Inhibition of DNA Repair in Cancer Therapy: Toward a Multi-Target Approach
Abstract
1. Introduction
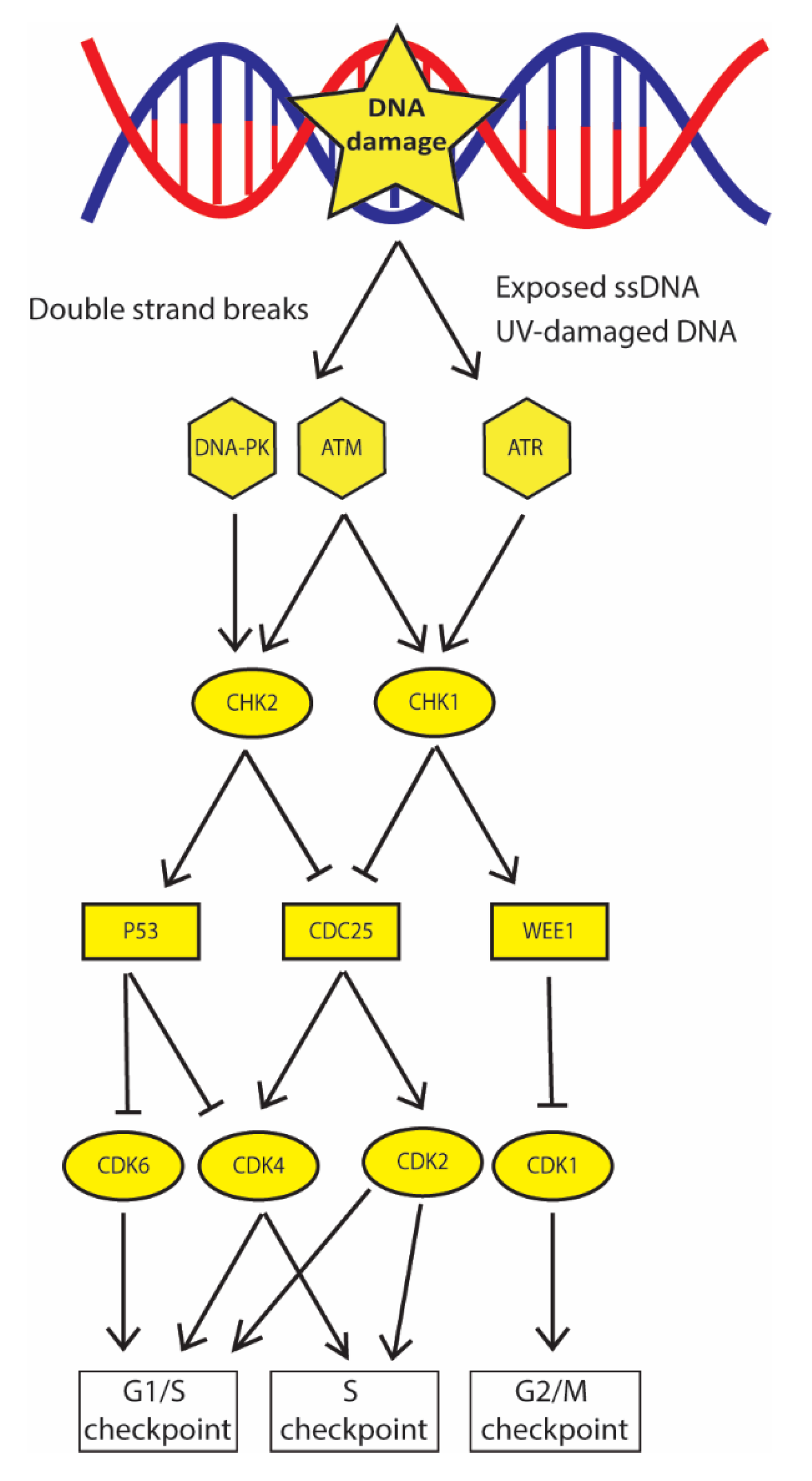
2. DNA Damage Checkpoint Promotes DNA Damage Recognition and Cell Cycle Arrest: The First Barrier to Cancer Progression
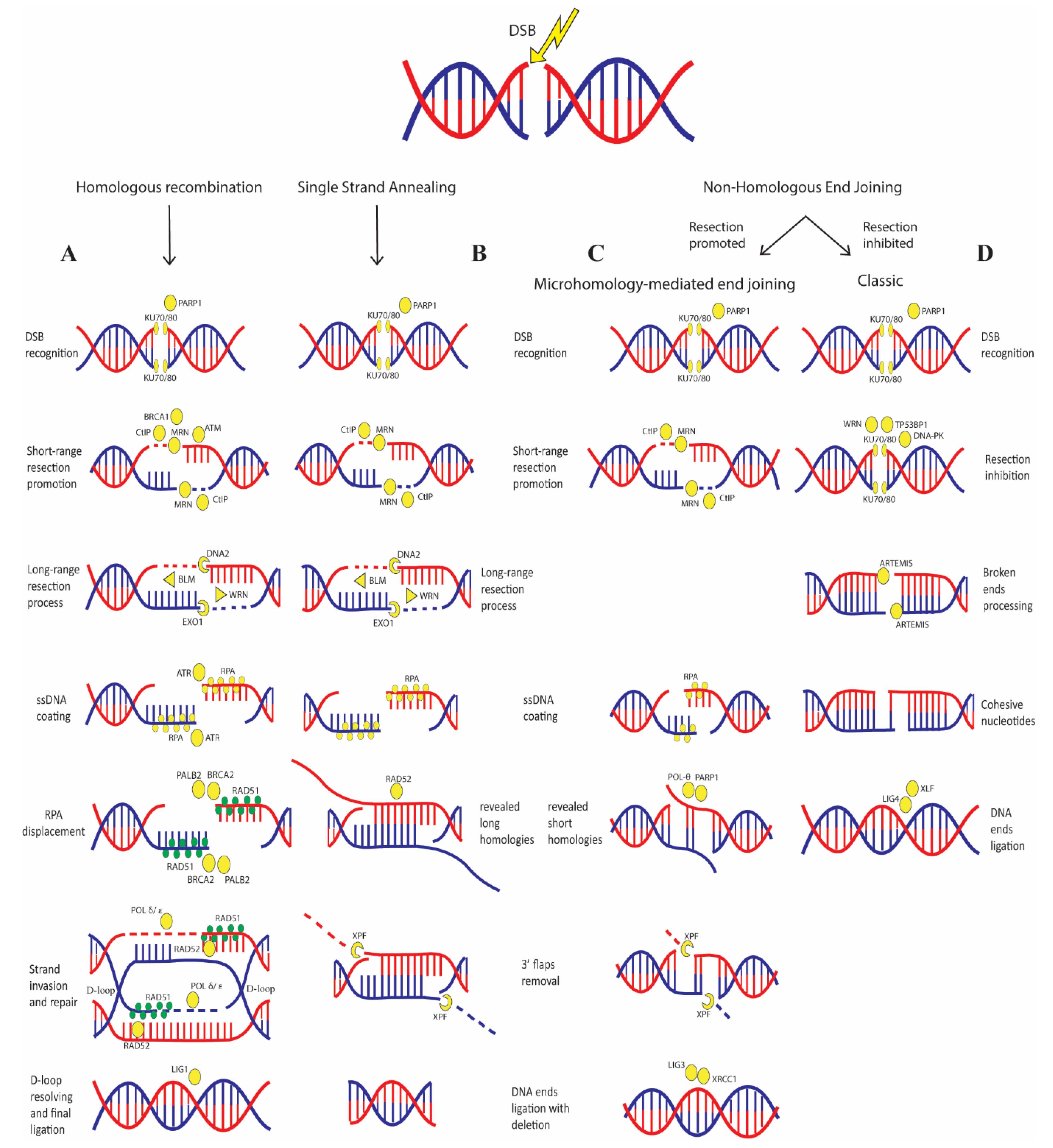
3. Defects in DSB Repair Greatly Promotes Cancer Development
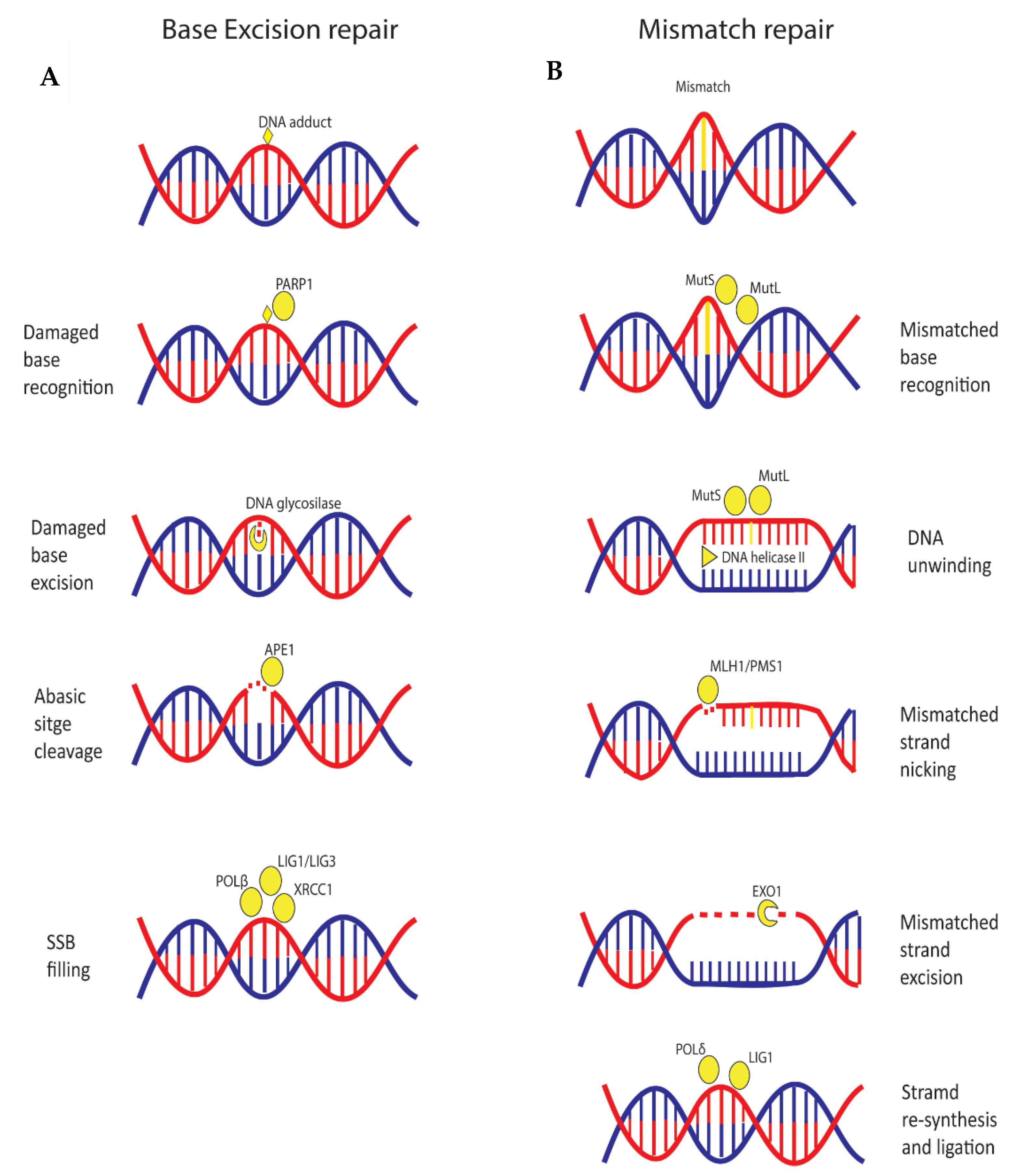
4. Single Base Variations and Mismatched Base Pairs Are Repaired by Specific Pathways Linked to Cancer
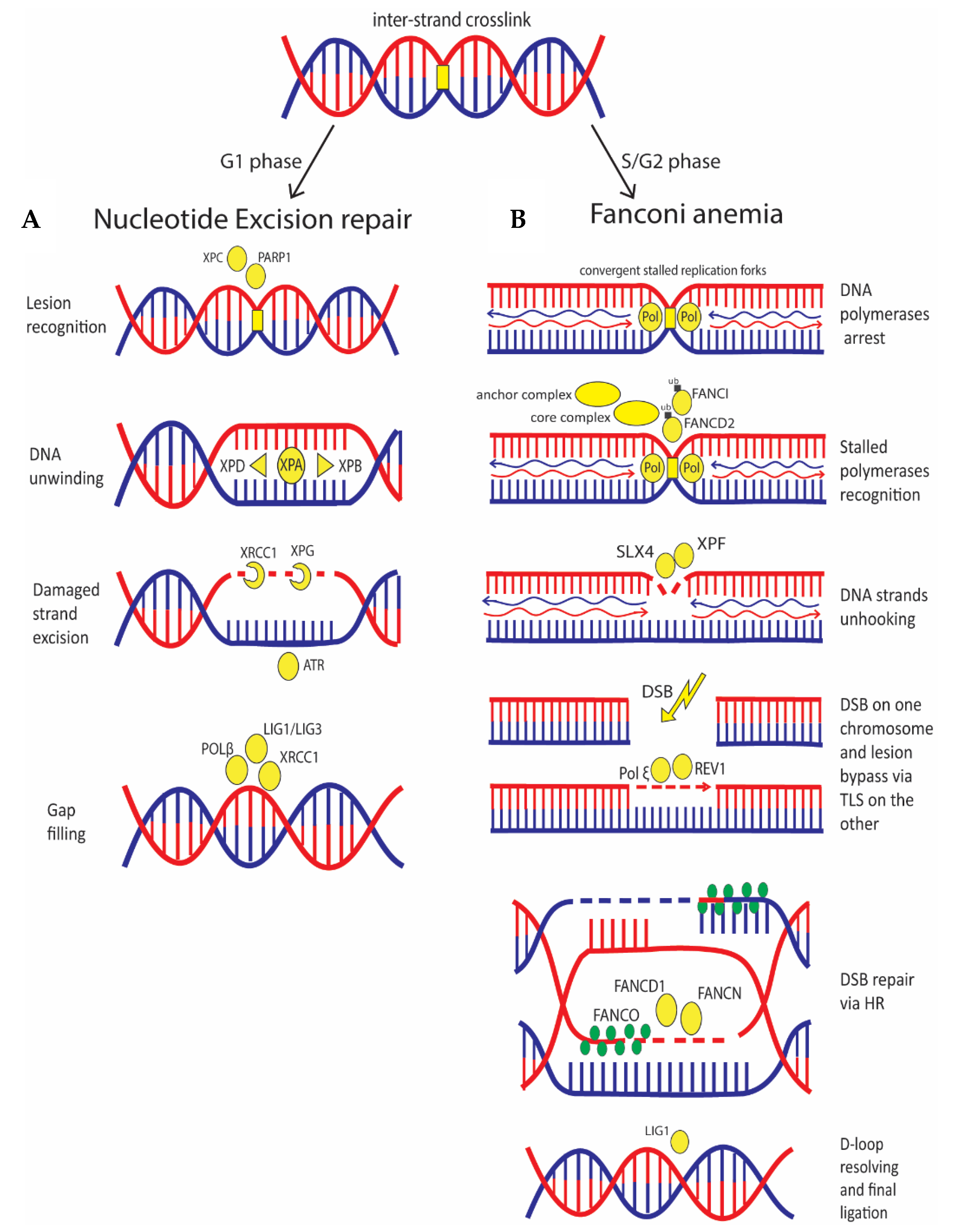
5. The DNA Crosslinks Repair Deals with NER and FA Pathways with Implication for Cancer
6. Classic Anti-Cancer Therapy Relies on DNA Damage Induction to Which Cancer Cells Are More Sensitive
7. Targeting DDC Is One of the Most Efficient Ways to Tackle Cancer Cells
8. Combination of DSB Inducers and DSB-Repair Proteins Inhibition Is Greatly Effective
9. Targeting DNA Helicases Increases Sensitivity to Several Treatments
10. Targeting Repair of Single Base Lesions and DNA Crosslinks Improves Efficiency of Cancer Treatments
11. Targeting MMR and Synthetic Lethality Interactions: Possible New Combination Therapies
12. PARP1 Inhibition Gave the Most Important Results in the Synthetic Lethality Field Promoting Development of New Drugs and Treatments
13. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Beard, W.A.; Horton, J.K.; Prasad, R.; Wilson, S.H. Eukaryotic Base Excision Repair: New Approaches Shine Light on Mechanism. Annu. Rev. Biochem. 2019, 88, 137–162. [Google Scholar] [CrossRef] [PubMed]
- Spivak, G. Nucleotide excision repair in humans. DNA Repair. (Amst.) 2015, 36, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Ceccaldi, R.; Sarangi, P.; D’Andrea, A.D. The Fanconi anaemia pathway: New players and new functions. Nat. Rev. Mol. Cell Biol. 2016, 17, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Larrea, A.A.; Lujan, S.A.; Kunkel, T.A. SnapShot: DNA mismatch repair. Cell 2010, 141, 730.e1. [Google Scholar] [CrossRef]
- Waters, L.S.; Minesinger, B.K.; Wiltrout, M.E.; D’Souza, S.; Woodruff, R.V.; Walker, G.C. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 2009, 73, 134–154. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Guo, R.; Xu, D. Non-homologous end joining: Advances and frontiers. Acta Biochim. Biophys. Sin. (Shanghai) 2016, 48, 632–640. [Google Scholar] [CrossRef]
- Wright, W.D.; Shah, S.S.; Heyer, W.D. Homologous recombination and the repair of DNA double-strand breaks. J. Biol. Chem. 2018, 293, 10524–10535. [Google Scholar] [CrossRef] [PubMed]
- McVey, M.; Lee, S.E. MMEJ repair of double-strand breaks (director’s cut): Deleted sequences and alternative endings. Trends Genet. 2008, 24, 529–538. [Google Scholar] [CrossRef]
- Bhargava, R.; Onyango, D.O.; Stark, J.M. Regulation of Single-Strand Annealing and its Role in Genome Maintenance. Trends Genet. 2016, 32, 566–575. [Google Scholar] [CrossRef]
- Jeggo, P.A.; Lobrich, M. How cancer cells hijack DNA double-strand break repair pathways to gain genomic instability. Biochem. J. 2015, 471, 1–11. [Google Scholar] [CrossRef]
- O’Neil, N.J.; Bailey, M.L.; Hieter, P. Synthetic lethality and cancer. Nat. Rev. Genet. 2017, 18, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Hoadley, K.A.; Yau, C.; Hinoue, T.; Wolf, D.M.; Lazar, A.J.; Drill, E.; Shen, R.; Taylor, A.M.; Cherniack, A.D.; Thorsson, V.; et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 2018, 173, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Horlbeck, M.A.; Gilbert, L.A.; Villalta, J.E.; Adamson, B.; Pak, R.A.; Chen, Y.; Fields, A.P.; Park, C.Y.; Corn, J.E.; Kampmann, M.; et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. Elife 2016, 5, e19760. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Yu, H.; Hughes, N.W.; Liu, B.; Kendirli, A.; Klein, K.; Chen, W.W.; Lander, E.S.; Sabatini, D.M. Gene Essentiality Profiling Reveals Gene Networks and Synthetic Lethal Interactions with Oncogenic Ras. Cell 2017, 168, 890–903. [Google Scholar] [CrossRef]
- Blackford, A.N.; Jackson, S.P. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol. Cell 2017, 66, 801–817. [Google Scholar] [CrossRef]
- Burger, K.; Schlackow, M.; Gullerova, M. Tyrosine kinase c-Abl couples RNA polymerase II transcription to DNA double-strand breaks. Nucleic Acids Res. 2019, 47, 3467–3484. [Google Scholar] [CrossRef]
- Pong, S.K.; Gullerova, M. Noncanonical functions of microRNA pathway enzymes-Drosha, DGCR8, Dicer and Ago proteins. FEBS Lett. 2018, 592, 2973–2986. [Google Scholar] [CrossRef]
- Falck, J.; Coates, J.; Jackson, S.P. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 2005, 434, 605–611. [Google Scholar] [CrossRef]
- Zhang, Y.; Hunter, T. Roles of Chk1 in cell biology and cancer therapy. Int. J. Cancer 2014, 134, 1013–1023. [Google Scholar] [CrossRef]
- Zannini, L.; Delia, D.; Buscemi, G. CHK2 kinase in the DNA damage response and beyond. J. Mol. Cell Biol. 2014, 6, 442–457. [Google Scholar] [CrossRef]
- Wagner, S.A.; Oehler, H.; Voigt, A.; Dalic, D.; Freiwald, A.; Serve, H.; Beli, P. ATR inhibition rewires cellular signaling networks induced by replication stress. Proteomics 2016, 16, 402–416. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.; Kipps, T.; Kurzrock, R. ATM Mutations in Cancer: Therapeutic Implications. Mol. Cancer Ther. 2016, 15, 1781–1791. [Google Scholar] [CrossRef]
- Goodwin, J.F.; Knudsen, K.E. Beyond DNA repair: DNA-PK function in cancer. Cancer Discov. 2014, 4, 1126–1139. [Google Scholar] [CrossRef]
- Sishc, B.J.; Davis, A.J. The Role of the Core Non-Homologous End Joining Factors in Carcinogenesis and Cancer. Cancers (Basel) 2017, 9, 81. [Google Scholar] [CrossRef]
- Tanaka, A.; Weinel, S.; Nagy, N.; O’Driscoll, M.; Lai-Cheong, J.E.; Kulp-Shorten, C.L.; Knable, A.; Carpenter, G.; Fisher, S.A.; Hiragun, M.; et al. Germline mutation in ATR in autosomal- dominant oropharyngeal cancer syndrome. Am. J. Hum. Genet. 2012, 90, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.F.; Ruiz-Vega, R.; Vasudeva, P.; Espitia, F.; Krasieva, T.B.; de Feraudy, S.; Tromberg, B.J.; Huang, S.; Garner, C.P.; Wu, J.; et al. ATR Mutations Promote the Growth of Melanoma Tumors by Modulating the Immune Microenvironment. Cell Rep. 2017, 18, 2331–2342. [Google Scholar] [CrossRef] [PubMed]
- Ray Chaudhuri, A.; Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610–621. [Google Scholar] [CrossRef]
- Yang, G.; Liu, C.; Chen, S.H.; Kassab, M.A.; Hoff, J.D.; Walter, N.G.; Yu, X. Super-resolution imaging identifies PARP1 and the Ku complex acting as DNA double-strand break sensors. Nucleic Acids Res. 2018, 46, 3446–3457. [Google Scholar] [CrossRef] [PubMed]
- Mian, E.; Wiesmuller, L. Phenotypic Analysis of ATM Protein Kinase in DNA Double-Strand Break Formation and Repair. Methods Mol. Biol. 2017, 1599, 317–334. [Google Scholar] [CrossRef] [PubMed]
- Marini, F.; Rawal, C.C.; Liberi, G.; Pellicioli, A. Regulation of DNA Double Strand Breaks Processing: Focus on Barriers. Front. Mol. Biosci. 2019, 6, 55. [Google Scholar] [CrossRef]
- Zhao, X.; Wei, C.; Li, J.; Xing, P.; Li, J.; Zheng, S.; Chen, X. Cell cycle-dependent control of homologous recombination. Acta Biochim. Biophys. Sin. (Shanghai) 2017, 49, 655–668. [Google Scholar] [CrossRef]
- Gupta, A.; Hunt, C.R.; Chakraborty, S.; Pandita, R.K.; Yordy, J.; Ramnarain, D.B.; Horikoshi, N.; Pandita, T.K. Role of 53BP1 in the regulation of DNA double-strand break repair pathway choice. Radiat. Res. 2014, 181, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Peterson, S.E.; Li, Y.; Wu-Baer, F.; Chait, B.T.; Baer, R.; Yan, H.; Gottesman, M.E.; Gautier, J. Activation of DSB processing requires phosphorylation of CtIP by ATR. Mol. Cell 2013, 49, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Ismail, I.H.; Gagne, J.P.; Genois, M.M.; Strickfaden, H.; McDonald, D.; Xu, Z.; Poirier, G.G.; Masson, J.Y.; Hendzel, M.J. The RNF138 E3 ligase displaces Ku to promote DNA end resection and regulate DNA repair pathway choice. Nat. Cell Biol. 2015, 17, 1446–1457. [Google Scholar] [CrossRef] [PubMed]
- Reginato, G.; Cannavo, E.; Cejka, P. Physiological protein blocks direct the Mre11-Rad50-Xrs2 and Sae2 nuclease complex to initiate DNA end resection. Genes Dev. 2017, 31, 2325–2330. [Google Scholar] [CrossRef]
- Daley, J.M.; Jimenez-Sainz, J.; Wang, W.; Miller, A.S.; Xue, X.; Nguyen, K.A.; Jensen, R.B.; Sung, P. Enhancement of BLM-DNA2-Mediated Long-Range DNA End Resection by CtIP. Cell Rep. 2017, 21, 324–332. [Google Scholar] [CrossRef]
- Buisson, R.; Masson, J.Y. PALB2 self-interaction controls homologous recombination. Nucleic Acids Res. 2012, 40, 10312–10323. [Google Scholar] [CrossRef]
- Liu, J.; Doty, T.; Gibson, B.; Heyer, W.D. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 2010, 17, 1260–1262. [Google Scholar] [CrossRef]
- Honda, M.; Okuno, Y.; Yoo, J.; Ha, T.; Spies, M. Tyrosine phosphorylation enhances RAD52-mediated annealing by modulating its DNA binding. EMBO J. 2011, 30, 3368–3382. [Google Scholar] [CrossRef]
- Piazza, A.; Shah, S.S.; Wright, W.D.; Gore, S.K.; Koszul, R.; Heyer, W.D. Dynamic Processing of Displacement Loops during Recombinational DNA Repair. Mol. Cell 2019, 73, 1255–1266 e1254. [Google Scholar] [CrossRef]
- Ferrari, M.; Rawal, C.C.; Lodovichi, S.; Vietri, M.Y.; Pellicioli, A. Rad9/53BP1 promotes DNA repair via crossover recombination by limiting the Sgs1 and Mph1 helicases. Nat. Commun. 2020, 11, 3181. [Google Scholar] [CrossRef] [PubMed]
- Truong, L.N.; Li, Y.; Shi, L.Z.; Hwang, P.Y.; He, J.; Wang, H.; Razavian, N.; Berns, M.W.; Wu, X. Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl. Acad. Sci. USA 2013, 110, 7720–7725. [Google Scholar] [CrossRef]
- Rodgers, K.; McVey, M. Error-Prone Repair of DNA Double-Strand Breaks. J. Cell Physiol. 2016, 231, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Shamanna, R.A.; Lu, H.; de Freitas, J.K.; Tian, J.; Croteau, D.L.; Bohr, V.A. WRN regulates pathway choice between classical and alternative non-homologous end joining. Nat. Commun. 2016, 7, 13785. [Google Scholar] [CrossRef] [PubMed]
- Faridounnia, M.; Folkers, G.E.; Boelens, R. Function and Interactions of ERCC1-XPF in DNA Damage Response. Molecules 2018, 23, 3205. [Google Scholar] [CrossRef]
- Knijnenburg, T.A.; Wang, L.; Zimmermann, M.T.; Chambwe, N.; Gao, G.F.; Cherniack, A.D.; Fan, H.; Shen, H.; Way, G.P.; Greene, C.S.; et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018, 23, 239–254. [Google Scholar] [CrossRef]
- Heeke, A.L.; Pishvaian, M.J.; Lynce, F.; Xiu, J.; Brody, J.R.; Chen, W.J.; Baker, T.M.; Marshall, J.L.; Isaacs, C. Prevalence of Homologous Recombination-Related Gene Mutations Across Multiple Cancer Types. JCO Precis. Oncol. 2018, 2018. [Google Scholar] [CrossRef]
- Nielsen, F.C.; van Overeem Hansen, T.; Sorensen, C.S. Hereditary breast and ovarian cancer: New genes in confined pathways. Nat. Rev. Cancer 2016, 16, 599–612. [Google Scholar] [CrossRef]
- Kleibl, Z.; Kristensen, V.N. Women at high risk of breast cancer: Molecular characteristics, clinical presentation and management. Breast 2016, 28, 136–144. [Google Scholar] [CrossRef]
- Maresca, L.; Lodovichi, S.; Lorenzoni, A.; Cervelli, T.; Monaco, R.; Spugnesi, L.; Tancredi, M.; Falaschi, E.; Zavaglia, K.; Landucci, E.; et al. Functional Interaction Between BRCA1 and DNA Repair in Yeast May Uncover a Role of RAD50, RAD51, MRE11A, and MSH6 Somatic Variants in Cancer Development. Front. Genet. 2018, 9, 397. [Google Scholar] [CrossRef]
- Gachechiladze, M.; Skarda, J.; Soltermann, A.; Joerger, M. RAD51 as a potential surrogate marker for DNA repair capacity in solid malignancies. Int. J. Cancer 2017, 141, 1286–1294. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Kim, J.W.; Hwang, I.G.; Lee, H.S.; Kim, W.H. Expression of DNA Damage Response Markers in Early-Onset or Familial Gastric Cancers. Asian Pac. J. Cancer Prev. 2019, 20, 1369–1376. [Google Scholar] [CrossRef] [PubMed]
- Takagi, M.; Yoshida, M.; Nemoto, Y.; Tamaichi, H.; Tsuchida, R.; Seki, M.; Uryu, K.; Nishii, R.; Miyamoto, S.; Saito, M.; et al. Loss of DNA Damage Response in Neuroblastoma and Utility of a PARP Inhibitor. J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef] [PubMed]
- Simonetti, G.; Padella, A.; do Valle, I.F.; Fontana, M.C.; Fonzi, E.; Bruno, S.; Baldazzi, C.; Guadagnuolo, V.; Manfrini, M.; Ferrari, A.; et al. Aneuploid acute myeloid leukemia exhibits a signature of genomic alterations in the cell cycle and protein degradation machinery. Cancer 2019, 125, 712–725. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Sanz, P.; Trivino, J.C.; Mota, A.; Perez Lopez, M.; Colas, E.; Rojo-Sebastian, A.; Garcia, A.; Gatius, S.; Ruiz, M.; Prat, J.; et al. Chromatin remodelling and DNA repair genes are frequently mutated in endometrioid endometrial carcinoma. Int. J. Cancer 2017, 140, 1551–1563. [Google Scholar] [CrossRef]
- Zhen, J.T.; Syed, J.; Nguyen, K.A.; Leapman, M.S.; Agarwal, N.; Brierley, K.; Llor, X.; Hofstatter, E.; Shuch, B. Genetic testing for hereditary prostate cancer: Current status and limitations. Cancer 2018, 124, 3105–3117. [Google Scholar] [CrossRef]
- Kaluzna, E.M.; Rembowska, J.; Ziolkowska-Suchanek, I.; Swiatek-Koscielna, B.; Gabryel, P.; Dyszkiewicz, W.; Nowak, J.S. Heterozygous p.I171V mutation of the NBN gene as a risk factor for lung cancer development. Oncol. Lett. 2015, 10, 3300–3304. [Google Scholar] [CrossRef][Green Version]
- Hurley, R.M.; Wahner Hendrickson, A.E.; Visscher, D.W.; Ansell, P.; Harrell, M.I.; Wagner, J.M.; Negron, V.; Goergen, K.M.; Maurer, M.J.; Oberg, A.L.; et al. 53BP1 as a potential predictor of response in PARP inhibitor-treated homologous recombination-deficient ovarian cancer. Gynecol. Oncol. 2019, 153, 127–134. [Google Scholar] [CrossRef]
- Yao, J.; Huang, A.; Zheng, X.; Liu, T.; Lin, Z.; Zhang, S.; Yang, Q.; Zhang, T.; Ma, H. 53BP1 loss induces chemoresistance of colorectal cancer cells to 5-fluorouracil by inhibiting the ATM-CHK2-P53 pathway. J. Cancer Res. Clin. Oncol. 2017, 143, 419–431. [Google Scholar] [CrossRef]
- Mirza-Aghazadeh-Attari, M.; Mohammadzadeh, A.; Yousefi, B.; Mihanfar, A.; Karimian, A.; Majidinia, M. 53BP1: A key player of DNA damage response with critical functions in cancer. DNA Repair (Amst) 2019, 73, 110–119. [Google Scholar] [CrossRef]
- Reczek, C.R.; Shakya, R.; Miteva, Y.; Szabolcs, M.; Ludwig, T.; Baer, R. The DNA resection protein CtIP promotes mammary tumorigenesis. Oncotarget 2016, 7, 32172–32183. [Google Scholar] [CrossRef] [PubMed]
- Wood, R.D.; Doublie, S. DNA polymerase theta (POLQ), double-strand break repair, and cancer. DNA Repair (Amst) 2016, 44, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Brosh, R.M., Jr. DNA helicases involved in DNA repair and their roles in cancer. Nat. Rev. Cancer 2013, 13, 542–558. [Google Scholar] [CrossRef] [PubMed]
- Pawlowska, E.; Szczepanska, J.; Blasiak, J. DNA2-An Important Player in DNA Damage Response or Just Another DNA Maintenance Protein? Int. J. Mol. Sci. 2017, 18, 1562. [Google Scholar] [CrossRef]
- Moser, M.J.; Bigbee, W.L.; Grant, S.G.; Emond, M.J.; Langlois, R.G.; Jensen, R.H.; Oshima, J.; Monnat, R.J., Jr. Genetic instability and hematologic disease risk in Werner syndrome patients and heterozygotes. Cancer Res. 2000, 60, 2492–2496. [Google Scholar]
- Sallmyr, A.; Tomkinson, A.E.; Rassool, F.V. Up-regulation of WRN and DNA ligase IIIalpha in chronic myeloid leukemia: Consequences for the repair of DNA double-strand breaks. Blood 2008, 112, 1413–1423. [Google Scholar] [CrossRef]
- Lee, T.H.; Kang, T.H. DNA Oxidation and Excision Repair Pathways. Int. J. Mol. Sci. 2019, 20, 6092. [Google Scholar] [CrossRef]
- Breslin, C.; Hornyak, P.; Ridley, A.; Rulten, S.L.; Hanzlikova, H.; Oliver, A.W.; Caldecott, K.W. The XRCC1 phosphate-binding pocket binds poly (ADP-ribose) and is required for XRCC1 function. Nucleic Acids Res. 2015, 43, 6934–6944. [Google Scholar] [CrossRef]
- Hanzlikova, H.; Gittens, W.; Krejcikova, K.; Zeng, Z.; Caldecott, K.W. Overlapping roles for PARP1 and PARP2 in the recruitment of endogenous XRCC1 and PNKP into oxidized chromatin. Nucleic Acids Res. 2017, 45, 2546–2557. [Google Scholar] [CrossRef]
- Liu, D.; Keijzers, G.; Rasmussen, L.J. DNA mismatch repair and its many roles in eukaryotic cells. Mutat. Res. 2017, 773, 174–187. [Google Scholar] [CrossRef]
- Kunkel, T.A.; Erie, D.A. Eukaryotic Mismatch Repair in Relation to DNA Replication. Annu. Rev. Genet. 2015, 49, 291–313. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, U.; Alani, E. Understanding how mismatch repair proteins participate in the repair/anti-recombination decision. FEMS Yeast Res. 2016, 16, 1–12. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Farrington, S.M.; Tenesa, A.; Barnetson, R.; Wiltshire, A.; Prendergast, J.; Porteous, M.; Campbell, H.; Dunlop, M.G. Germline susceptibility to colorectal cancer due to base-excision repair gene defects. Am. J. Hum. Genet. 2005, 77, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Sahadevan, M.; Lee, O.; Muzzio, M.; Phan, B.; Jacobs, L.; Khouri, N.; Wang, J.; Hu, H.; Stearns, V.; Chatterton, R.T. The relationship of single-strand breaks in DNA to breast cancer risk and to tissue concentrations of oestrogens. Biomarkers 2017, 22, 689–697. [Google Scholar] [CrossRef]
- Wang, M.; Long, K.; Li, E.; Li, L.; Li, B.; Ci, S.; He, L.; Pan, F.; Hu, Z.; Guo, Z. DNA polymerase beta modulates cancer progression via enhancing CDH13 expression by promoter demethylation. Oncogene 2020, 39, 5507–5519. [Google Scholar] [CrossRef]
- Abdel-Fatah, T.M.; Russell, R.; Agarwal, D.; Moseley, P.; Abayomi, M.A.; Perry, C.; Albarakati, N.; Ball, G.; Chan, S.; Caldas, C.; et al. DNA polymerase beta deficiency is linked to aggressive breast cancer: A comprehensive analysis of gene copy number, mRNA and protein expression in multiple cohorts. Mol. Oncol 2014, 8, 520–532. [Google Scholar] [CrossRef]
- Alnajjar, K.S.; Negahbani, A.; Nakhjiri, M.; Krylov, I.S.; Kashemirov, B.A.; McKenna, C.E.; Goodman, M.F.; Sweasy, J.B. DNA Polymerase beta Cancer-Associated Variant I260M Exhibits Nonspecific Selectivity toward the beta-gamma Bridging Group of the Incoming dNTP. Biochemistry 2017, 56, 5449–5456. [Google Scholar] [CrossRef]
- Mahmoud, M.M.; Schechter, A.; Alnajjar, K.S.; Huang, J.; Towle-Weicksel, J.; Eckenroth, B.E.; Doublie, S.; Sweasy, J.B. Defective Nucleotide Release by DNA Polymerase beta Mutator Variant E288K Is the Basis of Its Low Fidelity. Biochemistry 2017, 56, 5550–5559. [Google Scholar] [CrossRef]
- Zhou, T.; Pan, F.; Cao, Y.; Han, Y.; Zhao, J.; Sun, H.; Zhou, X.; Wu, X.; He, L.; Hu, Z.; et al. R152C DNA Pol beta mutation impairs base excision repair and induces cellular transformation. Oncotarget 2016, 7, 6902–6915. [Google Scholar] [CrossRef]
- Malik, S.S.; Masood, N.; Asif, M.; Ahmed, P.; Shah, Z.U.; Khan, J.S. Expressional analysis of MLH1 and MSH2 in breast cancer. Curr. Probl. Cancer 2019, 43, 97–105. [Google Scholar] [CrossRef]
- Wang, S.M.; Jiang, B.; Deng, Y.; Huang, S.L.; Fang, M.Z.; Wang, Y. Clinical significance of MLH1/MSH2 for stage II/III sporadic colorectal cancer. World J. Gastrointest. Oncol. 2019, 11, 1065–1080. [Google Scholar] [CrossRef] [PubMed]
- Liccardo, R.; Della Ragione, C.; Mitilini, N.; De Rosa, M.; Izzo, P.; Duraturo, F. Novel variants of unknown significance in the PMS2 gene identified in patients with hereditary colon cancer. Cancer Manag. Res. 2019, 11, 6719–6725. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.E.; Jackson, S.A.; Susswein, L.R.; Zeinomar, N.; Ma, X.; Marshall, M.L.; Stettner, A.R.; Milewski, B.; Xu, Z.; Solomon, B.D.; et al. MSH6 and PMS2 germ-line pathogenic variants implicated in Lynch syndrome are associated with breast cancer. Genet. Med. 2018, 20, 1167–1174. [Google Scholar] [CrossRef] [PubMed]
- Maresca, L.; Spugnesi, L.; Lodovichi, S.; Cozzani, C.; Naccarato, A.G.; Tancredi, M.; Collavoli, A.; Falaschi, E.; Rossetti, E.; Aretini, P.; et al. MSH2 role in BRCA1-driven tumorigenesis: A preliminary study in yeast and in human tumors from BRCA1-VUS carriers. Eur. J. Med. Genet. 2015, 58, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, S.; Anai, H.; Hanada, K. Mechanisms of interstrand DNA crosslink repair and human disorders. Genes Environ. 2016, 38, 9. [Google Scholar] [CrossRef]
- Robu, M.; Shah, R.G.; Purohit, N.K.; Zhou, P.; Naegeli, H.; Shah, G.M. Poly(ADP-ribose) polymerase 1 escorts XPC to UV-induced DNA lesions during nucleotide excision repair. Proc. Natl. Acad. Sci. USA 2017, 114, E6847–E6856. [Google Scholar] [CrossRef]
- Sugitani, N.; Sivley, R.M.; Perry, K.E.; Capra, J.A.; Chazin, W.J. XPA: A key scaffold for human nucleotide excision repair. DNA Repair (Amst) 2016, 44, 123–135. [Google Scholar] [CrossRef]
- Park, J.M.; Kang, T.H. Transcriptional and Posttranslational Regulation of Nucleotide Excision Repair: The Guardian of the Genome against Ultraviolet Radiation. Int. J. Mol. Sci. 2016, 17, 1840. [Google Scholar] [CrossRef]
- Klein Douwel, D.; Boonen, R.A.; Long, D.T.; Szypowska, A.A.; Raschle, M.; Walter, J.C.; Knipscheer, P. XPF-ERCC1 acts in Unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol. Cell 2014, 54, 460–471. [Google Scholar] [CrossRef]
- Howlett, N.G.; Harney, J.A.; Rego, M.A.; Kolling, F.W., IV; Glover, T.W. Functional interaction between the Fanconi Anemia D2 protein and proliferating cell nuclear antigen (PCNA) via a conserved putative PCNA interaction motif. J. Biol. Chem. 2009, 284, 28935–28942. [Google Scholar] [CrossRef]
- Jo, U.; Kim, H. Exploiting the Fanconi Anemia Pathway for Targeted Anti-Cancer Therapy. Mol. Cells 2015, 38, 669–676. [Google Scholar] [CrossRef]
- Michl, J.; Zimmer, J.; Tarsounas, M. Interplay between Fanconi anemia and homologous recombination pathways in genome integrity. EMBO J. 2016, 35, 909–923. [Google Scholar] [CrossRef] [PubMed]
- Alter, B.P. Fanconi anemia and the development of leukemia. Best Pract. Res. Clin. Haematol. 2014, 27, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Niraj, J.; Farkkila, A.; D’Andrea, A.D. The Fanconi Anemia Pathway in Cancer. Annu. Rev. Cancer Biol. 2019, 3, 457–478. [Google Scholar] [CrossRef]
- Kondo, N.; Takahashi, A.; Ono, K.; Ohnishi, T. DNA damage induced by alkylating agents and repair pathways. J. Nucleic Acids 2010, 2010, 543531. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Wu, Q.; Luan, S.; Yin, Z.; He, C.; Yin, L.; Zou, Y.; Yuan, Z.; Li, L.; Song, X.; et al. A comprehensive review of topoisomerase inhibitors as anticancer agents in the past decade. Eur. J. Med. Chem. 2019, 171, 129–168. [Google Scholar] [CrossRef]
- Calderon-Montano, J.M.; Burgos-Moron, E.; Orta, M.L.; Lopez-Lazaro, M. Effect of DNA repair deficiencies on the cytotoxicity of drugs used in cancer therapy-a review. Curr. Med. Chem. 2014, 21, 3419–3454. [Google Scholar] [CrossRef]
- Peters, G.J. Novel developments in the use of antimetabolites. Nucleosides Nucleotides Nucleic Acids 2014, 33, 358–374. [Google Scholar] [CrossRef]
- Chen, H.H.W.; Kuo, M.T. Improving radiotherapy in cancer treatment: Promises and challenges. Oncotarget 2017, 8, 62742–62758. [Google Scholar] [CrossRef]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef]
- Vitale, I.; Galluzzi, L.; Castedo, M.; Kroemer, G. Mitotic catastrophe: A mechanism for avoiding genomic instability. Nat. Rev. Mol. Cell Biol. 2011, 12, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Sia, J.; Szmyd, R.; Hau, E.; Gee, H.E. Molecular Mechanisms of Radiation-Induced Cancer Cell Death: A Primer. Front. Cell Dev. Biol. 2020, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.X.; Zhou, P.K. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal. Transduct Target. Ther. 2020, 5, 60. [Google Scholar] [CrossRef]
- Balmus, G.; Pilger, D.; Coates, J.; Demir, M.; Sczaniecka-Clift, M.; Barros, A.C.; Woods, M.; Fu, B.; Yang, F.; Chen, E.; et al. ATM orchestrates the DNA-damage response to counter toxic non-homologous end-joining at broken replication forks. Nat. Commun. 2019, 10, 87. [Google Scholar] [CrossRef] [PubMed]
- Riches, L.C.; Trinidad, A.G.; Hughes, G.; Jones, G.N.; Hughes, A.M.; Thomason, A.G.; Gavine, P.; Cui, A.; Ling, S.; Stott, J.; et al. Pharmacology of the ATM Inhibitor AZD0156: Potentiation of Irradiation and Olaparib Responses Preclinically. Mol. Cancer Ther. 2020, 19, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Hong, R.; Ma, F.; Zhang, W.; Yu, X.; Li, Q.; Luo, Y.; Zhu, C.; Jiang, W.; Xu, B. 53BP1 depletion causes PARP inhibitor resistance in ATM-deficient breast cancer cells. BMC Cancer 2016, 16, 725. [Google Scholar] [CrossRef]
- Durant, S.T.; Zheng, L.; Wang, Y.; Chen, K.; Zhang, L.; Zhang, T.; Yang, Z.; Riches, L.; Trinidad, A.G.; Fok, J.H.L.; et al. The brain-penetrant clinical ATM inhibitor AZD1390 radiosensitizes and improves survival of preclinical brain tumor models. Sci. Adv. 2018, 4, eaat1719. [Google Scholar] [CrossRef]
- Jin, M.H.; Oh, D.Y. ATM in DNA repair in cancer. Pharmacol. Ther. 2019, 203, 107391. [Google Scholar] [CrossRef]
- Geng, W.; Tian, D.; Wang, Q.; Shan, S.; Zhou, J.; Xu, W.; Shan, H. DNAPKcs inhibitor increases the sensitivity of gastric cancer cells to radiotherapy. Oncol. Rep. 2019, 42, 561–570. [Google Scholar] [CrossRef]
- Mohiuddin, I.S.; Kang, M.H. DNA-PK as an Emerging Therapeutic Target in Cancer. Front. Oncol. 2019, 9, 635. [Google Scholar] [CrossRef]
- Wise, H.C.; Iyer, G.V.; Moore, K.; Temkin, S.M.; Gordon, S.; Aghajanian, C.; Grisham, R.N. Activity of M3814, an Oral DNA-PK Inhibitor, In Combination with Topoisomerase II Inhibitors in Ovarian Cancer Models. Sci. Rep. 2019, 9, 18882. [Google Scholar] [CrossRef]
- Timme, C.R.; Rath, B.H.; O’Neill, J.W.; Camphausen, K.; Tofilon, P.J. The DNA-PK Inhibitor VX-984 Enhances the Radiosensitivity of Glioblastoma Cells Grown In Vitro and as Orthotopic Xenografts. Mol. Cancer Ther. 2018, 17, 1207–1216. [Google Scholar] [CrossRef] [PubMed]
- Gavande, N.S.; VanderVere-Carozza, P.S.; Hinshaw, H.D.; Jalal, S.I.; Sears, C.R.; Pawelczak, K.S.; Turchi, J.J. DNA repair targeted therapy: The past or future of cancer treatment? Pharmacol. Ther. 2016, 160, 65–83. [Google Scholar] [CrossRef] [PubMed]
- Saldivar, J.C.; Cortez, D.; Cimprich, K.A. The essential kinase ATR: Ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 2017, 18, 622–636. [Google Scholar] [CrossRef] [PubMed]
- Rundle, S.; Bradbury, A.; Drew, Y.; Curtin, N.J. Targeting the ATR-CHK1 Axis in Cancer Therapy. Cancers (Basel) 2017, 9, 41. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; George, E.; Ragland, R.; Rafail, S.; Zhang, R.; Krepler, C.; Morgan, M.; Herlyn, M.; Brown, E.; Simpkins, F. Targeting the ATR/CHK1 Axis with PARP Inhibition Results in Tumor Regression in BRCA-Mutant Ovarian Cancer Models. Clin. Cancer Res. 2017, 23, 3097–3108. [Google Scholar] [CrossRef] [PubMed]
- Vendetti, F.P.; Lau, A.; Schamus, S.; Conrads, T.P.; O’Connor, M.J.; Bakkenist, C.J. The orally active and bioavailable ATR kinase inhibitor AZD6738 potentiates the anti-tumor effects of cisplatin to resolve ATM-deficient non-small cell lung cancer in vivo. Oncotarget 2015, 6, 44289–44305. [Google Scholar] [CrossRef]
- Kwok, M.; Davies, N.; Agathanggelou, A.; Smith, E.; Oldreive, C.; Petermann, E.; Stewart, G.; Brown, J.; Lau, A.; Pratt, G.; et al. ATR inhibition induces synthetic lethality and overcomes chemoresistance in TP53- or ATM-defective chronic lymphocytic leukemia cells. Blood 2016, 127, 582–595. [Google Scholar] [CrossRef]
- Jin, J.; Fang, H.; Yang, F.; Ji, W.; Guan, N.; Sun, Z.; Shi, Y.; Zhou, G.; Guan, X. Combined Inhibition of ATR and WEE1 as a Novel Therapeutic Strategy in Triple-Negative Breast Cancer. Neoplasia 2018, 20, 478–488. [Google Scholar] [CrossRef]
- Sanjiv, K.; Hagenkort, A.; Calderon-Montano, J.M.; Koolmeister, T.; Reaper, P.M.; Mortusewicz, O.; Jacques, S.A.; Kuiper, R.V.; Schultz, N.; Scobie, M.; et al. Cancer-Specific Synthetic Lethality between ATR and CHK1 Kinase Activities. Cell Rep. 2016, 14, 298–309. [Google Scholar] [CrossRef]
- Dillon, M.T.; Boylan, Z.; Smith, D.; Guevara, J.; Mohammed, K.; Peckitt, C.; Saunders, M.; Banerji, U.; Clack, G.; Smith, S.A.; et al. PATRIOT: A phase I study to assess the tolerability, safety and biological effects of a specific ataxia telangiectasia and Rad3-related (ATR) inhibitor (AZD6738) as a single agent and in combination with palliative radiation therapy in patients with solid tumours. Clin. Transl. Radiat. Oncol. 2018, 12, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Prevo, R.; Fokas, E.; Reaper, P.M.; Charlton, P.A.; Pollard, J.R.; McKenna, W.G.; Muschel, R.J.; Brunner, T.B. The novel ATR inhibitor VE-821 increases sensitivity of pancreatic cancer cells to radiation and chemotherapy. Cancer Biol. Ther. 2012, 13, 1072–1081. [Google Scholar] [CrossRef] [PubMed]
- Wengner, A.M.; Siemeister, G.; Lucking, U.; Lefranc, J.; Wortmann, L.; Lienau, P.; Bader, B.; Bomer, U.; Moosmayer, D.; Eberspacher, U.; et al. The Novel ATR Inhibitor BAY 1895344 Is Efficacious as Monotherapy and Combined with DNA Damage-Inducing or Repair-Compromising Therapies in Preclinical Cancer Models. Mol. Cancer Ther. 2020, 19, 26–38. [Google Scholar] [CrossRef]
- Dai, C.H.; Wang, Y.; Chen, P.; Jiang, Q.; Lan, T.; Li, M.Y.; Su, J.Y.; Wu, Y.; Li, J. Suppression of the FA pathway combined with CHK1 inhibitor hypersensitize lung cancer cells to gemcitabine. Sci. Rep. 2017, 7, 15031. [Google Scholar] [CrossRef]
- Trenner, A.; Sartori, A.A. Harnessing DNA Double-Strand Break Repair for Cancer Treatment. Front. Oncol. 2019, 9, 1388. [Google Scholar] [CrossRef] [PubMed]
- Jividen, K.; Kedzierska, K.Z.; Yang, C.S.; Szlachta, K.; Ratan, A.; Paschal, B.M. Genomic analysis of DNA repair genes and androgen signaling in prostate cancer. BMC Cancer 2018, 18, 960. [Google Scholar] [CrossRef]
- Petroni, M.; Sardina, F.; Infante, P.; Bartolazzi, A.; Locatelli, E.; Fabretti, F.; Di Giulio, S.; Capalbo, C.; Cardinali, B.; Coppa, A.; et al. MRE11 inhibition highlights a replication stress-dependent vulnerability of MYCN-driven tumors. Cell Death Dis. 2018, 9, 895. [Google Scholar] [CrossRef] [PubMed]
- Bian, L.; Meng, Y.; Zhang, M.; Li, D. MRE11-RAD50-NBS1 complex alterations and DNA damage response: Implications for cancer treatment. Mol. Cancer 2019, 18, 169. [Google Scholar] [CrossRef]
- Huang, F.; Mazin, A.V. A small molecule inhibitor of human RAD51 potentiates breast cancer cell killing by therapeutic agents in mouse xenografts. PLoS ONE 2014, 9, e100993. [Google Scholar] [CrossRef]
- Alagpulinsa, D.A.; Ayyadevara, S.; Shmookler Reis, R.J. A Small-Molecule Inhibitor of RAD51 Reduces Homologous Recombination and Sensitizes Multiple Myeloma Cells to Doxorubicin. Front. Oncol. 2014, 4, 289. [Google Scholar] [CrossRef]
- Wera, A.C.; Lobbens, A.; Stoyanov, M.; Lucas, S.; Michiels, C. Radiation-induced synthetic lethality: Combination of poly(ADP-ribose) polymerase and RAD51 inhibitors to sensitize cells to proton irradiation. Cell Cycle 2019, 18, 1770–1783. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Cai, D.; Li, M.; Wu, X. The homologous recombination protein RAD51 is a promising therapeutic target for cervical carcinoma. Oncol. Rep. 2017, 38, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Guan, J.; Zhang, Z.; Lv, J.; Wang, Y.; Liu, L.; Zhou, Q.; Mao, W. Inhibition of Rad51 sensitizes breast cancer cells with wild-type PTEN to olaparib. Biomed. Pharmacother. 2017, 94, 165–168. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Chen, H.; Guo, X.E.; Qiu, X.L.; Hu, C.M.; Chamberlin, A.R.; Lee, W.H. Synthesis, molecular modeling, and biological evaluation of novel RAD51 inhibitors. Eur. J. Med. Chem. 2015, 96, 196–208. [Google Scholar] [CrossRef]
- Roberti, M.; Schipani, F.; Bagnolini, G.; Milano, D.; Giacomini, E.; Falchi, F.; Balboni, A.; Manerba, M.; Farabegoli, F.; De Franco, F.; et al. Rad51/BRCA2 disruptors inhibit homologous recombination and synergize with olaparib in pancreatic cancer cells. Eur. J. Med. Chem. 2019, 165, 80–92. [Google Scholar] [CrossRef]
- Chen, C.C.; Feng, W.; Lim, P.X.; Kass, E.M.; Jasin, M. Homology-Directed Repair and the Role of BRCA1, BRCA2, and Related Proteins in Genome Integrity and Cancer. Annu. Rev. Cancer Biol. 2018, 2, 313–336. [Google Scholar] [CrossRef]
- Trenner, A.; Godau, J.; Sartori, A.A. A Short BRCA2-Derived Cell-Penetrating Peptide Targets RAD51 Function and Confers Hypersensitivity toward PARP Inhibition. Mol. Cancer Ther. 2018, 17, 1392–1404. [Google Scholar] [CrossRef]
- Pessetto, Z.Y.; Yan, Y.; Bessho, T.; Natarajan, A. Inhibition of BRCT(BRCA1)-phosphoprotein interaction enhances the cytotoxic effect of olaparib in breast cancer cells: A proof of concept study for synthetic lethal therapeutic option. Breast Cancer Res. Treat. 2012, 134, 511–517. [Google Scholar] [CrossRef][Green Version]
- Nogueira, A.; Fernandes, M.; Catarino, R.; Medeiros, R. RAD52 Functions in Homologous Recombination and Its Importance on Genomic Integrity Maintenance and Cancer Therapy. Cancers (Basel) 2019, 11, 1622. [Google Scholar] [CrossRef]
- Huang, F.; Goyal, N.; Sullivan, K.; Hanamshet, K.; Patel, M.; Mazina, O.M.; Wang, C.X.; An, W.F.; Spoonamore, J.; Metkar, S.; et al. Targeting BRCA1- and BRCA2-deficient cells with RAD52 small molecule inhibitors. Nucleic Acids Res. 2016, 44, 4189–4199. [Google Scholar] [CrossRef]
- Toma, M.; Sullivan-Reed, K.; Sliwinski, T.; Skorski, T. RAD52 as a Potential Target for Synthetic Lethality-Based Anticancer Therapies. Cancers (Basel) 2019, 11, 1561. [Google Scholar] [CrossRef] [PubMed]
- Sullivan-Reed, K.; Bolton-Gillespie, E.; Dasgupta, Y.; Langer, S.; Siciliano, M.; Nieborowska-Skorska, M.; Hanamshet, K.; Belyaeva, E.A.; Bernhardy, A.J.; Lee, J.; et al. Simultaneous Targeting of PARP1 and RAD52 Triggers Dual Synthetic Lethality in BRCA-Deficient Tumor Cells. Cell Rep. 2018, 23, 3127–3136. [Google Scholar] [CrossRef] [PubMed]
- Weterings, E.; Gallegos, A.C.; Dominick, L.N.; Cooke, L.S.; Bartels, T.N.; Vagner, J.; Matsunaga, T.O.; Mahadevan, D. A novel small molecule inhibitor of the DNA repair protein Ku70/80. DNA Repair (Amst) 2016, 43, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yu, M.; Fei, B.; Sun, J.; Wang, D. Identification Of Natural Compound Derivative For Inhibition Of XLF And Overcoming Chemoresistance In Colorectal Cancer Cells. Drug Des. Devel. Ther. 2019, 13, 3823–3834. [Google Scholar] [CrossRef] [PubMed]
- Kondo, N.; Takahashi, A.; Mori, E.; Ohnishi, K.; McKinnon, P.J.; Sakaki, T.; Nakase, H.; Ohnishi, T. DNA ligase IV as a new molecular target for temozolomide. Biochem. Biophys. Res. Commun. 2009, 387, 656–660. [Google Scholar] [CrossRef]
- Perfetti, M.T.; Baughman, B.M.; Dickson, B.M.; Mu, Y.; Cui, G.; Mader, P.; Dong, A.; Norris, J.L.; Rothbart, S.B.; Strahl, B.D.; et al. Identification of a fragment-like small molecule ligand for the methyl-lysine binding protein, 53BP1. ACS Chem. Biol. 2015, 10, 1072–1081. [Google Scholar] [CrossRef]
- Canny, M.D.; Moatti, N.; Wan, L.C.K.; Fradet-Turcotte, A.; Krasner, D.; Mateos-Gomez, P.A.; Zimmermann, M.; Orthwein, A.; Juang, Y.C.; Zhang, W.; et al. Inhibition of 53BP1 favors homology-dependent DNA repair and increases CRISPR-Cas9 genome-editing efficiency. Nat. Biotechnol. 2018, 36, 95–102. [Google Scholar] [CrossRef]
- Mohni, K.N.; Thompson, P.S.; Luzwick, J.W.; Glick, G.G.; Pendleton, C.S.; Lehmann, B.D.; Pietenpol, J.A.; Cortez, D. A Synthetic Lethal Screen Identifies DNA Repair Pathways that Sensitize Cancer Cells to Combined ATR Inhibition and Cisplatin Treatments. PLoS ONE 2015, 10, e0125482. [Google Scholar] [CrossRef]
- Patterson-Fortin, J.; D’Andrea, A.D. Exploiting the Microhomology-Mediated End-Joining Pathway in Cancer Therapy. Cancer Res. 2020. [Google Scholar] [CrossRef]
- Wang, Z.; Song, Y.; Li, S.; Kurian, S.; Xiang, R.; Chiba, T.; Wu, X. DNA polymerase theta (POLQ) is important for repair of DNA double-strand breaks caused by fork collapse. J. Biol. Chem. 2019, 294, 3909–3919. [Google Scholar] [CrossRef]
- Feng, W.; Simpson, D.A.; Carvajal-Garcia, J.; Price, B.A.; Kumar, R.J.; Mose, L.E.; Wood, R.D.; Rashid, N.; Purvis, J.E.; Parker, J.S.; et al. Genetic determinants of cellular addiction to DNA polymerase theta. Nat. Commun. 2019, 10, 4286. [Google Scholar] [CrossRef] [PubMed]
- Ceccaldi, R.; Liu, J.C.; Amunugama, R.; Hajdu, I.; Primack, B.; Petalcorin, M.I.; O’Connor, K.W.; Konstantinopoulos, P.A.; Elledge, S.J.; Boulton, S.J.; et al. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature 2015, 518, 258–262. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Gelot, C.; Pantelidou, C.; Li, A.; Yücel, H.; Davis, R.E.; Farkkila, A.; Kochupurakkal, B.; Syed, A.; Shapiro, G.I.; et al. Polymerase Theta Inhibition Kills Homologous Recombination Deficient Tumors. bioRxiv 2020. [Google Scholar] [CrossRef]
- Tripathi, V.; Agarwal, H.; Priya, S.; Batra, H.; Modi, P.; Pandey, M.; Saha, D.; Raghavan, S.C.; Sengupta, S. MRN complex-dependent recruitment of ubiquitylated BLM helicase to DSBs negatively regulates DNA repair pathways. Nat. Commun. 2018, 9, 1016. [Google Scholar] [CrossRef] [PubMed]
- Hengel, S.R.; Spies, M.A.; Spies, M. Small-Molecule Inhibitors Targeting DNA Repair and DNA Repair Deficiency in Research and Cancer Therapy. Cell Chem. Biol. 2017, 24, 1101–1119. [Google Scholar] [CrossRef]
- Wanrooij, P.H.; Burgers, P.M. Yet another job for Dna2: Checkpoint activation. DNA Repair (Amst) 2015, 32, 17–23. [Google Scholar] [CrossRef]
- Liu, W.; Zhou, M.; Li, Z.; Li, H.; Polaczek, P.; Dai, H.; Wu, Q.; Liu, C.; Karanja, K.K.; Popuri, V.; et al. A Selective Small Molecule DNA2 Inhibitor for Sensitization of Human Cancer Cells to Chemotherapy. EBioMedicine 2016, 6, 73–86. [Google Scholar] [CrossRef]
- Orlovetskie, N.; Serruya, R.; Abboud-Jarrous, G.; Jarrous, N. Targeted inhibition of WRN helicase, replication stress and cancer. Biochim. Biophys. Acta Rev. Cancer 2017, 1867, 42–48. [Google Scholar] [CrossRef]
- Moles, R.; Bai, X.T.; Chaib-Mezrag, H.; Nicot, C. WRN-targeted therapy using inhibitors NSC 19630 and NSC 617145 induce apoptosis in HTLV-1-transformed adult T-cell leukemia cells. J. Hematol. Oncol. 2016, 9, 121. [Google Scholar] [CrossRef]
- Aggarwal, M.; Banerjee, T.; Sommers, J.A.; Brosh, R.M., Jr. Targeting an Achilles’ heel of cancer with a WRN helicase inhibitor. Cell Cycle 2013, 12, 3329–3335. [Google Scholar] [CrossRef]
- Sommers, J.A.; Kulikowicz, T.; Croteau, D.L.; Dexheimer, T.; Dorjsuren, D.; Jadhav, A.; Maloney, D.J.; Simeonov, A.; Bohr, V.A.; Brosh, R.M., Jr. A high-throughput screen to identify novel small molecule inhibitors of the Werner Syndrome Helicase-Nuclease (WRN). PLoS ONE 2019, 14, e0210525. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, T.; Aggarwal, M.; Sommers, J.A.; Brosh, R.M., Jr. Biochemical and cell biological assays to identify and characterize DNA helicase inhibitors. Methods 2016, 108, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Malfatti, M.C.; Gerratana, L.; Dalla, E.; Isola, M.; Damante, G.; Di Loreto, C.; Puglisi, F.; Tell, G. APE1 and NPM1 protect cancer cells from platinum compounds cytotoxicity and their expression pattern has a prognostic value in TNBC. J. Exp. Clin. Cancer Res. 2019, 38, 309. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.L.; He, F.; Ye, J.Z.; Wu, H.N.; Zhang, J.Y.; Liu, Z.H.; Li, Y.Q.; Luo, X.L.; Lin, Y.; Liang, R. APE1 overexpression is associated with poor survival in patients with solid tumors: A meta-analysis. Oncotarget 2017, 8, 59720–59728. [Google Scholar] [CrossRef] [PubMed]
- Poletto, M.; Malfatti, M.C.; Dorjsuren, D.; Scognamiglio, P.L.; Marasco, D.; Vascotto, C.; Jadhav, A.; Maloney, D.J.; Wilson, D.M., 3rd; Simeonov, A.; et al. Inhibitors of the apurinic/apyrimidinic endonuclease 1 (APE1)/nucleophosmin (NPM1) interaction that display anti-tumor properties. Mol. Carcinog. 2016, 55, 688–704. [Google Scholar] [CrossRef]
- Codrich, M.; Comelli, M.; Malfatti, M.C.; Mio, C.; Ayyildiz, D.; Zhang, C.; Kelley, M.R.; Terrosu, G.; Pucillo, C.E.M.; Tell, G. Inhibition of APE1-endonuclease activity affects cell metabolism in colon cancer cells via a p53-dependent pathway. DNA Repair (Amst) 2019, 82, 102675. [Google Scholar] [CrossRef]
- Guerreiro, P.S.; Corvacho, E.; Costa, J.G.; Saraiva, N.; Fernandes, A.S.; Castro, M.; Miranda, J.P.; Oliveira, N.G. The APE1 redox inhibitor E3330 reduces collective cell migration of human breast cancer cells and decreases chemoinvasion and colony formation when combined with docetaxel. Chem. Biol. Drug Des. 2017, 90, 561–571. [Google Scholar] [CrossRef]
- Ma, X.; Dang, C.; Min, W.; Diao, Y.; Hui, W.; Wang, X.; Dai, Z.; Wang, X.; Kang, H. Downregulation of APE1 potentiates breast cancer cells to olaparib by inhibiting PARP-1 expression. Breast Cancer Res. Treat. 2019, 176, 109–117. [Google Scholar] [CrossRef]
- Nickoloff, J.A.; Jones, D.; Lee, S.H.; Williamson, E.A.; Hromas, R. Drugging the Cancers Addicted to DNA Repair. J. Natl. Cancer Inst. 2017, 109, 1–13. [Google Scholar] [CrossRef]
- Jaiswal, A.S.; Panda, H.; Law, B.K.; Sharma, J.; Jani, J.; Hromas, R.; Narayan, S. NSC666715 and Its Analogs Inhibit Strand-Displacement Activity of DNA Polymerase beta and Potentiate Temozolomide-Induced DNA Damage, Senescence and Apoptosis in Colorectal Cancer Cells. PLoS ONE 2015, 10, e0123808. [Google Scholar] [CrossRef]
- Song, X.; Wang, S.; Hong, X.; Li, X.; Zhao, X.; Huai, C.; Chen, H.; Gao, Z.; Qian, J.; Wang, J.; et al. Single nucleotide polymorphisms of nucleotide excision repair pathway are significantly associated with outcomes of platinum-based chemotherapy in lung cancer. Sci. Rep. 2017, 7, 11785. [Google Scholar] [CrossRef] [PubMed]
- Pajuelo-Lozano, N.; Bargiela-Iparraguirre, J.; Dominguez, G.; Quiroga, A.G.; Perona, R.; Sanchez-Perez, I. XPA, XPC, and XPD Modulate Sensitivity in Gastric Cisplatin Resistance Cancer Cells. Front. Pharmacol. 2018, 9, 1197. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.B.; Chen, Y.; Meng, X.D.; Yu, P.; He, X.; Li, J. Nucleotide Excision Repair Factor XPC Ameliorates Prognosis by Increasing the Susceptibility of Human Colorectal Cancer to Chemotherapy and Ionizing Radiation. Front. Oncol. 2018, 8, 290. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Wang, Y.; Chen, L.; Gan, Y.; Wu, Q. High ERCC1 expression is associated with platinum-resistance, but not survival in patients with epithelial ovarian cancer. Oncol. Lett. 2016, 12, 857–862. [Google Scholar] [CrossRef]
- Chabanon, R.M.; Muirhead, G.; Krastev, D.B.; Adam, J.; Morel, D.; Garrido, M.; Lamb, A.; Henon, C.; Dorvault, N.; Rouanne, M.; et al. PARP inhibition enhances tumor cell-intrinsic immunity in ERCC1-deficient non-small cell lung cancer. J. Clin. Investig. 2019, 129, 1211–1228. [Google Scholar] [CrossRef]
- Cheng, H.; Zhang, Z.; Borczuk, A.; Powell, C.A.; Balajee, A.S.; Lieberman, H.B.; Halmos, B. PARP inhibition selectively increases sensitivity to cisplatin in ERCC1-low non-small cell lung cancer cells. Carcinogenesis 2013, 34, 739–749. [Google Scholar] [CrossRef]
- Arora, S.; Heyza, J.; Zhang, H.; Kalman-Maltese, V.; Tillison, K.; Floyd, A.M.; Chalfin, E.M.; Bepler, G.; Patrick, S.M. Identification of small molecule inhibitors of ERCC1-XPF that inhibit DNA repair and potentiate cisplatin efficacy in cancer cells. Oncotarget 2016, 7, 75104–75117. [Google Scholar] [CrossRef]
- McNeil, E.M.; Astell, K.R.; Ritchie, A.M.; Shave, S.; Houston, D.R.; Bakrania, P.; Jones, H.M.; Khurana, P.; Wallace, C.; Chapman, T.; et al. Inhibition of the ERCC1-XPF structure-specific endonuclease to overcome cancer chemoresistance. DNA Repair (Amst) 2015, 31, 19–28. [Google Scholar] [CrossRef]
- Burkitt, K.; Ljungman, M. Phenylbutyrate interferes with the Fanconi anemia and BRCA pathway and sensitizes head and neck cancer cells to cisplatin. Mol. Cancer 2008, 7, 24. [Google Scholar] [CrossRef]
- Chen, C.C.; Taniguchi, T.; D’Andrea, A. The Fanconi anemia (FA) pathway confers glioma resistance to DNA alkylating agents. J. Mol. Med. (Berl) 2007, 85, 497–509. [Google Scholar] [CrossRef]
- Chirnomas, D.; Taniguchi, T.; de la Vega, M.; Vaidya, A.P.; Vasserman, M.; Hartman, A.R.; Kennedy, R.; Foster, R.; Mahoney, J.; Seiden, M.V.; et al. Chemosensitization to cisplatin by inhibitors of the Fanconi anemia/BRCA pathway. Mol. Cancer Ther. 2006, 5, 952–961. [Google Scholar] [CrossRef] [PubMed]
- Kee, Y.; Huang, M.; Chang, S.; Moreau, L.A.; Park, E.; Smith, P.G.; D’Andrea, A.D. Inhibition of the Nedd8 system sensitizes cells to DNA interstrand cross-linking agents. Mol. Cancer Res. 2012, 10, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Li, S.; Cui, X.; Han, K.; Wang, J.; Hou, X.; Cui, L.; He, S.; Xiao, J.; Yang, Y. Inhibition of Ubiquitin Specific Protease 1 Sensitizes Colorectal Cancer Cells to DNA-Damaging Chemotherapeutics. Front. Oncol. 2019, 9, 1406. [Google Scholar] [CrossRef]
- Liang, Q.; Dexheimer, T.S.; Zhang, P.; Rosenthal, A.S.; Villamil, M.A.; You, C.; Zhang, Q.; Chen, J.; Ott, C.A.; Sun, H.; et al. A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses. Nat. Chem. Biol. 2014, 10, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, R.D.; Chen, C.C.; Stuckert, P.; Archila, E.M.; De la Vega, M.A.; Moreau, L.A.; Shimamura, A.; D’Andrea, A.D. Fanconi anemia pathway-deficient tumor cells are hypersensitive to inhibition of ataxia telangiectasia mutated. J. Clin. Investig. 2007, 117, 1440–1449. [Google Scholar] [CrossRef] [PubMed]
- Kamaletdinova, T.; Fanaei-Kahrani, Z.; Wang, Z.Q. The Enigmatic Function of PARP1: From PARylation Activity to PAR Readers. Cells 2019, 8, 1625. [Google Scholar] [CrossRef]
- Chen, K.; Li, Y.; Xu, H.; Zhang, C.; Li, Z.; Wang, W.; Wang, B. An analysis of the gene interaction networks identifying the role of PARP1 in metastasis of non-small cell lung cancer. Oncotarget 2017, 8, 87263–87275. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Y.; Zhao, Y.; Gao, D.; Xing, J.; Liu, H. High PARP-1 expression is associated with tumor invasion and poor prognosis in gastric cancer. Oncol. Lett. 2016, 12, 3825–3835. [Google Scholar] [CrossRef]
- Giovannini, S.; Weller, M.C.; Repmann, S.; Moch, H.; Jiricny, J. Synthetic lethality between BRCA1 deficiency and poly(ADP-ribose) polymerase inhibition is modulated by processing of endogenous oxidative DNA damage. Nucleic Acids Res. 2019, 47, 9132–9143. [Google Scholar] [CrossRef]
- Awada, A.; Campone, M.; Varga, A.; Aftimos, P.; Frenel, J.S.; Bahleda, R.; Gombos, A.; Bourbouloux, E.; Soria, J.C. An open-label, dose-escalation study to evaluate the safety and pharmacokinetics of CEP-9722 (a PARP-1 and PARP-2 inhibitor) in combination with gemcitabine and cisplatin in patients with advanced solid tumors. Anticancer Drugs 2016, 27, 342–348. [Google Scholar] [CrossRef]
- Russo, A.L.; Kwon, H.C.; Burgan, W.E.; Carter, D.; Beam, K.; Weizheng, X.; Zhang, J.; Slusher, B.S.; Chakravarti, A.; Tofilon, P.J.; et al. In vitro and in vivo radiosensitization of glioblastoma cells by the poly (ADP-ribose) polymerase inhibitor E7016. Clin. Cancer Res. 2009, 15, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Jannetti, S.A.; Zeglis, B.M.; Zalutsky, M.R.; Reiner, T. Poly(ADP-Ribose)Polymerase (PARP) Inhibitors and Radiation Therapy. Front. Pharmacol 2020, 11, 170. [Google Scholar] [CrossRef] [PubMed]
- Pirovano, G.; Jannetti, S.A.; Carter, L.M.; Sadique, A.; Kossatz, S.; Guru, N.; Demetrio De Souza Franca, P.; Maeda, M.; Zeglis, B.M.; Lewis, J.S.; et al. Targeted Brain Tumor Radiotherapy Using an Auger Emitter. Clin. Cancer Res. 2020, 26, 2871–2881. [Google Scholar] [CrossRef] [PubMed]
- Makvandi, M.; Lee, H.; Puentes, L.N.; Reilly, S.W.; Rathi, K.S.; Weng, C.C.; Chan, H.S.; Hou, C.; Raman, P.; Martinez, D.; et al. Targeting PARP-1 with Alpha-Particles Is Potently Cytotoxic to Human Neuroblastoma in Preclinical Models. Mol. Cancer Ther. 2019, 18, 1195–1204. [Google Scholar] [CrossRef]
- Gadducci, A.; Guarneri, V.; Peccatori, F.A.; Ronzino, G.; Scandurra, G.; Zamagni, C.; Zola, P.; Salutari, V. Current strategies for the targeted treatment of high-grade serous epithelial ovarian cancer and relevance of BRCA mutational status. J. Ovarian Res. 2019, 12, 9. [Google Scholar] [CrossRef]
- Gadducci, A.; Guerrieri, M.E. PARP inhibitors alone and in combination with other biological agents in homologous recombination deficient epithelial ovarian cancer: From the basic research to the clinic. Crit. Rev. Oncol. Hematol. 2017, 114, 153–165. [Google Scholar] [CrossRef]
- D’Andrea, A.D. Mechanisms of PARP inhibitor sensitivity and resistance. DNA Repair (Amst) 2018, 71, 172–176. [Google Scholar] [CrossRef]
- La Ferla, M.; Mercatanti, A.; Rocchi, G.; Lodovichi, S.; Cervelli, T.; Pignata, L.; Caligo, M.A.; Galli, A. Expression of human poly (ADP-ribose) polymerase 1 in Saccharomyces cerevisiae: Effect on survival, homologous recombination and identification of genes involved in intracellular localization. Mutat. Res. 2015, 774, 14–24. [Google Scholar] [CrossRef]
- Lodovichi, S.; Mercatanti, A.; Cervelli, T.; Galli, A. Computational analysis of data from a genome-wide screening identifies new PARP1 functional interactors as potential therapeutic targets. Oncotarget 2019, 10, 2722–2737. [Google Scholar] [CrossRef]
- Antolin, A.A.; Ameratunga, M.; Banerji, U.; Clarke, P.A.; Workman, P.; Al-Lazikani, B. The kinase polypharmacology landscape of clinical PARP inhibitors. Sci. Rep. 2020, 10, 2585. [Google Scholar] [CrossRef]
- Knezevic, C.E.; Wright, G.; Rix, L.L.R.; Kim, W.; Kuenzi, B.M.; Luo, Y.; Watters, J.M.; Koomen, J.M.; Haura, E.B.; Monteiro, A.N.; et al. Proteome-wide Profiling of Clinical PARP Inhibitors Reveals Compound-Specific Secondary Targets. Cell Chem. Biol. 2016, 23, 1490–1503. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.R.; Corrales, L.; Gajewski, T.F. Innate immune recognition of cancer. Annu. Rev. Immunol. 2015, 33, 445–474. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Garraway, L.A.; Ashworth, A.; Weber, B. Synthetic lethality as an engine for cancer drug target discovery. Nat. Rev. Drug Discov. 2020, 19, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Dhanjal, J.K.; Radhakrishnan, N.; Sundar, D. Identifying synthetic lethal targets using CRISPR/Cas9 system. Methods 2017, 131, 66–73. [Google Scholar] [CrossRef] [PubMed]
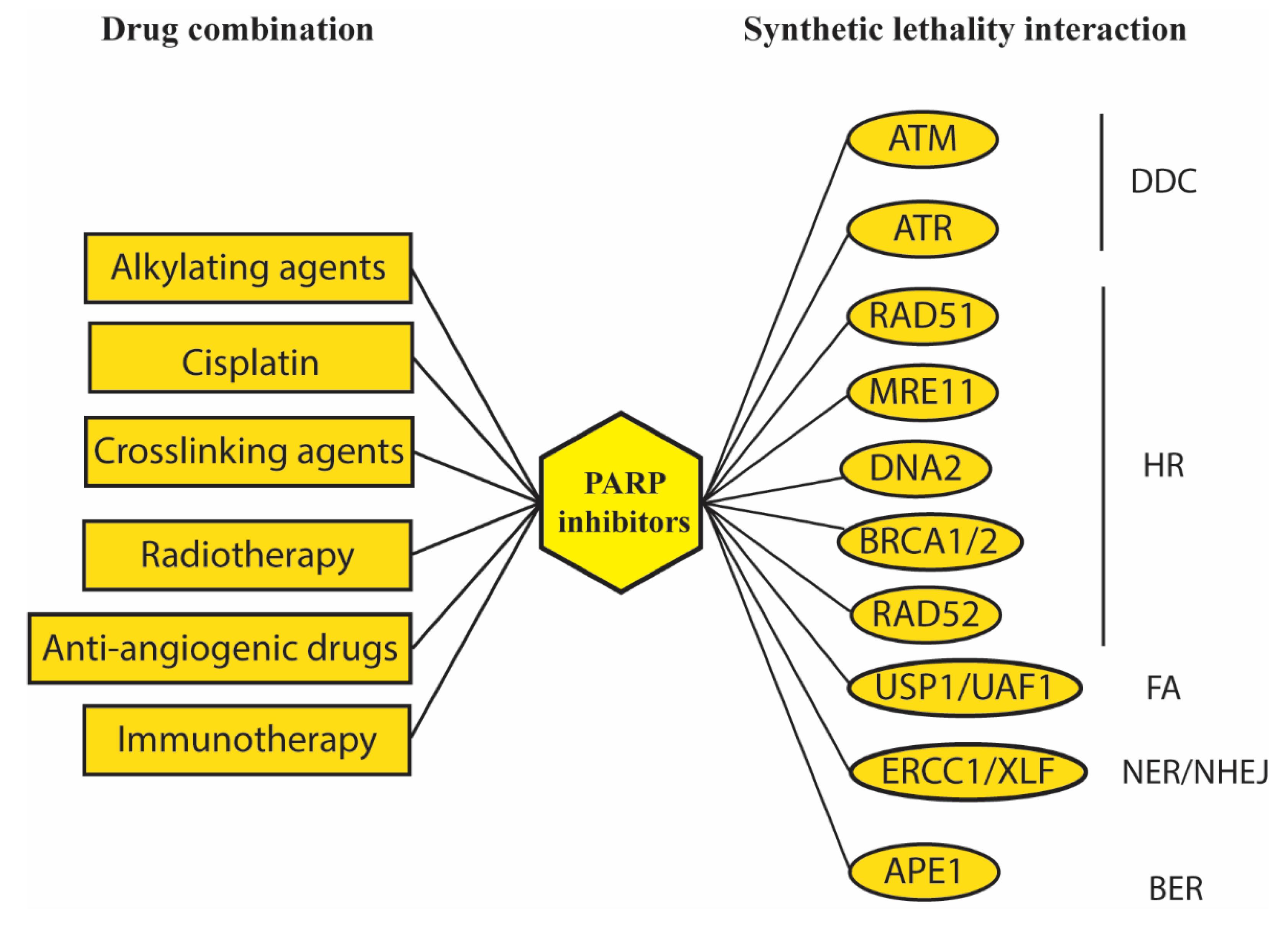
| DDC Pathway | |||||
|---|---|---|---|---|---|
| Protein | Drug Name | Combined Treatment | Possible Synthetic Lethal Interaction | Clinical Trial | References |
| ATM | AZD0156 | Ionizing radiation Topoisomerase I and II inhibitors | PARP1 inhibition | NCT02588105 | [105,106,107,108] |
| AZD1390 | NCT03423628 | ||||
| KU-55933 | |||||
| KU-60019 | |||||
| KU-59403 | |||||
| ATR | M6620 | Ionizing radiation Cisplatin | ATM inhibition | NCT02589522 NCT02567422 | [115,116,117,118,119,120,121,122,123] |
| AZD6738 | Ionizing radiation | ATM inhibition WEE1 inhibition | |||
| BAY1895344 | Ionizing radiation Cisplatin | ATM inhibition PARP1 inhibition | |||
| VE-821 | Ionizing radiation Anti-metabolites | ATM inhibition CHK1 inhibition | |||
| DNA-PK | M3814 | Topoisomerase I and II inhibitors Anti-metabolites Ionizing radiation | ATM inhibition | NCT03770689 | [109,110,111,112,113] |
| VX-984 | Ionizing radiation Topoisomerase I and II inhibitors | NCT02644278 | |||
| CC-115 | Ionizing radiation | NCT02516813 | |||
| MSC2490484A | Ionizing radiation | NCT02833883 | |||
| CHK1 | AZD7762 | Anti-metabolites | ATR inhibition | NCT00937664 | [120] |
| MK-8776 | FANCD2 inhibition | [124] | |||
| WEE1 | AZD1775 | Cisplatin Ionizing radiation | PARP1 inhibition | NCT03028766 | [119] |
| HR | |||||
|---|---|---|---|---|---|
| Protein | Drug Name | Combined Treatment | Possible Synthetic Lethal Interaction | Clinical Trial | References |
| MRE11 | Mirin | PARP1 inhibition | [126,127,128] | ||
| RAD51 | B02 | Ionizing radiation Topoisomerases I and II inhibitors | PARP1 inhibition | [129,130,131,132] | |
| RI-1 | Cisplatin Ionizing radiation | [132,133] | |||
| IBR120 | Ionizing radiation | [134,135] | |||
| CYT-0851 | NCT03997968 | [125] | |||
| BRCA1 | PARP1 inhibition | [24,137,138] | |||
| BRCA2 | |||||
| RAD52 | F79 6-OH-dopa D-I03 | Cisplatin | PARP1 inhibition BRCA2 inhibition BRCA1/PALB2 inhibition | [139,140,141,142] | |
| A5MP AICAR/ZMP | BRCA1 inhibition | ||||
| NP-004255 F779-0434 | BRCA2 inhibition | ||||
| NHEJ | ||||
|---|---|---|---|---|
| Protein | Drug Name | Combined Treatment | Possible Synthetic Lethal Interaction | References |
| TP53BP1 | i53 | Cisplatin | ATR inhibition | [146,147,148] |
| UNC2170 | ||||
| KU70/80 | Ionizing radiation | [143] | ||
| LIG4 | Alkyating agents | [145] | ||
| XLF | G3 | Cisplatin | PARP1 inhibition | [144] |
| Anti-metabolities | ||||
| Pol-θ | Novobicin | Topoisomerase I and II inhibitors | ATR inhibition | [150] |
| FANCD2 inhibition | [152,153] | |||
| Helicases/Nucleases | ||||
|---|---|---|---|---|
| Protein | Drug Name | Combined Treatment | Possible Synthetic Lethal Interaction | References |
| BLM | ML216 | [155] | ||
| DNA2 | C5 | Topoisomerase I and II inhibitors | PARP1 inhibition | [157] |
| WRN | NSC 19630 | [159,160,161] | ||
| NSC617145 | Mitomycin C | FANCD2/DNA-PK inhibition | ||
| BER/MMR | ||||
|---|---|---|---|---|
| Protein | Drug Name | Combined Treatment | Possible Synthetic Lethal Interaction | References |
| APE1 | E3330 | Bleomycin | PARP1 inhibition | [163,164,165,166,167,168] |
| POL-β | NSC666715 | Alkylating agents | MSH2/MLH1 inhibition | [169,170] |
| NER/FA | ||||
|---|---|---|---|---|
| Protein | Drug Name | Combined Treatment | Possible Synthetic Lethal Interaction | References |
| ERCC1 | NSC16168 | Cisplatin | PARP1 inhibition | [174,175,176,177,178] |
| FANCS | Phenylbutyrate | Cisplatin | [179] | |
| FANCD2 | Curcumin | Cisplatin | ATM inhibition CHK1 inhibition | [124] |
| MLN4924 | DNA cross-linking agents | [180,181,182] | ||
| USP1/UAF1 | ML323 | Topoisomerase I and II inhibitors | PARP1 inhibition | [183,184] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lodovichi, S.; Cervelli, T.; Pellicioli, A.; Galli, A. Inhibition of DNA Repair in Cancer Therapy: Toward a Multi-Target Approach. Int. J. Mol. Sci. 2020, 21, 6684. https://doi.org/10.3390/ijms21186684
Lodovichi S, Cervelli T, Pellicioli A, Galli A. Inhibition of DNA Repair in Cancer Therapy: Toward a Multi-Target Approach. International Journal of Molecular Sciences. 2020; 21(18):6684. https://doi.org/10.3390/ijms21186684
Chicago/Turabian StyleLodovichi, Samuele, Tiziana Cervelli, Achille Pellicioli, and Alvaro Galli. 2020. "Inhibition of DNA Repair in Cancer Therapy: Toward a Multi-Target Approach" International Journal of Molecular Sciences 21, no. 18: 6684. https://doi.org/10.3390/ijms21186684
APA StyleLodovichi, S., Cervelli, T., Pellicioli, A., & Galli, A. (2020). Inhibition of DNA Repair in Cancer Therapy: Toward a Multi-Target Approach. International Journal of Molecular Sciences, 21(18), 6684. https://doi.org/10.3390/ijms21186684






