Abstract
(1) Background: Obesity and mood disorders are considered as the most prevalent morbidities in many countries. We suppose that epigenetic mechanisms may induce higher rates of obesity in subjects who suffer from mood disorders. In this systematic review, we focused on the potential roles of DNA methylation on mood disorders and obesity development. (2) Methods: This systematic review was conducted in accordance with the PRISMA statement and registered in Prospero. A systematic search was conducted in MEDLINE, Scopus, Web of Science, Cochrane Central database, EMBASE, and CINHAL. We also conducted a Grey literature search, such as Google Scholar. (3) Results: After deduplication, we identified 198 potentially related citations. Finally, ten unique studies met our inclusion criteria. We have found three overlap genes that show significant DNA methylation changes, both in obesity and depression. Pathway analysis interaction for TAPBP, BDNF, and SORBS2 confirmed the relation of these genes in both obesity and mood disorders. (4) Conclusions: While mechanisms linking both obesity and mood disorders to epigenetic response are still unknown, we have already known chronic inflammation induces a novel epigenetic program. As the results of gene enrichment, pathways analysis showed that TAPBP, BDNF, and SORBS2 linked together by inflammatory pathways. Hypermethylation in these genes might play a crucial rule in the co-occurrence of obesity and mood disorders.
1. Introduction
Obesity and mood disorders are considered as the most prevalent morbidities in developed and developing countries [1,2,3]. The worldwide prevalence of obesity has nearly tripled since 1975. In 2016, more than 650 million adults were obese and 38 million children under the age of 5 were overweight or obese in 2019 [4]. The prevalence of mood disorders differs based on sex and disease. For example, the prevalence of major depressive disorder (MDD) and anxiety are 17.4% and 18.2% in men, 22.7% and 23.6 in women, respectively [5].
A combination of genetics and environmental factors affect the incidence and development of obesity and mood disorders [6,7]. The type and amount of food consumed during depression appeared to be significantly correlated and could affect the weight in a long time [8]. It has been well-established that 12% of the responsible genes for obesity are shared with depression [9], and changes in the mutual pathways of the shared genes could lead to altering the pathological eating behavior in patients with mood disorders. In addition, antidepressant drugs can alter body mass indexes [10,11,12].
One of the possible biological changes that could be responsible for the co-occurrence of these disorders might be epigenetic changes [13,14]. Epigenetics could legitimize modifications in the chromatin level, which alters the expression of genes involved in obesity and mood disorder [15,16]. Epigenetics could explain complex interactions between the genome and the environment. Epigenetic modifications, such as DNA methylation and histone modification, alter DNA accessibility and chromatin structure, thereby regulating patterns of gene expression [17]. For example, increasing the methylation of DNA reduces the expression of genes, while decreasing methylation is associated with increased gene expression [18]. DNA methylation occurs in the whole genome but could play an important role in repressing gene transcription when affecting the gene promoter (especially in the CpG islands, shore, and shelves) [19]. The relationship between DNA methylation and obesity and mood disorders has been reported previously [20,21,22,23,24].
The critical question is whether epigenetic changes in overlapped genes could cause obesity and mood disorders. In other words, mood disorders, especially depression, may result in obesity through DNA methylation of the shared genes, which could affect the body composition. We hypothesized that epigenetic mechanisms might induce higher rates of obesity in subjects who suffer from mood disorders. Therefore, in this article, we focused on the potential roles of DNA methylation on mood disorders and obesity development. To answer this question, we systematically reviewed the studies investigating the methylation regions in overlap genes in patients with obesity or mood disorders. Then, we discussed possible pathways that are impressed by DNA methylation in overlap genes and possible consequent changes.
2. Methods
This systematic review was conducted following the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) statement and was registered in a prospective international register of systematic reviews [PROSPERO (Prospective Register of Systematic Reviews). To find relevant articles, searches were made in MEDLINE via PubMed (www.pubmed.com; National Library of Medicine), Scopus (www.scopus.com), ISI Web of Science (www.thomsonreuters.com), Cochrane Central database, EMBASE, and CINHAL. We also searched Google Scholar (www.scholar.google.com) as Gray literature. There is no restriction regarding language, publication period, patient age (children or adult), or study design. The study identification also included manual search, based on the screening of the citations of the relevant studies.
2.1. Search Strategy
2.1.1. Step 1: Identification of Candidate Genes for Obesity
We carried out a systematic search of DNA methylation in epigenome-wide association study (EWAS) for obesity. We reviewed EWAS study papers published until November 2019 for obesity or body mass index (BMI). All EWAS significant information such as reported genes, author(s), PubMed ID, date of publication, journal, discovery, and replication sample sizes was searched. An obesity gene was considered as a candidate gene if (1) at least one CpG site within or nearby to the gene was identified; and (2) it was functionally relevant to influence at least one of the genes related to obesity.
2.1.2. Step 2: Exploration of the Role of Differentiated Methylated Obesity Genes in Mood Disorders
In the second systematic search, we conducted a literature search in the above-mentioned data-source for any epigenome-wide association with the candidate gene analysis when the study is published in the fields of mood disorders until January 2020.
2.1.3. Search Term
Three groups of medical subject headings (MeSH) and non-MeSH keywords were selected to search the databases, as follows: “Obesity AND (DNA Methylation, obesity, and depression, mood disorders, bipolar, suicide”.
2.2. Screening
Three independent reviewers (MG, NM, MD) initially scanned titles and abstracts to select potential full-text articles for further study. When any reviewer could not exclude the title and abstract, the full text of the article was obtained via Isfahan University of Medical Sciences library. Any differences in opinion were resolved through team discussion. Inclusion or exclusion of each study was determined by discussion and consensus between the two reviews. A reference list of related articles was also checked for any missing related articles. We included cohort and case-control studies.
Data on the author(s), year of publication, sample size, study design, study cohort, experimental methods, type of tissues, candidate genes or genome, DNA purification method, DNA methylation method, DNA methylation validation, genotyping, gene expression, experimental factors, statistical methods, and significant findings were extracted independently by two reviewers. For those studies with multiple reports, a single record denoted one study with the information extracted from multiple reports. All disagreements were resolved through discussions. The reviewers endeavored to contact the original authors of the studies for any missing information in order to gather complete and consistent study information. Open-ended questions were used to prevent misleading answers.
2.3. Inclusion Criteria
The following inclusion criteria used: (1) Studies using EWAS approach focusing on the global DNA methylation since we aimed to find overlap methylated genes in both obesity and depression; (2) we consider all types of a mood disorders such as depression, bipolar and suicide.
2.4. Exclusion Criteria
Review articles, randomized clinical trials, or any paper with no quantitative data was omitted.
2.5. The Following Outcome Measures Were Considered
The outcome of interest was obesity, depression, psychological disorder, and suicide. We anticipated that DNA methylation levels would be reported as either categorical (DNA is either hypo-, hyper- or normally methylated) or continuous data (i.e., percentage of methylated DNA). We also searched abstracts from relevant conference papers.
2.6. Types of Tissue Samples Included in the Review
We decided to include methylation data regardless of the source of the sample, e.g., peripheral blood, placenta, umbilical cord blood, or buccal mucosa.
2.7. Format of Data Input for Factors
Risk of bias and quality assessment of selected studies were assessed through a modified Downs and Black checklist for methodological quality assessment [25]. We chose to use this checklist for quality assessment used. Additionally, this checklist provides an overall quality index as well as four sub-scales of quality assessment (reporting, external quality, internal validity-bias, and internal validity-confounding). We did not exclude any study based on quality.
3. Results
After deduplication, we identified 198 potentially related citations. Based on the title and abstract, 123 studies were excluded because of inappropriate exposure (gene mutations, gene polymorphism, and microRNA), irrelevant outcomes (autoimmune diseases, cancer, and inflammation-related diseases such as asthma), or both. We also excluded investigations conducted in mice or rats. Finally, ten studies were deliberated for full-text assessment. Figure 1 shows the detailed information of the process of study selection.
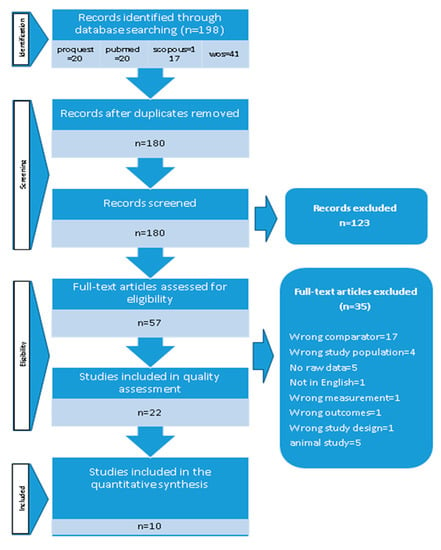
Figure 1.
Search strategy.
Table 1 presents a summary of the study characteristics of these selected studies. Most of the reviewed articles were published between 2014 and 2019, especially in the past four years. The selected studies mainly focused on both adults and adolescence. Most studies in this review were case-control or general population-based cohorts. There was a wide variety in terms of sample size, ranging from 5 to 115. Whole blood was the most commonly used biological sample analyzed by generally accepted DNA methylation methods, such as bisulfite conversion with pyrosequencing. Table 2 indicated the characteristics of the overlap genes. Table 3 displays a summary of biological pathways related to the TAPBP, BDNF, and SORBS2. Table 4 shows pathway analysis interaction for overlapped genes in obesity and mood disorders. Figure 2 demonstrate gene interaction between overlapped genes in obesity and mood disorders by the genemania software.

Table 1.
Genomic regions investigated in reviewed studies.

Table 2.
A summary of biological pathways related to the TAPBP, BDNF, and SORBS.

Table 3.
Pathway analysis interaction for overlap genes in obesity and mood disorder.

Table 4.
Report of Black and down score.
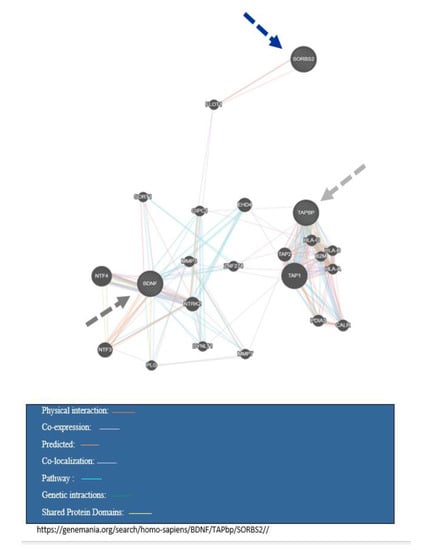
Figure 2.
Gene interaction between overlap genes in obesity and mood disorder.
4. Discussion
To the best of our knowledge, this is the first cross-disorder systematically review that assessed the role of DNA methylation in the overlapped genes and their affected biological pathways in mood disorders and obesity. Our results revealed three overlapped genes with different methylated patterns during obesity or mood disorders, which can assist us to understand better the molecular pathophysiology of these disorders. In the further step, we attempted to identify the possible pathways that could be involved in obesity and mood disorders through the overlap genes.
In the era of the increasing prevalence of obesity and mood disorders, especially in both developing and developed world, results from our systematic review suggest an interplay between genetic susceptibility, diet, epigenetics, metagenomics, and the environment [36,37].
Evidently, obesity was found to increase the risk of depression, and depression was found to be predictive of developing obesity. Remarkably, obese persons had a 55% increased risk of developing depression over time, whereas depressed persons had a 58% increased risk of becoming obese. Neuroendocrine disturbances may also lead to depression, which in turn would cause an increase in weight over time by dysregulated stress systems or through unhealthy lifestyles. It is also possible that obesity, by its adverse effects on self-image or somatic consequences, results in the development of depression over time [38]. So, scientists struggled to find responsible genes through genome-wide association studies (GWAS) to identify the risk associated with single nucleotide polymorphisms, which might also be responsible for the co-occurrence of two conditions.
In recent years, scientific documents proved that genes are not responsible for disease by themselves, and the interaction of genes and environment is better determinants for phenotypes. Accordingly, the latest researches are likely to focus on epigenome-wide association studies (EWAS). The advantages of EWAS is considering the interaction of both genes and environments. The information gained from GWAS and EWAS has potential applications in disease control and treatment. In this study, we merely focused on DNA methylation, which could cause alterations in gene expressions and changes in the pathophysiology of diseases. We found three overlapped genes between mood disorders and obesity “TAPBP, SORBS2, and BDNF.” As these genes were found through published results of EWAS, we will discuss canonical pathways that might be involved in co-occurrence mood disorders and obesity.
TAPBP: The TAPBP gene is located in chromosome 6 and encodes tapsin; a transmembrane glycoprotein that mediates the interaction between newly assembled major histocompatibility complex (MHC) class I molecules. MHC1 is a transporter associated with antigen processing (TAP), which is required for the transport of antigenic peptides across the ER membrane [39,40]. TAPBP-mutant mice have defects in the expression of MHC class I, antigen presentation, and immune responses. Remarkably, Cui et al. found that the expression levels of HLA-ABC were upregulated even in the TAPBP knock-out cells by the interferon treatment, and immune rejection was reduced in TAPBP-deficient hESC line. Potent inflammatory molecules such as eicosanoids are able to upregulate TAPBP [41,42].
The important role of TAPBP is not recognized in the past in both obesity and mood disorders, and just in recent years. The results of EWAS-approved methylation in this gene could play a crucial role in these conditions. Murphy et al. identified epigenetic changes such as differentiated methylated regions (DMR) located in the third intron of the TAPBP gene that is related to the major depressive disorder and suicide [27]. Another study demonstrated hypermethylated CpG sites observed in the promoter region of TAPBP in obese and overweight subjects. These results confirmed by NEST cohort results revealed differentially methylated CpGs of TAPBP gene is related to the maternal pre-pregnancy obesity [28]. In vitro experiments revealed higher methylation levels of TAPBP, such as those found in above-mentioned studies might decrease tapsin via reduced transcriptional activity, leading to impaired immune responses and lower CD8 + T-cell responses [43,44,45]. In mice, tapsin is activated by the cytokines like IFN-γ and IFN-β, and to a lesser extent, TNF-α [45].
These results were very thought-provoking and cited several times by others and unlocked doors to the diagnosis of pathophysiology and new treatments.
TAPBP is linked to both mood disorders and obesity through the JNK pathway. This pathway plays a vital role in the inflammatory response and oxidative stress [43]. Briefly, stress-induced JNK activation occurs in the adipose and liver tissue of obese mice, whether obesity is induced by a high-fat diet or genetically through leptin deficiency (obese/obese mice). Insulin resistance in obese mice through ER stress-mediated JNK pathway is induced by the phosphorylation of insulin receptor substrate 1 (IRS1), which impairs insulin action and causes insulin resistance [44].
Interestingly, in the different tissues of obese subjects, inflammatory factors can be observed to cause continuous activation of JNK. The activated JNK acts on nuclear factor-κB (NF-κB) and activator protein-1 (AP-1) to produce more inflammatory factors, further reducing the sensitivity of insulin target cells toward insulin, finally forming a vicious circle and aggravating insulin resistance. Moreover, a network framed by PPARγ, NF-κB, and PTP1B signaling pathways crossing with the JNK signaling pathway plays a crucial role in regulating insulin resistance [39].
We assumed that a better understanding of the JNK signaling pathway and its relationship with PPARγ, NF-κB, PTP1B signaling pathways are necessary for a new drug targeting the treatment of obesity and mood disorders [39].
SORBS2: The role of SORBS2 gene in obesity and mood disorders has been discovered recently by different genome-wide methylation studies [30]. This gene located on the 4q35. 1 encodes the Arg protein tyrosine kinase binding protein 2 (ArgBP2). SORBS2 is an RNA-binding protein, which is involved in the regulation of RNA metabolism [46]. SORBS2 is involved in several biological pathways (Table 2). Sorbin, the product of SORBS2, is an ArgBP2 protein and SH3 domain-containing protein 2 and might be involved in insulin-mediated translocation of GLUT4 and thereby might affect energy storage [47]. Previous research has highlighted the role of this functional protein in disease states [48,49,50,51]. Downregulation of this gene was reported to be associated with mood disorders [52]. Linear regression analyses revealed a strong association of methylation with BMI for SORBS2 in abdominal omental visceral adipose tissue [53]. There is enough data to provide functional evidence that promoter methylation in SORBS2 directly influences gene activity and thus contributes to the abiogenesis. We suggest that SORBS2 is related to obesity through the innate immunity and inflammation response by the Notch signaling pathway that plays a major role in adipogenic differentiation [54]. Increased Notch signaling in mice blocked the expansion of white adipose tissue, ectopic fat accumulation, and insulin resistance [55].
The genetic deletion of Sorbin in mice leads to mood disorders by a reduction in the average number of spines per dendrite [49]. Additionally, to the grapevine, SORBS2 is related to mood disorders through two different pathways; actin-related proteins and the Notch signaling pathway [56,57]. Notch signaling is important in regulating neural cell proliferation, differentiation, and neural cellular growth, and is considered as a contributor in adaptive and innate immune responses. Active Notch signaling has been observed under a variety of inflammatory conditions such as atherosclerosis [55,58]. Interestingly, prototypical proinflammatory cytokines positively regulate Notch signaling and its target gene expression. For example, TNF induces expression of Notch1, Notch4 [59]. In addition, IL-1β induces Notch target genes, and Interferon-γ (IFNγ) functions as a negative regulator of Notch pathway activation [60].
BDNF: This gene is located in the 11p14.1 and encodes a member of the nerve growth factor family of proteins [61]. Alternative splicing results in multiple transcripts, at least one of which encodes a preproprotein that is proteolytically processed to generate the mature protein. Binding of this protein to its cognate receptor promotes neuronal survival in the adult brain. BDNF gene structure is complex, regulated by nine functional promoters. Each promoter regulates the expression of this gene [62]. BDNF encompasses several biological pathways (Table 2) and has a complex regulation; the exact roles of BDNF and its transcripts are not fully understood. BDNF insufficiency or missense mutations in its receptor, TrkB, are associated with weight gain and obesity in humans and mouse models [63,64]. In line with these observations, both exogenous BDNF administration and BDNF gene transfer in mouse model support the concept of the BDNF deficit in the brain induces a metabotropic impairment leading to obesity. Essentially, it has been established that the hypothalamic reduction of BDNF modulates energy homeostasis affecting food intake and promoting an anorectic signal [65].
There are several pieces of evidence about the role of BDNF in brain function and mood disorders [66,67,68]. Previous studies indicated that the positive correlation between brain and circulating BDNF suggests that BDNF levels in the blood reflect the levels occurring in the central nervous system. Thus, circulating BDNF has been proposed as a potential biomarker for neuropsychiatric disorders and neurodegenerative diseases [69,70,71,72,73,74].
BDNF is one of the major neurotrophic factors, plays an important role in the maintenance and survival of neurons and in synaptic plasticity. Several lines of evidence suggest that BDNF is involved in depression and plays an important role in the maintenance and survival of neurons and in synaptic plasticity. Recent documents demonstrated that the expression of BDNF is decreased in depressed patients [75]. BDNF has a multifaceted role from its neurotrophic activity to inflammation, metabolism, and cardiovascular diseases [76,77,78].
Methylation of the BDNF gene was analyzed at CpG sites in upstream of exon I. It is also possible that the hypomethylation promotor is located in exon I, which could cause altered BDNF expression, leading to abnormal eating behaviors [35,79]. Gardner et al. displayed different methylation in the promoter of BDNF related to obesity [35]. Interestingly, three of the obesity-associated CpGs were located within two of the numerous promoters of BDNF, and differential BDNF transcripts are expressed at different time points and in different cellular compartments [80,81]. Carriers of the risk allele at rs10767664 had higher methylation in the pII promoter of BDNF, and lower methylation in the pVI promoter of BDNF [31]. Januar et al. have revealed that late-life depression is associated with elevated BDNF methylation of specific CpG sites within promoters I and IV, with all associations remaining after adjustment for a range of covariates [33].
Furthermore, recent studies reported an increased BDNF methylation is associated with depression in animal models [82] and in humans [83]. Decreased BDNF may relate to the reduced function of the BDNF gene in promoting neural growth and repair in depression. Thus, among depressive patients, those with a higher BDNF methylation status are at a greater risk of suicidal behavior [84]. Hypermethylation in Exon I, in the promoter region, reduced BDNF levels in the plasma and post-mortem hippocampus of depressed individuals [85,86,87,88]. Another post-stroke cohort indicated that higher BDNF promoter methylation status was independently associated with depressive symptoms over one year after the onset of stroke, although not associated with baseline depressive symptom severity [84,89,90]. The methylation state of CpG sites within mouse promoter/exon IV is correlated with the expression of BDNF in the developing mouse forebrain, and similar associations were found with chronic depression, and these effects were not driven by antidepressant treatment [69]. For example, Jin et al. using the Sequenom Mass Array platform, demonstrated in mice model that fluoxetine can downregulate the expression of BDNF by the methylation of 11 CpG sites in promoter IV [91].
Strangely, BDNF has leading biological roles in inflammation and apoptosis; consequently, it is a crucial neurotrophic factor for preserving normal nervous system function. Moreover, BDNF is an associated member of the neurotrophic factor family that is mainly secreted by neuron or glial cells [92].
Sources of chronic inflammation or non-resolving inflammation may originate from either pathophysiological (e.g., inflammatory diseases, immune-based disorders, T cell dysfunction) or non-pathological conditions, including aging and obesity. Interestingly, BDNF has main biological roles in inflammation and apoptosis; thus, it is a crucial neurotrophic factor for preserving normal nervous system function [92].
Additionally, BDNF has a multifaceted role from its neurotrophic activity to inflammation, metabolism, and cardiovascular diseases. BDNF is considered as a potential modulator/mediator with anti-inflammatory effects [86].
BDNF-related neuroprotective effects are elicited by activation of extracellular signal-related kinase (ERK) and phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT)-signaling pathways. Production of inflammatory cytokines can regulate by complex signaling pathways, especially nuclear factor-κb (NF-κB) and inflammatory response signal pathway (BDNF-TrkB-MEK-ERK-NF-κB pathway) [93,94,95].
5. Limitation
This study strengthens the novel findings related to the overlap genes in obesity and mood disorders but is limited in accessing row epigenome data to do gene enrichment analysis. None of the authors of the included studies were interested in responding to our inquiry to share their raw data to do a meta-analysis.
6. Conclusions
While mechanisms linking both obesity and mood disorders to epigenetic response are still unknown, it is well-known that chronic inflammation induces a novel epigenetic program. As the results of gene enrichment pathways analysis exhibited that TAPBP, BDNF, and SRBP2 are related together by inflammatory pathways, we hypothesis that hypermethylation in these genes might play a crucial role in the co-occurrence of obesity and mood disorders due to the inflammation process. Our results shed light on our understanding of such associations. Future studies should focus on the molecular pathophysiology of these disorders in the hope of opening new approaches for target treatment.
Author Contributions
M.G.: conceived of the presented idea, supervised the findings, contributed data and analysis tools, drafting, proof reading, perform gene enrichment analysis, M.B.: conceived of the presented idea, and proofreading, J.S.: conceived of the presented idea, N.E.: collected the data, reviewing title and abstract, L.S.: Pathway searching, rechecking references, rechecking black and down score, M.D.: reviewing title and abstract, performed the analysis, N.S.: conceived of the presented idea, S.J.: Conducting and supervising meta-analysis and systematic review, proofreading, editing. All authors contributed to the final version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Abbreviations
| ArgBP2 | Arg/c-Abl kinase binding protein 2 |
| BMI | body mass index |
| DMR | differentiated methylated regions |
| ER | endoplasmic reticulum |
| EWAS | Epigenome wide association study |
| IRS1 | insulin receptor substrate 1 |
| MDD | Major Depressive Disorder |
| MHC | major histocompatibility complex |
| OVAT | omental visceral adipose tissue |
| PRISMA | Preferred Reporting Items for Systematic Reviews and Meta-Analyses |
| PROSPERO | Prospective Register of Systematic Reviews |
| SOHOs | orbin homology |
| TAP | transporter associated with antigen processing |
References
- Popkin, B.M.; Adair, L.S.; Ng, S.W. Global nutrition transition and the pandemic of obesity in developing countries. Nutr. Rev. 2012, 70, 3–21. [Google Scholar] [CrossRef] [PubMed]
- Kelly, T.; Yang, W.; Chen, C.-S.; Reynolds, K.; He, J. Global burden of obesity in 2005 and projections to 2030. Int. J. Obes. 2008, 32, 1431–1437. [Google Scholar] [CrossRef] [PubMed]
- Kessler, R.C.; Aguilar-Gaxiola, S.; Alonso, J.; Chatterji, S.; Lee, S.; Ormel, J.; Üstün, T.B.; Wang, P.S. The global burden of mental disorders: An update from the WHO World Mental Health (WMH) surveys. Epidemiologia e Psichiatr. Soc. 2009, 18, 23–33. [Google Scholar] [CrossRef] [PubMed]
- WHO. Obesity and Overweight; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Veisani, Y.; Mohamadian, F.; Delpisheh, A. Prevalence and comorbidity of common mental disorders and associations with suicidal ideation in the adult population. Epidemiol. Heal. 2017, 39, e2017031. [Google Scholar] [CrossRef] [PubMed]
- Palou, A.; Serra, F.; Bonet, M.; Picó, C. Obesity: Molecular bases of a multifactorial problem. Eur. J. Nutr. 2000, 39, 127–144. [Google Scholar] [CrossRef] [PubMed]
- Akiskal, H.S. New insights into the nature and heterogeneity of mood disorders. J. Clin. Psychiatry 1989, 50, 50. [Google Scholar]
- Morris, W.N.; Reilly, N.P. Toward the self-regulation of mood: Theory and research. Motiv. Emot. 1987, 11, 215–249. [Google Scholar] [CrossRef]
- Afari, N.; Noonan, C.; Goldberg, J.; Roy-Byrne, P.; Schur, E.; Golnari, G.; Buchwald, D. Depression and obesity: Do shared genes explain the relationship? Depression Anxiety 2010, 27, 799–806. [Google Scholar] [CrossRef]
- Khan, A.; Schwartz, K.A.; Kolts, R.L.; Brown, W.A. BMI, sex, and antidepressant response. J. Affect. Disord. 2007, 99, 101–106. [Google Scholar] [CrossRef]
- Hinze-Selch, D.; Schuld, A.; Kraus, T.; Kühn, M.; Uhr, M.; Haack, M.; Pollmacher, T. Effects of Antidepressants on Weight and on the Plasma Levels of Leptin, TNF-α and Soluble TNF Receptors A Longitudinal Study in Patients Treated with Amitriptyline or Paroxetine. Neuropsychopharmacol 2000, 23, 13–19. [Google Scholar] [CrossRef]
- Gafoor, R.; Booth, H.P.; Gulliford, M.C. Antidepressant utilisation and incidence of weight gain during 10 years’ follow-up: Population based cohort study. BMJ 2018, 361, k1951. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, S.J.; EpiSCOPE, M.O.; Molloy, P.L.; Varinli, H.; Morrison, J.L.; Muhlhausler, B.S. Epigenetics and human obesity. Int. J. Obes. 2014, 39, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Fass, D.M.; Schroeder, F.A.; Perlis, R.H.; Haggarty, S.J. Epigenetic mechanisms in mood disorders: Targeting neuroplasticity. Neuroscience 2013, 264, 112–130. [Google Scholar] [CrossRef] [PubMed]
- Schones, D.E.; Leung, A.; Natarajan, R. Chromatin Modifications Associated with Diabetes and Obesity. Arter. Thromb. Vasc. Boil. 2015, 35, 1557–1561. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, K.; Molina-Márquez, A.M.; Saavedra, N.; Zambrano, T.; Salazar, L.A. Epigenetic Modifications of Major Depressive Disorder. Int. J. Mol. Sci. 2016, 17, 1279. [Google Scholar] [CrossRef]
- Alegría-Torres, J.A.; Baccarelli, A.A.; Bollati, V. Epigenetics and lifestyle. Epigenomics 2011, 3, 267–277. [Google Scholar] [CrossRef]
- Attwood, J.; Yung, R.; Richardson, B. DNA methylation and the regulation of gene transcription. Cell. Mol. Life Sci. 2002, 59, 241–257. [Google Scholar] [CrossRef]
- Kouzmenko, A.; Ohtake, F.; Fujiki, R.; Kato, S. Hormonal gene regulation through DNA methylation and demethylation. Epigenomics 2010, 2, 765–774. [Google Scholar] [CrossRef]
- Sayols-Baixeras, S.; Subirana, I.; Fernández-Sanlés, A.; Sentí, M.; Lluís-Ganella, C.; Marrugat, J.; Elosua, R. DNA methylation and obesity traits: An epigenome-wide association study. The REGICOR study. Epigenetics 2017, 12, 909–916. [Google Scholar] [CrossRef]
- Sonne, S.B.; Yadav, R.; Yin, G.; Dalgaard, M.D.; Myrmel, L.S.; Gupta, R.; Wang, J.; Madsen, L.; Kajimura, S.; Kristiansen, K. Obesity is associated with depot-specific alterations in adipocyte DNA methylation and gene expression. Adipocyte 2017, 6, 124–133. [Google Scholar] [CrossRef]
- De Mello, V.D.; Pulkkinen, L.; Lalli, M.; Kolehmainen, M.; Pihlajamäki, J.; Uusitupa, M. DNA methylation in obesity and type 2 diabetes. Ann. Med. 2014, 46, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Kader, F.; Ghai, M.; Maharaj, L. The effects of DNA methylation on human psychology. Behav. Brain Res. 2018, 346, 47–65. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Meng, L.; Pei, F.; Zheng, Y.; Leng, J. A review of DNA methylation in depression. J. Clin. Neurosci. 2017, 43, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Downs, S.H.; Black, N. The feasibility of creating a checklist for the assessment of the methodological quality both of randomised and non-randomised studies of health care interventions. J. Epidemiol. Community Heal. 1998, 52, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Monteiro, C.; Matos, A.; You, J.; Fraga, A.; Pereira, C.; Catalán, V.; Rodríguez, A.; Gómez-Ambrosi, J.; Frühbeck, G.; et al. Epigenome-wide DNA methylation profiling of periprostatic adipose tissue in prostate cancer patients with excess adiposity—A pilot study. Clin. Epigenetics 2018, 10, 54. [Google Scholar] [CrossRef]
- Murphy, T.M.; Crawford, B.; Dempster, E.L.; Hannon, E.; Burrage, J.; Turecki, G.; Kaminsky, Z.; Mill, J. Methylomic profiling of cortex samples from completed suicide cases implicates a role for PSORS1C3 in major depression and suicide. Transl. Psychiatry 2017, 7, e989. [Google Scholar] [CrossRef]
- Martin, C.L.; Jima, D.; Sharp, G.C.; McCullough, L.E.; Park, S.S.; Gowdy, K.M.; Skaar, D.; Cowley, M.; Maguire, R.L.; Fuemmeler, B.; et al. Maternal pre-pregnancy obesity, offspring cord blood DNA methylation, and offspring cardiometabolic health in early childhood: An epigenome-wide association study. Epigenetics 2019, 14, 325–340. [Google Scholar] [CrossRef]
- Rhee, J.K.; Lee, J.H.; Yang, H.K.; Kim, T.M.; Yoon, K.H. DNA Methylation profiles of blood cells are distinct between early-onset obese and control individuals. Genom. Inform. 2017, 15, 28. [Google Scholar] [CrossRef]
- Zhu, Y.; Strachan, E.; Fowler, E.; Bacus, T.; Roy-Byrne, P.; Zhao, J. Genome-Wide profiling of DNA methylome and transcriptome in peripheral blood monocytes for major depression: A Monozygotic Discordant Twin Study. Transl. Psychiatry 2019, 9, 215. [Google Scholar] [CrossRef]
- Keller, S.; Sarchiapone, M.; Zarrilli, F.; Videtic, A.; Ferraro, A.; Carli, V.; Sacchetti, S.; Lembo, F.; Angiolillo, A.; Jovanović, N.; et al. Increased BDNF Promoter Methylation in the Wernicke Area of Suicide Subjects. Arch. Gen. Psychiatry 2010, 67, 258–267. [Google Scholar] [CrossRef]
- Perroud, N.; Salzmann, A.; Prada, P.; Nicastro, R.; Hoeppli, M.E.; Furrer, S.; Ardu, S.; Krejci, I.; Karege, F.; Malafosse, A. Response to psychotherapy in borderline personality disorder and methylation status of the BDNF gene. Transl. Psychiatry 2013, 3, e207. [Google Scholar] [CrossRef] [PubMed]
- Januar, V.; Ancelin, M.-L.; Ritchie, K.; Saffery, R.; Ryan, J. BDNF promoter methylation and genetic variation in late-life depression. Transl. Psychiatry 2015, 5, e619. [Google Scholar] [CrossRef] [PubMed]
- Voisin, S.; Almén, M.S.; Zheleznyakova, G.Y.; Lundberg, L.; Zarei, S.; Castillo, S.; Eriksson, F.E.; Nilsson, E.K.; Blüher, M.; Böttcher, Y.; et al. Many obesity-associated SNPs strongly associate with DNA methylation changes at proximal promoters and enhancers. Genome Med. 2015, 7, 1–6. [Google Scholar]
- Gardner, K.; Sapienza, C.; Fisher, J.J.P. Genetic and epigenetic associations to obesity-related appetite phenotypes among A frican–A merican children. Pediatric Obesity 2015, 10, 476–482. [Google Scholar] [CrossRef]
- Chapman, D.P.; Perry, G.S. Peer reviewed: Depression as a major component of public health for older adults. Prev. Chron. Dis. 2008, 5, 1. [Google Scholar]
- Nigatu, Y.T.; Reijneveld, S.A.; De Jonge, P.; Van Rossum, E.; Bültmann, U. The Combined Effects of Obesity, Abdominal Obesity and Major Depression/Anxiety on Health-Related Quality of Life: The LifeLines Cohort Study. PLoS ONE 2016, 11, e0148871. [Google Scholar] [CrossRef]
- Chen, K.-W.; Chen, L. Epigenetic Regulation of BDNF Gene during Development and Diseases. Int. J. Mol. Sci. 2017, 18, 571. [Google Scholar] [CrossRef]
- Mayer, W.E.; Klein, J. Is tapasin a modified Mhc class I molecule? Immunogenetics 2001, 53, 719–723. [Google Scholar] [CrossRef]
- Herberg, J.A.; Sgouros, J.; Jones, T.; Copeman, J.; Humphray, S.J.; Sheer, D. Genomic analysis of the Tapasin gene, located close to the TAP loci in the MHC. Eur. J. Immunol. 1998, 28, 459–467. [Google Scholar] [CrossRef]
- Montfort, A.; Martin, P.G.; Levade, T.; Benoist, H.; Ségui, B. FAN (factor associated with neutral sphingomyelinase activation), a moonlighting protein in TNF-R1 signaling. J. Leukoc. Boil. 2010, 88, 897–903. [Google Scholar] [CrossRef]
- Cui, D.; Wang, J.; Zeng, Y.; Rao, L.; Chen, H.; Li, W.; Li, Y.; Li, H.; Cui, C.; Xiao, L. Generating hESCs with reduced immunogenicity by disrupting TAP1 or TAPBP. Biosci. Biotechnol. Biochem. 2016, 80, 1484–1491. [Google Scholar] [CrossRef] [PubMed]
- Grootjans, J.; Kaser, A.; Kaufman, R.J.; Blumberg, R.S. The unfolded protein response in immunity and inflammation. Nat. Rev. Immunol. 2016, 16, 469–484. [Google Scholar] [CrossRef] [PubMed]
- Ozcan, U. Endoplasmic Reticulum Stress Links Obesity, Insulin Action, and Type 2 Diabetes. Science 2004, 306, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Abarca-Heidemann, K.; Friederichs, S.; Klamp, T.; Boehm, U.; Guethlein, L.A.; Ortmann, B. Regulation of the expression of mouse TAP-associated glycoprotein (tapasin) by cytokines. Immunol. Lett. 2002, 83, 197–207. [Google Scholar] [CrossRef]
- Lukong, K.E.; Chang, K.-W.; Khandjian, E.W.; Richard, S. RNA-Binding proteins in human genetic disease. Trends Genet. 2008, 24, 416–425. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, T.; Tanaka, S.; Ishii, A.; Watanabe, M.; Fujitani, N.; Sugeo, A. A brain-specific Grb2-associated regulator of extracellular signal-regulated kinase (Erk)/mitogen-activated protein kinase (MAPK)(GAREM) subtype, GAREM2, contributes to neurite outgrowth of neuroblastoma cells by regulating Erk signaling. J. Cell Sci. 2013, 288, 29934. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Bei, Y.; Shen, S.; Huang, P.; Shi, J.; Zhang, J.; Sun, Q.; Chen, Y.; Yang, Y.; Xu, T.; et al. miR-21-3p controls sepsis-associated cardiac dysfunction via regulating SORBS2. J. Mol. Cell. Cardiol. 2016, 94, 43–53. [Google Scholar] [CrossRef]
- Zhang, Q.; Gao, X.; Li, C.; Feliciano, C.; Wang, N.; Zhou, D.; Mei, Y.; Monteiro, P.; Anand, M.; Itohara, S.; et al. Impaired Dendritic Development and Memory in Sorbs2 Knock-Out Mice. J. Neurosci. 2016, 36, 2247–2260. [Google Scholar] [CrossRef]
- Lu, P.; Qiao, J.; He, W.; Wang, J.; Jia, Y.; Sun, Y.; Tang, S.; Fu, L.; Qin, Y. Genome-Wide Gene Expression Profile Analyses Identify CTTN as a Potential Prognostic Marker in Esophageal Cancer. PLoS ONE 2014, 9, e88918. [Google Scholar] [CrossRef]
- Cheli, S.; Francois, S.; Bodega, B.; Ferrari, F.; Tenedini, E.; Roncaglia, E.; Ferrari, S.; Ginelli, E.; Meneveri, R. Expression Profiling of FSHD-1 and FSHD-2 Cells during Myogenic Differentiation Evidences Common and Distinctive Gene Dysregulation Patterns. PLoS ONE 2011, 6, e20966. [Google Scholar] [CrossRef]
- McCarthy, M.J.; Leckband, S.G.; Kelsoe, J.R. Pharmacogenetics of lithium response in bipolar disorder. Pharmacogenomics 2010, 11, 1439–1465. [Google Scholar] [CrossRef] [PubMed]
- Keller, M.; Hopp, L.; Liu, X.; Wohland, T.; Rohde, K.; Cancello, R.; Klös, M.; Bacos, K.; Kern, M.; Eichelmann, F.; et al. Genome-wide DNA promoter methylation and transcriptome analysis in human adipose tissue unravels novel candidate genes for obesity. Mol. Metab. 2017, 6, 86–100. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Lorente, R.; Bejar, M.T.; Badimon, L. Notch Signaling Pathway Activation in Normal and Hyperglycemic Rats Differs in the Stem Cells of Visceral and Subcutaneous Adipose Tissue. Stem Cells Dev. 2014, 23, 3034–3048. [Google Scholar] [CrossRef] [PubMed]
- Fung, E.; Tang, S.-M.T.; Canner, J.P.; Morishige, K.; Arboleda-Velasquez, J.F.; Cardoso, A.A.; Carlesso, N.; Aster, J.C.; Aikawa, E. Delta-Like 4 Induces Notch Signaling in Macrophages. Circulation 2007, 115, 2948–2956. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Zhao, B.; Deng, Y.; Shangguan, S.; Zhou, F.; Zhou, W.; Li, X.; Li, Y.; Chen, G. Notch signaling in cerebrovascular diseases (Review). Mol. Med. Rep. 2016, 14, 2883–2898. [Google Scholar] [CrossRef] [PubMed]
- Ahearn, E.P.; Speer, M.C.; Chen, Y.; Steffens, D.C.; Cassidy, F.; Van Meter, S.; Provenzale, J.M.; Weisler, R.H.; Krishnan, K.R.R. Investigation of Notch3 as a candidate gene for bipolar disorder using brain hyperintensities as an endophenotype. Am. J. Med. Genet. 2002, 114, 652–658. [Google Scholar] [CrossRef]
- Aoyama, T.; Takeshita, K.; Kikuchi, R.; Yamamoto, K.; Cheng, X.W.; Liao, J.K.; Murohara, T. γ-Secretase inhibitor reduces diet-induced atherosclerosis in apolipoprotein E-deficient mice. Biochem. Biophys. Res. Commun. 2009, 383, 216–221. [Google Scholar] [CrossRef]
- Ando, K.; Kanazawa, S.; Tetsuka, T.; Ohta, S.; Jiang, X.; Tada, T.; Kobayashi, M.; Matsui, N.; Okamoto, T. Induction of Notch signaling by tumor necrosis factor in rheumatoid synovial fibroblasts. Oncogene 2003, 22, 7796–7803. [Google Scholar] [CrossRef]
- Hu, X.; Chung, A.Y.; Wu, I.; Foldi, J.; Chen, J.; Ji, J.D.; Tateya, T.; Kang, Y.J.; Han, J.; Gessler, M.; et al. Integrated Regulation of Toll-like Receptor Responses by Notch and Interferon-γ Pathways. Immunity 2008, 29, 691–703. [Google Scholar] [CrossRef]
- Hanson, I.M.; Seawright, A.; Van Heyningen, V. The human BDNF gene maps between FSHB and HVBS1 at the boundary of 11p13–p14. Genomics 1992, 13, 1331–1333. [Google Scholar] [CrossRef]
- Pruunsild, P.; Kazantseva, A.; Aid, T.; Palm, K.; Timmusk, T. Dissecting the human BDNF locus: Bidirectional transcription, complex splicing, and multiple promoters. Genomics 2007, 90, 397–406. [Google Scholar] [CrossRef]
- Gray, J.; Yeo, G.S.H.; Cox, J.J.; Morton, J.; Adlam, A.; Keogh, J.M.; A Yanovski, J.; El Gharbawy, A.; Han, J.C.; Tung, Y.L.; et al. Hyperphagia, severe obesity, impaired cognitive function, and hyperactivity associated with functional loss of one copy of the brain-derived neurotrophic factor (BDNF) gene. Diabetes 2006, 55, 3366–3371. [Google Scholar] [CrossRef] [PubMed]
- Bonaccorso, S.; Sodhi, M.; Li, J.; Bobo, W.V.; Chen, Y.; Tumuklu, M.; Theleritis, C.; Jayathilake, K.; Meltzer, H.Y. The brain-derived neurotrophic factor (BDNF) Val66Met polymorphism is associated with increased body mass index and insulin resistance measures in bipolar disorder and schizophrenia. Bipolar. Disord. 2015, 17, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Stanley, S.; Wynne, K.; McGowan, B.; Bloom, S.R. Hormonal Regulation of Food Intake. Physiol. Rev. 2005, 85, 1131–1158. [Google Scholar] [CrossRef] [PubMed]
- Ray, M.T.; Weickert, C.S.; Wyatt, E.; Webster, M.J. Decreased BDNF, trkB-TK+ and GAD67 mRNA expression in the hippocampus of individuals with schizophrenia and mood disorders. J. Psychiatry Neurosci. 2011, 36, 195–203. [Google Scholar] [CrossRef]
- Martinowich, K.; Manji, H.; Lu, B. New insights into BDNF function in depression and anxiety. Nat. Neurosci. 2007, 10, 1089–1093. [Google Scholar] [CrossRef]
- Jin, Y.; Sun, L.H.; Yang, W.; Cui, R.J.; Xu, S.B. The Role of BDNF in the Neuroimmune Axis Regulation of Mood Disorders. Front. Neurol. 2019, 10. [Google Scholar] [CrossRef]
- Polyakova, M.; Stuke, K.; Schuemberg, K.; Mueller, K.; Schoenknecht, P.; Schroeter, M.L. BDNF as a biomarker for successful treatment of mood disorders: A systematic & quantitative meta-analysis. J. Affect. Disord. 2015, 174, 432–440. [Google Scholar] [CrossRef]
- Fernandes, B.S.; Molendijk, M.L.; Köhler, C.A.; Soares, J.C.; Leite, C.M.G.S.; Machado-Vieira, R.; Ribeiro, T.L.; Silva, J.C.; Sales, P.M.G.; Quevedo, J.; et al. Peripheral brain-derived neurotrophic factor (BDNF) as a biomarker in bipolar disorder: A meta-analysis of 52 studies. BMC Med. 2015, 13, 289. [Google Scholar] [CrossRef]
- Sanada, K.; Zorrilla, I.; Iwata, Y.; Bermúdez-Ampudia, C.; Graff-Guerrero, A.; Martínez-Cengotitabengoa, M. The efficacy of non-pharmacological interventions on brain-derived neurotrophic factor in schizophrenia: A systematic review and meta-analysis. Int. J. Mol. Sci. 2016, 17, 1766. [Google Scholar] [CrossRef]
- Green, M.J.; Matheson, S.L.; Shepherd, A.; Weickert, C.S.; Carr, V.J. Brain-Derived neurotrophic factor levels in schizophrenia: A systematic review with meta-analysis. Mol. Psychiatry 2010, 16, 960–972. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Jin, Y.; Wang, J.; Weng, X.; Li, C. Serum brain-derived neurotrophic factor (BDNF) levels in schizophrenia: A systematic review. Shanghai Arch. Psychiatry 2012, 24, 250–261. [Google Scholar] [PubMed]
- Fernandes, B.S.; Steiner, J.; Berk, M.; Molendijk, M.L.; Gonzàlez-Pinto, A.; Turck, C.W.; Nardin, P.; Gonçalves, C.-A. Peripheral brain-derived neurotrophic factor in schizophrenia and the role of antipsychotics: Meta-Analysis and implications. Mol. Psychiatry 2014, 20, 1108–1119. [Google Scholar] [CrossRef] [PubMed]
- Papathanassoglou, E.D.; Miltiadous, P.; Karanikola, M.N. May BDNF Be Implicated in the Exercise-Mediated Regulation of Inflammation? Critical Review and Synthesis of Evidence. Boil. Res. Nurs. 2014, 17, 521–539. [Google Scholar] [CrossRef]
- Zhang, J.-C.; Yao, W.; Hashimoto, K. Brain-Derived Neurotrophic Factor (BDNF)-TrkB Signaling in Inflammation-related Depression and Potential Therapeutic Targets. Curr. Neuropharmacol. 2016, 14, 721–731. [Google Scholar] [CrossRef]
- Chaldakov, G.N.; Fiore, M.; Stankulov, I.S.; Manni, L.; Hristova, M.G.; Antonelli, A.; Ghenev, P.I.; Aloe, L. Neurotrophin presence in human coronary atherosclerosis and metabolic syndrome: A role for NGF and BDNF in cardiovascular disease? In Progress in Brain Research; Elsevier BV: Amsterdam, The Netherlands, 2004; Volume 146, pp. 279–289. [Google Scholar]
- Sandrini, L.; Di Minno, G.; Amadio, P.; Ieraci, A.; Tremoli, E.; Barbieri, S.S. Association between Obesity and Circulating Brain-Derived Neurotrophic Factor (BDNF) Levels: Systematic Review of Literature and Meta-Analysis. Int. J. Mol. Sci. 2018, 19, 2281. [Google Scholar] [CrossRef]
- Silviera, M.L.; Smith, B.P.; Powell, J.; Sapienza, C. Epigenetic differences in normal colon mucosa of cancer patients suggest altered dietary metabolic pathways. Cancer Prev. Res. 2012, 5, 374–384. [Google Scholar] [CrossRef]
- Marosi, K.; Mattson, M.P. BDNF mediates adaptive brain and body responses to energetic challenges. Trends Endocrinol. Metab. 2014, 25, 89–98. [Google Scholar] [CrossRef]
- Martínez-Levy, G.A.; Cruz-Fuentes, C.S. Genetic and Epigenetic Regulation of the Brain-Derived Neurotrophic Factor in the Central Nervous System. Yale J. Boil. Med. 2014, 87, 173–186. [Google Scholar]
- Tsankova, N.M.; Berton, O.; Renthal, W.; Kumar, A.; Neve, R.L.; Nestler, E.J. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat. Neurosci. 2006, 9, 519–525. [Google Scholar] [CrossRef]
- Fuchikami, M.; Morinobu, S.; Segawa, M.; Okamoto, Y.; Yamawaki, S.; Ozaki, N.; Inoue, T.; Kusumi, I.; Koyama, T.; Tsuchiyama, K.; et al. DNA Methylation Profiles of the Brain-Derived Neurotrophic Factor (BDNF) Gene as a Potent Diagnostic Biomarker in Major Depression. PLoS ONE 2011, 6, e23881. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.-J.; Kim, J.-M.; Lee, J.-Y.; Kim, S.-Y.; Bae, K.-Y.; Kim, S.-W.; Shin, I.-S.; Kim, H.-R.; Shin, M.-G.; Yoon, J.-S. BDNF promoter methylation and suicidal behavior in depressive patients. J. Affect. Disord. 2013, 151, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Chiavetto, L.B.; Bagnardi, V.; Zanardini, R.; Molteni, R.; Nielsen, M.G.; Placentino, A.; Giovannini, C.; Rillosi, L.; Ventriglia, M.; Riva, M.A.; et al. Serum and plasma BDNF levels in major depression: A replication study and meta-analyses. World J. Boil. Psychiatry 2010, 11, 763–773. [Google Scholar] [CrossRef] [PubMed]
- Kurita, M.; Nishino, S.; Kato, M.; Numata, Y.; Sato, T. Plasma Brain-Derived Neurotrophic Factor Levels Predict the Clinical Outcome of Depression Treatment in a Naturalistic Study. PLoS ONE 2012, 7, e39212. [Google Scholar] [CrossRef]
- Lee, B.-H.; Kim, H.; Park, S.-H.; Kim, Y.-K. Decreased plasma BDNF level in depressive patients. J. Affect. Disord. 2007, 101, 239–244. [Google Scholar] [CrossRef]
- Tripp, A.; Oh, H.; Guilloux, J.-P.; Martinowich, K.; Lewis, D.A.; Sibille, E. Brain-Derived Neurotrophic Factor Signaling and Subgenual Anterior Cingulate Cortex Dysfunction in Major Depressive Disorder. Am. J. Psychiatry 2012, 169, 1194–1202. [Google Scholar] [CrossRef]
- Kim, J.-M.; Stewart, R.J.; Glozier, N.; Prince, M.; Kim, S.-W.; Yang, S.-J.; Shin, I.-S.; Yoon, J.-S. Physical health, depression and cognitive function as correlates of disability in an older Korean population. Int. J. Geriatr. Psychiatry 2005, 20, 160–167. [Google Scholar] [CrossRef]
- Kim, J.-M.; Stewart, R.J.; Kang, H.-J.; Kim, S.Y.; Kim, S.-W.; Shin, I.-S.; Park, M.-S.; Kim, H.-R.; Shin, M.-G.; Cho, K.-H.; et al. A longitudinal study of BDNF promoter methylation and genotype with poststroke depression. J. Affect. Disord. 2013, 149, 93–99. [Google Scholar] [CrossRef]
- Jin, H.-J.; Pei, L.; Li, Y.-N.; Zheng, H.; Yang, S.; Wan, Y.; Mao, L.; Xia, Y.-P.; He, Q.-W.; Li, M.; et al. Alleviative effects of fluoxetine on depressive-like behaviors by epigenetic regulation of BDNF gene transcription in mouse model of post-stroke depression. Sci. Rep. 2017, 7, 14926. [Google Scholar] [CrossRef]
- Bathina, S.; Das, U.N. Brain-Derived neurotrophic factor and its clinical implications. Arch. Med. Sci. 2015, 11, 1164–1178. [Google Scholar] [CrossRef]
- Crowder, R.J.; Freeman, R.S. Phosphatidylinositol 3-Kinase and Akt Protein Kinase Are Necessary and Sufficient for the Survival of Nerve Growth Factor-Dependent Sympathetic Neurons. J. Neurosci. 1998, 18, 2933–2943. [Google Scholar] [CrossRef] [PubMed]
- Han, B.H.; Holtzman, D.M. BDNF Protects the Neonatal Brain from Hypoxic-Ischemic Injury In Vivo via the ERK Pathway. J. Neurosci. 2000, 20, 5775–5781. [Google Scholar] [CrossRef] [PubMed]
- Makar, T.K.; Trisler, D.; Sura, K.; Sultana, S.; Patel, N.; Bever, C.T. Brain derived neurotrophic factor treatment reduces inflammation and apoptosis in experimental allergic encephalomyelitis. J. Neurol. Sci. 2008, 270, 70–76. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).