Human Mesenchymal Stromal Cells Unveil an Unexpected Differentiation Potential toward the Dopaminergic Neuronal Lineage
Abstract
1. Introduction
2. Results
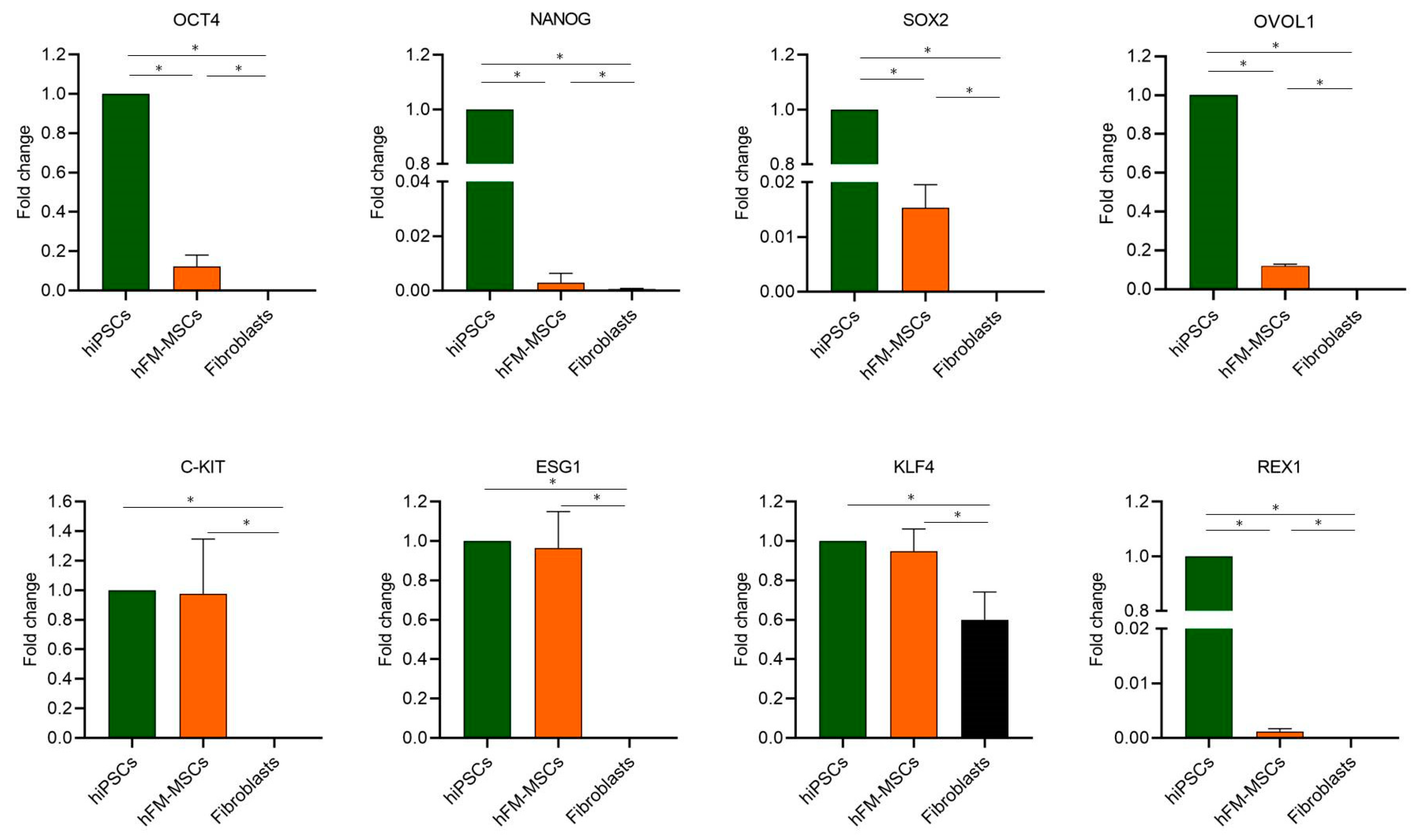
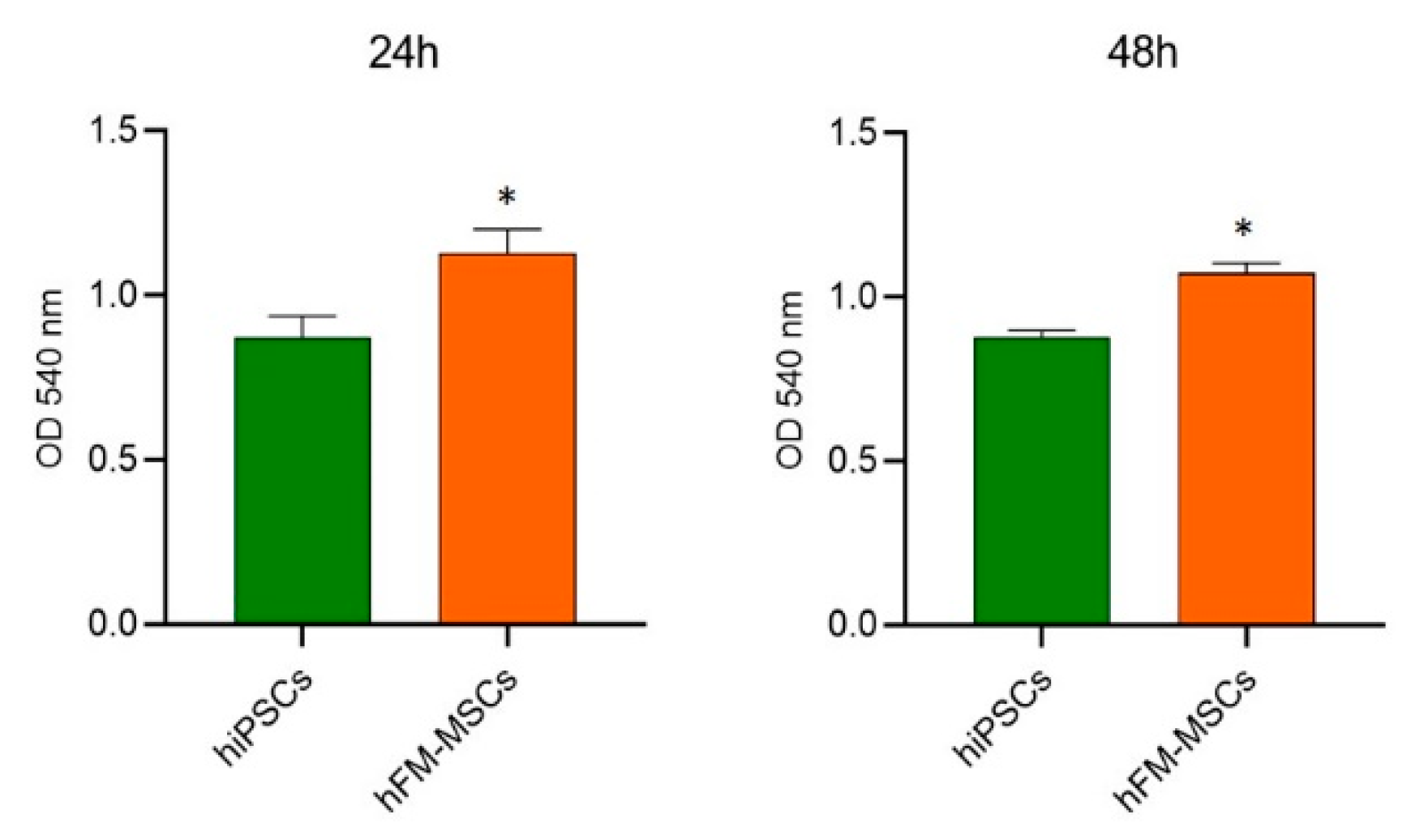
2.1. hFM-MSCs Exhibit Some Molecular and Biological Characteristics of Pluripotent Stem Cells
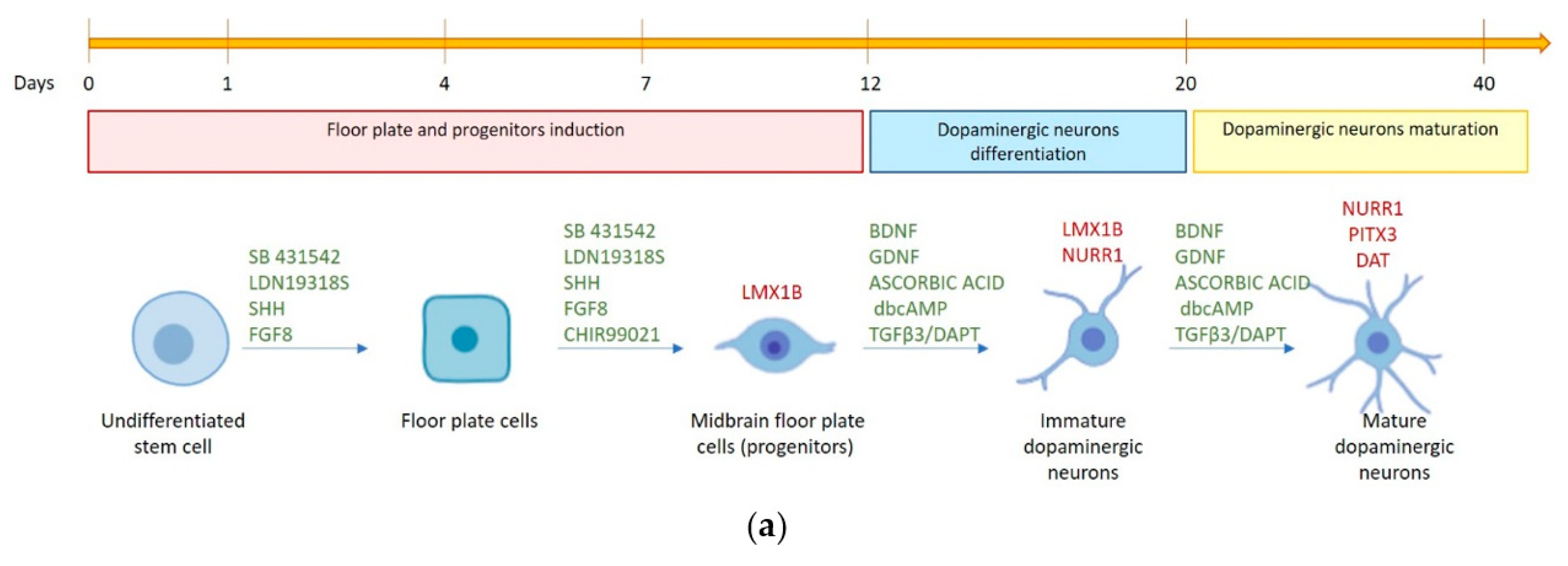
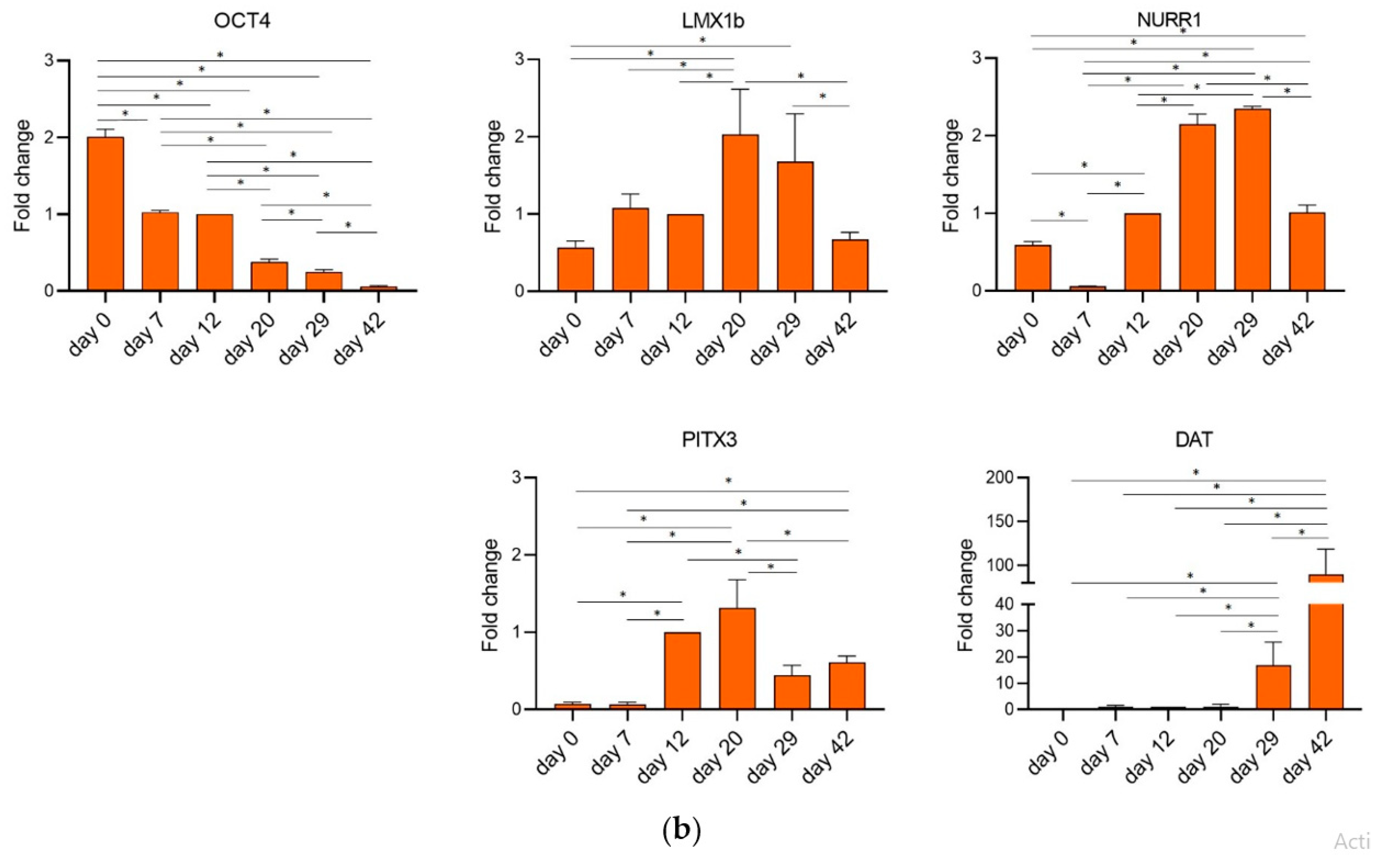
2.2. hFM-MSCs Can Be Driven toward a DA Neuronal Fate
2.3. hFM-MSCs Differentiated Cells Resemble the Shape of Primary Midbrain Dopaminergic Neurons and Express Dopaminergic Markers
3. Discussion
4. Materials and Methods
4.1. Culture of Human Cell
4.2. MTT Assay
4.3. Dopaminergic Differentiation
4.4. RNA Extraction and Reverse Transcription
4.5. Real Time PCR (qPCR)
4.6. Immunofluorescence
4.7. Immunoblot Analysis
4.8. Flow Cytometry
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tiklová, K.; Björklund, Å.K.; Lahti, L.; Fiorenzano, A.; Nolbrant, S.; Gillberg, L.; Volakakis, N.; Yokota, C.; Hilscher, M.M.; Hauling, T. Single-cell RNA sequencing reveals midbrain dopamine neuron diversity emerging during mouse brain development. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Xi, J.; Liu, Y.; Liu, H.; Chen, H.; Emborg, M.E.; Zhang, S.-C. Specification of midbrain dopamine neurons from primate pluripotent stem cells. Stem Cells 2012, 30, 1655–1663. [Google Scholar] [CrossRef] [PubMed]
- Ang, S.-L. Transcriptional control of midbrain dopaminergic neuron development. Development 2006, 133, 3499–3506. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Song, J.-J.; Puspita, L.; Valiulahi, P.; Shim, J.; Lee, S.-H. In vitro generation of mature midbrain-type dopamine neurons by adjusting exogenous Nurr1 and Foxa2 expressions to their physiologic patterns. Exp. Mol. Med. 2017, 49, e300. [Google Scholar] [CrossRef]
- Daubner, S.C.; Le, T.; Wang, S. Tyrosine hydroxylase and regulation of dopamine synthesis. Arch. Biochem. Biophys. 2011, 508, 1–12. [Google Scholar] [CrossRef]
- Vernier, P. The degeneration of dopamine neurons in Parkinson’s Disease: Insights from embryology and evolution of the mesostriatocortical system. Ann. N. Y. Acad. Sci. 2004, 1035, 231–249. [Google Scholar] [CrossRef]
- Lindvall, O. Dopaminergic neurons for Parkinson’s therapy. Nat. Biotechnol. 2012, 30, 56–58. [Google Scholar] [CrossRef]
- Mahajani, S.; Raina, A.; Fokken, C.; Kügler, S.; Bähr, M. Homogenous generation of dopaminergic neurons from multiple hiPSC lines by transient expression of transcription factors. Cell Death Dis. 2019, 10. [Google Scholar] [CrossRef]
- Di Baldassarre, A.; Cimetta, E.; Bollini, S.; Gaggi, G.; Ghinassi, B. Human-induced pluripotent stem cell technology and cardiomyocyte generation: Progress and clinical applications. Cells 2018, 7, 48. [Google Scholar] [CrossRef]
- Gaggi, G.; Izzicupo, P.; Di Credico, A.; Sancilio, S.; Di Baldassarre, A.; Ghinassi, B. spare parts from discarded materials: Fetal annexes in regenerative medicine. Int. J. Mol. Sci. 2019, 20, 1573. [Google Scholar] [CrossRef]
- Gaggi, G.; Di Credico, A.; Izzicupo, P.; Antonucci, I.; Crescioli, C.; Di Giacomo, V.; Di Ruscio, A.; Amabile, G.; Alviano, F.; Di Baldassarre, A.; et al. Epigenetic features of human perinatal stem cells redefine their stemness potential. Cells 2020, 9, 1304. [Google Scholar] [CrossRef] [PubMed]
- Shyh-Chang, N.; Daley, G.Q.; Cantley, L.C. Stem cell metabolism in tissue development and aging. Development 2013, 140, 2535–2547. [Google Scholar] [CrossRef]
- Pauklin, S.; Vallier, L. The cell-cycle state of stem cells determines cell fate propensity. Cell 2013, 155, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Ghule, P.N.; Medina, R.; Lengner, C.J.; Mandeville, M.; Qiao, M.; Dominski, Z.; Lian, J.B.; Stein, J.L.; van Wijnen, A.J.; Stein, G.S. Reprogramming the pluripotent cell cycle: Restoration of an abbreviated G1 phase in human induced pluripotent stem (iPS) cells. J. Cell. Physiol. 2011, 226, 1149–1156. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Ocampo, A.; Belmonte, J.C.I. Cellular metabolism and induced pluripotency. Cell 2016, 166, 1371–1385. [Google Scholar] [CrossRef]
- Papathanou, M.; Dumas, S.; Pettersson, H.; Olson, L.; Wallén-Mackenzie, Å. Off-target effects in transgenic mice: Characterization of Dopamine Transporter (DAT)-cre transgenic mouse lines exposes multiple non-dopaminergic neuronal clusters available for selective targeting within limbic neurocircuitry. Eneuro 2019, 6. [Google Scholar] [CrossRef]
- Harkness, L.; Chen, X.; Gillard, M.; Gray, P.P.; Davies, A.M. Media composition modulates human embryonic stem cell morphology and may influence preferential lineage differentiation potential. PLoS ONE 2019, 14, e0213678. [Google Scholar] [CrossRef]
- Rodríguez-Traver, E.; Solís, O.; Díaz-Guerra, E.; Ortiz, Ó.; Vergaño-Vera, E.; Méndez-Gómez, H.R.; García-Sanz, P.; Moratalla, R.; Vicario-Abejón, C. Role of nurr1 in the generation and differentiation of dopaminergic neurons from stem cells. Neurotox Res. 2016, 30, 14–31. [Google Scholar] [CrossRef]
- Masato, A.; Plotegher, N.; Boassa, D.; Bubacco, L. Impaired dopamine metabolism in Parkinson’s disease pathogenesis. Mol. Neurodegener. 2019, 14. [Google Scholar] [CrossRef]
- Papanikolaou, T.; Lennington, J.B.; Betz, A.; Figueiredo, C.; Salamone, J.D.; Conover, J.C. In vitro generation of dopaminergic neurons from adult subventricular zone neural progenitor cells. Stem Cells Dev. 2008, 17, 157–172. [Google Scholar] [CrossRef]
- Alizadeh, R.; Bagher, Z.; Kamrava, S.K.; Falah, M.; Ghasemi Hamidabadi, H.; Eskandarian Boroujeni, M.; Mohammadi, F.; Khodaverdi, S.; Zare-Sadeghi, A.; Olya, A.; et al. Differentiation of human mesenchymal stem cells (MSC) to dopaminergic neurons: A comparison between Wharton’s Jelly and olfactory mucosa as sources of MSCs. J. Chem. Neuroanat. 2019, 96, 126–133. [Google Scholar] [CrossRef]
- Melikian, H.E. Neurotransmitter transporter trafficking: Endocytosis, recycling, and regulation. Pharmacol. Ther. 2004, 104, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Maraldi, T.; Bertoni, L.; Riccio, M.; Zavatti, M.; Carnevale, G.; Resca, E.; Guida, M.; Beretti, F.; La Sala, G.B.; De Pol, A. Human amniotic fluid stem cells: Neural differentiation in vitro and in vivo. Cell Tissue Res. 2014, 357, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, A.E.; Cai, J.; Yang, M.; Iacovitti, L. Human amniotic fluid stem cells do not differentiate into dopamine neurons in vitro or after transplantation in vivo. Stem Cells Dev. 2009, 18, 1003–1012. [Google Scholar] [CrossRef] [PubMed]
- Paldino, E.; Cenciarelli, C.; Giampaolo, A.; Milazzo, L.; Pescatori, M.; Hassan, H.J.; Casalbore, P. Induction of dopaminergic neurons from human Wharton’s jelly mesenchymal stem cell by Forskolin. J. Cell. Physiol. 2014, 229, 232–244. [Google Scholar] [CrossRef]
- Chen, R.; Furman, C.A.; Gnegy, M.E. Dopamine transporter trafficking: Rapid response on demand. Future Neurol. 2010, 5, 123–134. [Google Scholar] [CrossRef]
- Zhang, P.; Andrianakos, R.; Yang, Y.; Liu, C.; Lu, W. Kruppel-like factor 4 (Klf4) prevents embryonic stem (es) cell differentiation by regulating nanog gene expression. J. Biol. Chem. 2010, 285, 9180–9189. [Google Scholar] [CrossRef]
- Renaud, S.J.; Chakraborty, D.; Mason, C.W.; Rumi, M.A.K.; Vivian, J.L.; Soares, M.J. OVO-like 1 regulates progenitor cell fate in human trophoblast development. Proc. Natl. Acad. Sci. USA 2015, 112, E6175–E6184. [Google Scholar] [CrossRef]
- Qian, X.; Kim, J.K.; Tong, W.; Villa-Diaz, L.G.; Krebsbach, P.H. DPPA5 supports pluripotency and reprogramming by regulating NANOG turnover: Role of DPPA5 in human pluripotent stem cells. Stem Cells 2016, 34, 588–600. [Google Scholar] [CrossRef]
- Filardi, T.; Ghinassi, B.; Di Baldassarre, A.; Tanzilli, G.; Morano, S.; Lenzi, A.; Basili, S.; Crescioli, C. Cardiomyopathy Associated with Diabetes: The Central Role of the Cardiomyocyte. Int. J. Mol. Sci. 2019, 20, 3299. [Google Scholar] [CrossRef]
- Yi, S.-H.; He, X.-B.; Rhee, Y.-H.; Park, C.-H.; Takizawa, T.; Nakashima, K.; Lee, S.-H. Foxa2 acts as a co-activator potentiating expression of the Nurr1-induced DA phenotype via epigenetic regulation. Development 2014, 141, 761–772. [Google Scholar] [CrossRef] [PubMed][Green Version]
- García-Yagüe, Á.J.; Rada, P.; Rojo, A.I.; Lastres-Becker, I.; Cuadrado, A. Nuclear import and export signals control the subcellular localization of nurr1 protein in response to oxidative stress. J. Biol. Chem. 2013, 288, 5506–5517. [Google Scholar] [CrossRef]
- Eriksen, J.; Rasmussen, S.G.F.; Rasmussen, T.N.; Vaegter, C.B.; Cha, J.H.; Zou, M.-F.; Newman, A.H.; Gether, U. Visualization of dopamine transporter trafficking in live neurons by use of fluorescent cocaine analogs. J. Neurosci. 2009, 29, 6794–6808. [Google Scholar] [CrossRef] [PubMed]
- Zahniser, N.R.; Sorkin, A. Trafficking of dopamine transporters in psychostimulant actions. Semin. Cell Dev. Biol. 2009, 20, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Eriksen, J.; Bjørn-Yoshimoto, W.E.; Jørgensen, T.N.; Newman, A.H.; Gether, U. Postendocytic sorting of constitutively internalized dopamine transporter in cell lines and dopaminergic neurons. J. Biol. Chem. 2010, 285, 27289–27301. [Google Scholar] [CrossRef]
- Rao, A.; Simmons, D.; Sorkin, A. Differential subcellular distribution of endosomal compartments and the dopamine transporter in dopaminergic neurons. Mol. Cell. Neurosci. 2011, 46, 148–158. [Google Scholar] [CrossRef]
- Block, E.R.; Nuttle, J.; Balcita-Pedicino, J.J.; Caltagarone, J.; Watkins, S.C.; Sesack, S.R.; Sorkin, A. Brain region-specific trafficking of the dopamine transporter. J. Neurosci. 2015, 35, 12845–12858. [Google Scholar] [CrossRef]
- Pickel, V.M.; Joh, T.H.; Field, P.M.; Becker, C.G.; Reis, D.J. Cellular localization of tyrosine hydroxylase by immunohistochemistry. J. Histochem. Cytochem. 1975, 23, 1–12. [Google Scholar] [CrossRef]
- Amabile, G.; Welner, R.S.; Nombela-Arrieta, C.; D’Alise, A.M.; Di Ruscio, A.; Ebralidze, A.K.; Kraytsberg, Y.; Ye, M.; Kocher, O.; Neuberg, D.S.; et al. In vivo generation of transplantable human hematopoietic cells from induced pluripotent stem cells. Blood 2013, 121, 1255–1264. [Google Scholar] [CrossRef]
- Chatgilialoglu, A.; Rossi, M.; Alviano, F.; Poggi, P.; Zannini, C.; Marchionni, C.; Ricci, F.; Tazzari, P.L.; Taglioli, V.; Calder, P.C.; et al. Restored in vivo-like membrane lipidomics positively influence in vitro features of cultured mesenchymal stromal/stem cells derived from human placenta. Stem Cell Res. Ther. 2017, 8. [Google Scholar] [CrossRef]
- Leoni, V.; Gatta, V.; Palladini, A.; Nicoletti, G.; Ranieri, D.; Dall’Ora, M.; Grosso, V.; Rossi, M.; Alviano, F.; Bonsi, L.; et al. Systemic delivery of HER2-retargeted oncolytic-HSV by mesenchymal stromal cells protects from lung and brain metastases. Oncotarget 2015, 6, 34774–34787. [Google Scholar] [CrossRef] [PubMed]
- D’amico, M.A.; Ghinassi, B.; Izzicupo, P.; Di Ruscio, A.; Di Baldassarre, A. IL-6 Activates PI3K and PKCζ signaling and determines cardiac differentiation in rat embryonic H9c2 cells: IL-6 and cardiac differentiation of H9c2 cells. J. Cell. Physiol. 2016, 231, 576–586. [Google Scholar] [CrossRef] [PubMed]
- Ghinassi, B.; D’Addazio, G.; Di Baldassarre, A.; Femminella, B.; Di Vincenzo, G.; Piattelli, M.; Gaggi, G.; Sinjari, B. Immunohistochemical results of soft tissues around a new implant healing-abutment surface: A human study. JCM 2020, 9, 1009. [Google Scholar] [CrossRef]
- Kriks, S.; Shim, J.-W.; Piao, J.; Ganat, Y.M.; Wakeman, D.R.; Xie, Z.; Carrillo-Reid, L.; Auyeung, G.; Antonacci, C.; Buch, A.; et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature 2011, 480, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Shimojo, D.; Onodera, K.; Doi-Torii, Y.; Ishihara, Y.; Hattori, C.; Miwa, Y.; Tanaka, S.; Okada, R.; Ohyama, M.; Shoji, M.; et al. Rapid, efficient, and simple motor neuron differentiation from human pluripotent stem cells. Mol. Brain 2015, 8. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Jakovcevski, M.; Bharadwaj, R.; Connor, C.; Schroeder, F.A.; Lin, C.L.; Straubhaar, J.; Martin, G.; Akbarian, S. Setdb1 histone methyltransferase regulates mood-related behaviors and expression of the NMDA Receptor Subunit NR2B. J. Neurosci. 2010, 30, 7152–7167. [Google Scholar] [CrossRef]
- Aasen, T.; Raya, A.; Barrero, M.J.; Garreta, E.; Consiglio, A.; Gonzalez, F.; Vassena, R.; Bilić, J.; Pekarik, V.; Tiscornia, G.; et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat. Biotechnol. 2008, 26, 1276–1284. [Google Scholar] [CrossRef]
- Antonucci, I.; Di Pietro, R.; Alfonsi, M.; Centurione, M.A.; Centurione, L.; Sancilio, S.; Pelagatti, F.; D’amico, M.A.; Di Baldassarre, A.; Piattelli, A.; et al. Human second trimester amniotic fluid cells are able to create embryoid body-like structures in vitro and to show typical expression profiles of embryonic and primordial germ cells. Cell Transplant. 2014, 23, 1501–1515. [Google Scholar] [CrossRef]
- Volpicelli, F.; De Gregorio, R.; Pulcrano, S.; Perrone-Capano, C.; di Porzio, U.; Bellenchi, G.C. Direct regulation of pitx3 expression by nurr1 in culture and in developing mouse midbrain. PLoS ONE 2012, 7, e30661. [Google Scholar] [CrossRef]
- Di Baldassarre, A.; D’Amico, M.A.; Izzicupo, P.; Gaggi, G.; Guarnieri, S.; Mariggiò, M.A.; Antonucci, I.; Corneo, B.; Sirabella, D.; Stuppia, L.; et al. Cardiomyocytes derived from human cardiopoieticamniotic fluids. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]

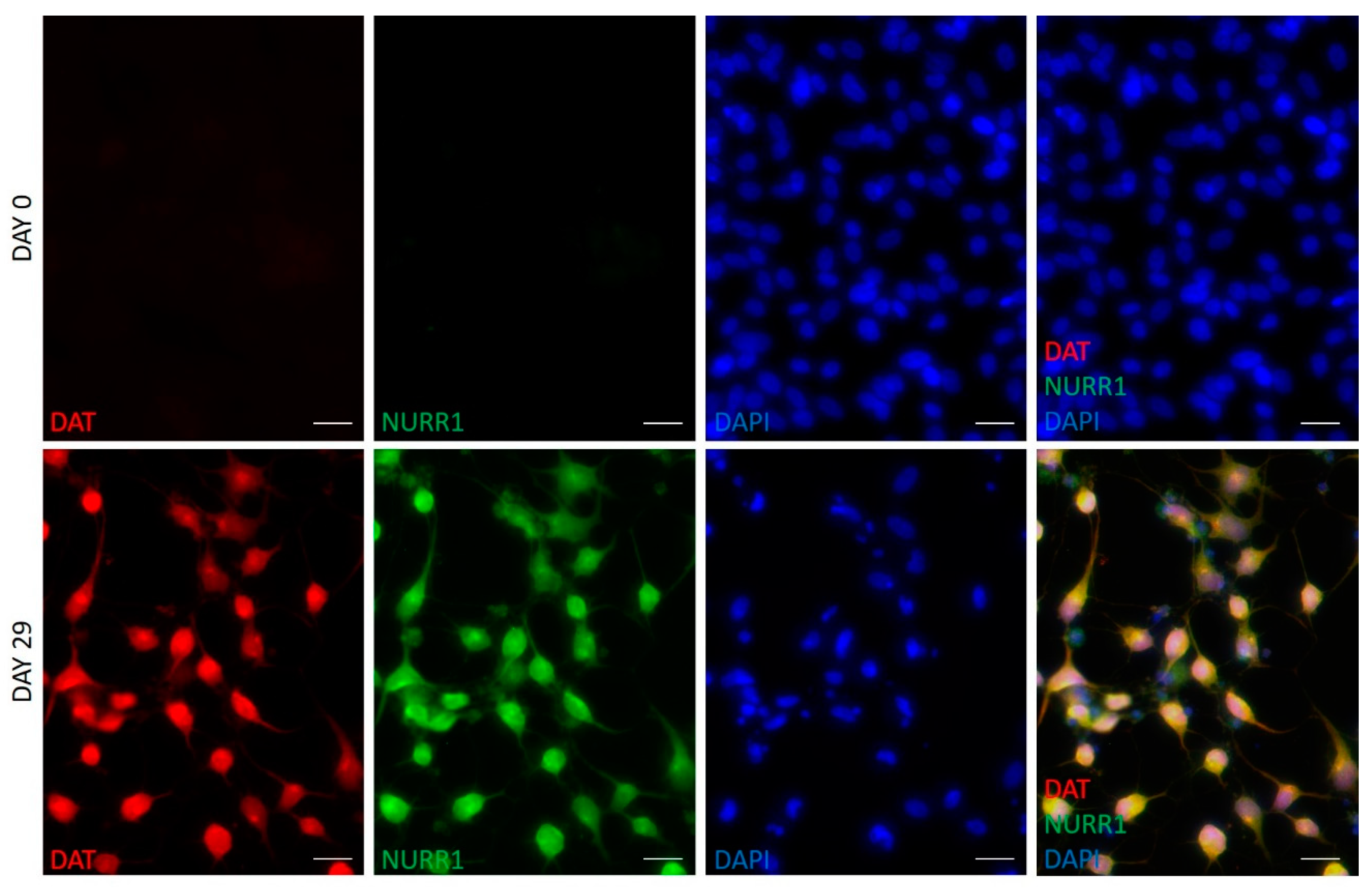
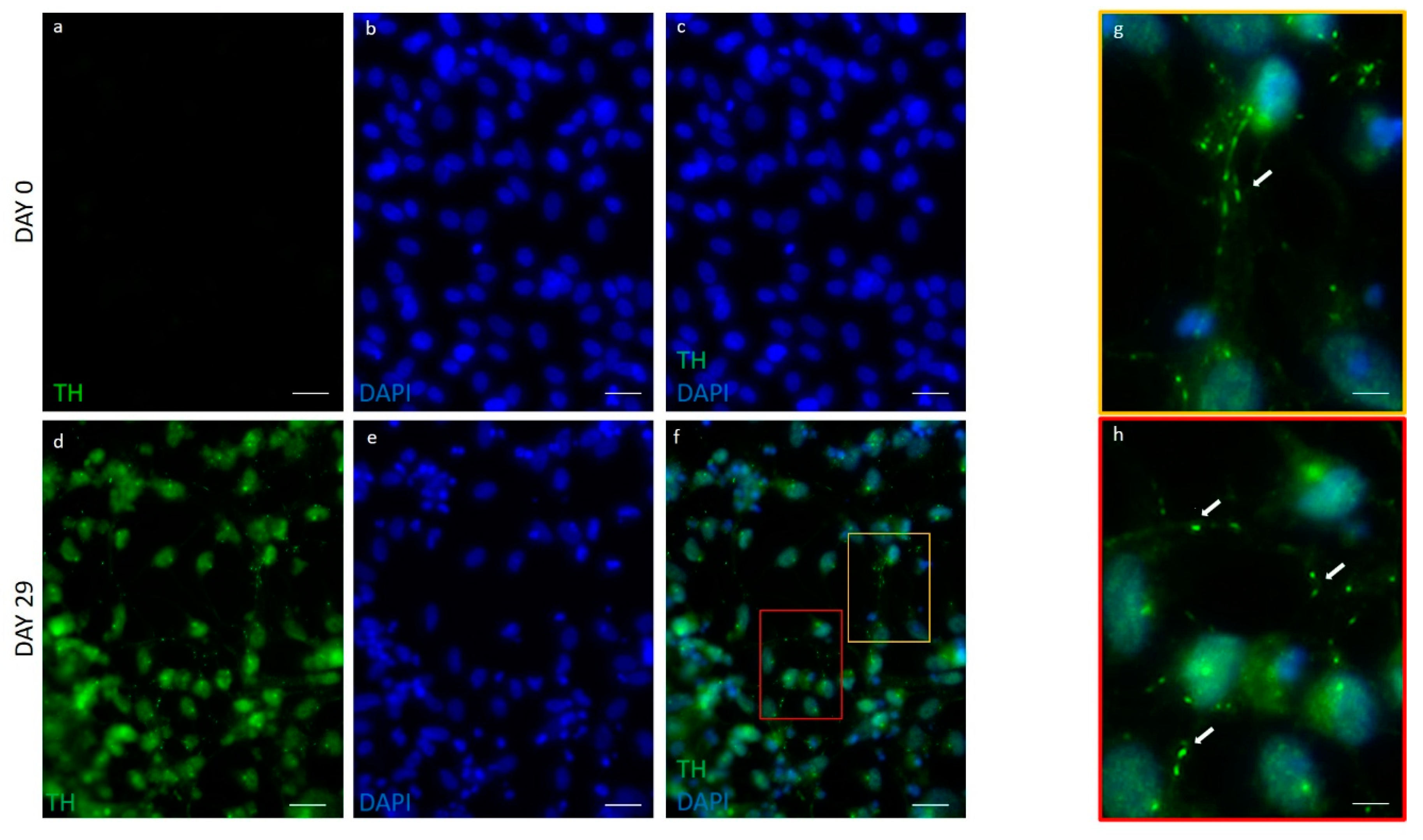
| DATpos Cells | NURR1pos Cells | Datposnurr1poscells | THpos Cells | |
|---|---|---|---|---|
| Undifferentiated FM-MSCs (day 0) | None | None | None | None |
| hFM-MSCs-derived DA neuron-like cells (day 29) | 78.5 ± 9.7% * | 73.8 ± 8.4% * | 75.6 ± 8.4% * | 79.2 ± 11.8% * |
| Gene | Sequence (5′ to 3′) |
|---|---|
| 18S FW [46] | CATGGCCGTTCTTAGTTGGT |
| 18S RW [46] | CGCTGAGCCAGTCAGTGTAG |
| NANOG-FW [11] | CCAGACCCAGAACATCCAGTC |
| NANOG-RW [11] | CACTGGCAGGAGAATTTGGC |
| Endo-OCT4-FW [47] | GGGTTTTTGGGATTAAGTTCTTCA |
| Endo-OCT4-RW [47] | GCCCCCACCCTTTGTGTT |
| Endo-SOX2-FW [47] | CAAAAATGGCCATGCAGGTT |
| Endo-SOX2-RW [47] | AGTTGGGATCGAACAAAAGCTATT |
| Endo-KLF4-FW [47] | AGCCTAAATGATGGTGCTTGGT |
| Endo-KLF4-RW [47] | TTGAAAACTTTGGCTTCCTTGTT |
| C-KIT-FW [11] | CCACACCCTGTTCACTCCTT |
| C-KIT-RW [11] | TTCTGGGAAACTCCCATTTGTG |
| REX1-FW | GCGCAATCGCTTGTCCTCAG |
| REX1-RW | CACATTCCGCACAGACGTGG |
| OVOL1-FW [48] | AGAGCAGAGACCATGGCTTC |
| OVOL1-RW [48] | GACGTGTCTCTTGAGGTCGA |
| ESG1-FW | CCATGAATGCCCTCGAACTAGG |
| ESG1-RW | CCTTAACTCTTTAGGCTGGAGCA |
| LMX1b-FW | GCTGCATGGAGAAGATCGCC |
| LMX1b-RW | CTTGCGTAGCTGCCGTTCA |
| PITX3-FW | CCAACCTTAGTCCGTGCCAG |
| PITX3-RW | AGCCAGTCAAAATGACCCCA |
| DAT-FW | ACGTAGATCTGTGCAGCGAG |
| DAT-RW | CTCAGCAGGTGCGTCTACAA |
| NURR1-FW [49] | CAACTACAGCACAGGCTACGA |
| NURR1-RW [49] | GCATCTGAATGTCTTCTACCTTAATG |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaggi, G.; Di Credico, A.; Izzicupo, P.; Alviano, F.; Di Mauro, M.; Di Baldassarre, A.; Ghinassi, B. Human Mesenchymal Stromal Cells Unveil an Unexpected Differentiation Potential toward the Dopaminergic Neuronal Lineage. Int. J. Mol. Sci. 2020, 21, 6589. https://doi.org/10.3390/ijms21186589
Gaggi G, Di Credico A, Izzicupo P, Alviano F, Di Mauro M, Di Baldassarre A, Ghinassi B. Human Mesenchymal Stromal Cells Unveil an Unexpected Differentiation Potential toward the Dopaminergic Neuronal Lineage. International Journal of Molecular Sciences. 2020; 21(18):6589. https://doi.org/10.3390/ijms21186589
Chicago/Turabian StyleGaggi, Giulia, Andrea Di Credico, Pascal Izzicupo, Francesco Alviano, Michele Di Mauro, Angela Di Baldassarre, and Barbara Ghinassi. 2020. "Human Mesenchymal Stromal Cells Unveil an Unexpected Differentiation Potential toward the Dopaminergic Neuronal Lineage" International Journal of Molecular Sciences 21, no. 18: 6589. https://doi.org/10.3390/ijms21186589
APA StyleGaggi, G., Di Credico, A., Izzicupo, P., Alviano, F., Di Mauro, M., Di Baldassarre, A., & Ghinassi, B. (2020). Human Mesenchymal Stromal Cells Unveil an Unexpected Differentiation Potential toward the Dopaminergic Neuronal Lineage. International Journal of Molecular Sciences, 21(18), 6589. https://doi.org/10.3390/ijms21186589










