High-Temperature-Responsive Poplar lncRNAs Modulate Target Gene Expression via RNA Interference and Act as RNA Scaffolds to Enhance Heat Tolerance
Abstract
1. Introduction
2. Results
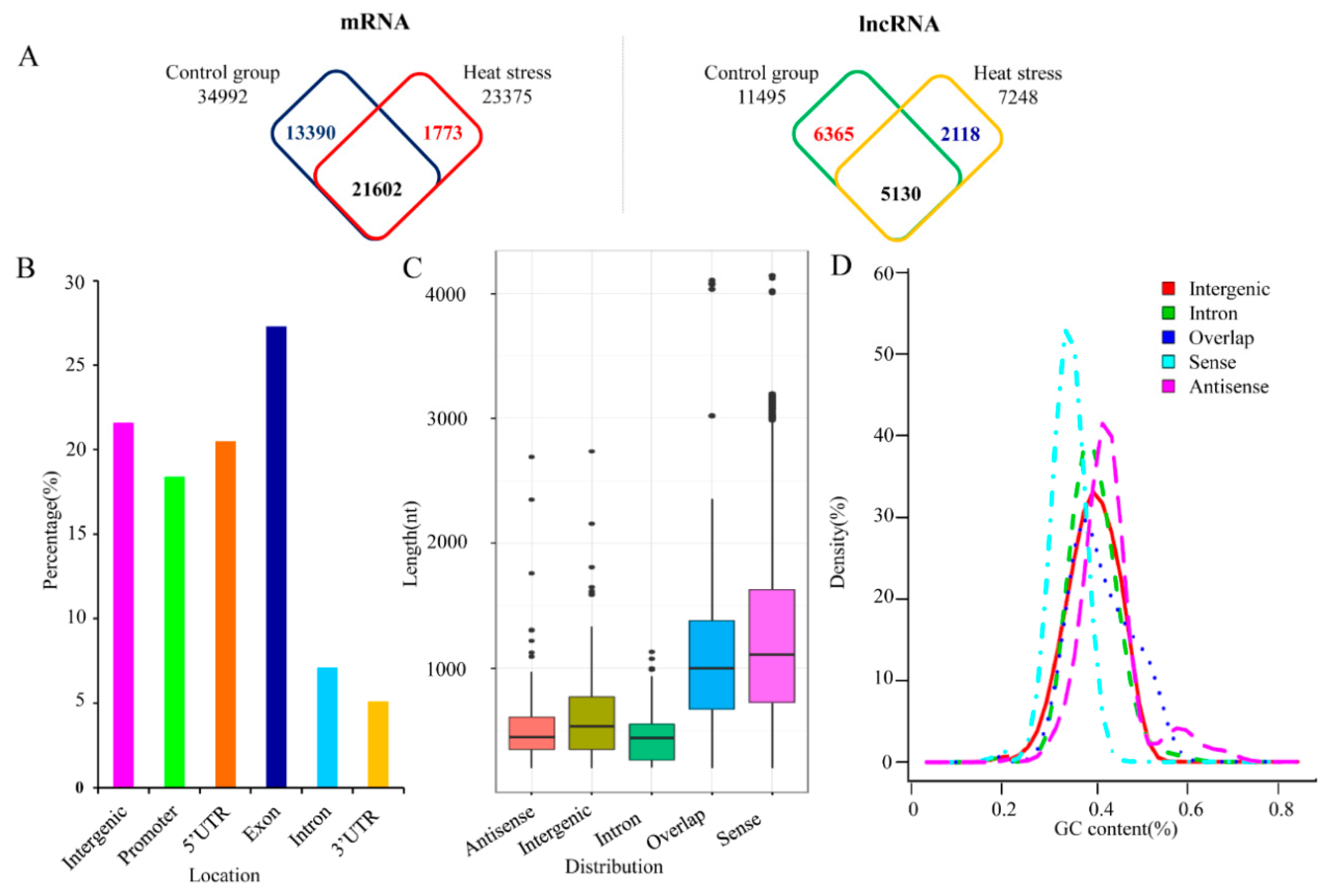
2.1. Characterization of High-Temperature-Responsive lncRNAs in P. simonii
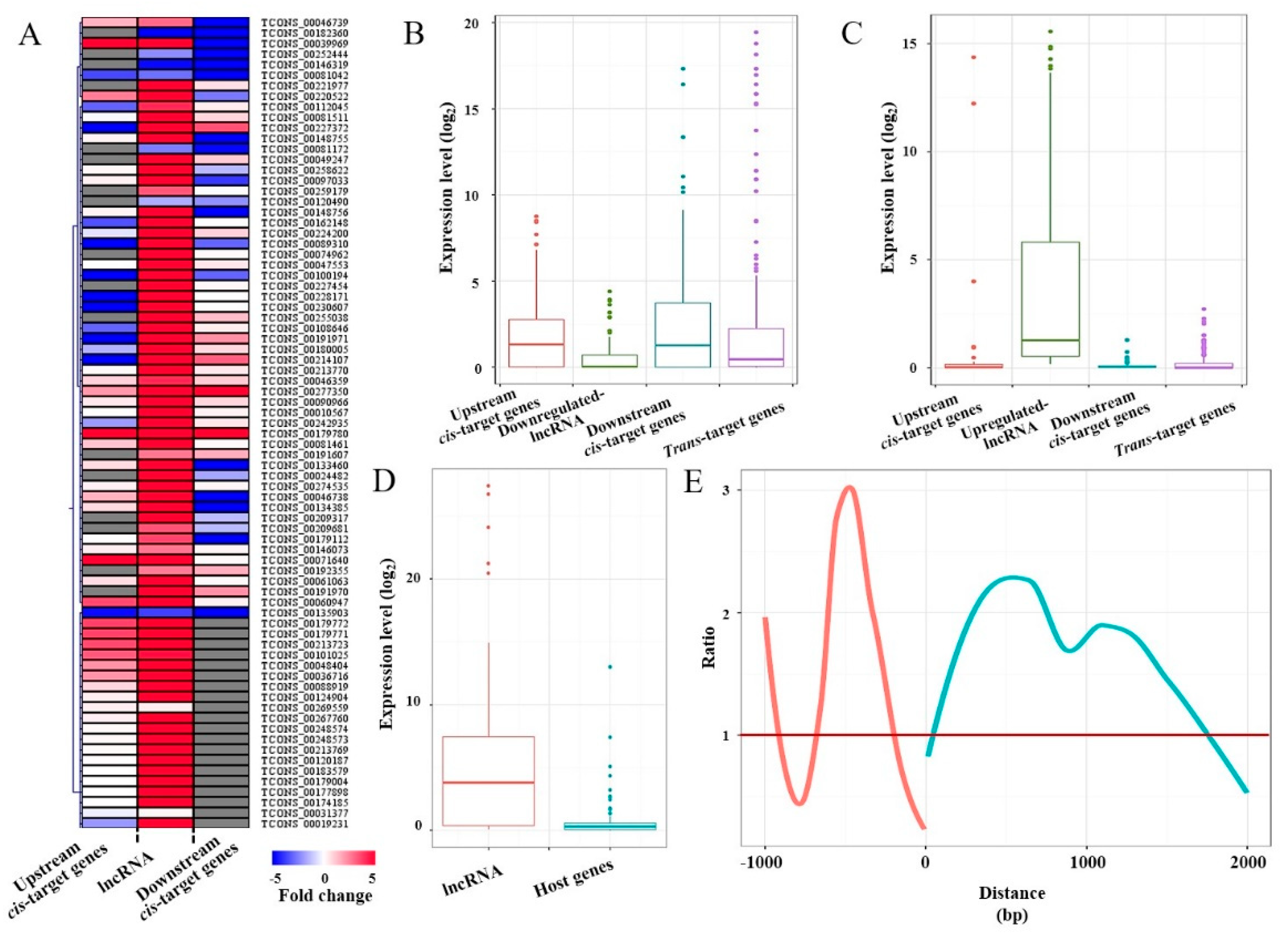
2.2. Identification of High-Temperature-Responsive lncRNAs and Their Potential Targets in P. simonii
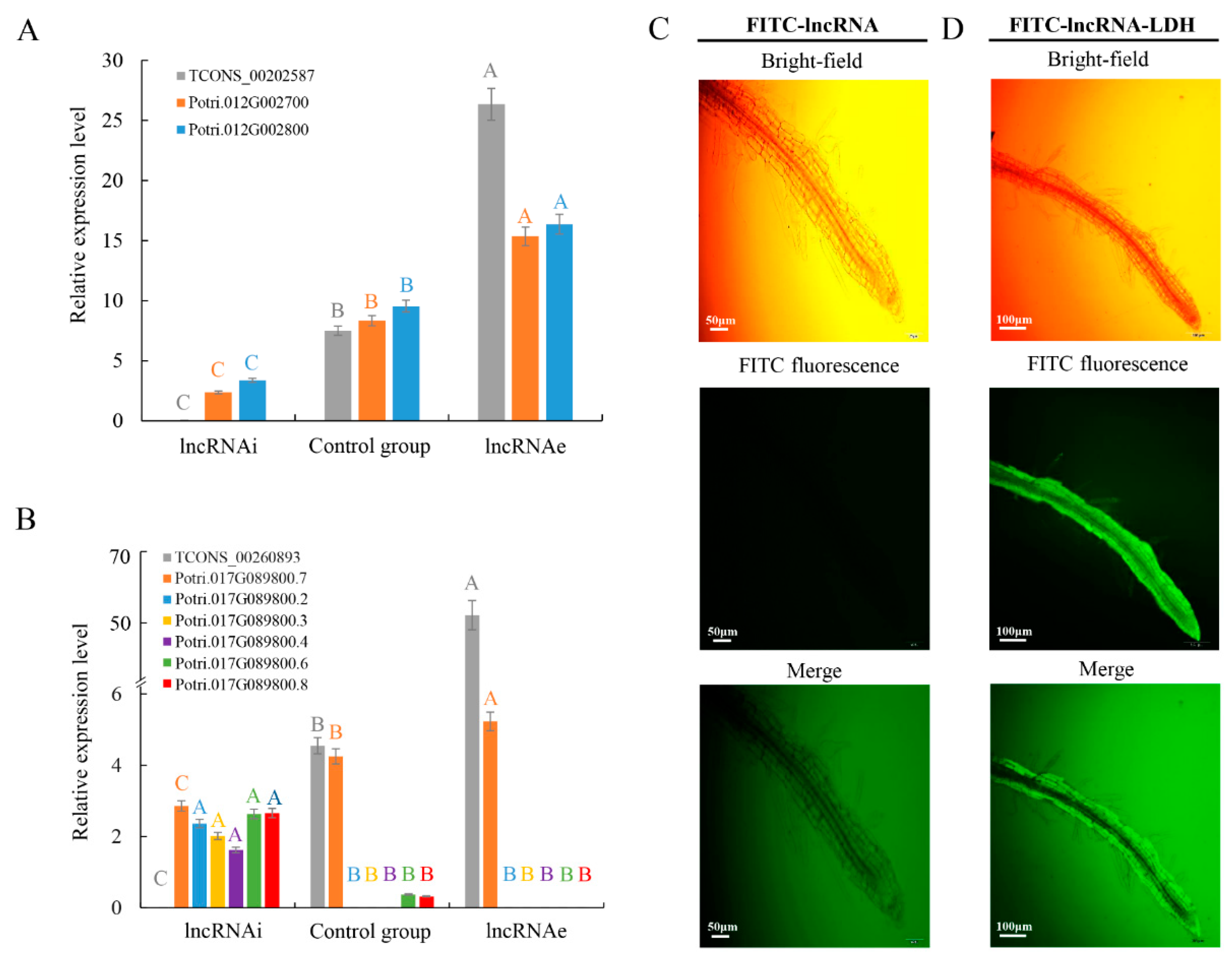
2.3. Effects of High-Temperature-Responsive lncRNAs on Target Gene Expression
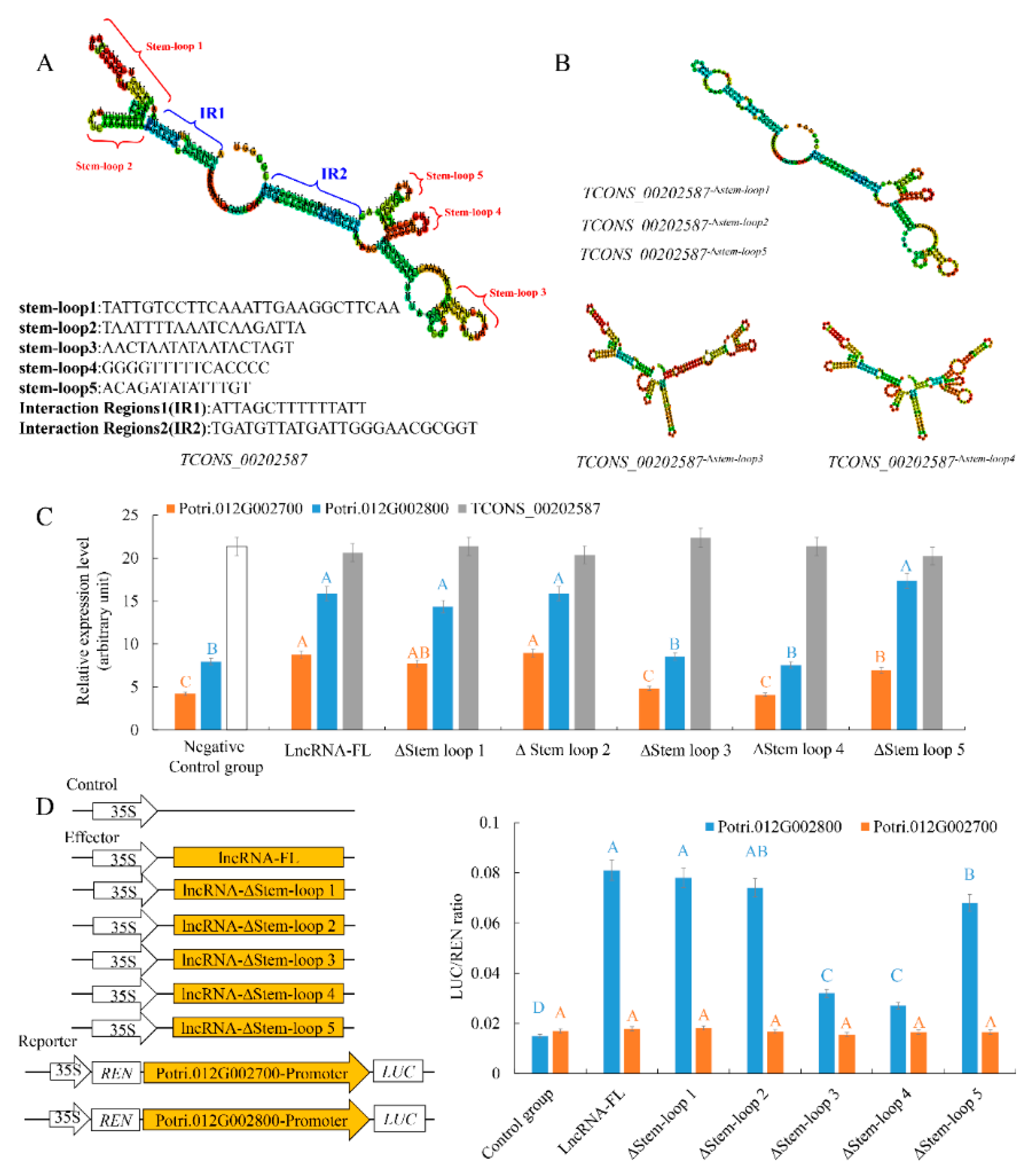
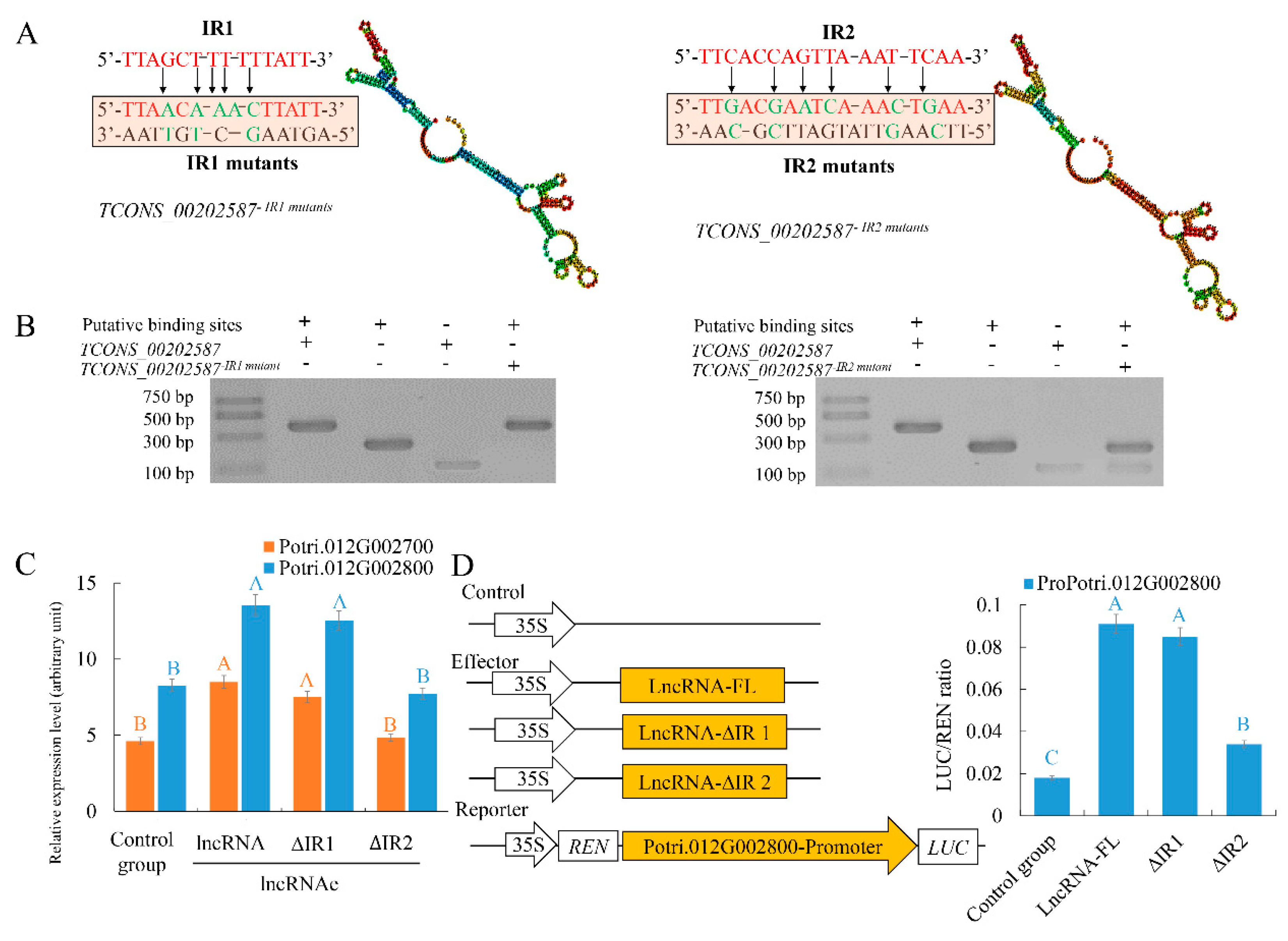
2.4. LncRNAs TCONS_00202587 Transcribed from Proximal Promoter Sequences of a Downstream Gene Can Regulate Transcription by Acting as an RNA Scaffold
2.5. High-Temperature-Induced lncRNA TCONS_00260893 Affects the Expression of Specific Isoforms of Potri.017G089800 Transcripts
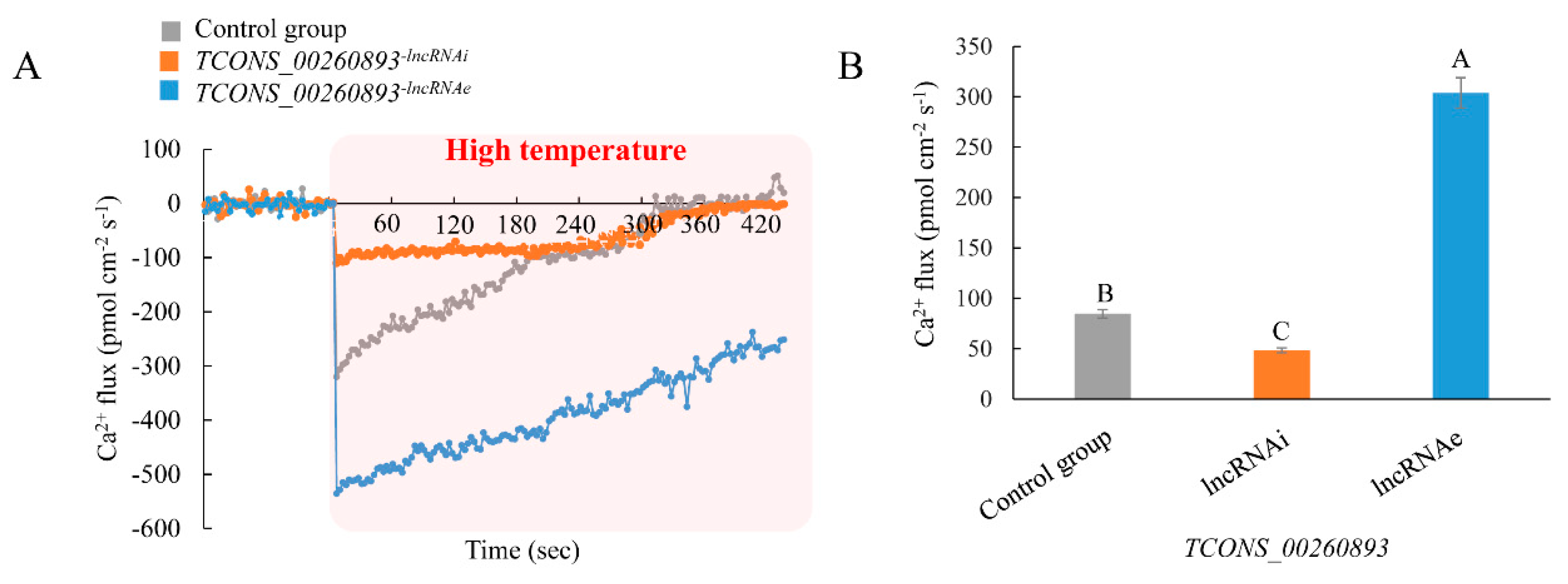
2.6. TCONS_00260893 Enhances Ca2+ Influx under High-Temperature Treatment
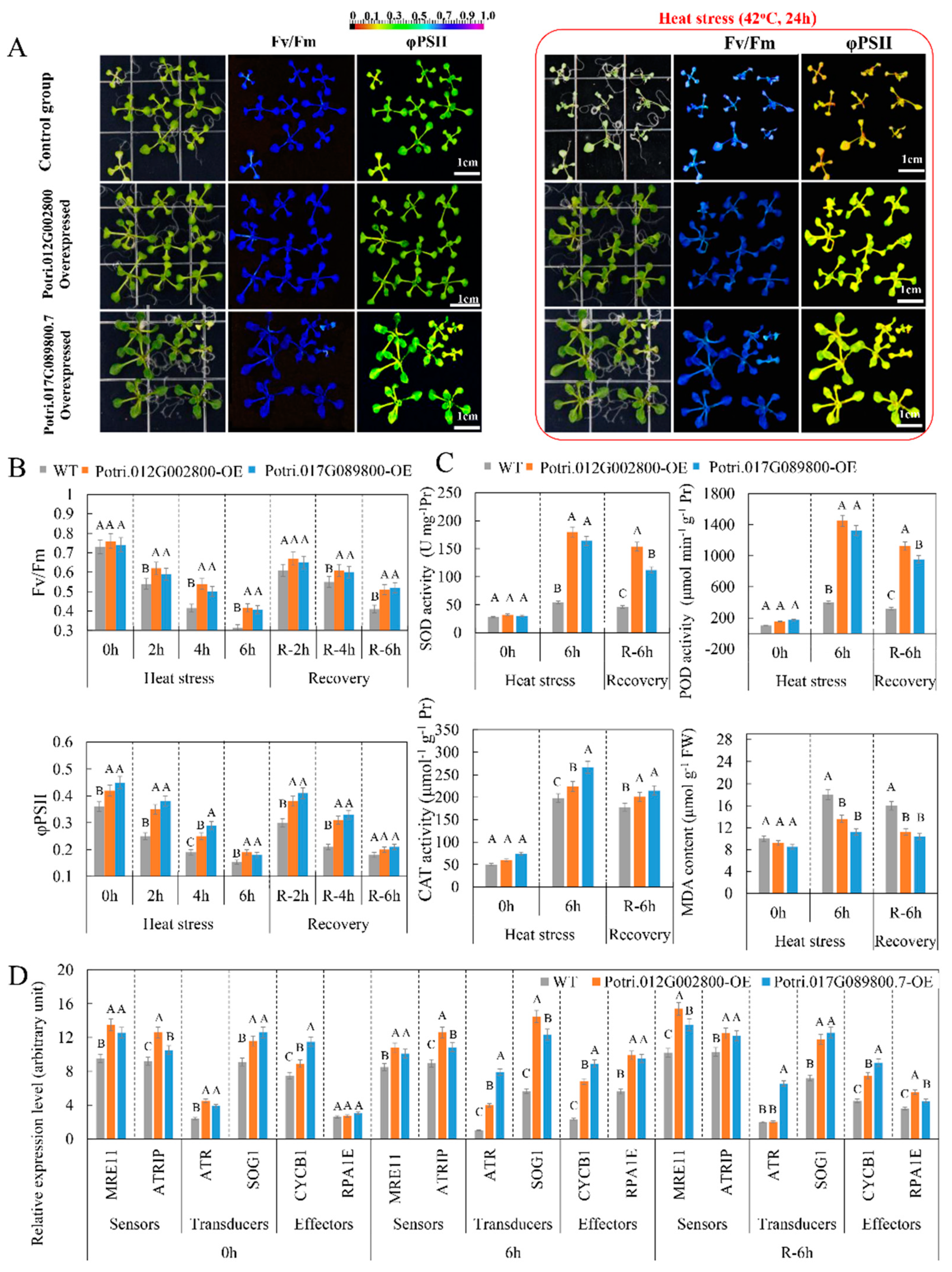
2.7. Overexpressed lncRNA Targets Enhance Heat Tolerance of Arabidopsis
3. Discussion
3.1. LncRNAs Function as Regulators of Temperature-Responsive Transcription
3.2. Specific Secondary Structures of lncRNAs Are Essential for Transcript Regulation
3.3. Temperature-Responsive lncRNAs TCONS_00260893 Mediate Ca2+ Signaling Transduction in Poplar
4. Materials and Methods
4.1. Plant Materials and Treatment
4.2. RNA Sequencing and Prediction of lncRNAs
4.3. Predication of Small Interfering RNA
4.4. Structural Motifs and Target Prediction for Heat-Responsive lncRNAs
4.5. Quantitative PCR Analysis
4.6. Differential Expression Analysis
4.7. Treatment with LDH–lncRNA Conjugates
4.8. Dual Luciferase Assay
4.9. Extracellular Ca2+ Flux Monitoring
4.10. Overexpressed Heat-Responsive lncRNA Targets in A. thaliana
4.11. Measurement of Chlorophyll Fluorescence, Physiological, and Biochemical Characteristics of Transgenic Arabidopsis
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bonan, G.B. Forests and climate change: Forcings, feedbacks, and the climate benefits of forests. Science 2008, 320, 1444–1449. [Google Scholar] [CrossRef] [PubMed]
- Whitham, T.G.; Difazio, S.P.; Schweitzer, J.A.; Shuster, S.M.; Allan, G.J.; Bailey, J.K.; Woolbright, S.A. Extending genomics to natural communities and ecosystems. Science 2008, 320, 492–495. [Google Scholar] [CrossRef]
- Harper, J.; Breton, G.; Harmon, A. Decoding Ca2+ signals through plant protein kinases. Annu. Rev. Plant Biol. 2004, 55, 263–288. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, T.; Shinozaki, K. Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant J. 2010, 61, 1041–1052. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Yang, L.; Chen, L.L. The Diversity of Long Noncoding RNAs and Their Generation. Trends Genet. 2017, 33, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, H.; Chua, N.H. Long noncoding RNA transcriptome of plants. Plant Biotechnol. J. 2015, 13, 319–328. [Google Scholar] [CrossRef]
- Wang, P.; Xue, Y.; Han, Y.; Lin, L.; Wu, C.; Xu, S.; Jiang, Z.; Xu, J.; Liu, Q.; Cao, X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 2014, 344, 310–313. [Google Scholar] [CrossRef]
- Lai, F.; Orom, U.A.; Cesaroni, M.; Beringer, M.; Taatjes, D.J.; Blobel, G.A.; Shiekhattar, R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 2013, 494, 497–501. [Google Scholar] [CrossRef]
- Xu, W.; Xu, H.; Li, K.; Fan, Y.; Liu, Y.; Yang, X.; Sun, Q. The R-loop is a common chromatin feature of the Arabidopsis genome. Nat. Plants 2017, 3, 704–714. [Google Scholar] [CrossRef]
- Cunningham, F.J.; Goh, N.S.; Demirer, G.S.; Matos, J.L.; Landry, M.P. Nanoparticle-Mediated Delivery towards Advancing Plant Genetic Engineering. Trends Biotechnol. 2018, 36, 882–897. [Google Scholar] [CrossRef]
- Kwak, S.Y.; Lew, T.T.S.; Sweeney, C.J.; Koman, V.B.; Wong, M.H.; Bohmert-Tatarev, K.; Snell, K.D.; Seo, J.S.; Chua, N.H.; Strano, M.S. Chloroplast-selective gene delivery and expression in planta using chitosan-complexed single-walled carbon nanotube carriers. Nat. Nanotechnol. 2019, 14, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jung, C.; Xu, J.; Wang, H.; Deng, S.; Bernad, L. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 2012, 24, 4333–4345. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Xie, S.; Liu, Y.; Yi, F.; Yu, J. Genome-wide annotation of genes and noncoding RNAs of foxtail millet in response to simulated drought stress by deep sequencing. Plant Mol. Biol. 2013, 83, 459–473. [Google Scholar] [CrossRef]
- Wang, T.Z.; Liu, M.; Zhao, M.G.; Chen, R.; Zhang, W.H. Identification and characterization of long non-coding RNAs involved in osmotic and salt stress in Medicago truncatula using genome-wide high-throughput sequencing. BMC Plant Biol. 2015, 15, 131. [Google Scholar] [CrossRef] [PubMed]
- Borsani, O.; Zhu, J.; Verslues, P.E.; Sunkar, R.; Zhu, J.K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 2005, 123, 1279–1291. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Ding, C.; Chu, Y.; Zhang, W.; Guo, G.; Chen, J.; Su, X. Pln24NT: A web resource for plant 24-nt siRNA producing loci. Bioinformatics 2017, 33, 2065–2067. [Google Scholar] [CrossRef]
- Mitter, N.; Worrall, E.A.; Robinson, K.E.; Li, P.; Jain, R.G.; Taochy, C.; Fletcher, S.J.; Carroll, B.J.; Lu, G.Q.; Xu, Z.P. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants 2017, 3, 16207. [Google Scholar] [CrossRef]
- Arazi, T.; Kaplan, B.; Sunkar, R.; Fromm, H. Cyclic-nucleotide- and Ca2+/calmodulin-regulated channels in plants: Targets for manipulating heavy-metal tolerance, and possible physiological roles. Biochem. Soc. Trans. 2000, 28, 471–475. [Google Scholar] [CrossRef]
- DeFalco, T.A.; Moeder, W.; Yoshioka, K. Opening the Gates: Insights into Cyclic Nucleotide-Gated Channel-Mediated Signaling. Trends Plant Sci. 2016, 21, 903–906. [Google Scholar] [CrossRef]
- Jha, S.K.; Sharma, M.; Pandey, G.K. Role of Cyclic Nucleotide Gated Channels in Stress Management in Plants. Curr. Genom. 2016, 17, 315–329. [Google Scholar] [CrossRef]
- Bourbousse, C.; Vegesna, N.; Law, J.A. SOG1 Activator and MYB3R Repressors Regulate a Complex DNA Damage Network in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E12453–E12462. [Google Scholar] [CrossRef]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef] [PubMed]
- Mathews, D.H.; Disney, M.D.; Childs, J.L.; Schroeder, S.J.; Zuker, M.; Turner, D.H. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA 2004, 101, 7287–7292. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.P.; Xuan, A.R.; Bu, C.H.; Ci, D.; Tian, M.; Zhang, D.Q. Osmotic stress-responsive promoter upstream transcripts (PROMPTs) act as carriers of MYB transcription factors to induce the expression of target genes in Populus simonii. Plant Biotechnol. 2019, 17, 164–177. [Google Scholar] [CrossRef] [PubMed]
- Kortmann, J.; Narberhaus, F. Bacterial RNA thermometers: Molecular zippers and switches. Nat. Rev. Microbiol. 2012, 10, 255–265. [Google Scholar] [CrossRef]
- Cruz, J.A.; Westhof, E. The dynamic landscapes of RNA architecture. Cell 2009, 136, 604–609. [Google Scholar] [CrossRef]
- Di, C.; Yuan, J.; Wu, Y.; Li, J.; Lin, H.; Hu, L.; Tang, T.; Qi, Y.J.; Gerstein, M.B.; Guo, Y.; et al. Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J. 2014, 80, 848–861. [Google Scholar] [CrossRef]
- Ray, D.; Kazan, H.; Cook, K.B.; Weirauch, M.T.; Najafabadi, H.S.; Li, X.; Hughes, T.R. A compendium of RNA-binding motifs for decoding gene regulation. Nature 2013, 499, 172–177. [Google Scholar] [CrossRef]
- Vandivier, L.E.; Anderson, S.J.; Foley, S.W.; Gregory, B.D. The conservation and function of RNA secondary structure in plants. Annu. Rev. Plant Biol. 2016, 67, 463–488. [Google Scholar] [CrossRef]
- Gong, J.; Liu, W.; Zhang, J.Y.; Miao, X.P.; Guo, A.Y. lncRNASNP: A database of SNPs in lncRNAs and their potential functions in human and mouse. Nucleic Acids Res. 2015, 43, D181–D186. [Google Scholar] [CrossRef]
- Vu, L.D.; Gevaert, K.; De Smet, I. Feeling the Heat: Searching for Plant Thermosensors. Trends Plant Sci. 2019, 24, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Thor, K. Calcium-Nutrient and Messenger. Front. Plant Sci. 2019, 10, 440. [Google Scholar] [CrossRef]
- Demidchik, V.; Bowen, H.C.; Maathuis, F.J.M.; Shabala, S.N.; Tester, M.A.; White, P.J.; Davies, J.M. Arabidopsis thaliana root non-selective cation channels mediate calcium uptake and are involved in growth. Plant J. 2002, 32, 799–808. [Google Scholar] [CrossRef] [PubMed]
- White, P.J. The pathways of calcium movement to the xylem. J. Exp. Bot. 2001, 52, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Finka, A.; Cuendet, A.F.; Maathuis, F.J.; Saidi, Y.; Goloubinoff, P. Plasma membrane cyclic nucleotide gated calcium channels control land plant thermal sensing and acquired thermotolerance. Plant Cell 2012, 24, 3333–3348. [Google Scholar] [CrossRef] [PubMed]
- Tress, M.L.; Abascal, F.; Valencia, A. Alternative splicing may not be the key to proteome complexity. Trends Biochem. Sci. 2017, 42, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Coulombe-Huntington, J.; Kang, S.; Sheynkman, G.M.; Hao, T.; Richardson, A.; Sun, S.; Yang, F.; Shen, Y.A.; Murray, R.R.; et al. Widespread expansion of protein interaction capabilities by alternative splicing. Cell 2016, 164, 805–817. [Google Scholar] [CrossRef]
- Knight, C.A.; Ackerly, D.D. An ecological and evolutionary analysis of photosynthetic thermotolerance using the temperature-dependent increase in fluorescence. Oecologia 2002, 130, 505–514. [Google Scholar] [CrossRef]
- Levin, J.Z.; Yassour, M.; Adiconis, X.; Nusbaum, C.; Thompson, D.A.; Friedman, N.; Gnirke, A.; Regev, A. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods 2010, 7, 709–715. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzbergm, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Min, X.J.; Butler, G.; Storms, R.; Tsang, A. OrfPredictor: Predicting protein-coding regions in EST-derived sequences. Nucleic Acids Res. 2005, 33, W677–W680. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.N.; Chen, R.S.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- El Baidouri, M.; Kim, K.D.; Abernathy, B.; Arikit, S.; Maumus, F.; Panaud, O.; Meyers, B.C.; Jackson, S.A. A new approach for annotation of transposable elements using small RNA mapping. Nucleic Acids Res. 2015, 43, e84. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Rabani, M.; Kertesz, M.; Segal, E. Computational prediction of RNA structural motifs involved in posttranscriptional regulatory processes. Proc. Natl. Acad. Sci. USA 2008, 105, 14885–14890. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Osak, M.; Bogu, G.K.; Stanton, L.W.; Johnson, R.; Lipovich, L. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA 2010, 16, 1478–1487. [Google Scholar] [CrossRef]
- Tafer, H.; Hofacker, I.L. RNAplex: A fast tool for RNA-RNA interaction search. Bioinformatics 2008, 24, 2657–2663. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple hypothesis testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Howe, E.; Holton, K.; Nair, S.; Schlauch, D.; Sinha, R.; Quackenbush, J. MeV: MultiExperiment Viewer. In Biomedical Informatics for Cancer Research; Ochs, M., Casagrande, J., Davuluri, R., Eds.; Springer: Boston, MA, USA, 2010; pp. 102–105. [Google Scholar]
- Sundell, D.; Mannapperuma, C.; Netotea, S.; Delhomme, N.; Lin, Y.C.; Sjödin, A.; Van de Peer, Y.; Jansson, S.; Hvidsten, T.R.; Street, N.R. The Plant Genome Integrative Explorer Resource: PlantGenIE.org. New Phytol. 2015, 208, 1149–1156. [Google Scholar] [CrossRef] [PubMed]
- Bao, W.; Wang, J.; Wang, Q.; O’Hare, D.; Wan, Y. Layered double hydroxide nanotransporter for molecule delivery to intact plant cells. Sci. Rep. 2016, 6, 26738. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Li, H.; Luo, J.; Ma, C.; Li, S.; Qu, L.; Gai, Y.; Jiang, X.J.; Janz, D.; Polle, A.; et al. A transcriptomic network underlies microstructural and physiological responses to cadmium in Populus × canescens. Plant Physiol. 2013, 162, 424–439. [Google Scholar] [CrossRef]
- Sun, J.; Wang, M.J.; Ding, M.Q.; Deng, S.R.; Liu, M.Q.; Lu, C.F.; Zhou, X.Y.; Shen, X.; Zheng, X.J.; Zhang, Z.K.; et al. H2O2 and cytosolic Ca2+ signals triggered by the PM H-coupled transport system mediate K+/Na+ homeostasis in NaCl-stressed Populus euphratica cells. Plant Cell Environ. 2010, 6, 943–958. [Google Scholar] [CrossRef]
- Zhang, Y.; Sa, G.; Zhang, Y.; Zhu, Z.; Deng, S.; Sun, J.; Li, N.; Li, J.; Yao, J.; Zhao, N.; et al. Paxillus involutus-Facilitated Cd2+ Influx through Plasma Membrane Ca2+-Permeable Channels Is Stimulated by H2O2 and H+-ATPase in Ectomycorrhizal Populus × canescens under Cadmium Stress. Front. Plant Sci. 2017, 7, 1975. [Google Scholar] [CrossRef]
- Song, Y.P.; Ci, D.; Tian, M.; Zhang, D.Q. Comparison of the physiological effects and transcriptome responses of Populus simonii under different abiotic stresses. Plant Mol. Biol. 2014, 86, 139–156. [Google Scholar] [CrossRef]
- Zhao, X.Y.; Zheng, T.C.; Shao, L.T.; Xiao, Z.H.; Wang, F.W.; Li, S.C.; Zang, L.; Zheng, M.; Li, Y.; Qu, G.Z. Variation analysis of physiological traits in Betula platyphylla overexpressing TaLEA-ThbZIP gene under salt stress. PLoS ONE 2016, 11, e0164820. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.P.; Chen, P.F.; Liu, P.; Bu, C.H.; Zhang, D.Q. High temperature-responsive poplar lncRNAs modulate target gene expression by RNA interference or acting as RNA scaffolds enhancing heat tolerance. Authorea 2020, 20, 5573. [Google Scholar] [CrossRef]
| Locus Name | Location | Targets | Annotation |
|---|---|---|---|
| TCONS_00260893 | Chr17:10685952-10690606 (Antisense strand) | Potri.017G089800 | Cyclic nucleotide- regulated ion channel family protein |
| TCONS_00202587 | Chr12: 251176-251434 (Sense strand) | Potri.012G002700 | Highly ABA-induced PP2C gene 3 |
| Potri.012G002800 | Protein kinase superfamily protein |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, Y.; Chen, P.; Liu, P.; Bu, C.; Zhang, D. High-Temperature-Responsive Poplar lncRNAs Modulate Target Gene Expression via RNA Interference and Act as RNA Scaffolds to Enhance Heat Tolerance. Int. J. Mol. Sci. 2020, 21, 6808. https://doi.org/10.3390/ijms21186808
Song Y, Chen P, Liu P, Bu C, Zhang D. High-Temperature-Responsive Poplar lncRNAs Modulate Target Gene Expression via RNA Interference and Act as RNA Scaffolds to Enhance Heat Tolerance. International Journal of Molecular Sciences. 2020; 21(18):6808. https://doi.org/10.3390/ijms21186808
Chicago/Turabian StyleSong, Yuepeng, Panfei Chen, Peng Liu, Chenhao Bu, and Deqiang Zhang. 2020. "High-Temperature-Responsive Poplar lncRNAs Modulate Target Gene Expression via RNA Interference and Act as RNA Scaffolds to Enhance Heat Tolerance" International Journal of Molecular Sciences 21, no. 18: 6808. https://doi.org/10.3390/ijms21186808
APA StyleSong, Y., Chen, P., Liu, P., Bu, C., & Zhang, D. (2020). High-Temperature-Responsive Poplar lncRNAs Modulate Target Gene Expression via RNA Interference and Act as RNA Scaffolds to Enhance Heat Tolerance. International Journal of Molecular Sciences, 21(18), 6808. https://doi.org/10.3390/ijms21186808




