Enhanced Antiproliferative Effect of Combined Treatment with Calcitriol and All-Trans Retinoic Acid in Relation to Vitamin D Receptor and Retinoic Acid Receptor α Expression in Osteosarcoma Cell Lines
Abstract
1. Introduction
2. Results
2.1. Calcitriol Slightly Increases the Antiproliferative Effect of ATRA in the Saos-2 Reference Cell Line
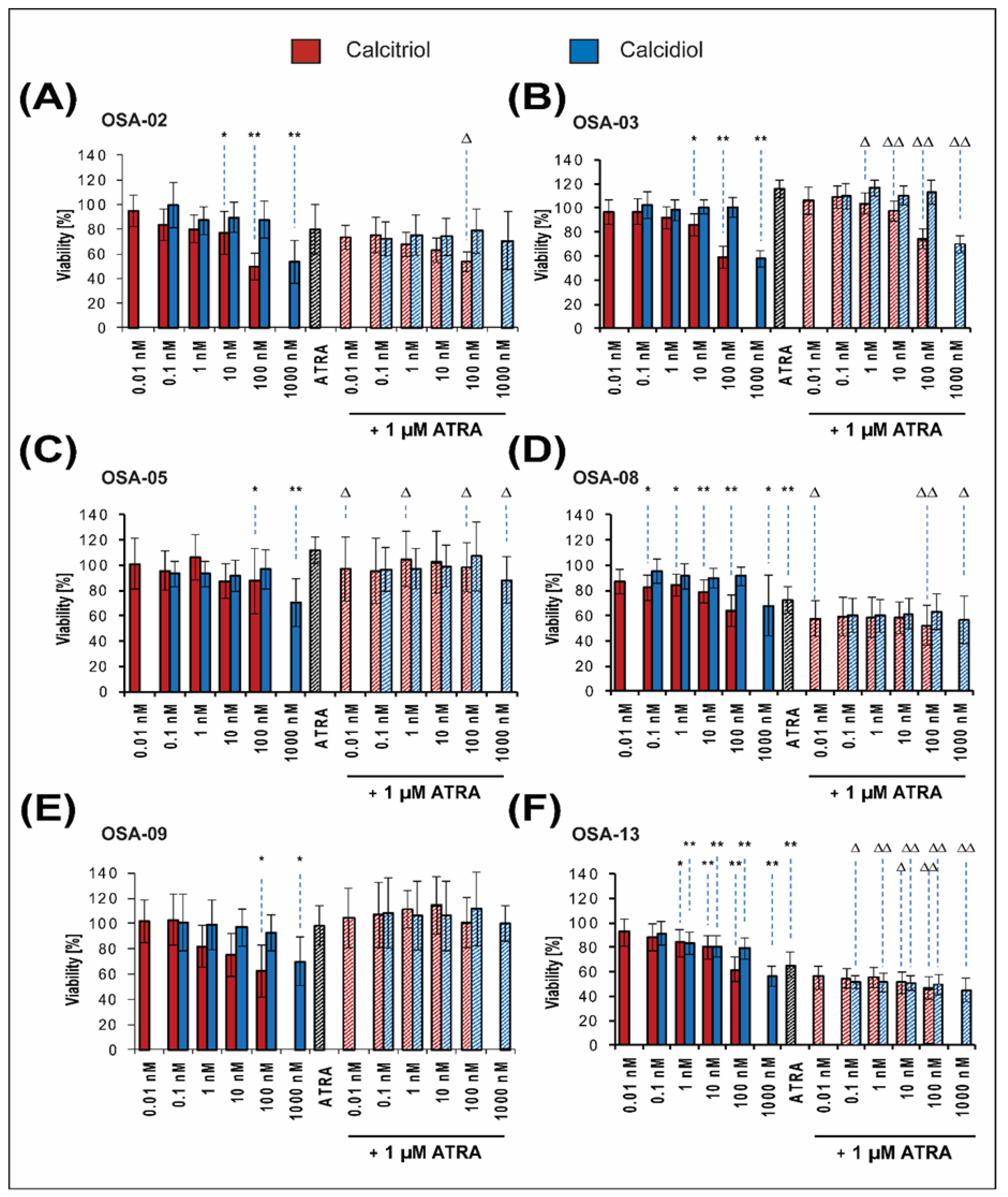
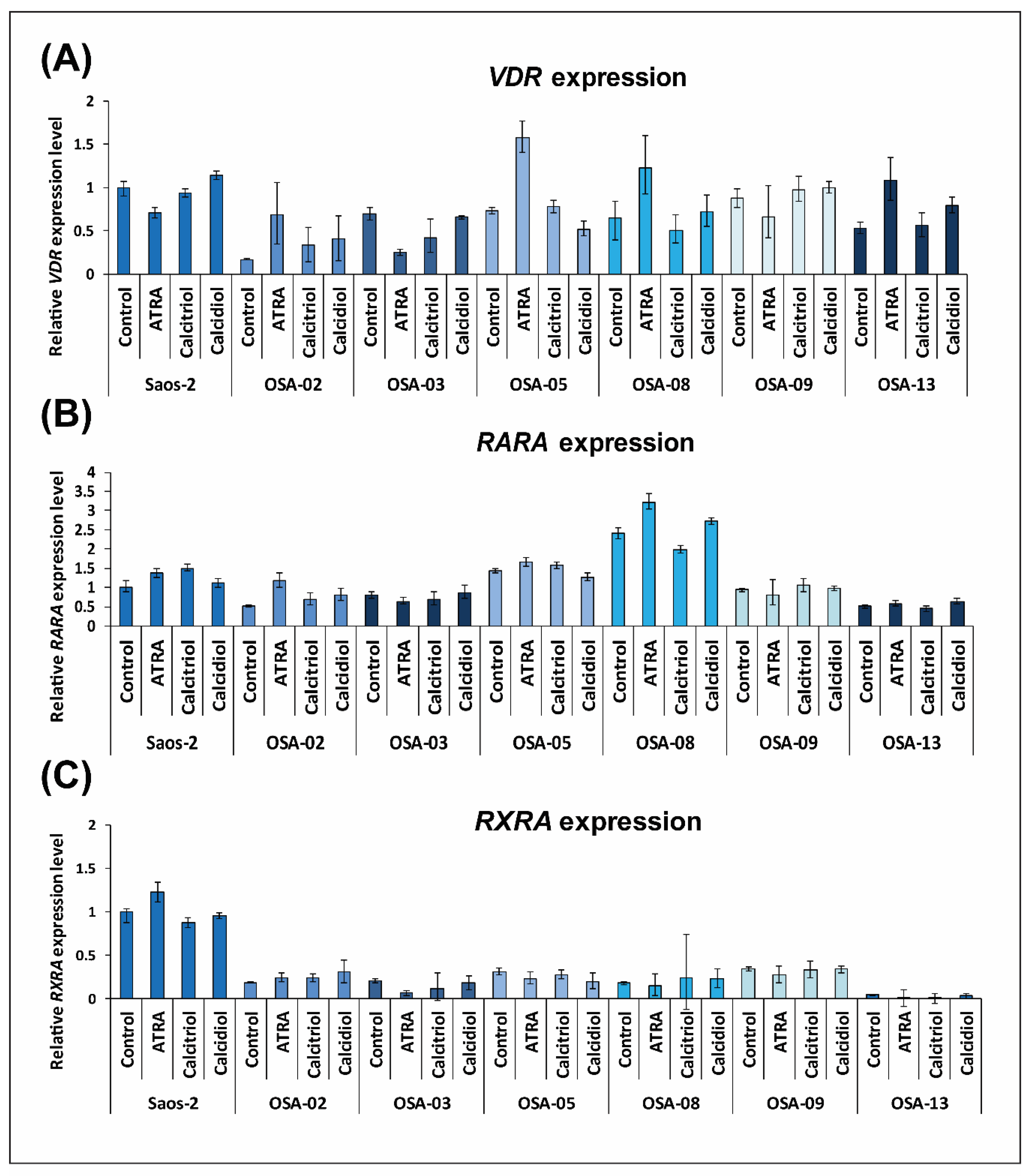
2.2. Patient-Derived Osteosarcoma Cell Lines Show Various Levels of Sensitivity to Calcitriol, Calcidiol, and Their Combinations with ATRA
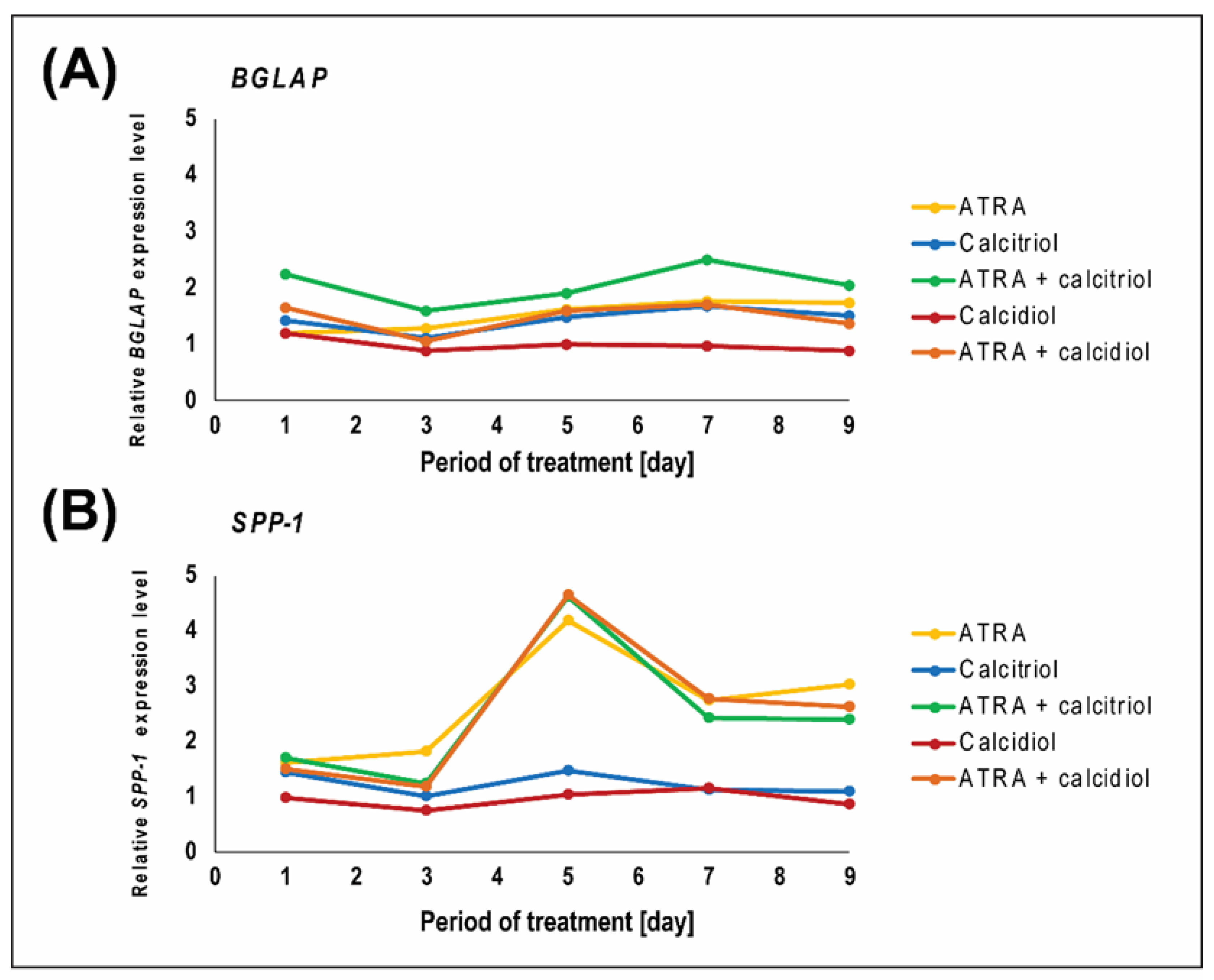
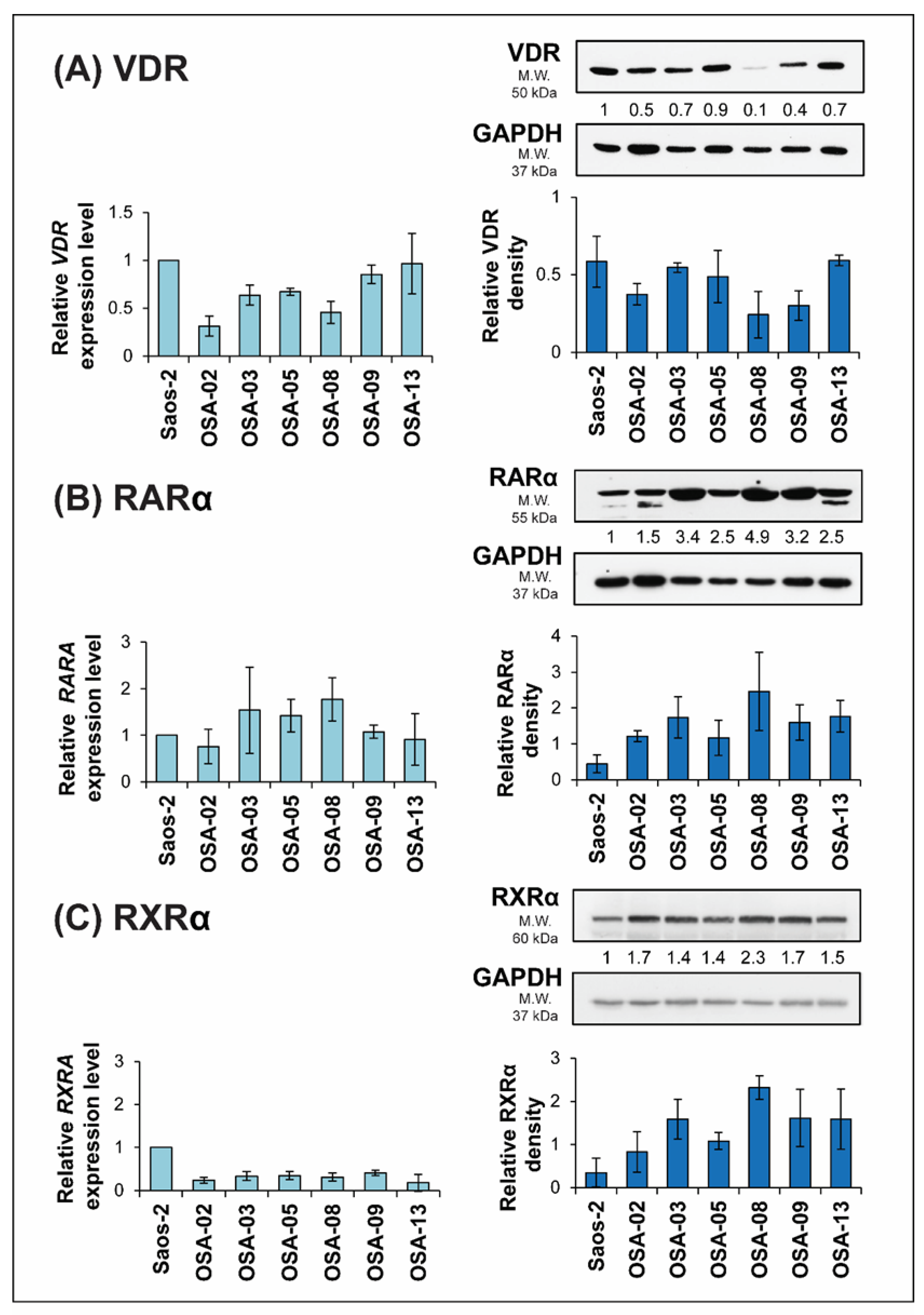
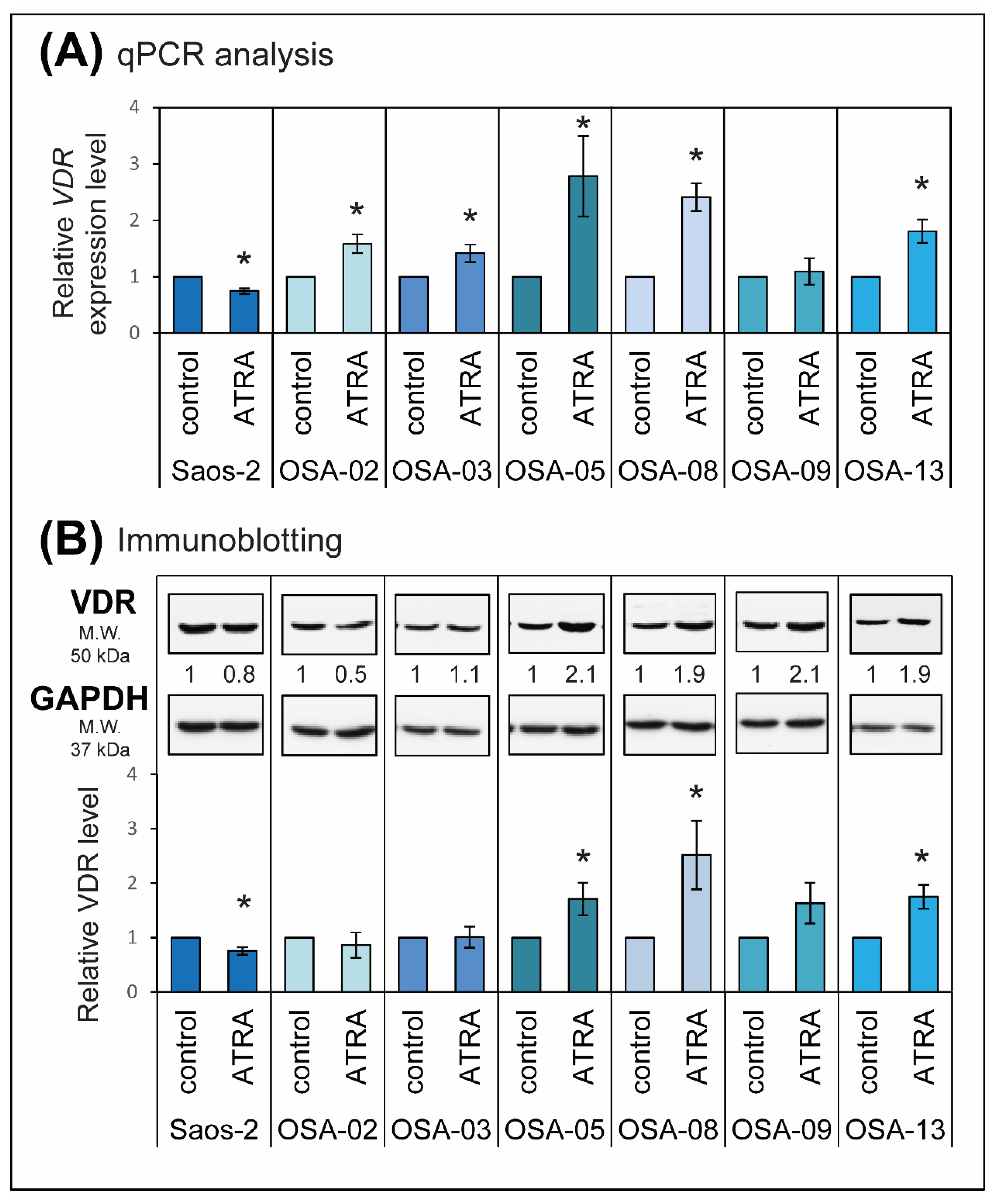
2.3. ATRA Influences VDR Expression
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Chemicals
4.3. Treatment
4.4. Cell Viability
4.5. RT-PCR
4.6. Immunoblotting
4.7. Statistics
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ATRA | All-trans retinoic acid |
| COX | Cyclooxygenase |
| DMEM | Dulbecco’s modified Eagle’s medium |
| DMSO | Dimethyl sulfoxide |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| LOX | Lipoxygenase |
| MTT | 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide |
| RARα | Retinoic acid receptor α |
| RARE | Retinoic acid response element |
| RXRα | Retinoid X receptor α |
| VDR | Vitamin D receptor |
| VDRE | Vitamin D response element |
| VDRE-BP | Vitamin D response element-binding protein |
Appendix A
| Calcitriol (Ya) | ATRA 1 µM (Yb) | Yab,P | Yab,O | Calcitriol (Ya) | ATRA 1 µM (Yb) | Yab,P | Yab,O | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Saos-2 Day 3 | 0.01 nM | 0.03 | 0.27 | 0.29 | 0.35 | OSA-05 | 0.01 nM | −0.01 | −0.12 | −0.13 | 0.03 |
| 0.1 nM | −0.01 | 0.27 | 0.26 | 0.33 | 0.1 nM | 0.04 | −0.12 | −0.07 | 0.04 | ||
| 1 nM | 0.03 | 0.27 | 0.29 | 0.33 | 1 nM | −0.06 | −0.12 | −0.19 | −0.04 | ||
| 10 nM | 0.05 | 0.27 | 0.30 | 0.40 | 10 nM | 0.13 | −0.12 | 0.02 | −0.03 | ||
| 100 nM | 0.03 | 0.27 | 0.28 | 0.38 | 100 nM | 0.13 | −0.12 | 0.02 | 0.02 | ||
| Saos-2 Day 7 | 0.01 nM | −0.01 | 0.35 | 0.34 | 0.48 | OSA-08 | 0.01 nM | 0.13 | 0.28 | 0.37 | 0.43 |
| 0.1 nM | −0.03 | 0.35 | 0.34 | 0.48 | 0.1 nM | 0.18 | 0.28 | 0.41 | 0.41 | ||
| 1 nM | 0.00 | 0.35 | 0.35 | 0.47 | 1 nM | 0.16 | 0.28 | 0.39 | 0.41 | ||
| 10 nM | 0.03 | 0.35 | 0.37 | 0.49 | 10 nM | 0.21 | 0.28 | 0.43 | 0.42 | ||
| 100 nM | 0.02 | 0.35 | 0.37 | 0.48 | 100 nM | 0.36 | 0.28 | 0.54 | 0.48 | ||
| OSA-02 | 0.01 nM | 0.05 | 0.20 | 0.24 | 0.27 | OSA-09 | 0.01 nM | −0.02 | 0.02 | 0.00 | −0.04 |
| 0.1 nM | 0.17 | 0.20 | 0.33 | 0.25 | 0.1 nM | −0.03 | 0.02 | −0.01 | −0.07 | ||
| 1 nM | 0.20 | 0.20 | 0.36 | 0.32 | 1 nM | 0.18 | 0.02 | 0.19 | −0.11 | ||
| 10 nM | 0.23 | 0.20 | 0.38 | 0.37 | 10 nM | 0.25 | 0.02 | 0.26 | −0.15 | ||
| 100 nM | 0.51 | 0.20 | 0.60 | 0.47 | 100 nM | 0.38 | 0.02 | 0.38 | 0.00 | ||
| OSA-03 | 0.01 nM | 0.03 | −0.15 | −0.12 | −0.06 | OSA-13 | 0.01 nM | 0.07 | 0.35 | 0.40 | 0.43 |
| 0.1 nM | 0.03 | −0.15 | −0.12 | −0.09 | 0.1 nM | 0.12 | 0.35 | 0.43 | 0.46 | ||
| 1 nM | 0.08 | −0.15 | −0.06 | −0.04 | 1 nM | 0.16 | 0.35 | 0.45 | 0.45 | ||
| 10 nM | 0.14 | −0.15 | 0.01 | 0.03 | 10 nM | 0.20 | 0.35 | 0.48 | 0.49 | ||
| 100 nM | 0.41 | −0.15 | 0.32 | 0.26 | 100 nM | 0.39 | 0.35 | 0.60 | 0.54 | ||
| Calcidiol (Ya) | ATRA 1 µM (Yb) | Yab,P | Yab,O | Calcidiol (Ya) | ATRA 1 µM (Yb) | Yab,P | Yab,O | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Saos-2 Day 3 | 0.1 nM | −0.02 | 0.27 | 0.25 | 0.30 | OSA-05 | 0.1 nM | 0.07 | −0.12 | −0.05 | 0.04 |
| 1 nM | 0.01 | 0.27 | 0.28 | 0.29 | 1 nM | 0.07 | −0.12 | −0.05 | 0.03 | ||
| 10 nM | 0.04 | 0.27 | 0.29 | 0.31 | 10 nM | 0.08 | −0.12 | −0.03 | 0.01 | ||
| 100 nM | 0.03 | 0.27 | 0.29 | 0.32 | 100 nM | 0.03 | −0.12 | −0.09 | −0.07 | ||
| 1000 nM | −0.03 | 0.27 | 0.24 | 0.28 | 1000 nM | 0.29 | −0.12 | 0.21 | 0.12 | ||
| Saos-2 Day 7 | 0.1 nM | 0.03 | 0.35 | 0.37 | 0.24 | OSA-08 | 0.1 nM | 0.05 | 0.28 | 0.31 | 0.40 |
| 1 nM | 0.04 | 0.35 | 0.38 | 0.26 | 1 nM | 0.09 | 0.28 | 0.34 | 0.40 | ||
| 10 nM | 0.03 | 0.35 | 0.37 | 0.26 | 10 nM | 0.10 | 0.28 | 0.35 | 0.39 | ||
| 100 nM | 0.03 | 0.35 | 0.37 | 0.26 | 100 nM | 0.09 | 0.28 | 0.34 | 0.37 | ||
| 1000 nM | 0.08 | 0.35 | 0.40 | 0.29 | 1000 nM | 0.32 | 0.28 | 0.51 | 0.44 | ||
| OSA-02 | 0.1 nM | 0.00 | 0.20 | 0.20 | 0.28 | OSA-09 | 0.1 nM | −0.01 | 0.02 | 0.01 | −0.08 |
| 1 nM | 0.13 | 0.20 | 0.30 | 0.25 | 1 nM | 0.01 | 0.02 | 0.02 | −0.06 | ||
| 10 nM | 0.10 | 0.20 | 0.28 | 0.26 | 10 nM | 0.03 | 0.02 | 0.04 | −0.06 | ||
| 100 nM | 0.12 | 0.20 | 0.30 | 0.21 | 100 nM | 0.07 | 0.02 | 0.09 | −0.12 | ||
| 1000 nM | 0.47 | 0.20 | 0.57 | 0.29 | 1000 nM | 0.30 | 0.02 | 0.31 | 0.00 | ||
| OSA-03 | 0.1 nM | −0.02 | −0.15 | −0.18 | −0.10 | OSA-13 | 0.1 nM | 0.09 | 0.35 | 0.41 | 0.48 |
| 1 nM | 0.02 | −0.15 | −0.13 | −0.16 | 1 nM | 0.16 | 0.35 | 0.46 | 0.49 | ||
| 10 nM | −0.01 | −0.15 | −0.16 | −0.10 | 10 nM | 0.19 | 0.35 | 0.47 | 0.50 | ||
| 100 nM | 0.00 | −0.15 | −0.16 | −0.13 | 100 nM | 0.21 | 0.35 | 0.49 | 0.50 | ||
| 1000 nM | 0.42 | −0.15 | 0.33 | 0.31 | 1000 nM | 0.44 | 0.35 | 0.64 | 0.55 | ||
| BGLAP Relative Expression | ||||||
| DAYS | ATRA | CALCITRIOL | ATRA + CALCITRIOL | CALCIDIOL | ATRA + CALCIDIOL | |
| 1 | MEAN | 1.198 | 1.426 | 2.255 | 1.185 | 1.645 |
| SD | 0.027 | 0.040 | 0.230 | 0.168 | 0.882 | |
| STATISTICS | * | * | * | |||
| 3 | MEAN | 1.292 | 1.124 | 1.604 | 0.888 | 1.067 |
| SD | 0.094 | 0.220 | 0.320 | 0.085 | 0.220 | |
| STATISTICS | * | |||||
| 5 | MEAN | 1.633 | 1.472 | 1.908 | 1.010 | 1.587 |
| SD | 0.182 | 0.344 | 0.370 | 0.208 | 0.384 | |
| STATISTICS | * | * | ||||
| 7 | MEAN | 1.749 | 1.681 | 2.497 | 0.964 | 1.710 |
| SD | 0.218 | 0.325 | 0.680 | 0.182 | 0.494 | |
| STATISTICS | * | * | * | |||
| 9 | MEAN | 1.728 | 1.505 | 2.054 | 1.710 | 1.366 |
| SD | 0.430 | 0.244 | 0.022 | 0.108 | 0.464 | |
| STATISTICS | * | * | ||||
| SPP1 Relative Expression | ||||||
| DAYS | ATRA | CALCITRIOL | ATRA + CALCITRIOL | CALCIDIOL | ATRA + CALCIDIOL | |
| 1 | MEAN | 1.619 | 1.432 | 1.705 | 0.972 | 1.495 |
| SD | 0.284 | 0.167 | 0.383 | 0.223 | 0.341 | |
| STATISTICS | ||||||
| 3 | MEAN | 1.828 | 1.000 | 1.238 | 0.754 | 1.191 |
| SD | 0.182 | 0.351 | 0.097 | 0.188 | 0.422 | |
| STATISTICS | * | |||||
| 5 | MEAN | 4.181 | 1.481 | 4.613 | 1.047 | 4.646 |
| SD | 0.506 | 0.247 | 0.192 | 0.198 | 0.787 | |
| STATISTICS | * | * | * | |||
| 7 | MEAN | 2.730 | 1.132 | 2.411 | 1.163 | 2.760 |
| SD | 0.354 | 0.067 | 0.631 | 0.038 | 0.794 | |
| STATISTICS | * | * | * | |||
| 9 | MEAN | 3.022 | 1.105 | 2.378 | 0.850 | 2.634 |
| SD | 0.575 | 0.412 | 0.771 | 0.174 | 0.820 | |
| STATISTICS | * | * | ||||
References
- Durfee, R.A.; Mohammed, M.; Luu, H.H. Review of Osteosarcoma and Current Management. Rheumatol. Ther. 2016, 3, 221–243. [Google Scholar] [CrossRef] [PubMed]
- Damron, T.A.; Ward, W.G.; Stewart, A. Osteosarcoma, Chondrosarcoma, and Ewing’s Sarcoma: National Cancer Data Base Report. Clin. Orthop. Relat. Res. 2007, 459, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Weiss, A.; Gill, J.; Goldberg, J.; Lagmay, J.; Spraker-Perlman, H.; Venkatramani, R.; Reed, D. Advances in Therapy for Pediatric Sarcomas. Curr. Oncol. Rep. 2014, 16, 395. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Tsuchiya, H.; Yamamoto, N.; Shirai, T.; Nishida, H.; Takeuchi, A.; Kimura, H.; Miwa, S.; Inatani, H.; Okamoto, H.; et al. Diagnosis and treatment of low-grade osteosarcoma: Experience with nine cases. Int. J. Clin. Oncol. 2014, 19, 731–738. [Google Scholar] [CrossRef]
- Hundsdoerfer, P.; Albrecht, M.; Rühl, U.; Fengler, R.; Kulozik, A.E.; Henze, G. Long-term outcome after polychemotherapy and intensive local radiation therapy of high-grade osteosarcoma. Eur. J. Cancer 2009, 45, 2447–2451. [Google Scholar] [CrossRef]
- Eilber, F.R.; Rosen, G. Adjuvant chemotherapy for osteosarcoma. Semin. Oncol. 1989, 16, 312–322. [Google Scholar]
- Ferrari, S.; Smeland, S.; Mercuri, M.; Bertoni, F.; Longhi, A.; Ruggieri, P.; Alvegard, T.A.; Picci, P.; Capanna, R.; Bernini, G.; et al. Neoadjuvant chemotherapy with high-dose Ifosfamide, high-dose methotrexate, cisplatin, and doxorubicin for patients with localized osteosarcoma of the extremity: A joint study by the Italian and Scandinavian Sarcoma Groups. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2005, 23, 8845–8852. [Google Scholar] [CrossRef]
- Yu, D.; Zhang, S.; Feng, A.; Xu, D.; Zhu, Q.; Mao, Y.; Zhao, Y.; Lv, Y.; Han, C.; Liu, R.; et al. Methotrexate, doxorubicin, and cisplatinum regimen is still the preferred option for osteosarcoma chemotherapy: A meta-analysis and clinical observation. Medicine 2019, 98, e15582. [Google Scholar] [CrossRef]
- Allison, D.C.; Carney, S.C.; Ahlmann, E.R.; Hendifar, A.; Chawla, S.; Fedenko, A.; Angeles, C.; Menendez, L.R. A meta-analysis of osteosarcoma outcomes in the modern medical era. Sarcoma 2012, 2012, 704872. [Google Scholar] [CrossRef]
- Bielack, S.S.; Smeland, S.; Whelan, J.S.; Marina, N.; Jovic, G.; Hook, J.M.; Krailo, M.D.; Gebhardt, M.; Pápai, Z.; Meyer, J.; et al. Methotrexate, Doxorubicin, and Cisplatin (MAP) Plus Maintenance Pegylated Interferon Alfa-2b Versus MAP Alone in Patients With Resectable High-Grade Osteosarcoma and Good Histologic Response to Preoperative MAP: First Results of the EURAMOS-1 Good Response Randomized Controlled Trial. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2015, 33, 2279–2287. [Google Scholar] [CrossRef]
- Lewis, I.J.; Nooij, M.A.; Whelan, J.; Sydes, M.R.; Grimer, R.; Hogendoorn, P.C.W.; Memon, M.A.; Weeden, S.; Uscinska, B.M.; van Glabbeke, M.; et al. Improvement in histologic response but not survival in osteosarcoma patients treated with intensified chemotherapy: A randomized phase III trial of the European Osteosarcoma Intergroup. J. Natl. Cancer Inst. 2007, 99, 112–128. [Google Scholar] [CrossRef] [PubMed]
- Marina, N.M.; Smeland, S.; Bielack, S.S.; Bernstein, M.; Jovic, G.; Krailo, M.D.; Hook, J.M.; Arndt, C.; van den Berg, H.; Brennan, B.; et al. Comparison of MAPIE versus MAP in patients with a poor response to preoperative chemotherapy for newly diagnosed high-grade osteosarcoma (EURAMOS-1): An open-label, international, randomised controlled trial. Lancet Oncol. 2016, 17, 1396–1408. [Google Scholar] [CrossRef]
- Nowak, D.; Stewart, D.; Koeffler, H.P. Differentiation therapy of leukemia: 3 decades of development. Blood 2009, 113, 3655–3665. [Google Scholar] [CrossRef] [PubMed]
- Enane, F.O.; Saunthararajah, Y.; Korc, M. Differentiation therapy and the mechanisms that terminate cancer cell proliferation without harming normal cells. Cell Death Dis. 2018, 9, 912. [Google Scholar] [CrossRef] [PubMed]
- Sakashita, A.; Kizaki, M.; Pakkala, S.; Schiller, G.; Tsuruoka, N.; Tomosaki, R.; Cameron, J.F.; Dawson, M.I.; Koeffler, H.P. 9-cis-retinoic acid: Effects on normal and leukemic hematopoiesis in vitro. Blood 1993, 81, 1009–1016. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.A.; Villablanca, J.G.; Reynolds, C.P.; Avramis, V.I. Pharmacokinetic studies of 13-cis-retinoic acid in pediatric patients with neuroblastoma following bone marrow transplantation. Cancer Chemother. Pharmacol. 1996, 39, 34–41. [Google Scholar] [CrossRef]
- Van heusden, J.; Wouters, W.; Ramaekers, F.C.; Krekels, M.D.; Dillen, L.; Borgers, M.; Smets, G. All-trans-retinoic acid metabolites significantly inhibit the proliferation of MCF-7 human breast cancer cells in vitro. Br. J. Cancer 1998, 77, 26–32. [Google Scholar] [CrossRef][Green Version]
- Reynolds, C.P. Differentiating agents in pediatric malignancies: Retinoids in neuroblastoma. Curr. Oncol. Rep. 2000, 2, 511–518. [Google Scholar] [CrossRef]
- Iskakova, M.; Karbyshev, M.; Piskunov, A.; Rochette-Egly, C. Nuclear and extranuclear effects of vitamin A. Can. J. Physiol. Pharmacol. 2015, 93, 1065–1075. [Google Scholar] [CrossRef]
- Patatanian, E.; Thompson, D.F. Retinoic acid syndrome: A review. J. Clin. Pharm. Ther. 2008, 33, 331–338. [Google Scholar] [CrossRef]
- Schultze, E.; Collares, T.; Lucas, C.G.; Seixas, F.K. Synergistic and additive effects of ATRA in combination with different anti-tumor compounds. Chem. Biol. Interact. 2018, 285, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Chlapek, P.; Redova, M.; Zitterbart, K.; Hermanova, M.; Sterba, J.; Veselska, R. Enhancement of ATRA-induced differentiation of neuroblastoma cells with LOX/COX inhibitors: An expression profiling study. J. Exp. Clin. Cancer Res. 2010, 29, 45. [Google Scholar] [CrossRef] [PubMed]
- Chlapek, P.; Neradil, J.; Redova, M.; Zitterbart, K.; Sterba, J.; Veselska, R. The ATRA-induced differentiation of medulloblastoma cells is enhanced with LOX/COX inhibitors: An analysis of gene expression. Cancer Cell Int. 2014, 14, 51. [Google Scholar] [CrossRef] [PubMed]
- Redova, M.; Chlapek, P.; Loja, T.; Zitterbart, K.; Hermanova, M.; Sterba, J.; Veselska, R. Influence of LOX/COX inhibitors on cell differentiation induced by all-trans retinoic acid in neuroblastoma cell lines. Int. J. Mol. Med. 2010, 25, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Krzyzankova, M.; Chovanova, S.; Chlapek, P.; Radsetoulal, M.; Neradil, J.; Zitterbart, K.; Sterba, J.; Veselska, R. LOX/COX inhibitors enhance the antineoplastic effects of all-trans retinoic acid in osteosarcoma cell lines. Tumor Biol. 2014, 35, 7617–7627. [Google Scholar] [CrossRef]
- Satake, K.; Takagi, E.; Ishii, A.; Kato, Y.; Imagawa, Y.; Kimura, Y.; Tsukuda, M. Anti-tumor effect of vitamin A and D on head and neck squamous cell carcinoma. Auris. Nasus. Larynx 2003, 30, 403–412. [Google Scholar] [CrossRef]
- Elmaci, I.; Ozpinar, A.; Ozpinar, A.; Perez, J.L.; Altinoz, M.A. From epidemiology and neurometabolism to treatment: Vitamin D in pathogenesis of glioblastoma Multiforme (GBM) and a proposal for Vitamin D + all-trans retinoic acid + Temozolomide combination in treatment of GBM. Metab. Brain Dis. 2019, 34, 687–704. [Google Scholar] [CrossRef]
- Peehl, D.M.; Feldman, D. Interaction of nuclear receptor ligands with the Vitamin D signaling pathway in prostate cancer. J. Steroid Biochem. Mol. Biol. 2004, 92, 307–315. [Google Scholar] [CrossRef]
- Jeon, S.-M.; Shin, E.-A. Exploring vitamin D metabolism and function in cancer. Exp. Mol. Med. 2018, 50, 20. [Google Scholar] [CrossRef]
- Montenegro, K.R.; Cruzat, V.; Carlessi, R.; Newsholme, P. Mechanisms of vitamin D action in skeletal muscle. Nutr. Res. Rev. 2019, 32, 192–204. [Google Scholar] [CrossRef]
- Norman, A.W. Sunlight, season, skin pigmentation, vitamin D, and 25-hydroxyvitamin D: Integral components of the vitamin D endocrine system. Am. J. Clin. Nutr. 1998, 67, 1108–1110. [Google Scholar] [CrossRef] [PubMed]
- Carlberg, C.; Seuter, S. A genomic perspective on vitamin D signaling. Anticancer Res. 2009, 29, 3485–3493. [Google Scholar] [PubMed]
- Keum, N.; Lee, D.H.; Greenwood, D.C.; Manson, J.E.; Giovannucci, E. Vitamin D supplementation and total cancer incidence and mortality: A meta-analysis of randomized controlled trials. Ann. Oncol. 2019, 30, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Li, H.; Chan, A.T.; Hollis, B.W.; Lee, I.-M.; Stampfer, M.J.; Wu, K.; Giovannucci, E.; Ma, J. Circulating Levels of Vitamin D and Colon and Rectal Cancer: The Physicians’ Health Study and a Meta-analysis of Prospective Studies. Cancer Prev. Res. 2011, 4, 735–743. [Google Scholar] [CrossRef]
- Deschasaux, M.; Souberbielle, J.-C.; Latino-Martel, P.; Sutton, A.; Charnaux, N.; Druesne-Pecollo, N.; Galan, P.; Hercberg, S.; Clerc, S.L.; Kesse-Guyot, E.; et al. A prospective study of plasma 25-hydroxyvitamin D concentration and prostate cancer risk. Br. J. Nutr. 2016, 115, 305–314. [Google Scholar] [CrossRef]
- Chlebowski, R.T.; Johnson, K.C.; Kooperberg, C.; Pettinger, M.; Wactawski-Wende, J.; Rohan, T.; Rossouw, J.; Lane, D.; O’Sullivan, M.J.; Yasmeen, S.; et al. Calcium Plus Vitamin D Supplementation and the Risk of Breast Cancer. JNCI J. Natl. Cancer Inst. 2008, 100, 1581–1591. [Google Scholar] [CrossRef]
- De Smedt, J.; Van Kelst, S.; Boecxstaens, V.; Stas, M.; Bogaerts, K.; Vanderschueren, D.; Aura, C.; Vandenberghe, K.; Lambrechts, D.; Wolter, P.; et al. Vitamin D supplementation in cutaneous malignant melanoma outcome (ViDMe): A randomized controlled trial. BMC Cancer 2017, 17, 562. [Google Scholar] [CrossRef]
- Tokuumi, Y. Correlation between the concentration of 1, 25 alpha dihydroxyvitamin D3 receptors and growth inhibition, and differentiation of human osteosarcoma cells induced by vitamin D3. Nihon Seikeigeka Gakkai Zasshi 1995, 69, 181–190. [Google Scholar]
- Auken, M.V.; Buckley, D.; Ray, R.; Holick, M.F.; Baran, D.T. Effects of the vitamin D3 analog 1α, 25-dihydroxyvitamin D3-3β-bromoacetate on rat osteosarcoma cells: Comparison with 1α, 25-dihydroxyvitamin D3. J. Cell. Biochem. 1996, 63, 302–310. [Google Scholar] [CrossRef]
- Thompson, L.; Wang, S.; Tawfik, O.; Templeton, K.; Tancabelic, J.; Pinson, D.; Anderson, H.C.; Keighley, J.; Garimella, R. Effect of 25-hydroxyvitamin D3 and 1α, 25 dihydroxyvitamin D3 on differentiation and apoptosis of human osteosarcoma cell lines. J. Orthop. Res. 2012, 30, 831–844. [Google Scholar] [CrossRef]
- Shimizu, T.; Kamel, W.A.; Yamaguchi-Iwai, S.; Fukuchi, Y.; Muto, A.; Saya, H. Calcitriol exerts an anti-tumor effect in osteosarcoma by inducing the endoplasmic reticulum stress response. Cancer Sci. 2017, 108, 1793–1802. [Google Scholar] [CrossRef] [PubMed]
- Garimella, R.; Tadikonda, P.; Tawfik, O.; Gunewardena, S.; Rowe, P.; Van Veldhuizen, P. Vitamin D Impacts the Expression of Runx2 Target Genes and Modulates Inflammation, Oxidative Stress and Membrane Vesicle Biogenesis Gene Networks in 143B Osteosarcoma Cells. Int. J. Mol. Sci. 2017, 18, 642. [Google Scholar] [CrossRef] [PubMed]
- Engel, N.; Adamus, A.; Schauer, N.; Kühn, J.; Nebe, B.; Seitz, G.; Kraft, K. Synergistic Action of Genistein and Calcitriol in Immature Osteosarcoma MG-63 Cells by SGPL1 Up-Regulation. PLoS ONE 2017, 12, e0169742. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Hu, W.; Lu, L.; Zhao, Y.; Zhou, Y.; Xiao, Z.; Zhang, L.; Zhang, H.; Li, X.; Li, W.; et al. Repurposing vitamin D for treatment of human malignancies via targeting tumor microenvironment. Acta Pharm. Sin. B 2019, 9, 203–219. [Google Scholar] [CrossRef]
- Saggese, G.; Federico, G.; Cinquanta, L. In vitro effects of growth hormone and other hormones on chondrocytes and osteoblast-like cells. Acta Paediatr. 1993, 82, 54–59. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, X.; Zanello, L.P. 1α, 25-Dihydroxyvitamin D3 antiproliferative actions involve vitamin D receptor-mediated activation of MAPK pathways and AP-1/p21waf1 upregulation in human osteosarcoma. Cancer Lett. 2007, 254, 75–86. [Google Scholar] [CrossRef]
- Skoda, J.; Nunukova, A.; Loja, T.; Zambo, I.; Neradil, J.; Mudry, P.; Zitterbart, K.; Hermanova, M.; Hampl, A.; Sterba, J.; et al. Cancer stem cell markers in pediatric sarcomas: Sox2 is associated with tumorigenicity in immunodeficient mice. Tumor Biol. 2016, 37, 9535–9548. [Google Scholar] [CrossRef]
- Al Tanoury, Z.; Piskunov, A.; Rochette-Egly, C. Vitamin A and retinoid signaling: Genomic and nongenomic effects. J. Lipid Res. 2013, 54, 1761–1775. [Google Scholar] [CrossRef]
- Bover, J.; Egido, J.; Fernández-Giráldez, E.; Praga, M.; Solozábal-Campos, C.; Torregrosa, J.V.; Martínez-Castelao, A. Vitamin D, vitamin D receptor and the importance of its activation in patients with chronic kidney disease. Nefrol. Pub. Off. Soc. Espanola Nefrol. 2015, 35, 28–41. [Google Scholar] [CrossRef]
- Petkovich, P.M.; Heersche, J.N.; Tinker, D.O.; Jones, G. Retinoic acid stimulates 1, 25-dihydroxyvitamin D3 binding in rat osteosarcoma cells. J. Biol. Chem. 1984, 259, 8274–8280. [Google Scholar]
- Petkovich, P.M.; Heersche, J.N.; Aubin, J.E.; Grigoriadis, A.E.; Jones, G. Retinoic acid-induced changes in 1 alpha, 25-dihydroxyvitamin D3 receptor levels in tumor and nontumor cells derived from rat bone. J. Natl. Cancer Inst. 1987, 78, 265–270. [Google Scholar] [PubMed]
- Lee, K.-L.; Petkovich, P.M.; Heersche, J.N.M. The Effects of Sodium Butyrate on the Retinoic Acid-Induced Changes in 1, 25-Dihydroxyvitamin D3 Receptors in Tumorigenic and Nontumorigenic Bone Derived Cell Lines. Endocrinology 1988, 122, 2399–2406. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Koga, M.; Takaoka, K.; Ono, K.; Sato, B. Effects of retinoic acid on steroid and vitamin D3 receptors in cultured mouse osteosarcoma cells. Bone 1993, 14, 7–12. [Google Scholar] [CrossRef]
- Nakajima, H.; Kizaki, M.; Ueno, H.; Muto, A.; Takayama, N.; Matsushita, H.; Sonoda, A.; Ikeda, Y. All-trans and 9-cis retinoic acid enhance 1, 25-dihydroxyvitamin D3-induced monocytic differentiation of U937 cells. Leuk. Res. 1996, 20, 665–676. [Google Scholar] [CrossRef]
- Folgueira, M.A.A.K.; Federico, M.H.H.; Katayama, M.L.H.; Silva, M.R.P.; Brentani, M.M. Expression of vitamin D receptor (VDR) in HL-60 cells is differentially regulated during the process of differentiation induced by phorbol ester, retinoic acid or interferon-γ. J. Steroid Biochem. Mol. Biol. 1998, 66, 193–201. [Google Scholar] [CrossRef]
- Gocek, E.; Marchwicka, A.; Baurska, H.; Chrobak, A.; Marcinkowska, E. Opposite regulation of vitamin D receptor by ATRA in AML cells susceptible and resistant to vitamin D-induced differentiation. J. Steroid Biochem. Mol. Biol. 2012, 132, 220–226. [Google Scholar] [CrossRef]
- Marchwicka, A.; Cebrat, M.; Łaszkiewicz, A.; Śnieżewski, Ł.; Brown, G.; Marcinkowska, E. Regulation of vitamin D receptor expression by retinoic acid receptor alpha in acute myeloid leukemia cells. J. Steroid Biochem. Mol. Biol. 2016, 159, 121–130. [Google Scholar] [CrossRef]
- Janik, S.; Nowak, U.; Łaszkiewicz, A.; Satyr, A.; Majkowski, M.; Marchwicka, A.; Śnieżewski, Ł.; Berkowska, K.; Gabryś, M.; Cebrat, M.; et al. Diverse Regulation of Vitamin D Receptor Gene Expression by 1, 25-Dihydroxyvitamin D and ATRA in Murine and Human Blood Cells at Early Stages of Their Differentiation. Int. J. Mol. Sci. 2017, 18, 1323. [Google Scholar] [CrossRef]
- Uchida, H.; Hasegawa, Y.; Takahashi, H.; Makishima, M. 1α-Dihydroxyvitamin D3 and Retinoic Acid Increase Nuclear Vitamin D Receptor Expression in Monocytic THP-1 Cells. Anticancer Res. 2016, 36, 6297–6301. [Google Scholar] [CrossRef]
- Miyamoto, K.; Kesterson, R.A.; Yamamoto, H.; Taketani, Y.; Nishiwaki, E.; Tatsumi, S.; Inoue, Y.; Morita, K.; Takeda, E.; Pike, J.W. Structural Organization of the Human Vitamin D Receptor Chromosomal Gene and Its Promoter. Mol. Endocrinol. 1997, 11, 1165–1179. [Google Scholar] [CrossRef]
- Chen, H.; Hu, B.; Allegretto, E.A.; Adams, J.S. The Vitamin D Response Element-binding Protein a Novel Dominant-negative Regulator of Vitamin D-directed Transactivation. J. Biol. Chem. 2000, 275, 35557–35564. [Google Scholar] [CrossRef] [PubMed]
- Veselska, R.; Kuglik, P.; Cejpek, P.; Svachova, H.; Neradil, J.; Loja, T.; Relichova, J. Nestin expression in the cell lines derived from glioblastoma multiforme. BMC Cancer 2006, 6, 32. [Google Scholar] [CrossRef] [PubMed]
- Sramek, M.; Neradil, J.; Sterba, J.; Veselska, R. Non-DHFR-mediated effects of methotrexate in osteosarcoma cell lines: Epigenetic alterations and enhanced cell differentiation. Cancer Cell Int. 2016, 16, 14. [Google Scholar] [CrossRef] [PubMed]
- Veselska, R.; Hermanova, M.; Loja, T.; Chlapek, P.; Zambo, I.; Vesely, K.; Zitterbart, K.; Sterba, J. Nestin expression in osteosarcomas and derivation of nestin/CD133 positive osteosarcoma cell lines. BMC Cancer 2008, 8, 300. [Google Scholar] [CrossRef]
- Zhao, W.; Sachsenmeier, K.; Zhang, L.; Sult, E.; Hollingsworth, R.E.; Yang, H. A New Bliss Independence Model to Analyze Drug Combination Data. J. Biomol. Screen. 2014, 19, 817–821. [Google Scholar] [CrossRef]
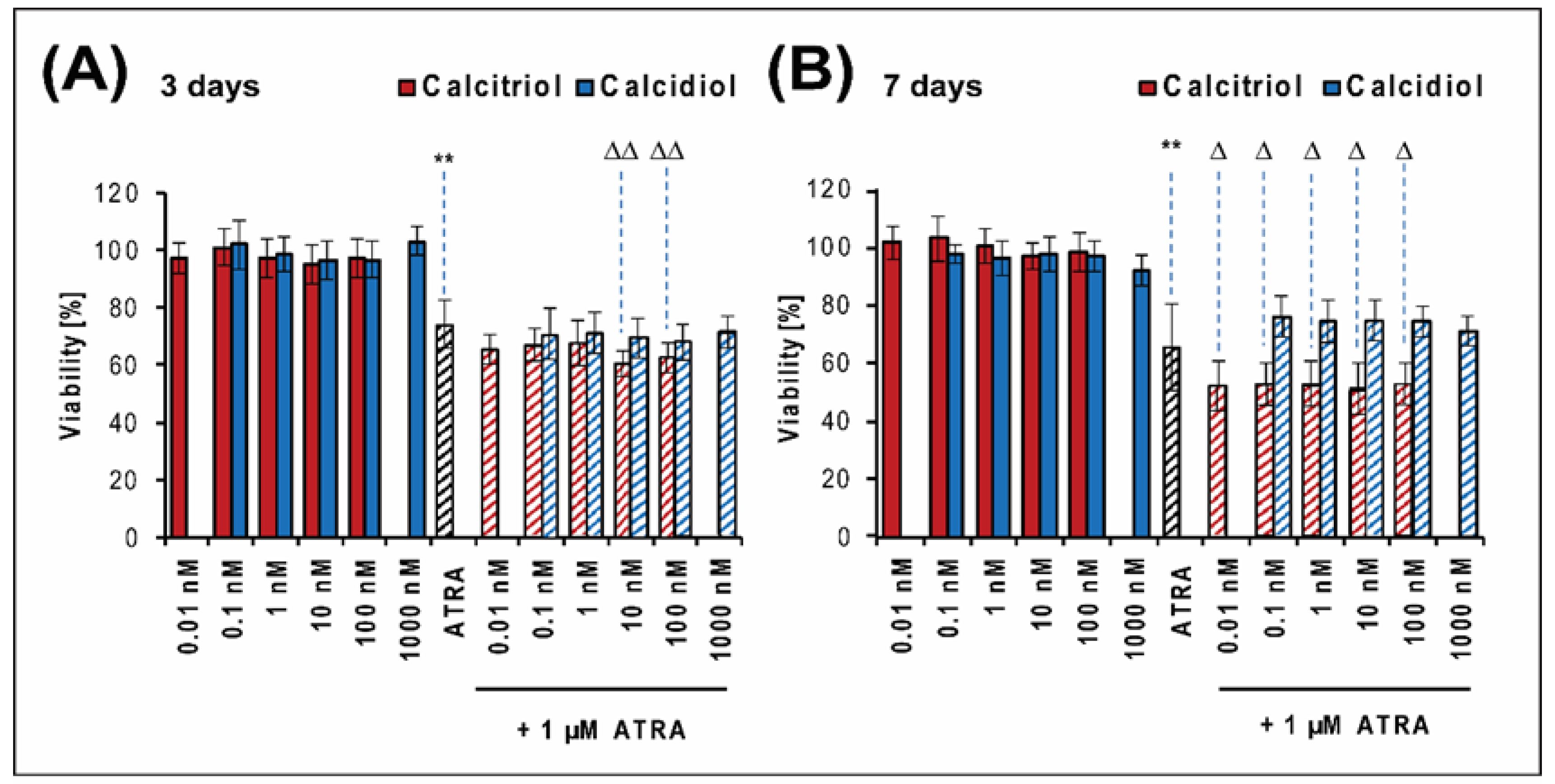
| Combination of Drugs | Saos-2 Cells | Combination of Drugs | Saos-2 Cells | ||||
|---|---|---|---|---|---|---|---|
| Calcitriol | ATRA | Day 3 | Day 7 | Calcidiol | ATRA | Day 3 | Day 7 |
| 0.01 nM | 1 µM | 0.1 nM | 1 µM | ||||
| 0.1 nM | 1 µM | 1 nM | 1 µM | ||||
| 1 nM | 1 µM | 10 nM | 1 µM | ||||
| 10 nM | 1 µM | 100 nM | 1 µM | ||||
| 100 nM | 1 µM | 1000 nM | 1 µM | ||||
| Day 1 | Day 3 | Day 5 | Day 7 | Day 9 | |
|---|---|---|---|---|---|
| Up-regulation of BGLAP expression after combined treatment with ATRA and calcitriol [%] | |||||
| Predicted effect | 0.62 | 0.42 | 1.10 | 1.43 | 1.23 |
| Observed effect | 1.26 | 0.60 | 0.91 | 1.50 | 1.05 |
| Up-regulation of BGLAP expression after combined treatment with ATRA and calcidiol [%] | |||||
| Predicted effect | 0.38 | 0.18 | 0.64 | 0.71 | 0.60 |
| Observed effect | 0.65 | 0.07 | 0.59 | 0.71 | 0.37 |
| Up-regulation of SPP-1 expression after combined treatment with ATRA and calcitriol [%] | |||||
| Predicted effect | 1.05 | 0.83 | 3.66 | 1.86 | 2.13 |
| Observed effect | 0.70 | 0.24 | 3.61 | 1.41 | 1.38 |
| Up-regulation of SPP-1 expression after combined treatment with ATRA and calcidiol [%] | |||||
| Predicted effect | 0.59 | 0.58 | 3.23 | 1.89 | 1.87 |
| Observed effect | 0.49 | 0.19 | 3.65 | 1.76 | 1.63 |
| Concentrations of Drugs | Patient-Derived Cell Lines | ||||||
|---|---|---|---|---|---|---|---|
| Calcitriol | ATRA | OSA-02 | OSA-03 | OSA-05 | OSA-08 | OSA-09 | OSA-13 |
| 0.01 nM | 1 µM | ||||||
| 0.1 nM | 1 µM | ||||||
| 1 nM | 1 µM | ||||||
| 10 nM | 1 µM | ||||||
| 100 nM | 1 µM | ||||||
| Concentrations of Drugs | Patient-Derived Cell Lines | ||||||
|---|---|---|---|---|---|---|---|
| Calcidiol | ATRA | OSA-02 | OSA-03 | OSA-05 | OSA-08 | OSA-09 | OSA-13 |
| 0.1 nM | 1 µM | ||||||
| 1 nM | 1 µM | ||||||
| 10 nM | 1 µM | ||||||
| 100 nM | 1 µM | ||||||
| 1000 nM | 1 µM | ||||||
| Gene | Primer Sequence | Product Length (bp) |
|---|---|---|
| BGLAP | F: 5′-GAG GGC AGC GAG GTA GTG AA-3′ | 152 |
| R: 5′-TCC TGA AAG CCG ATG TGG TC-3′ | ||
| SPP1 | F: 5′-GCC GAG GTG ATA GTG TGG TT-3′ | 242 |
| R: 5′-GTG GGT TTC AGC ACT CTG GT-3′ | ||
| HSP90AB1 | F: 5′-CGC ATG AAG GAG ACA CAG AA-3′ | 169 |
| R: 5′-TCC CAT CAA ATT CCT TGA GC-3′ | ||
| RARA | F: 5’-CGA CCG AAA CAA GAA GAA GAA GG-3´ | 166 |
| R: 5´-TTC TGA GCT GTT GTT CGT AGT GT-3´ | ||
| RXRA | F: 5´-CTC AAT GGC GTC CTC AAG GT-3´ | 111 |
| R: 5´-CAC TCC ATA GTG CTT GCC TGA-3´ | ||
| VDR | F: 5´-AGC CTC AAT GAG GAG CAC TCC AAG-3´ | 206 |
| R: 5´-ACG GGT GAG GAG GGC TGC TGA GTA-3´ | ||
| GAPDH | F: 5´-AGC CAC ATC GCT CAG ACA CC-3´ | 302 |
| R: 5´-GTA CTC AGC GCC AGC ATC G-3´ |
| Primary Antibodies | |||||
| Antigen | Type/Host | Clone | Catalog No. | Manufacturer | Dilution |
| RARα | Mono/Mo | H1920 | ab41934 | Abcam | 1:1000 |
| RXRα | Mono/Rb | D6H10 | 3085 | Cell Signaling | 1:1000 |
| VDR | Mono/Rb | EPR4552 | ab109234 | Abcam | 1: 2000 |
| GAPDH | Mono/Rb | 14C10 | 2118S | Cell Signaling | 1:10,000 |
| Secondary Antibodies | |||||
| Host | Specificity | Conjugate | Catalog No. | Manufacturer | Dilution |
| Goat | anti-Rb IgG | HRP | 7074 | Cell Signaling | 1:5000 |
| Horse | anti-Mo IgG | HRP | 7076 | Cell Signaling | 1:5000 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paukovcekova, S.; Valik, D.; Sterba, J.; Veselska, R. Enhanced Antiproliferative Effect of Combined Treatment with Calcitriol and All-Trans Retinoic Acid in Relation to Vitamin D Receptor and Retinoic Acid Receptor α Expression in Osteosarcoma Cell Lines. Int. J. Mol. Sci. 2020, 21, 6591. https://doi.org/10.3390/ijms21186591
Paukovcekova S, Valik D, Sterba J, Veselska R. Enhanced Antiproliferative Effect of Combined Treatment with Calcitriol and All-Trans Retinoic Acid in Relation to Vitamin D Receptor and Retinoic Acid Receptor α Expression in Osteosarcoma Cell Lines. International Journal of Molecular Sciences. 2020; 21(18):6591. https://doi.org/10.3390/ijms21186591
Chicago/Turabian StylePaukovcekova, Silvia, Dalibor Valik, Jaroslav Sterba, and Renata Veselska. 2020. "Enhanced Antiproliferative Effect of Combined Treatment with Calcitriol and All-Trans Retinoic Acid in Relation to Vitamin D Receptor and Retinoic Acid Receptor α Expression in Osteosarcoma Cell Lines" International Journal of Molecular Sciences 21, no. 18: 6591. https://doi.org/10.3390/ijms21186591
APA StylePaukovcekova, S., Valik, D., Sterba, J., & Veselska, R. (2020). Enhanced Antiproliferative Effect of Combined Treatment with Calcitriol and All-Trans Retinoic Acid in Relation to Vitamin D Receptor and Retinoic Acid Receptor α Expression in Osteosarcoma Cell Lines. International Journal of Molecular Sciences, 21(18), 6591. https://doi.org/10.3390/ijms21186591





