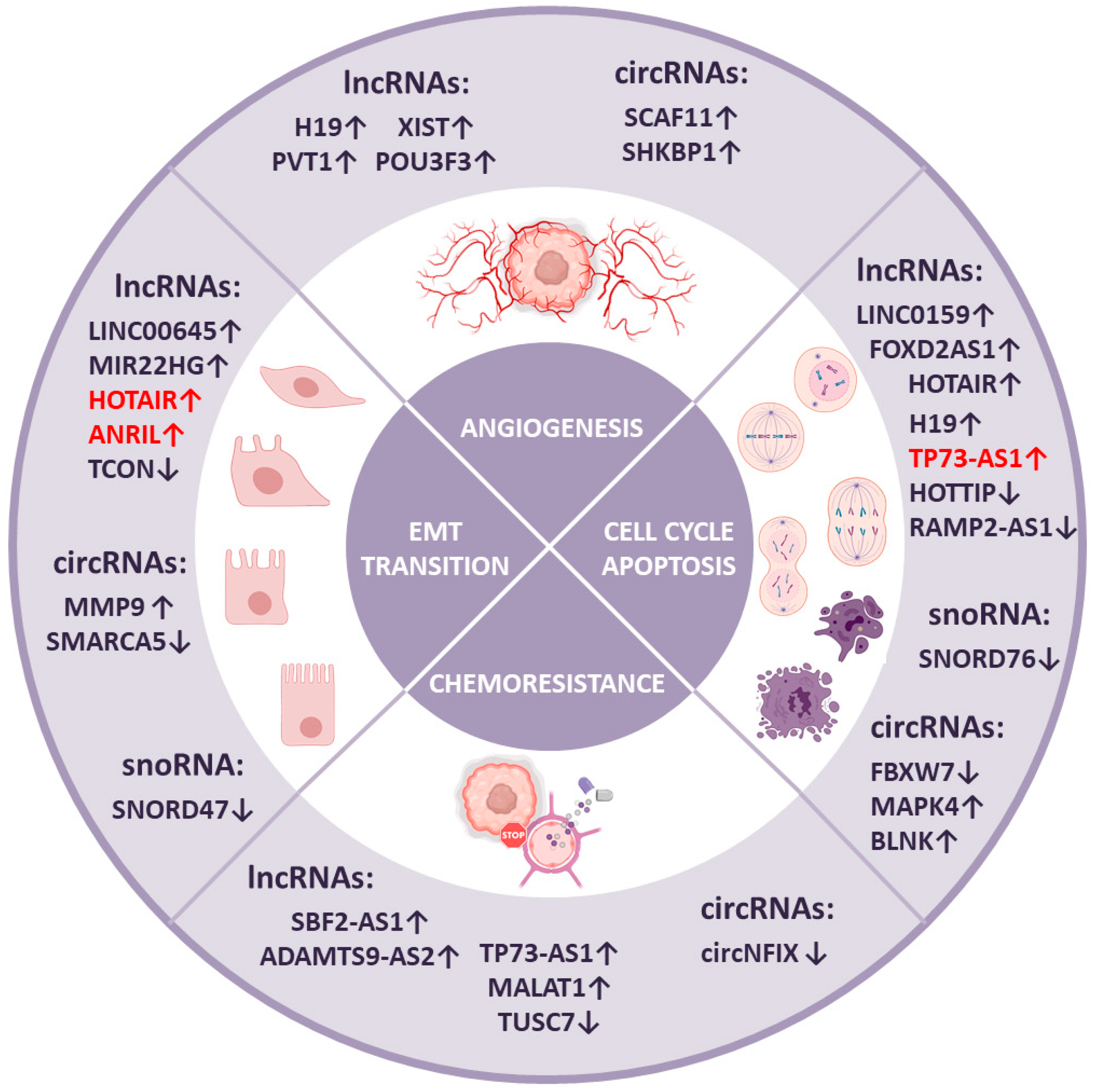
4. NcRNAs in Brain Tumor Progression
4.1. Cell Cycle and Apoptosis
4.2. Angiogenesis
4.3. Epithelial-to-Mesenchymal Transition
4.4. Chemoresistance
4.5. Regulation of the Immune System
5. Non-Coding RNAs in Glioma Stem Cells
6. Diagnostic and Therapeutic Potential of ncRNAs
6.1. Diagnostic Potential
6.2. Therapeutic Relevance
6.2.1. ncRNAs as Therapeutic Targets
6.2.2. Therapeutic Approaches Toward ncRNAs—Present State and Perspectives
7. Conclusions
Funding
Conflicts of Interest
References
- Miranda-Filho, A.; Pineros, M.; Soerjomataram, I.; Deltour, I.; Bray, F. Cancers of the brain and CNS: Global patterns and trends in incidence. Neuro Oncol. 2017, 19, 270–280. [Google Scholar] [CrossRef] [PubMed]
- Brain, G.B.D.; Other, C.N.S.C.C. Global, regional, and national burden of brain and other CNS cancer, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019, 18, 376–393. [Google Scholar] [CrossRef]
- Ferris, S.P.; Hofmann, J.W.; Solomon, D.A.; Perry, A. Characterization of gliomas: From morphology to molecules. Virchows Arch. 2017, 471, 257–269. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Brinkman, T.M.; Krasin, M.J.; Liu, W.; Armstrong, G.T.; Ojha, R.P.; Sadighi, Z.S.; Gupta, P.; Kimberg, C.; Srivastava, D.; Merchant, T.E.; et al. Long-Term Neurocognitive Functioning and Social Attainment in Adult Survivors of Pediatric CNS Tumors: Results From the St Jude Lifetime Cohort Study. J. Clin. Oncol. 2016, 34, 1358–1367. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. The Microenvironmental Landscape of Brain Tumors. Cancer Cell 2017, 31, 326–341. [Google Scholar] [CrossRef] [PubMed]
- Mackay, A.; Burford, A.; Carvalho, D.; Izquierdo, E.; Fazal-Salom, J.; Taylor, K.R.; Bjerke, L.; Clarke, M.; Vinci, M.; Nandhabalan, M.; et al. Integrated Molecular Meta-Analysis of 1000 Pediatric High-Grade and Diffuse Intrinsic Pontine Glioma. Cancer Cell 2017, 32, 520–537 e525. [Google Scholar] [CrossRef]
- Gilbertson, R.J. Mapping cancer origins. Cell 2011, 145, 25–29. [Google Scholar] [CrossRef]
- Ling, H.; Vincent, K.; Pichler, M.; Fodde, R.; Berindan-Neagoe, I.; Slack, F.J.; Calin, G.A. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene 2015, 34, 5003–5011. [Google Scholar] [CrossRef]
- Wei, J.W.; Huang, K.; Yang, C.; Kang, C.S. Non-coding RNAs as regulators in epigenetics (Review). Oncol. Rep. 2017, 37, 3–9. [Google Scholar] [CrossRef]
- Mattick, J.S.; Makunin, I.V. Non-coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef] [PubMed]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding RNA networks in cancer. Nat. Rev. Cancer 2018, 18, 5–18. [Google Scholar] [CrossRef] [PubMed]
- De Windt, L.J.; Giacca, M. Non-coding RNA function in stem cells and Regenerative Medicine. Noncoding RNA Res. 2018, 3, 39–41. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, N.; Honma, R.; Sekino, Y.; Goto, K.; Sentani, K.; Ishikawa, A.; Oue, N.; Yasui, W. Non-coding RNAs are promising targets for stem cell-based cancer therapy. Noncoding RNA Res. 2017, 2, 83–87. [Google Scholar] [CrossRef]
- Turner, J.D.; Williamson, R.; Almefty, K.K.; Nakaji, P.; Porter, R.; Tse, V.; Kalani, M.Y. The many roles of microRNAs in brain tumor biology. Neurosurg. Focus 2010, 28, E3. [Google Scholar] [CrossRef]
- Nicoloso, M.S.; Calin, G.A. MicroRNA involvement in brain tumors: From bench to bedside. Brain Pathol. 2008, 18, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; Wick, W.; Aldape, K.; Brada, M.; Berger, M.; Pfister, S.M.; Nishikawa, R.; Rosenthal, M.; Wen, P.Y.; Stupp, R.; et al. Glioma. Nat. Rev. Dis. Primers 2015, 1, 15017. [Google Scholar] [CrossRef]
- Lakhan, S.E.; Harle, L. Difficult diagnosis of brainstem glioblastoma multiforme in a woman: A case report and review of the literature. J. Med. Case Rep. 2009, 3, 87. [Google Scholar] [CrossRef]
- Weller, M.; van den Bent, M.; Tonn, J.C.; Stupp, R.; Preusser, M.; Cohen-Jonathan-Moyal, E.; Henriksson, R.; Le Rhun, E.; Balana, C.; Chinot, O.; et al. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol. 2017, 18, e315–e329. [Google Scholar] [CrossRef]
- Chaumeil, M.M.; Lupo, J.M.; Ronen, S.M. Magnetic Resonance (MR) Metabolic Imaging in Glioma. Brain Pathol. 2015, 25, 769–780. [Google Scholar] [CrossRef]
- Wrensch, M.; Minn, Y.; Chew, T.; Bondy, M.; Berger, M.S. Epidemiology of primary brain tumors: Current concepts and review of the literature. Neuro Oncol. 2002, 4, 278–299. [Google Scholar] [CrossRef] [PubMed]
- Walid, M.S. Prognostic factors for long-term survival after glioblastoma. Perm. J. 2008, 12, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Karcher, S.; Steiner, H.H.; Ahmadi, R.; Zoubaa, S.; Vasvari, G.; Bauer, H.; Unterberg, A.; Herold-Mende, C. Different angiogenic phenotypes in primary and secondary glioblastomas. Int. J. Cancer 2006, 118, 2182–2189. [Google Scholar] [CrossRef] [PubMed]
- Lieberman, F. Glioblastoma update: Molecular biology, diagnosis, treatment, response assessment, and translational clinical trials. F1000Research 2017, 6, 1892. [Google Scholar] [CrossRef]
- Chamberlain, M.C. Radiographic patterns of relapse in glioblastoma. J. Neurooncol. 2011, 101, 319–323. [Google Scholar] [CrossRef]
- Gladson, C.L.; Prayson, R.A.; Liu, W.M. The pathobiology of glioma tumors. Annu. Rev. Pathol. 2010, 5, 33–50. [Google Scholar] [CrossRef]
- Gusyatiner, O.; Hegi, M.E. Glioma epigenetics: From subclassification to novel treatment options. Semin. Cancer Biol. 2018, 51, 50–58. [Google Scholar] [CrossRef]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef]
- Ohgaki, H.; Kleihues, P. Genetic pathways to primary and secondary glioblastoma. Am. J. Pathol. 2007, 170, 1445–1453. [Google Scholar] [CrossRef]
- Purkait, S.; Jha, P.; Sharma, M.C.; Suri, V.; Sharma, M.; Kale, S.S.; Sarkar, C. CDKN2A deletion in pediatric versus adult glioblastomas and predictive value of p16 immunohistochemistry. Neuropathology 2013, 33, 405–412. [Google Scholar] [CrossRef]
- Wang, Y.; Leung, F.C. An evaluation of new criteria for CpG islands in the human genome as gene markers. Bioinformatics 2004, 20, 1170–1177. [Google Scholar] [CrossRef] [PubMed]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Reon, B.J.; Anaya, J.; Zhang, Y.; Mandell, J.; Purow, B.; Abounader, R.; Dutta, A. Expression of lncRNAs in Low-Grade Gliomas and Glioblastoma Multiforme: An In Silico Analysis. PLoS Med. 2016, 13, e1002192. [Google Scholar] [CrossRef]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef] [PubMed]
- Jue, T.R.; McDonald, K.L. The challenges associated with molecular targeted therapies for glioblastoma. J. Neurooncol. 2016, 127, 427–434. [Google Scholar] [CrossRef]
- Cheng, J.; Meng, J.; Zhu, L.; Peng, Y. Exosomal noncoding RNAs in Glioma: Biological functions and potential clinical applications. Mol. Cancer 2020, 19, 66. [Google Scholar] [CrossRef]
- Malbari, F.; Lindsay, H. Genetics of Common Pediatric Brain Tumors. Pediatr. Neurol. 2020, 104, 3–12. [Google Scholar] [CrossRef]
- Sturm, D.; Pfister, S.M.; Jones, D.T.W. Pediatric Gliomas: Current Concepts on Diagnosis, Biology, and Clinical Management. J. Clin. Oncol. 2017, 35, 2370–2377. [Google Scholar] [CrossRef]
- Gajjar, A.; Bowers, D.C.; Karajannis, M.A.; Leary, S.; Witt, H.; Gottardo, N.G. Pediatric Brain Tumors: Innovative Genomic Information Is Transforming the Diagnostic and Clinical Landscape. J. Clin. Oncol. 2015, 33, 2986–2998. [Google Scholar] [CrossRef]
- Packer, R.J.; Pfister, S.; Bouffet, E.; Avery, R.; Bandopadhayay, P.; Bornhorst, M.; Bowers, D.C.; Ellison, D.; Fangusaro, J.; Foreman, N.; et al. Pediatric low-grade gliomas: Implications of the biologic era. Neuro Oncol. 2017, 19, 750–761. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.W.; Kieran, M.W.; Bouffet, E.; Alexandrescu, S.; Bandopadhayay, P.; Bornhorst, M.; Ellison, D.; Fangusaro, J.; Fisher, M.J.; Foreman, N.; et al. Pediatric low-grade gliomas: Next biologically driven steps. Neuro Oncol. 2018, 20, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Crawford, J.R.; MacDonald, T.J.; Packer, R.J. Medulloblastoma in childhood: New biological advances. Lancet Neurol. 2007, 6, 1073–1085. [Google Scholar] [CrossRef]
- Ramaswamy, V.; Nor, C.; Taylor, M.D. p53 and Meduloblastoma. Cold Spring Harb. Perspect. Med. 2015, 6, a026278. [Google Scholar] [CrossRef] [PubMed]
- Northcott, P.A.; Korshunov, A.; Witt, H.; Hielscher, T.; Eberhart, C.G.; Mack, S.; Bouffet, E.; Clifford, S.C.; Hawkins, C.E.; French, P.; et al. Medulloblastoma comprises four distinct molecular variants. J. Clin. Oncol. 2011, 29, 1408–1414. [Google Scholar] [CrossRef]
- Northcott, P.A.; Shih, D.J.; Peacock, J.; Garzia, L.; Morrissy, A.S.; Zichner, T.; Stutz, A.M.; Korshunov, A.; Reimand, J.; Schumacher, S.E.; et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature 2012, 488, 49–56. [Google Scholar] [CrossRef]
- Taylor, M.D.; Northcott, P.A.; Korshunov, A.; Remke, M.; Cho, Y.J.; Clifford, S.C.; Eberhart, C.G.; Parsons, D.W.; Rutkowski, S.; Gajjar, A.; et al. Molecular subgroups of medulloblastoma: The current consensus. Acta Neuropathol. 2012, 123, 465–472. [Google Scholar] [CrossRef]
- Holley, R.W.; Apgar, J.; Everett, G.A.; Madison, J.T.; Marquisee, M.; Merrill, S.H.; Penswick, J.R.; Zamir, A. STRUCTURE OF A RIBONUCLEIC ACID. Science 1965, 147, 1462–1465. [Google Scholar] [CrossRef]
- Eddy, S.R. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001, 2, 919–929. [Google Scholar] [CrossRef]
- Cao, J. The functional role of long non-coding RNAs and epigenetics. Biol. Proced. Online 2014, 16, 11. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs-microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Kung, J.T.; Colognori, D.; Lee, J.T. Long noncoding RNAs: Past, present, and future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef] [PubMed]
- Beermann, J.; Piccoli, M.T.; Viereck, J.; Thum, T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef] [PubMed]
- Hacisuleyman, E.; Goff, L.A.; Trapnell, C.; Williams, A.; Henao-Mejia, J.; Sun, L.; McClanahan, P.; Hendrickson, D.G.; Sauvageau, M.; Kelley, D.R.; et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 2014, 21, 198–206. [Google Scholar] [CrossRef]
- Lin, A.; Li, C.; Xing, Z.; Hu, Q.; Liang, K.; Han, L.; Wang, C.; Hawke, D.H.; Wang, S.; Zhang, Y.; et al. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat. Cell Biol. 2016, 18, 213–224. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Dahariya, S.; Paddibhatla, I.; Kumar, S.; Raghuwanshi, S.; Pallepati, A.; Gutti, R.K. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Mol. Immunol. 2019, 112, 82–92. [Google Scholar] [CrossRef]
- Marchese, F.P.; Raimondi, I.; Huarte, M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017, 18, 206. [Google Scholar] [CrossRef]
- Cai, B.; Song, X.Q.; Cai, J.P.; Zhang, S. HOTAIR: A cancer-related long non-coding RNA. Neoplasma 2014, 61, 379–391. [Google Scholar] [CrossRef]
- Tano, K.; Mizuno, R.; Okada, T.; Rakwal, R.; Shibato, J.; Masuo, Y.; Ijiri, K.; Akimitsu, N. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010, 584, 4575–4580. [Google Scholar] [CrossRef]
- Leveille, N.; Melo, C.A.; Rooijers, K.; Diaz-Lagares, A.; Melo, S.A.; Korkmaz, G.; Lopes, R.; Moqadam, F.A.; Maia, A.R.; Wijchers, P.J.; et al. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat. Commun. 2015, 6, 6520. [Google Scholar] [CrossRef] [PubMed]
- Greene, J.; Baird, A.M.; Brady, L.; Lim, M.; Gray, S.G.; McDermott, R.; Finn, S.P. Circular RNAs: Biogenesis, Function and Role in Human Diseases. Front. Mol. Biosci. 2017, 4, 38. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Z. Efficient backsplicing produces translatable circular mRNAs. RNA 2015, 21, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Barrett, S.P.; Wang, P.L.; Salzman, J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife 2015, 4, e07540. [Google Scholar] [CrossRef]
- Okholm, T.L.H.; Nielsen, M.M.; Hamilton, M.P.; Christensen, L.L.; Vang, S.; Hedegaard, J.; Hansen, T.B.; Kjems, J.; Dyrskjot, L.; Pedersen, J.S. Circular RNA expression is abundant and correlated to aggressiveness in early-stage bladder cancer. NPJ Genom. Med. 2017, 2, 36. [Google Scholar] [CrossRef]
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Lasda, E.; Parker, R. Circular RNAs: Diversity of form and function. RNA 2014, 20, 1829–1842. [Google Scholar] [CrossRef]
- Patop, I.L.; Wust, S.; Kadener, S. Past, present, and future of circRNAs. EMBO J. 2019, 38, e100836. [Google Scholar] [CrossRef]
- Wang, Z.; Lei, X.; Wu, F.X. Identifying Cancer-Specific circRNA-RBP Binding Sites Based on Deep Learning. Molecules 2019, 24, 4035. [Google Scholar] [CrossRef]
- Abe, N.; Matsumoto, K.; Nishihara, M.; Nakano, Y.; Shibata, A.; Maruyama, H.; Shuto, S.; Matsuda, A.; Yoshida, M.; Ito, Y.; et al. Rolling Circle Translation of Circular RNA in Living Human Cells. Sci. Rep. 2015, 5, 16435. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Zhou, H.; Feng, Z.; Xu, Z.; Tang, Y.; Li, P.; Wu, M. CircRNA: Functions and properties of a novel potential biomarker for cancer. Mol. Cancer 2017, 16, 94. [Google Scholar] [CrossRef] [PubMed]
- Scotti, M.M.; Swanson, M.S. RNA mis-splicing in disease. Nat. Rev. Genet. 2016, 17, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Chen, Q.; Yao, T.; Li, T.; Ying, S.; Hu, Y.; Guo, J. Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma. Oncotarget 2017, 8, 43878–43888. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Lyu, Z.; Pan, L.; Zeng, G.; Randhawa, P. Defining housekeeping genes suitable for RNA-seq analysis of the human allograft kidney biopsy tissue. BMC Med. Genom. 2019, 12, 86. [Google Scholar] [CrossRef]
- Yin, Q.F.; Yang, L.; Zhang, Y.; Xiang, J.F.; Wu, Y.W.; Carmichael, G.G.; Chen, L.L. Long noncoding RNAs with snoRNA ends. Mol. Cell 2012, 48, 219–230. [Google Scholar] [CrossRef]
- Filipowicz, W.; Pogacic, V. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 2002, 14, 319–327. [Google Scholar] [CrossRef]
- Falaleeva, M.; Stamm, S. Processing of snoRNAs as a new source of regulatory non-coding RNAs: snoRNA fragments form a new class of functional RNAs. Bioessays 2013, 35, 46–54. [Google Scholar] [CrossRef]
- Grzechnik, P.; Szczepaniak, S.A.; Dhir, S.; Pastucha, A.; Parslow, H.; Matuszek, Z.; Mischo, H.E.; Kufel, J.; Proudfoot, N.J. Nuclear fate of yeast snoRNA is determined by co-transcriptional Rnt1 cleavage. Nat. Commun. 2018, 9, 1783. [Google Scholar] [CrossRef]
- Kishore, S.; Khanna, A.; Zhang, Z.; Hui, J.; Balwierz, P.J.; Stefan, M.; Beach, C.; Nicholls, R.D.; Zavolan, M.; Stamm, S. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum. Mol. Genet. 2010, 19, 1153–1164. [Google Scholar] [CrossRef]
- Kishore, S.; Stamm, S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science 2006, 311, 230–232. [Google Scholar] [CrossRef]
- Lafontaine, D.L. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat. Struct. Mol. Biol. 2015, 22, 11–19. [Google Scholar] [CrossRef] [PubMed]
- McMahon, M.; Contreras, A.; Ruggero, D. Small RNAs with big implications: New insights into H/ACA snoRNA function and their role in human disease. Wiley Interdiscip. Rev. RNA 2015, 6, 173–189. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Sawyer, I.A.; Sung, M.H.; Sturgill, D.; Shevtsov, S.P.; Pegoraro, G.; Hakim, O.; Baek, S.; Hager, G.L.; Dundr, M. Cajal bodies are linked to genome conformation. Nat. Commun. 2016, 7, 10966. [Google Scholar] [CrossRef] [PubMed]
- Abel, Y.; Rederstorff, M. SnoRNAs and the emerging class of sdRNAs: Multifaceted players in oncogenesis. Biochimie 2019, 164, 17–21. [Google Scholar] [CrossRef]
- Taft, R.J.; Glazov, E.A.; Lassmann, T.; Hayashizaki, Y.; Carninci, P.; Mattick, J.S. Small RNAs derived from snoRNAs. RNA 2009, 15, 1233–1240. [Google Scholar] [CrossRef]
- Ender, C.; Krek, A.; Friedlander, M.R.; Beitzinger, M.; Weinmann, L.; Chen, W.; Pfeffer, S.; Rajewsky, N.; Meister, G. A human snoRNA with microRNA-like functions. Mol. Cell 2008, 32, 519–528. [Google Scholar] [CrossRef]
- Scott, M.S.; Avolio, F.; Ono, M.; Lamond, A.I.; Barton, G.J. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput. Biol. 2009, 5, e1000507. [Google Scholar] [CrossRef]
- Brameier, M.; Herwig, A.; Reinhardt, R.; Walter, L.; Gruber, J. Human box C/D snoRNAs with miRNA like functions: Expanding the range of regulatory RNAs. Nucleic Acids Res. 2011, 39, 675–686. [Google Scholar] [CrossRef]
- Dong, X.Y.; Rodriguez, C.; Guo, P.; Sun, X.; Talbot, J.T.; Zhou, W.; Petros, J.; Li, Q.; Vessella, R.L.; Kibel, A.S.; et al. SnoRNA U50 is a candidate tumor-suppressor gene at 6q14.3 with a mutation associated with clinically significant prostate cancer. Hum. Mol. Genet. 2008, 17, 1031–1042. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.Y.; Guo, P.; Boyd, J.; Sun, X.; Li, Q.; Zhou, W.; Dong, J.T. Implication of snoRNA U50 in human breast cancer. J. Genet. Genom. 2009, 36, 447–454. [Google Scholar] [CrossRef]
- Xu, G.; Yang, F.; Ding, C.L.; Zhao, L.J.; Ren, H.; Zhao, P.; Wang, W.; Qi, Z.T. Small nucleolar RNA 113-1 suppresses tumorigenesis in hepatocellular carcinoma. Mol. Cancer 2014, 13, 216. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.P.; Liao, J.P.; Shen, J.; Yu, L.; Liu, B.L.; Liu, L.; Li, R.Y.; Ji, L.; Dorsey, S.G.; Jiang, Z.R.; et al. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene 2012, 31, 2794–2804. [Google Scholar] [CrossRef]
- Valleron, W.; Laprevotte, E.; Gautier, E.F.; Quelen, C.; Demur, C.; Delabesse, E.; Agirre, X.; Prosper, F.; Kiss, T.; Brousset, P. Specific small nucleolar RNA expression profiles in acute leukemia. Leukemia 2012, 26, 2052–2060. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef]
- Zhu, J.; Ye, J.; Zhang, L.; Xia, L.; Hu, H.; Jiang, H.; Wan, Z.; Sheng, F.; Ma, Y.; Li, W.; et al. Differential Expression of Circular RNAs in Glioblastoma Multiforme and Its Correlation with Prognosis. Transl. Oncol. 2017, 10, 271–279. [Google Scholar] [CrossRef]
- Cavaille, J.; Buiting, K.; Kiefmann, M.; Lalande, M.; Brannan, C.I.; Horsthemke, B.; Bachellerie, J.P.; Brosius, J.; Huttenhofer, A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. USA 2000, 97, 14311–14316. [Google Scholar] [CrossRef]
- Chen, L.; Han, L.; Wei, J.; Zhang, K.; Shi, Z.; Duan, R.; Li, S.; Zhou, X.; Pu, P.; Zhang, J.; et al. SNORD76, a box C/D snoRNA, acts as a tumor suppressor in glioblastoma. Sci. Rep. 2015, 5, 8588. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Liu, S.; Mitra, R.; Zhao, M.M.; Fan, W.; Eischen, C.M.; Yin, F.; Zhao, Z. The Potential Roles of Long Noncoding RNAs (lncRNA) in Glioblastoma Development. Mol. Cancer Ther. 2016, 15, 2977–2986. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.; Xie, M. LINC01579 promotes cell proliferation by acting as a ceRNA of miR-139-5p to upregulate EIF4G2 expression in glioblastoma. J. Cell. Physiol. 2019, 234, 23658–23666. [Google Scholar] [CrossRef]
- Wang, J.; Li, B.; Wang, C.; Luo, Y.; Zhao, M.; Chen, P. Long noncoding RNA FOXD2-AS1 promotes glioma cell cycle progression and proliferation through the FOXD2-AS1/miR-31/CDK1 pathway. J. Cell. Biochem. 2019, 120, 19784–19795. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Sun, X.; Zhou, X.; Han, L.; Chen, L.; Shi, Z.; Zhang, A.; Ye, M.; Wang, Q.; Liu, C.; et al. Long non-coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget 2015, 6, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Ke, J.; Yao, Y.L.; Zheng, J.; Wang, P.; Liu, Y.H.; Ma, J.; Li, Z.; Liu, X.B.; Li, Z.Q.; Wang, Z.H.; et al. Knockdown of long non-coding RNA HOTAIR inhibits malignant biological behaviors of human glioma cells via modulation of miR-326. Oncotarget 2015, 6, 21934–21949. [Google Scholar] [CrossRef]
- Zhao, H.; Peng, R.; Liu, Q.; Liu, D.; Du, P.; Yuan, J.; Peng, G.; Liao, Y. The lncRNA H19 interacts with miR-140 to modulate glioma growth by targeting iASPP. Arch. Biochem. Biophys. 2016, 610, 1–7. [Google Scholar] [CrossRef]
- Xu, L.M.; Chen, L.; Li, F.; Zhang, R.; Li, Z.Y.; Chen, F.F.; Jiang, X.D. Over-expression of the long non-coding RNA HOTTIP inhibits glioma cell growth by BRE. J. Exp. Clin. Cancer Res. 2016, 35, 162. [Google Scholar] [CrossRef]
- Yang, Y.; Gao, X.; Zhang, M.; Yan, S.; Sun, C.; Xiao, F.; Huang, N.; Yang, X.; Zhao, K.; Zhou, H.; et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J. Natl. Cancer Inst. 2018, 110. [Google Scholar] [CrossRef]
- Liu, Y.; Ma, C.; Qin, X.; Yu, H.; Shen, L.; Jin, H. Circular RNA circ_001350 regulates glioma cell proliferation, apoptosis, and metastatic properties by acting as a miRNA sponge. J. Cell. Biochem. 2019, 120, 15280–15287. [Google Scholar] [CrossRef]
- He, J.; Huang, Z.; He, M.; Liao, J.; Zhang, Q.; Wang, S.; Xie, L.; Ouyang, L.; Koeffler, H.P.; Yin, D.; et al. Circular RNA MAPK4 (circ-MAPK4) inhibits cell apoptosis via MAPK signaling pathway by sponging miR-125a-3p in gliomas. Mol. Cancer 2020, 19, 17. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Ye, M.H.; Lv, S.G.; Wang, Q.X.; Wu, M.J.; Xiao, B.; Kang, C.S.; Zhu, X.G. SNORD47, a box C/D snoRNA, suppresses tumorigenesis in glioblastoma. Oncotarget 2017, 8, 43953–43966. [Google Scholar] [CrossRef] [PubMed]
- Varon, M.; Levy, T.; Mazor, G.; Ben David, H.; Marciano, R.; Krelin, Y.; Prasad, M.; Elkabets, M.; Pauck, D.; Ahmadov, U.; et al. The long noncoding RNA TP73-AS1 promotes tumorigenicity of medulloblastoma cells. Int. J. Cancer 2019, 145, 3402–3413. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhu, Y.; Wang, H.; Ji, X. Targeting Long Noncoding RNA in Glioma: A Pathway Perspective. Mol. Ther. Nucleic Acids 2018, 13, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Lugano, R.; Ramachandran, M.; Dimberg, A. Tumor angiogenesis: Causes, consequences, challenges and opportunities. Cell. Mol. Life Sci. 2020, 77, 1745–1770. [Google Scholar] [CrossRef] [PubMed]
- Jia, P.; Cai, H.; Liu, X.; Chen, J.; Ma, J.; Wang, P.; Liu, Y.; Zheng, J.; Xue, Y. Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer Lett. 2016, 381, 359–369. [Google Scholar] [CrossRef]
- Yu, H.; Xue, Y.; Wang, P.; Liu, X.; Ma, J.; Zheng, J.; Li, Z.; Li, Z.; Cai, H.; Liu, Y. Knockdown of long non-coding RNA XIST increases blood-tumor barrier permeability and inhibits glioma angiogenesis by targeting miR-137. Oncogenesis 2017, 6, e303. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, P.; Xue, Y.; Qu, C.; Zheng, J.; Liu, X.; Ma, J.; Liu, Y. PVT1 affects growth of glioma microvascular endothelial cells by negatively regulating miR-186. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2017, 39, 1010428317694326. [Google Scholar] [CrossRef]
- Lang, H.L.; Hu, G.W.; Chen, Y.; Liu, Y.; Tu, W.; Lu, Y.M.; Wu, L.; Xu, G.H. Glioma cells promote angiogenesis through the release of exosomes containing long non-coding RNA POU3F3. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 959–972. [Google Scholar]
- Meng, Q.; Li, S.; Liu, Y.; Zhang, S.; Jin, J.; Zhang, Y.; Guo, C.; Liu, B.; Sun, Y. Circular RNA circSCAF11 Accelerates the Glioma Tumorigenesis through the miR-421/SP1/VEGFA Axis. Mol. Ther. Nucleic Acids 2019, 17, 669–677. [Google Scholar] [CrossRef]
- He, Q.; Zhao, L.; Liu, Y.; Liu, X.; Zheng, J.; Yu, H.; Cai, H.; Ma, J.; Liu, L.; Wang, P.; et al. circ-SHKBP1 Regulates the Angiogenesis of U87 Glioma-Exposed Endothelial Cells through miR-544a/FOXP1 and miR-379/FOXP2 Pathways. Mol. Ther. Nucleic Acids 2018, 10, 331–348. [Google Scholar] [CrossRef] [PubMed]
- Xin, S.; Huang, K.; Zhu, X.G. Non-coding RNAs: Regulators of glioma cell epithelial-mesenchymal transformation. Pathol. Res. Pract. 2019, 215, 152539. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zheng, H.; Hou, W.; Bao, H.; Xiong, J.; Che, W.; Gu, Y.; Sun, H.; Liang, P. Long non-coding RNA linc00645 promotes TGF-beta-induced epithelial-mesenchymal transition by regulating miR-205-3p-ZEB1 axis in glioma. Cell Death Dis. 2019, 10, 717. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Wang, Y.; Zhang, L.; Wang, J.; Wang, W.; Han, X.; Mu, C.; Gao, D. Identification of novel LncRNA targeting Smad2/PKCalpha signal pathway to negatively regulate malignant progression of glioblastoma. J. Cell. Physiol. 2020, 235, 3835–3848. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Wang, S.; Fritah, S.; Wang, X.; Zhou, W.; Yang, N.; Ni, S.; Huang, B.; Chen, A.; Li, G.; et al. Interfering with long non-coding RNA MIR22HG processing inhibits glioblastoma progression through suppression of Wnt/beta-catenin signalling. Brain 2019, 143, 512–530. [Google Scholar] [CrossRef]
- Xue, Q.; Cao, L.; Chen, X.Y.; Zhao, J.; Gao, L.; Li, S.Z.; Fei, Z. High expression of MMP9 in glioma affects cell proliferation and is associated with patient survival rates. Oncol. Lett. 2017, 13, 1325–1330. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, S.; Chen, X.; Li, N.; Li, J.; Jia, R.; Pan, Y.; Liang, H. EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol. Cancer 2018, 17, 166. [Google Scholar] [CrossRef]
- Barbagallo, D.; Caponnetto, A.; Cirnigliaro, M.; Brex, D.; Barbagallo, C.; D’Angeli, F.; Morrone, A.; Caltabiano, R.; Barbagallo, G.M.; Ragusa, M.; et al. CircSMARCA5 Inhibits Migration of Glioblastoma Multiforme Cells by Regulating a Molecular Axis Involving Splicing Factors SRSF1/SRSF3/PTB. Int. J. Mol. Sci. 2018, 19, 480. [Google Scholar] [CrossRef]
- Zhang, J.; Li, N.; Fu, J.; Zhou, W. Long noncoding RNA HOTAIR promotes medulloblastoma growth, migration and invasion by sponging miR-1/miR-206 and targeting YY1. Biomed. Pharmacother. 2020, 124, 109887. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, X.; Chen, X. Potential Role of Long Non-Coding RNA ANRIL in Pediatric Medulloblastoma Through Promotion on Proliferation and Migration by Targeting miR-323. J. Cell. Biochem. 2017, 118, 4735–4744. [Google Scholar] [CrossRef]
- Sarkaria, J.N.; Kitange, G.J.; James, C.D.; Plummer, R.; Calvert, H.; Weller, M.; Wick, W. Mechanisms of chemoresistance to alkylating agents in malignant glioma. Clin. Cancer Res. 2008, 14, 2900–2908. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yin, J.; Lu, C.; Wei, Y.; Zeng, A.; You, Y. Exosomal transfer of long non-coding RNA SBF2-AS1 enhances chemoresistance to temozolomide in glioblastoma. J. Exp. Clin. Cancer Res. 2019, 38, 166. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Xu, Z.; Chen, X.; Wang, X.; Zeng, S.; Zhao, Z.; Qian, L.; Li, Z.; Wei, J.; Huo, L.; et al. Novel Function of lncRNA ADAMTS9-AS2 in Promoting Temozolomide Resistance in Glioblastoma via Upregulating the FUS/MDM2 Ubiquitination Axis. Front. Cell Dev. Biol. 2019, 7, 217. [Google Scholar] [CrossRef] [PubMed]
- Mazor, G.; Levin, L.; Picard, D.; Ahmadov, U.; Caren, H.; Borkhardt, A.; Reifenberger, G.; Leprivier, G.; Remke, M.; Rotblat, B. The lncRNA TP73-AS1 is linked to aggressiveness in glioblastoma and promotes temozolomide resistance in glioblastoma cancer stem cells. Cell Death Dis. 2019, 10, 246. [Google Scholar] [CrossRef] [PubMed]
- Cai, T.; Liu, Y.; Xiao, J. Long noncoding RNA MALAT1 knockdown reverses chemoresistance to temozolomide via promoting microRNA-101 in glioblastoma. Cancer Med. 2018, 7, 1404–1415. [Google Scholar] [CrossRef] [PubMed]
- Shang, C.; Tang, W.; Pan, C.; Hu, X.; Hong, Y. Long non-coding RNA TUSC7 inhibits temozolomide resistance by targeting miR-10a in glioblastoma. Cancer Chemother. Pharmacol. 2018, 81, 671–678. [Google Scholar] [CrossRef]
- Ding, C.; Yi, X.; Wu, X.; Bu, X.; Wang, D.; Wu, Z.; Zhang, G.; Gu, J.; Kang, D. Exosome-mediated transfer of circRNA CircNFIX enhances temozolomide resistance in glioma. Cancer Lett. 2020, 479, 1–12. [Google Scholar] [CrossRef]
- Nicolini, A.; Ferrari, P.; Rossi, G.; Carpi, A. Tumour growth and immune evasion as targets for a new strategy in advanced cancer. Endocr. Relat. Cancer 2018, 25, R577–R604. [Google Scholar] [CrossRef]
- Jethwa, K.; Wei, J.; McEnery, K.; Heimberger, A.B. miRNA-mediated immune regulation and immunotherapeutic potential in glioblastoma. Clin. Investig. (Lond.) 2011, 1, 1637–1650. [Google Scholar] [CrossRef]
- Razavi, S.M.; Lee, K.E.; Jin, B.E.; Aujla, P.S.; Gholamin, S.; Li, G. Immune Evasion Strategies of Glioblastoma. Front. Surg. 2016, 3, 11. [Google Scholar] [CrossRef]
- Zhang, Y.; Feng, J.; Fu, H.; Liu, C.; Yu, Z.; Sun, Y.; She, X.; Li, P.; Zhao, C.; Liu, Y.; et al. Coagulation Factor X Regulated by CASC2c Recruited Macrophages and Induced M2 Polarization in Glioblastoma Multiforme. Front. Immunol. 2018, 9, 1557. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhao, Z.; Yang, F.; Wang, H.; Wu, F.; Liang, T.; Yan, X.; Li, J.; Lan, Q.; Wang, J.; et al. An immune-related lncRNA signature for patients with anaplastic gliomas. J. Neurooncol. 2018, 136, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Meng, Y. Survival analysis of immune-related lncRNA in low-grade glioma. BMC Cancer 2019, 19, 813. [Google Scholar] [CrossRef]
- Wei, J.; Nduom, E.K.; Kong, L.Y.; Hashimoto, Y.; Xu, S.; Gabrusiewicz, K.; Ling, X.; Huang, N.; Qiao, W.; Zhou, S.; et al. MiR-138 exerts anti-glioma efficacy by targeting immune checkpoints. Neuro Oncol. 2016, 18, 639–648. [Google Scholar] [CrossRef]
- He, Z.; Ruan, X.; Liu, X.; Zheng, J.; Liu, Y.; Liu, L.; Ma, J.; Shao, L.; Wang, D.; Shen, S.; et al. FUS/circ_002136/miR-138-5p/SOX13 feedback loop regulates angiogenesis in Glioma. J. Exp. Clin. Cancer Res. 2019, 38, 65. [Google Scholar] [CrossRef]
- Xu, H.; Zhang, Y.; Qi, L.; Ding, L.; Jiang, H.; Yu, H. NFIX Circular RNA Promotes Glioma Progression by Regulating miR-34a-5p via Notch Signaling Pathway. Front. Mol. Neurosci. 2018, 11, 225. [Google Scholar] [CrossRef]
- Najbauer, J.; Kraljik, N.; Nemeth, P. Glioma stem cells: Markers, hallmarks and therapeutic targeting by metformin. Pathol. Oncol. Res. 2014, 20, 789–797. [Google Scholar] [CrossRef]
- Ren, Y.; Zhou, X.; Mei, M.; Yuan, X.B.; Han, L.; Wang, G.X.; Jia, Z.F.; Xu, P.; Pu, P.Y.; Kang, C.S. MicroRNA-21 inhibitor sensitizes human glioblastoma cells U251 (PTEN-mutant) and LN229 (PTEN-wild type) to taxol. BMC Cancer 2010, 10, 27. [Google Scholar] [CrossRef]
- Rani, S.B.; Rathod, S.S.; Karthik, S.; Kaur, N.; Muzumdar, D.; Shiras, A.S. MiR-145 functions as a tumor-suppressive RNA by targeting Sox9 and adducin 3 in human glioma cells. Neuro Oncol. 2013, 15, 1302–1316. [Google Scholar] [CrossRef]
- Zheng, J.; Li, X.D.; Wang, P.; Liu, X.B.; Xue, Y.X.; Hu, Y.; Li, Z.; Li, Z.Q.; Wang, Z.H.; Liu, Y.H. CRNDE affects the malignant biological characteristics of human glioma stem cells by negatively regulating miR-186. Oncotarget 2015, 6, 25339–25355. [Google Scholar] [CrossRef]
- Jiang, X.; Yan, Y.; Hu, M.; Chen, X.; Wang, Y.; Dai, Y.; Wu, D.; Wang, Y.; Zhuang, Z.; Xia, H. Increased level of H19 long noncoding RNA promotes invasion, angiogenesis, and stemness of glioblastoma cells. J. Neurosurg. 2016, 2016, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhao, B.S.; Zhou, A.; Lin, K.; Zheng, S.; Lu, Z.; Chen, Y.; Sulman, E.P.; Xie, K.; Bogler, O.; et al. m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 2017, 31, 591–606 e596. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Yu, W.; Zhu, S.; Cheng, K.; Xu, H.; Lv, Y.; Long, X.; Ma, L.; Huang, J.; Sun, S.; et al. Long noncoding RNA GAS5 regulates the proliferation, migration, and invasion of glioma cells by negatively regulating miR-18a-5p. J. Cell. Physiol. 2018, 234, 757–768. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, T.; Zhu, Y.; Li, Y.; Zhang, Y.; Wang, Y.; Li, X.; Xie, X.; Wang, J.; Huang, M.; et al. circPTN sponges miR-145-5p/miR-330-5p to promote proliferation and stemness in glioma. J. Exp. Clin. Cancer Res. 2019, 38, 398. [Google Scholar] [CrossRef] [PubMed]
- Orringer, D.A.; Golby, A.; Jolesz, F. Neuronavigation in the surgical management of brain tumors: Current and future trends. Expert Rev. Med. Devices 2012, 9, 491–500. [Google Scholar] [CrossRef]
- Rodriguez, F.J.; Vizcaino, M.A.; Lin, M.T. Recent Advances on the Molecular Pathology of Glial Neoplasms in Children and Adults. J. Mol. Diagn. 2016, 18, 620–634. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, S.; Pu, J.K.; Tsang, A.C.; Lee, D.; Man, V.O.; Lui, W.M.; Wong, S.T.; Leung, G.K. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol. Dis. 2012, 48, 1–8. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, J.; Liu, Y.; Zhang, W.; Zhou, J.; Duan, R.; Pu, P.; Kang, C.; Han, L. A novel cell cycle-associated lncRNA, HOXA11-AS, is transcribed from the 5-prime end of the HOXA transcript and is a biomarker of progression in glioma. Cancer Lett. 2016, 373, 251–259. [Google Scholar] [CrossRef]
- Jing, S.Y.; Lu, Y.Y.; Yang, J.K.; Deng, W.Y.; Zhou, Q.; Jiao, B.H. Expression of long non-coding RNA CRNDE in glioma and its correlation with tumor progression and patient survival. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3992–3996. [Google Scholar]
- Zhang, J.X.; Han, L.; Bao, Z.S.; Wang, Y.Y.; Chen, L.Y.; Yan, W.; Yu, S.Z.; Pu, P.Y.; Liu, N.; You, Y.P.; et al. HOTAIR, a cell cycle-associated long noncoding RNA and a strong predictor of survival, is preferentially expressed in classical and mesenchymal glioma. Neuro Oncol. 2013, 15, 1595–1603. [Google Scholar] [CrossRef]
- Li, M.; Long, S.; Hu, J.; Wang, Z.; Geng, C.; Ou, S. Systematic identification of lncRNA-based prognostic biomarkers for glioblastoma. Aging (Albany NY) 2019, 11, 9405–9423. [Google Scholar] [CrossRef] [PubMed]
- Xian, J.; Zhang, Q.; Guo, X.; Liang, X.; Liu, X.; Feng, Y. A prognostic signature based on three non-coding RNAs for prediction of the overall survival of glioma patients. FEBS Open Bio 2019, 9, 682–692. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhang, C.; Cai, J.; Yang, F.; Liang, T.; Yan, X.; Wang, H.; Wang, W.; Chen, J.; Jiang, T. Upregulation of long noncoding RNA HOXA-AS3 promotes tumor progression and predicts poor prognosis in glioma. Oncotarget 2017, 8, 53110–53123. [Google Scholar] [CrossRef]
- Min, W.; Dai, D.; Wang, J.; Zhang, D.; Zhang, Y.; Han, G.; Zhang, L.; Chen, C.; Li, X.; Li, Y.; et al. Long Noncoding RNA miR210HG as a Potential Biomarker for the Diagnosis of Glioma. PLoS ONE 2016, 11, e0160451. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Qian, J.; Wang, Y.Y.; Zhang, J.X.; You, Y.P. Long Noncoding RNA Profiles Reveal Three Molecular Subtypes in Glioma. CNS Neurosci. Ther. 2014, 20, 339–343. [Google Scholar] [CrossRef]
- Joshi, P.; Jallo, G.; Perera, R.J. In silico analysis of long non-coding RNAs in medulloblastoma and its subgroups. Neurobiol. Dis. 2020, 141, 1–43. [Google Scholar] [CrossRef]
- Gao, R.; Zhang, R.; Zhang, C.; Zhao, L.; Zhang, Y. Long noncoding RNA CCAT1 promotes cell proliferation and metastasis in human medulloblastoma via MAPK pathway. Tumori J. 2018, 104, 43–50. [Google Scholar] [CrossRef]
- Jin, P.; Huang, Y.; Zhu, P.; Zou, Y.; Shao, T.; Wang, O. CircRNA circHIPK3 serves as a prognostic marker to promote glioma progression by regulating miR-654/IGF2BP3 signaling. Biochem. Biophys. Res. Commun. 2018, 503, 1570–1574. [Google Scholar] [CrossRef]
- Yang, M.; Li, G.; Fan, L.; Zhang, G.; Xu, J.; Zhang, J. Circular RNA circ_0034642 elevates BATF3 expression and promotes cell proliferation and invasion through miR-1205 in glioma. Biochem. Biophys. Res. Commun. 2019, 508, 980–985. [Google Scholar] [CrossRef]
- Peng, H.; Qin, C.; Zhang, C.; Su, J.; Xiao, Q.; Xiao, Y.; Xiao, K.; Liu, Q. circCPA4 acts as a prognostic factor and regulates the proliferation and metastasis of glioma. J. Cell. Mol. Med. 2019, 23, 6658–6665. [Google Scholar] [CrossRef]
- Duan, X.; Liu, D.; Wang, Y.; Chen, Z. Circular RNA hsa_circ_0074362 Promotes Glioma Cell Proliferation, Migration, and Invasion by Attenuating the Inhibition of miR-1236-3p on HOXB7 Expression. DNA Cell Biol. 2018, 37, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Ma, K.; Sun, M.; Shi, S. Identification of the tumor-suppressive function of circular RNA ITCH in glioma cells through sponging miR-214 and promoting linear ITCH expression. Am. J. Transl. Res. 2018, 10, 1373–1386. [Google Scholar] [PubMed]
- Wang, Y.; Sui, X.; Zhao, H.; Cong, L.; Li, Y.; Xin, T.; Guo, M.; Hao, W. Decreased circular RNA hsa_circ_0001649 predicts unfavorable prognosis in glioma and exerts oncogenic properties in vitro and in vivo. Gene 2018, 676, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Wang, J.; He, Y.; Meng, N.; Yan, G.R. Peptides/Proteins Encoded by Non-coding RNA: A Novel Resource Bank for Drug Targets and Biomarkers. Front. Pharmacol. 2018, 9, 1295. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Huang, N.; Yang, X.; Luo, J.; Yan, S.; Xiao, F.; Chen, W.; Gao, X.; Zhao, K.; Zhou, H.; et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 2018, 37, 1805–1814. [Google Scholar] [CrossRef]
- Lv, T.; Miao, Y.F.; Jin, K.; Han, S.; Xu, T.Q.; Qiu, Z.L.; Zhang, X.H. Dysregulated circular RNAs in medulloblastoma regulate proliferation and growth of tumor cells via host genes. Cancer Med. 2018, 7, 6147–6157. [Google Scholar] [CrossRef]
- Pastori, C.; Kapranov, P.; Penas, C.; Peschansky, V.; Volmar, C.H.; Sarkaria, J.N.; Bregy, A.; Komotar, R.; St Laurent, G.; Ayad, N.G.; et al. The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc. Natl. Acad. Sci. USA 2015, 112, 8326–8331. [Google Scholar] [CrossRef]
- Sarma, K.; Levasseur, P.; Aristarkhov, A.; Lee, J.T. Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc. Natl. Acad. Sci. USA 2010, 107, 22196–22201. [Google Scholar] [CrossRef]
- Zhou, K.; Zhang, C.; Yao, H.; Zhang, X.; Zhou, Y.; Che, Y.; Huang, Y. Knockdown of long non-coding RNA NEAT1 inhibits glioma cell migration and invasion via modulation of SOX2 targeted by miR-132. Mol. Cancer 2018, 17, 105. [Google Scholar] [CrossRef]
- Du, P.; Zhao, H.; Peng, R.; Liu, Q.; Yuan, J.; Peng, G.; Liao, Y. LncRNA-XIST interacts with miR-29c to modulate the chemoresistance of glioma cell to TMZ through DNA mismatch repair pathway. Biosci. Rep. 2017, 37. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, T.; Wang, S.; Xiong, Y.; Zhang, R.; Zhang, X.; Zhao, J.; Yang, A.G.; Wang, L.; Jia, L. Nkx2-2as Suppression Contributes to the Pathogenesis of Sonic Hedgehog Medulloblastoma. Cancer Res. 2018, 78, 962–973. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Han, L.M.; Gao, Q.; Sun, Y. Long non-coding RNA CRNDE promotes tumor growth in medulloblastoma. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 2588–2597. [Google Scholar] [PubMed]
- Zhou, T.; Kim, Y.; MacLeod, A.R. Targeting Long Noncoding RNA with Antisense Oligonucleotide Technology as Cancer Therapeutics. Methods Mol. Biol. 2016, 1402, 199–213. [Google Scholar] [CrossRef] [PubMed]
- Katsushima, K.; Natsume, A.; Ohka, F.; Shinjo, K.; Hatanaka, A.; Ichimura, N.; Sato, S.; Takahashi, S.; Kimura, H.; Totoki, Y.; et al. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat. Commun. 2016, 7, 13616. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef]
- Barrett, S.P.; Salzman, J. Circular RNAs: Analysis, expression and potential functions. Development 2016, 143, 1838–1847. [Google Scholar] [CrossRef]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell 2017, 66, 9–21.e27. [Google Scholar] [CrossRef]
- Meganck, R.M.; Borchardt, E.K.; Castellanos Rivera, R.M.; Scalabrino, M.L.; Wilusz, J.E.; Marzluff, W.F.; Asokan, A. Tissue-Dependent Expression and Translation of Circular RNAs with Recombinant AAV Vectors In Vivo. Mol. Ther. Nucleic Acids 2018, 13, 89–98. [Google Scholar] [CrossRef]
- Wesselhoeft, R.A.; Kowalski, P.S.; Anderson, D.G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 2018, 9, 2629. [Google Scholar] [CrossRef]
- Nair, K.G.S.; Ramaiyan, V.; Sukumaran, S.K. Enhancement of drug permeability across blood brain barrier using nanoparticles in meningitis. Inflammopharmacology 2018, 26, 675–684. [Google Scholar] [CrossRef]
- Fareh, M.; Almairac, F.; Turchi, L.; Burel-Vandenbos, F.; Paquis, P.; Fontaine, D.; Lacas-Gervais, S.; Junier, M.P.; Chneiweiss, H.; Virolle, T. Cell-based therapy using miR-302-367 expressing cells represses glioblastoma growth. Cell Death Dis. 2017, 8, e2713. [Google Scholar] [CrossRef] [PubMed]
| ncRNA | Tumor | Target Name | Mechanism of Action | References |
|---|---|---|---|---|
| Cell cycle and apoptosis | ||||
| onco-ncRNAs | ||||
| lncRNA LINC01579 ↑ | Glioblastoma | miR-139-5p | Upregulation of EIF4G2 | [103] |
| lncRNA FOXD2-AS1 ↑ | Glioma | miR-31 | Upregulation of CDK1 | [104] |
| lncRNA HOTAIR ↑ | Glioma | miR-326 | PI3K/AKT and MEK1/2 pathways activation | [106] |
| lncRNA HOTAIR ↑ | Glioblastoma | PRC2 | Cell cycle arrest evading | [105] |
| lncRNA H19 ↑ | Glioma | miR-140 | Upregulation of iASPP | [107] |
| circBLNK ↑ | Glioma | miR-1236 | Possible upregulation of HOXB7 | [110] |
| circMAPK4 ↑ | Glioma | miR-125a-3p | Downregulation of phosphorylation of p38/MAPK | [111] |
| lncRNA TP73-AS1 ↑ | Medulloblastoma | Unknown | Unknown | [113] |
| antionco-ncRNAs | ||||
| lncRNA HOTTIP ↓ | Glioma | BRE | Downregulation of cyclin A and CDK2; upregulation of P53 | [108] |
| lncRNA RAMP2-AS1 ↓ | Glioma | NOTCH3 | Interacting with tumor promoter leading to silencing NOTCH3 | [114] |
| circRNA FBXW7 ↓ | Glioma | c-Myc | Encoding functional protein FBXW7-188aa | [109] |
| SNORD76 ↓ | Glioblastoma | Unknown | Upregulation of pRb; downregulation of Cyclin B1, CDK1, CDC25C | [112] |
| ncRNA | Tumor | Target Name | Mechanism of Action | References |
|---|---|---|---|---|
| Angiogenesis | ||||
| onco-ncRNAs | ||||
| lncRNA H19 ↑ | Glioma | miR-29 | Upregulation of VASH2 | [116] |
| lncRNA XIST ↑ | Glioma | miR-137 | Upregulation of FOXC1 and CXCR7 | [117] |
| lncRNA PVT1 ↑ | Glioma | miR-186 | Upregulation of Atg7 and Beclin1 | [118] |
| lncRNA POU3F3 ↑ | Glioma | Unknown | Upregulation of bFGF, bFGFR, and VEGFA | [119] |
| circRNA SCAF11 ↑ | Glioma | miR-421 | Upregulation of TF SP1 leading to increased expression of VEGF | [120] |
| circRNA SHKBP1 ↑ | Glioma | miR-379 and miR-544a | Upregulation of FOXP1 and FOXP2 | [121] |
| ncRNA | Tumor | Target Name | Mechanism of Action | References |
|---|---|---|---|---|
| Epithelial-to-mesenchymal transition | ||||
| onco-ncRNAs | ||||
| lncRNA LINC00645 ↑ | Glioma | miR-205-3p | Upregulation of ZEB1 | [123] |
| lncRNA MIR22HG ↑ | Glioblastoma | SFRP2 and PCDH15 | Producing miR-22-3p and miR-22-5p | [125] |
| lncRNA HOTAIR ↑ | Medulloblastoma | miR-206 | Upregulation of YY1 | [129] |
| circRNA MMP9 ↑ | Glioblastoma | miR-124 | Upregulation of CDK4 and AURKA | [127] |
| lncRNA ANRIL ↑ | Medulloblastoma | miR-323 | BRI3/CDK6 | [130] |
| antionco-ncRNAs | ||||
| lncRNA TCON ↓ | Glioblastoma | Smad2 and PKCα | Downregulation of Smad2 and PKCα possibly via complementary base pairing in mRNA | [124] |
| circRNA SMARCA5 ↓ | Glioblastoma | SRSF1 | Binds RNA binding proteins involved in splicing (e.g., SRSF1) | [128] |
| snoRNA SNORD47 ↓ | Glioblastoma | Unknown | Downregulation of N-cadherin | [112] |
| ncRNA | Tumor | Target Name | Mechanism of Action | References |
|---|---|---|---|---|
| Chemoresistance | ||||
| onco-ncRNAs | ||||
| lncRNA SBF2-AS1 ↑ | Glioblastoma | miR-151a-3p | Upregulation of XRCC4 | [132] |
| lncRNA ADAMTS9-AS2 ↑ | Glioblastoma | FUS | Inhibition of MDM2-medicated FUS K48 ubiquitination | [133] |
| lncRNA TP73-AS1 ↑ | Glioblastoma | Unknown | Upregulation of ALDH1A1 | [134] |
| lncRNA MALAT1 ↑ | Glioblastoma | miR-101 | Upregulation of GSK3β and MGMT | [135] |
| circNFIX ↑ | Glioma | miR-132 | Modulation of ABCG2 expression | [137] |
| antionco-ncRNAs | ||||
| lncRNA TUSC7↓ | Glioblastoma | miR-10a | Downregulation of MDR1 | [136] |
| ncRNA | Cancer Type | Therapeutical Strategy | References |
|---|---|---|---|
| ncRNAs as a target for molecular therapy | |||
| lncRNA HOTAIR | Bromodomain and extra-terminal (BET) domain proteins | [177] | |
| IRX3-80 (CRNDE) | MB | shRNA | [182] |
| lncRNA TUG1 | GBM | Anti-TUG1 ASOs | [184] |
| lncRNA XIST | Inhibition by miRNAs | [117] | |
| ncRNAs as a tool for molecular therapy | |||
| snoRNA SNORD47 | GBM | Lentiviral-based overexpression | [112] |
| miR-302-367 | GBM | Lentiviral-based overexpression | [191] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Latowska, J.; Grabowska, A.; Zarębska, Ż.; Kuczyński, K.; Kuczyńska, B.; Rolle, K. Non-Coding RNAs in Brain Tumors, the Contribution of lncRNAs, circRNAs, and snoRNAs to Cancer Development—Their Diagnostic and Therapeutic Potential. Int. J. Mol. Sci. 2020, 21, 7001. https://doi.org/10.3390/ijms21197001
Latowska J, Grabowska A, Zarębska Ż, Kuczyński K, Kuczyńska B, Rolle K. Non-Coding RNAs in Brain Tumors, the Contribution of lncRNAs, circRNAs, and snoRNAs to Cancer Development—Their Diagnostic and Therapeutic Potential. International Journal of Molecular Sciences. 2020; 21(19):7001. https://doi.org/10.3390/ijms21197001
Chicago/Turabian StyleLatowska, Julia, Adriana Grabowska, Żaneta Zarębska, Konrad Kuczyński, Bogna Kuczyńska, and Katarzyna Rolle. 2020. "Non-Coding RNAs in Brain Tumors, the Contribution of lncRNAs, circRNAs, and snoRNAs to Cancer Development—Their Diagnostic and Therapeutic Potential" International Journal of Molecular Sciences 21, no. 19: 7001. https://doi.org/10.3390/ijms21197001
APA StyleLatowska, J., Grabowska, A., Zarębska, Ż., Kuczyński, K., Kuczyńska, B., & Rolle, K. (2020). Non-Coding RNAs in Brain Tumors, the Contribution of lncRNAs, circRNAs, and snoRNAs to Cancer Development—Their Diagnostic and Therapeutic Potential. International Journal of Molecular Sciences, 21(19), 7001. https://doi.org/10.3390/ijms21197001





