The Malignant Role of Exosomes as Nanocarriers of Rare RNA Species
Abstract
1. Introduction
2. Exosomal Loading of Rare RNAs in Cancer
2.1. Exosomal Loading of Rare RNAs in Breast Cancer and Their Roles in Development and Progression
2.2. Exosomal Loading of Rare RNAs in Lung Tumors and Their Roles in Development and Progression
2.3. Exosomal Loading of Rare RNAs in Oral Tumors and Their Roles in Development and Progression
2.4. Exosomal Loading of Rare RNAs in Gastrointestinal Tumors and Their Roles in Development and Progression
2.5. Exosomal Loading of Rare RNAs in Billary Tract Tumors and Their Roles in Development and Progression
2.6. Exosomal Loading of Rare RNAs in Brain Tumors and Their Roles in Development and Progression
2.7. Exosomal Loading of Rare RNAs in Melanoma and Their Roles in Development and Progression
2.8. Exosomal Loading of Rare RNAs in Bone Cancer and Their Roles in Development and Progression
2.9. Exosomal Loading of Rare RNAs in Hematological Malignancies and Their Roles in Development and Progression
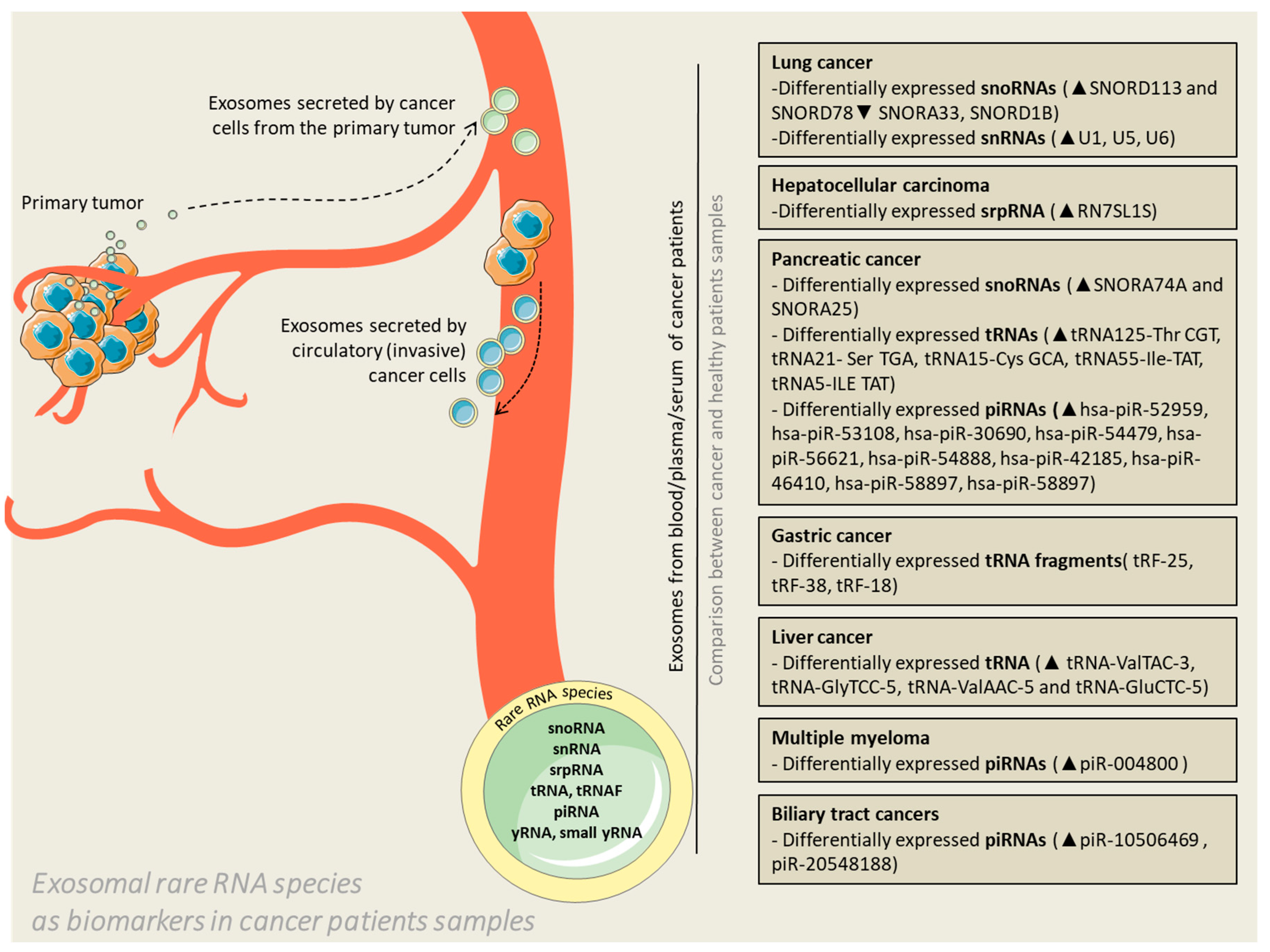
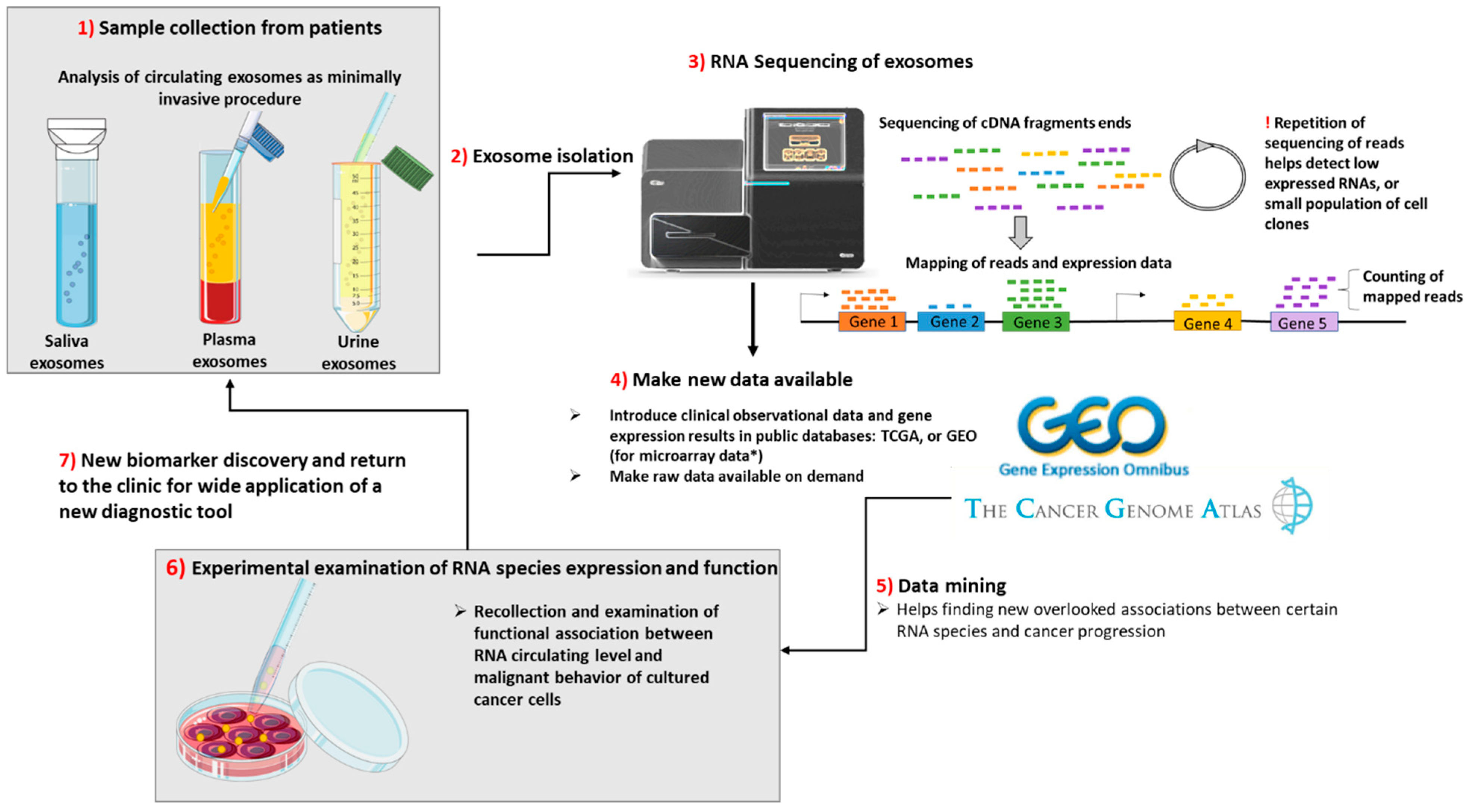
3. The Clinical Impact for New Biomarker and Therapeutic Target Discovery—Importance of Deep Sequencing and Data Mining
4. Final Remarks and Conclusions
| Type of RNA | Sequence Length (nt) | Region of the DNA | Type of Polymerase | Degree of Conservation across Species | Function | Biological Role | Type of Cancer | Cellular Localization | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| yRNA | 84, 98, 101, 112 | From specific DNA regions named hY1, hY3, hY4 and hY5; there are also pseudogenes of these YDNA regions | RNA Pol III | High degree of conservation | DNA replication; RNA stabilization (when bound to Ro60 protein) | Cell proliferation; cell cycle progression | Lung cancer, prostate cancer, colon cancer, renal cancer, cervical cancer, bladder cancer | Nucleus, cytoplasm | [90,91,92] |
| small yRNA | 19–60 | Represent fragments of yRNAs, especially from 5′ end of yRNA | RNA Pol III | High degree of conservation | Translation repression | Cell death, inflammation | Oral cancer, breast cancer | Cytoplasm | [11,16,93,94,95,96] |
| tRF | 14–30 | It has 446 specific genes in humans (for the full-length tRNAs) | RNA Pol III | Full length transcripts have high degree of conservation | Epigenetic control through heterochromatin modulation; competes with Dicer for miRNA binding; translation repression of mRNA; involved in RNA degradation and control of RNA stability | Cell cycle progression, cell proliferation, change in metabolic pathways, metastasis formation | Bladder cancer, lung cancer, renal cancer, colon cancer, breast cancer, ovarian cancer, liver cancer, lymphoma, chronic lymphocytic leukemia | Cytoplasm | [97,98,99,100,101,102] |
| tiRNA | 31–40 | 446 specific genes in humans (for the full-length tRNAs) | RNA Pol III | Full length transcripts have high degree of conservation | Chromatin/epigenetic modifications, translation repression, miRNA-like function; involved in RNA degradation and control of RNA stability | Cell cycle progression, cell proliferation | Breast cancer, prostate cancer, lung cancer, renal cancer | Cytoplasm | [98,99,102,103] |
| snoRNA | 60–300 | It is generally encoded by the introns or protein coding sequences of the DNA, but there it can also be encoded by DNA regions, where the snoRNA host genes (SNHGs) are located | RNA POL II or RNA POL III | snoRNAs in the intronic region show high degree of conservation | rRNA processing (2′-O-methylation or pseudouridylation, pre-rRNA cleavage) (H/ACA box snoRNA); mRNA splicing and mRNA processing (C/D box snoRNA), small Cajal body RNA (scaRNA), snoRNAs are origin for miRNAs | Activated in oxidative stress; involved in glucose metabolism; cell death, proliferation, invasion and metastasis | Lung cancer, prostate cancer, breast cancer, acute myeloid leukemia, acute promyelocytic leukemia, colorectal cancer, hepatocellular carcinoma, glioblastoma, osteosarcoma, pancreatic cancer | Nucleus (nucleolus), cytoplasm (only during cellular stress) | [20,104,105,106] |
| snRNA | 100–300 | Encoded by 5 DNA regions: U1, U2, U4, U5, and U6 | RNA POL III (U6), RNA POL II (U1-U5) | High degree of conservation | pre-mRNA splicing | Cell cycle progression, Tumorigenesis, oncogenic development | Breast cancer, lung cancer | Nucleus (main function), can also be exported into the cytoplasm (U6) | [107,108,109,110,111,112] |
| vtRNA | 88–100 | Special sequence from the DNA, vault DNA sequences are located on chromosome 5 | RNA POL III | High degree of conservation | Associated with specific proteins vault proteins in the cytoplasm, forming vault RNPs in the cytoplasm | Autophagy, intracellular and membrane trafficking, multidrug resistance (drug export from the cytoplasm) | Breast cancer, lymphoma, lung cancer, multiple myeloma | Cytoplasm | [22,113,114,115,116] |
| Alu-element RNA | around 250 | Alu-repeat containing transposable elements, comprises around 10% of human genome | RNA POL II, RNA POL III | The main genes containing ALU repeats are conserved, but there are also a number of evolved pseudogenes through single base substitution | They are involved in protein translation, are the ancestors of epigenetic enhancers, they offer new binding sites for transcription factors, impair mRNA or miRNA transcription | Increased expression in stress conditions, involved in the epithelial-to-mesenchymal transition, cell cycle progression | Breast cancer, colorectal cancer | Nucleus | [23,24,25,26] |
| piRNA | 21–35 | Encoded by protein-coding genes (untranslated regions of messenger RNAs), sequences form intergenic regions (long intergenic regions) | RNA POL III | High degree of conservation | It has a close interaction with piwi protein and together are involved in RNA cleavage. It also has epigenetic functions through heterochromatin regulation and induced changes in DNA methylation pattern. | Protects against germline genome stability and DNA integrity; maintains cancer stemness, apoptosis impairment, involved in telomerase activity, cell cycle progression and metastasis | Gastric cancer, lung cancer, cervical cancer, hepatocellular cancer, breast cancer, colorectal cancer, ovarian cancer | Cytoplasm | [117,118,119,120] |
Funding
Acknowledgments
Conflicts of Interest
References
- Nagai, H.; Kim, Y.H. Cancer prevention from the perspective of global cancer burden patterns. J. Thorac. Dis. 2017, 9, 448–451. [Google Scholar] [CrossRef]
- Jayaseelan, V.P. Emerging role of exosomes as promising diagnostic tool for cancer. Cancer Gene Ther. 2020, 27, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Gulei, D.; Petrut, B.; Tigu, A.B.; Onaciu, A.; Fischer-Fodor, E.; Atanasov, A.G.; Ionescu, C.; Berindan-Neagoe, I. Exosomes at a glance—Common nominators for cancer hallmarks and novel diagnosis tools. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.; Camussi, G.; Valadi, H.; Nazarenko, I.; Ekström, K.; Wang, X.; Principe, S.; Shah, N.; Ashraf, N.M.; Fatima, F.; et al. The emerging role of extracellular vesicles as biomarkers for urogenital cancers. Nat. Rev. Urol. 2014, 11, 688–701. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Maugeri, M.; Garre, E.; Nawaz, M.; Wahlgren, J.; Papadimitriou, A.; Lundqvist, C.; Lindfors, L.; Collén, A.; Sunnerhagen, P.; et al. Identification of RNA-binding proteins in exosomes capable of interacting with different types of RNA: RBP-facilitated transport of RNAs into exosomes. PLoS ONE 2018, 13, e0195969. [Google Scholar] [CrossRef] [PubMed]
- Fatima, F.; Nawaz, M. Vesiculated Long Non-Coding RNAs: Offshore Packages Deciphering Trans-Regulation between Cells, Cancer Progression and Resistance to Therapies. Noncoding RNA 2017, 3, 10. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.; Melo, S.A. Non-coding RNAs in Exosomes: New Players in Cancer Biology. Curr. Genom. 2015, 16, 295–303. [Google Scholar] [CrossRef]
- Xie, Y.; Dang, W.; Zhang, S.; Yue, W.; Yang, L.; Zhai, X.; Yan, Q.; Lu, J. The role of exosomal noncoding RNAs in cancer. Mol. Cancer 2019, 18, 37. [Google Scholar] [CrossRef]
- Braicu, C.; Zimta, A.A.; Harangus, A.; Iurca, I.; Irimie, A.; Coza, O.; Berindan-Neagoe, I. The Function of Non-Coding RNAs in Lung Cancer Tumorigenesis. Cancers 2019, 11, 605. [Google Scholar] [CrossRef]
- Irimie, A.I.; Zimta, A.A.; Ciocan, C.; Mehterov, N.; Dudea, D.; Braicu, C.; Berindan-Neagoe, I. The Unforeseen Non-Coding RNAs in Head and Neck Cancer. Genes 2018, 9, 134. [Google Scholar] [CrossRef]
- Dhahbi, J.; Nunez Lopez, Y.O.; Schneider, A.; Victoria, B.; Saccon, T.; Bharat, K.; McClatchey, T.; Atamna, H.; Scierski, W.; Golusinski, P.; et al. Profiling of tRNA Halves and YRNA Fragments in Serum and Tissue From Oral Squamous Cell Carcinoma Patients Identify Key Role of 5′ tRNA-Val-CAC-2-1 Half. Front. Oncol. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Yu, H.; Wang, J.; Sheng, Q.; Zhao, S.; Zhao, Y.Y.; Lehmann, B.D. The Landscape of Small Non-Coding RNAs in Triple-Negative Breast Cancer. Genes 2018, 9, 29. [Google Scholar] [CrossRef]
- Zhang, D.; Zhou, J.; Gao, J.; Wu, R.Y.; Huang, Y.L.; Jin, Q.W.; Chen, J.S.; Tang, W.Z.; Yan, L.H. Targeting snoRNAs as an emerging method of therapeutic development for cancer. Am. J. Cancer Res. 2019, 9, 1504–1516. [Google Scholar] [PubMed]
- Wang, X.; Xu, M.; Yan, Y.; Kuang, Y.; Li, P.; Zheng, W.; Liu, H.; Jia, B. Identification of Eight Small Nucleolar RNAs as Survival Biomarkers and Their Clinical Significance in Gastric Cancer. Front. Oncol. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, M.P.; Krude, T. Functional roles of non-coding Y RNAs. Int. J. Biochem. Cell Biol. 2015, 66, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Hizir, Z.; Bottini, S.; Grandjean, V.; Trabucchi, M.; Repetto, E. RNY (YRNA)-derived small RNAs regulate cell death and inflammation in monocytes/macrophages. Cell Death Dis. 2017, 8, e2530. [Google Scholar] [CrossRef]
- Anderson, P.; Ivanov, P. tRNA fragments in human health and disease. FEBS Lett. 2014, 588, 4297–4304. [Google Scholar] [CrossRef]
- Valadkhan, S.; Gunawardane, L.S. Role of small nuclear RNAs in eukaryotic gene expression. Essays Biochem. 2013, 54, 79–90. [Google Scholar] [CrossRef]
- Kiss, T. Small Nucleolar RNAs: An Abundant Group of Noncoding RNAs with Diverse Cellular Functions. Cell 2002, 109, 145–148. [Google Scholar] [CrossRef]
- Liang, J.; Wen, J.; Huang, Z.; Chen, X.P.; Zhang, B.X.; Chu, L. Small Nucleolar RNAs: Insight Into Their Function in Cancer. Front. Oncol. 2019, 9. [Google Scholar] [CrossRef]
- Horos, R.; Büscher, M.; Kleinendorst, R.; Alleaume, A.-M.; Tarafder, A.K.; Schwarzl, T.; Dziuba, D.; Tischer, C.; Zielonka, E.M.; Adak, A.; et al. The Small Non-coding Vault RNA1-1 Acts as a Riboregulator of Autophagy. Cell 2019, 176, 1054–1067.e12. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; OuYang, H.; An, X.; Liu, S. Vault RNAs partially induces drug resistance of human tumor cells MCF-7 by binding to the RNA/DNA-binding protein PSF and inducing oncogene GAGE6. PLoS ONE 2018, 13, e0191325. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-L.; Yang, L. ALUternative Regulation for Gene Expression. Trends Cell Biol. 2017, 27, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Caudron-Herger, M.; Pankert, T.; Seiler, J.; Németh, A.; Voit, R.; Grummt, I.; Rippe, K. Alu element-containing RNAs maintain nucleolar structure and function. EMBO J. 2015, 34, 2758–2774. [Google Scholar] [CrossRef] [PubMed]
- Di Ruocco, F.; Basso, V.; Rivoire, M.; Mehlen, P.; Ambati, J.; De Falco, S.; Tarallo, V. Alu RNA accumulation induces epithelial-to-mesenchymal transition by modulating miR-566 and is associated with cancer progression. Oncogene 2018, 37, 627–637. [Google Scholar] [CrossRef] [PubMed]
- Cantarella, S.; Carnevali, D.; Morselli, M.; Conti, A.; Pellegrini, M.; Montanini, B.; Dieci, G. Alu RNA Modulates the Expression of Cell Cycle Genes in Human Fibroblasts. Int. J. Mol. Sci. 2019, 20, 3315. [Google Scholar] [CrossRef] [PubMed]
- Tóth, K.F.; Pezic, D.; Stuwe, E.; Webster, A. The piRNA Pathway Guards the Germline Genome Against Transposable Elements. Adv. Exp. Med. Biol. 2016, 886, 51–77. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, S.; Matushansky, I. Piwis and piwi-interacting RNAs in the epigenetics of cancer. J. Cell. Biochem. 2012, 113, 373–380. [Google Scholar] [CrossRef]
- Cheng, Y.; Wang, Q.; Jiang, W.; Bian, Y.; Zhou, Y.; Gou, A.; Zhang, W.; Fu, K.; Shi, W. Emerging roles of piRNAs in cancer: Challenges and prospects. Aging 2019, 11, 9932–9946. [Google Scholar] [CrossRef]
- Pink, R.C.; Wicks, K.; Caley, D.P.; Punch, E.K.; Jacobs, L.; Carter, D.R.F. Pseudogenes: Pseudo-functional or key regulators in health and disease? RNA (N.Y.) 2011, 17, 792–798. [Google Scholar] [CrossRef]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed]
- King, M.-C.; Marks, J.H.; Mandell, J.B. Breast and Ovarian Cancer Risks Due to Inherited Mutations in BRCA1 and BRCA2. Science 2003, 302, 643–646. [Google Scholar] [CrossRef] [PubMed]
- Zimta, A.A.; Tigu, A.B.; Muntean, M.; Cenariu, D.; Slaby, O.; Berindan-Neagoe, I. Molecular Links between Central Obesity and Breast Cancer. Int. J. Mol. Sci. 2019, 20, 5364. [Google Scholar] [CrossRef] [PubMed]
- Tosar, J.P.; Gámbaro, F.; Sanguinetti, J.; Bonilla, B.; Witwer, K.W.; Cayota, A. Assessment of small RNA sorting into different extracellular fractions revealed by high-throughput sequencing of breast cell lines. Nucleic Acids Res. 2015, 43, 5601–5616. [Google Scholar] [CrossRef] [PubMed]
- Hashim, A.; Rizzo, F.; Marchese, G.; Ravo, M.; Tarallo, R.; Nassa, G.; Giurato, G.; Santamaria, G.; Cordella, A.; Cantarella, C.; et al. RNA sequencing identifies specific PIWI-interacting small non-coding RNA expression patterns in breast cancer. Oncotarget 2014, 5, 9901–9910. [Google Scholar] [CrossRef]
- Fu, A.; Jacobs, D.I.; Hoffman, A.E.; Zheng, T.; Zhu, Y. PIWI-interacting RNA 021285 is involved in breast tumorigenesis possibly by remodeling the cancer epigenome. Carcinogenesis 2015, 36, 1094–1102. [Google Scholar] [CrossRef]
- Gámbaro, F.; Li Calzi, M.; Fagúndez, P.; Costa, B.; Greif, G.; Mallick, E.; Lyons, S.; Ivanov, P.; Witwer, K.; Cayota, A.; et al. Stable tRNA halves can be sorted into extracellular vesicles and delivered to recipient cells in a concentration-dependent manner. RNA Biol. 2019, 1–15. [Google Scholar] [CrossRef]
- Ellis, P.M.; Vandermeer, R. Delays in the diagnosis of lung cancer. J. Thorac. Dis. 2011, 3, 183–188. [Google Scholar] [CrossRef]
- Savelyeva, A.V.; Kuligina, E.V.; Bariakin, D.N.; Kozlov, V.V.; Ryabchikova, E.I.; Richter, V.A.; Semenov, D.V. Variety of RNAs in Peripheral Blood Cells, Plasma, and Plasma Fractions. Biomed. Res. Int. 2017, 2017, 7404912. [Google Scholar] [CrossRef]
- Kumar, M.; Nanavati, R.; Modi, T.G.; Dobariya, C. Oral cancer: Etiology and risk factors: A review. J. Cancer Res. Ther. 2016, 12, 458–463. [Google Scholar] [CrossRef]
- Auguste, A.; Deloumeaux, J.; Joachim, C.; Gaete, S.; Michineau, L.; Herrmann-Storck, C.; Duflo, S.; Luce, D. Joint effect of tobacco, alcohol, and oral HPV infection on head and neck cancer risk in the French West Indies. Cancer Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, Y.; Taketomi, Y.; Murakami, M.; Tsujimoto, M.; Yanoshita, R. Small RNA transcriptomes of two types of exosomes in human whole saliva determined by next generation sequencing. Biol. Pharm. Bull. 2013, 36, 66–75. [Google Scholar] [CrossRef]
- Bahn, J.H.; Zhang, Q.; Li, F.; Chan, T.M.; Lin, X.; Kim, Y.; Wong, D.T.; Xiao, X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem. 2015, 61, 221–230. [Google Scholar] [CrossRef]
- Arnold, M.; Abnet, C.C.; Neale, R.E.; Vignat, J.; Giovannucci, E.L.; McGlynn, K.A.; Bray, F. Global Burden of 5 Major Types of Gastrointestinal Cancer. Gastroenterology 2020, 159, 335–349.e15. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Cao, J.; Chen, L.; Xi, X.; Wang, S.; Zhu, Y.; Yang, L.; Ma, L.; Wang, D.; Yin, J.; et al. Noncoding RNAs Serve as Diagnosis and Prognosis Biomarkers for Hepatocellular Carcinoma. Clin. Chem. 2019, 65, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, N.; Walter, P. The signal recognition particle (SRP) RNA links conformational changes in the SRP to protein targeting. Mol. Biol. Cell 2007, 18, 2728–2734. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Li, J.; Gong, Y.; Wu, Q.; Tan, S.; Sun, D.; Xu, X.; Zuo, Y.; Zhao, Y.; Wei, Y.Q.; et al. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol. Cancer 2019, 18, 74. [Google Scholar] [CrossRef]
- Kitagawa, T.; Taniuchi, K.; Tsuboi, M.; Sakaguchi, M.; Kohsaki, T.; Okabayashi, T.; Saibara, T. Circulating pancreatic cancer exosomal RNAs for detection of pancreatic cancer. Mol. Oncol. 2019, 13, 212–227. [Google Scholar] [CrossRef]
- Kumar, S.R.; Kimchi, E.T.; Manjunath, Y.; Gajagowni, S.; Stuckel, A.J.; Kaifi, J.T. RNA cargos in extracellular vesicles derived from blood serum in pancreas associated conditions. Sci. Rep. 2020, 10, 2800. [Google Scholar] [CrossRef]
- Lin, C.; Zheng, L.; Huang, R.; Yang, G.; Chen, J.; Li, H. tRFs as Potential Exosome tRNA-Derived Fragment Biomarkers for Gastric Carcinoma. Clin. Lab. 2020, 66. [Google Scholar] [CrossRef]
- Ren, J.; Zhou, Q.; Li, H.; Li, J.; Pang, L.; Su, L.; Gu, Q.; Zhu, Z.; Liu, B. Characterization of exosomal RNAs derived from human gastric cancer cells by deep sequencing. Tumour Biol. 2017, 39, 1010428317695012. [Google Scholar] [CrossRef] [PubMed]
- Fonseca Cabral, G.; Azevedo Dos Santos Pinheiro, J.; Vidal, A.F.; Santos, S.; Ribeiro-Dos-Santos, Â. piRNAs in Gastric Cancer: A New Approach Towards Translational Research. Int. J. Mol. Sci. 2020, 21, 2126. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Li, T.; Shen, Y.; Yu, X.; Xiao, B.; Guo, J. Using tRNA halves as novel biomarkers for the diagnosis of gastric cancer. Cancer Biomark. 2019, 25, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Xu, R.; Ji, H.; Greening, D.W.; Rai, A.; Izumikawa, K.; Ishikawa, H.; Takahashi, N.; Simpson, R.J. Transcriptome and long noncoding RNA sequencing of three extracellular vesicle subtypes released from the human colon cancer LIM1863 cell line. Sci. Rep. 2016, 6, 38397. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Wang, C.; Deng, H.; Qing, C.; Liu, R.; Liu, S.; Xue, X. Exosomal piRNA profiling revealed unique circulating piRNA signatures of cholangiocarcinoma and gallbladder carcinoma. Acta Biochim. Biophys. Sin. 2020, 52, 475–484. [Google Scholar] [CrossRef] [PubMed]
- Bosma, I.; Vos, M.J.; Heimans, J.J.; Taphoorn, M.J.B.; Aaronson, N.K.; Postma, T.J.; van der Ploeg, H.M.; Muller, M.; Vandertop, W.P.; Slotman, B.J.; et al. The course of neurocognitive functioning in high-grade glioma patients. Neuro Oncol. 2007, 9, 53–62. [Google Scholar] [CrossRef] [PubMed]
- van Kessel, E.; Baumfalk, A.E.; van Zandvoort, M.J.E.; Robe, P.A.; Snijders, T.J. Tumor-related neurocognitive dysfunction in patients with diffuse glioma: A systematic review of neurocognitive functioning prior to anti-tumor treatment. J. Neurooncol. 2017, 134, 9–18. [Google Scholar] [CrossRef]
- Li, C.C.; Eaton, S.A.; Young, P.E.; Lee, M.; Shuttleworth, R.; Humphreys, D.T.; Grau, G.E.; Combes, V.; Bebawy, M.; Gong, J.; et al. Glioma microvesicles carry selectively packaged coding and non-coding RNAs which alter gene expression in recipient cells. RNA Biol. 2013, 10, 1333–1344. [Google Scholar] [CrossRef]
- de Mooij, T.; Peterson, T.E.; Evans, J.; McCutcheon, B.; Parney, I.F. Short non-coding RNA sequencing of glioblastoma extracellular vesicles. J. Neurooncol. 2020, 146, 253–263. [Google Scholar] [CrossRef]
- Wei, Z.; Batagov, A.O.; Schinelli, S.; Wang, J.; Wang, Y.; El Fatimy, R.; Rabinovsky, R.; Balaj, L.; Chen, C.C.; Hochberg, F.; et al. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat. Commun. 2017, 8, 1145. [Google Scholar] [CrossRef]
- Kore, R.A.; Edmondson, J.L.; Jenkins, S.V.; Jamshidi-Parsian, A.; Dings, R.P.M.; Reyna, N.S.; Griffin, R.J. Hypoxia-derived exosomes induce putative altered pathways in biosynthesis and ion regulatory channels in glioblastoma cells. Biochem. Biophys. Rep. 2018, 14, 104–113. [Google Scholar] [CrossRef] [PubMed]
- van Balkom, B.W.; Eisele, A.S.; Pegtel, D.M.; Bervoets, S.; Verhaar, M.C. Quantitative and qualitative analysis of small RNAs in human endothelial cells and exosomes provides insights into localized RNA processing, degradation and sorting. J. Extracell. Vesicles 2015, 4, 26760. [Google Scholar] [CrossRef] [PubMed]
- Balaj, L.; Lessard, R.; Dai, L.; Cho, Y.J.; Pomeroy, S.L.; Breakefield, X.O.; Skog, J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2011, 2, 180. [Google Scholar] [CrossRef] [PubMed]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef]
- Lunavat, T.R.; Cheng, L.; Einarsdottir, B.O.; Olofsson Bagge, R.; Veppil Muralidharan, S.; Sharples, R.A.; Lässer, C.; Gho, Y.S.; Hill, A.F.; Nilsson, J.A.; et al. BRAFV600 inhibition alters the microRNA cargo in the vesicular secretome of malignant melanoma cells. Proc. Natl. Acad. Sci. USA 2017. [Google Scholar] [CrossRef]
- Luetke, A.; Meyers, P.A.; Lewis, I.; Juergens, H. Osteosarcoma treatment—Where do we stand? A state of the art review. Cancer Treat. Rev. 2014, 40, 523–532. [Google Scholar] [CrossRef]
- Liu, D.D.; Kang, Y. Ets2 anchors the prometastatic function of mutant p53 in osteosarcoma. Genes Dev. 2017, 31, 1823–1824. [Google Scholar] [CrossRef]
- Evdokimova, V.; Ruzanov, P.; Gassmann, H.; Zaidi, S.H.; Peltekova, V.; Heisler, L.E.; McPherson, J.D.; Orlic-Milacic, M.; Specht, K.; Steiger, K.; et al. Exosomes transmit retroelement RNAs to drive inflammation and immunosuppression in Ewing Sarcoma. bioRxiv 2019, 806851. [Google Scholar] [CrossRef]
- Vardiman, J.W.; Thiele, J.; Arber, D.A.; Brunning, R.D.; Borowitz, M.J.; Porwit, A.; Harris, N.L.; Le Beau, M.M.; Hellström-Lindberg, E.; Tefferi, A.; et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: Rationale and important changes. Blood 2009, 114, 937–951. [Google Scholar] [CrossRef]
- Baba AI, C.C. Chapter 17, TUMORS OF HEMATOPOIETIC AND LYMPHOID TISSUES. In Comparative Oncology; The Publishing House of the Romanian Academy: Bucharest, Romania, 2007. [Google Scholar]
- Ireland, R. Haematological malignancies: The rationale for integrated haematopathology services, key elements of organization and wider contribution to patient care. Histopathology 2011, 58, 145–154. [Google Scholar] [CrossRef]
- Lovisa, F.; Di Battista, P.; Gaffo, E.; Damanti, C.C.; Garbin, A.; Gallingani, I.; Carraro, E.; Pillon, M.; Biffi, A.; Bortoluzzi, S.; et al. RNY4 in Circulating Exosomes of Patients With Pediatric Anaplastic Large Cell Lymphoma: An Active Player? Front. Oncol. 2020, 10, 238. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Wang, H.; Tian, F.; Zhong, Y.; Liu, Z.; Liao, A. PIWI-Interacting RNA-004800 Is Regulated by S1P Receptor Signaling Pathway to Keep Myeloma Cell Survival. Front. Oncol. 2020, 10, 438. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Liu, J.; Zhuang, X.; Yu, S.; Zhu, S.; Liu, Y.; Chen, X. Identification and Comparison of piRNA Expression Profiles of Exosomes Derived from Human Stem Cells from the Apical Papilla and Bone Marrow Mesenchymal Stem Cells. Stem Cells Dev. 2020, 29, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Qiu, W.; Xu, X.; Kang, J.; Wang, J.; Wen, Y.; Tang, X.; Yan, Y.; Qian, H.; Zhang, X.; et al. Exosomes derived from human umbilical cord mesenchymal stem cells alleviate inflammatory bowel disease in mice through ubiquitination. Am. J. Transl. Res. 2018, 10, 2026–2036. [Google Scholar] [PubMed]
- He, J.G.; Xie, Q.L.; Li, B.B.; Zhou, L.; Yan, D. Exosomes Derived from IDO1-Overexpressing Rat Bone Marrow Mesenchymal Stem Cells Promote Immunotolerance of Cardiac Allografts. Cell Transplant. 2018, 27, 1657–1683. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Wang, J.; Zhou, X.; Xiong, Z.; Zhao, J.; Yu, R.; Huang, F.; Zhang, H.; Chen, L. Exosomes derived from human adipose mensenchymal stem cells accelerates cutaneous wound healing via optimizing the characteristics of fibroblasts. Sci. Rep. 2016, 6, 32993. [Google Scholar] [CrossRef]
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.; Jacob, H.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M.; et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genom. 2013, 14, 319. [Google Scholar] [CrossRef]
- Baglio, S.R.; Rooijers, K.; Koppers-Lalic, D.; Verweij, F.J.; Pérez Lanzón, M.; Zini, N.; Naaijkens, B.; Perut, F.; Niessen, H.W.; Baldini, N.; et al. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res. Ther. 2015, 6, 127. [Google Scholar] [CrossRef]
- Fan, G.; Li, J. Regions identity between the genome of vertebrates and non-retroviral families of insect viruses. Virol. J. 2011, 8, 511. [Google Scholar] [CrossRef]
- Wu, Z.; Sun, H.; Wang, C.; Liu, W.; Liu, M.; Zhu, Y.; Xu, W.; Jin, H.; Li, J. Mitochondrial Genome-Derived circRNA mc-COX2 Functions as an Oncogene in Chronic Lymphocytic Leukemia. Mol. Ther. Nucleic Acids 2020, 20, 801–811. [Google Scholar] [CrossRef]
- Sims, D.; Sudbery, I.; Ilott, N.E.; Heger, A.; Ponting, C.P. Sequencing depth and coverage: Key considerations in genomic analyses. Nat. Rev. Genet. 2014, 15, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Goldman, D.; Domschke, K. Making sense of deep sequencing. Int. J. Neuropsychopharmacol. 2014, 17, 1717–1725. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hwang, B.; Lee, J.H.; Bang, D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 2018, 50, 96. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Umbach, N.; Beißbarth, T.; Bleckmann, A.; Duttge, G.; Flatau, L.; König, A.; Kuhn, J.; Perera-Bel, J.; Roschauer, J.; Schulze, T.G.; et al. Clinical application of genomic high-throughput data: Infrastructural, ethical, legal and psychosocial aspects. Eur. Neuropsychopharmacol. J. Eur. Coll. Neuropsychopharmacol. 2020, 31, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.G.; Zeng, J.H.; Lin, P.; Mo, W.J.; Li, Q.; Feng, Z.B.; Luo, D.Z.; Yang, H.; Chen, G.; Zeng, J.J. Prognostic value of small nuclear RNAs (snRNAs) for digestive tract pan- adenocarcinomas identified by RNA sequencing data. Pathol. Res. Pract. 2019, 215, 414–426. [Google Scholar] [CrossRef]
- Dhahbi, J.M.; Atamna, H.; Selth, L.A. Data Mining of Small RNA-Seq Suggests an Association Between Prostate Cancer and Altered Abundance of 5′ Transfer RNA Halves in Seminal Fluid and Prostatic Tissues. Biomark. Cancer 2018, 10, 1179299x18759545. [Google Scholar] [CrossRef]
- Yao, D.; Sun, X.; Zhou, L.; Amanullah, M.; Pan, X.; Liu, Y.; Liang, M.; Liu, P.; Lu, Y. OncotRF: An online resource for exploration of tRNA-derived fragments in human cancers. RNA Biol. 2020, 1–11. [Google Scholar] [CrossRef]
- Wolin, S.L.; Steitz, J.A. Genes for two small cytoplasmic Ro RNAs are adjacent and appear to be single-copy in the human genome. Cell 1983, 32, 735–744. [Google Scholar] [CrossRef]
- Sacks, D.; Baxter, B.; Campbell, B.C.V.; Carpenter, J.S.; Cognard, C.; Dippel, D.; Eesa, M.; Fischer, U.; Hausegger, K.; Hirsch, J.A.; et al. Multisociety Consensus Quality Improvement Revised Consensus Statement for Endovascular Therapy of Acute Ischemic Stroke. Int. J. Stroke 2018, 13, 612–632. [Google Scholar] [CrossRef]
- Christov, C.P.; Trivier, E.; Krude, T. Noncoding human Y RNAs are overexpressed in tumours and required for cell proliferation. Br. J. Cancer 2008, 98, 981–988. [Google Scholar] [CrossRef] [PubMed]
- Sim, S.; Wolin, S.L. Emerging roles for the Ro 60-kDa autoantigen in noncoding RNA metabolism. Wiley Interdiscip. Rev. RNA 2011, 2, 686–699. [Google Scholar] [CrossRef]
- Dhahbi, J.M.; Spindler, S.R.; Atamna, H.; Boffelli, D.; Mote, P.; Martin, D.I.K. 5′-YRNA fragments derived by processing of transcripts from specific YRNA genes and pseudogenes are abundant in human serum and plasma. Physiol. Genomics 2013, 45, 990–998. [Google Scholar] [CrossRef]
- Gulìa, C.; Signore, F.; Gaffi, M.; Gigli, S.; Votino, R.; Nucciotti, R.; Bertacca, L.; Zaami, S.; Baffa, A.; Santini, E.; et al. Y RNA: An Overview of Their Role as Potential Biomarkers and Molecular Targets in Human Cancers. Cancers 2020, 12, 1238. [Google Scholar] [CrossRef]
- Dhahbi, J.M.; Spindler, S.R.; Atamna, H.; Boffelli, D.; Martin, D.I. Deep Sequencing of Serum Small RNAs Identifies Patterns of 5′ tRNA Half and YRNA Fragment Expression Associated with Breast Cancer. Biomark. Cancer 2014, 6, 37–47. [Google Scholar] [CrossRef]
- Boskovic, A.; Bing, X.Y.; Kaymak, E.; Rando, O.J. Control of noncoding RNA production and histone levels by a 5′ tRNA fragment. Genes Dev. 2020, 34, 118–131. [Google Scholar] [CrossRef]
- Goodenbour, J.M.; Pan, T. Diversity of tRNA genes in eukaryotes. Nucleic Acids Res. 2006, 34, 6137–6146. [Google Scholar] [CrossRef]
- Xie, Y.; Yao, L.; Yu, X.; Ruan, Y.; Li, Z.; Guo, J. Action mechanisms and research methods of tRNA-derived small RNAs. Signal Transduct. Target. Ther. 2020, 5, 109. [Google Scholar] [CrossRef]
- Kutsuna, T.; Hiyama, Y.; Kusaka, S.; Kusumoto, Y.; Tsuchiya, J.; Umeda, M.; Takahashi, T. The effect of short-term health promotion intervention on motor function in community-dwelling older adults. Aging Clin. Exp. Res. 2019, 31, 475–481. [Google Scholar] [CrossRef]
- Telonis, A.G.; Loher, P.; Magee, R.; Pliatsika, V.; Londin, E.; Kirino, Y.; Rigoutsos, I. tRNA Fragments Show Intertwining with mRNAs of Specific Repeat Content and Have Links to Disparities. Cancer Res. 2019, 79, 3034–3049. [Google Scholar] [CrossRef]
- Zhu, P.; Yu, J.; Zhou, P. Role of tRNA-derived fragments in cancer: Novel diagnostic and therapeutic targets tRFs in cancer. Am. J. Cancer Res. 2020, 10, 393–402. [Google Scholar]
- Lee, Y.S.; Shibata, Y.; Malhotra, A.; Dutta, A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009, 23, 2639–2649. [Google Scholar] [CrossRef]
- Holley, C.L.; Li, M.W.; Scruggs, B.S.; Matkovich, S.J.; Ory, D.S.; Schaffer, J.E. Cytosolic accumulation of small nucleolar RNAs (snoRNAs) is dynamically regulated by NADPH oxidase. J. Biol. Chem. 2015, 290, 11741–11748. [Google Scholar] [CrossRef]
- Scott, M.S.; Ono, M. From snoRNA to miRNA: Dual function regulatory non-coding RNAs. Biochimie 2011, 93, 1987–1992. [Google Scholar] [CrossRef]
- Hoeppner, M.P.; White, S.; Jeffares, D.C.; Poole, A.M. Evolutionarily stable association of intronic snoRNAs and microRNAs with their host genes. Genome Biol. Evol. 2009, 1, 420–428. [Google Scholar] [CrossRef]
- Kiss, T. Biogenesis of small nuclear RNPs. J. Cell Sci. 2004, 117, 5949–5951. [Google Scholar] [CrossRef]
- Jády, B.E.; Darzacq, X.; Tucker, K.E.; Matera, A.G.; Bertrand, E.; Kiss, T. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J. 2003, 22, 1878–1888. [Google Scholar] [CrossRef]
- Matera, A.G.; Terns, R.M.; Terns, M.P. Non-coding RNAs: Lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007, 8, 209–220. [Google Scholar] [CrossRef]
- Roy, K.; Chanfreau, G.F. Chapter Ten—The Diverse Functions of Fungal RNase III Enzymes in RNA Metabolism. In The Enzymes; Chanfreau, G.F., Tamanoi, F., Eds.; Academic Press: Cambridge, MA, USA, 2012; Volume 31, pp. 213–235. [Google Scholar]
- Dvinge, H.; Guenthoer, J.; Porter, P.L.; Bradley, R.K. RNA components of the spliceosome regulate tissue- and cancer-specific alternative splicing. Genome Res. 2019, 29, 1591–1604. [Google Scholar] [CrossRef]
- Oh, J.-M.; Venters, C.C.; Di, C.; Pinto, A.M.; Wan, L.; Younis, I.; Cai, Z.; Arai, C.; So, B.R.; Duan, J.; et al. U1 snRNP regulates cancer cell migration and invasion in vitro. Nat. Commun. 2020, 11, 1–8. [Google Scholar] [CrossRef]
- Büscher, M.; Horos, R.; Hentze, M.W. ‘High vault-age’: Non-coding RNA control of autophagy. Open Biol. 2020, 10, 190307. [Google Scholar] [CrossRef]
- Amort, M.; Nachbauer, B.; Tuzlak, S.; Kieser, A.; Schepers, A.; Villunger, A.; Polacek, N. Expression of the vault RNA protects cells from undergoing apoptosis. Nat. Commun. 2015, 6, 7030. [Google Scholar] [CrossRef]
- van Zon, A.; Mossink, M.H.; Houtsmuller, A.B.; Schoester, M.; Scheffer, G.L.; Scheper, R.J.; Sonneveld, P.; Wiemer, E.A. Vault mobility depends in part on microtubules and vaults can be recruited to the nuclear envelope. Exp. Cell Res. 2006, 312, 245–255. [Google Scholar] [CrossRef]
- Labuda, D.; Striker, G. Sequence conservation in Alu evolution. Nucleic Acids Res. 1989, 17, 2477–2491. [Google Scholar] [CrossRef]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef]
- Sun, T.; Han, X. The disease-related biological functions of PIWI-interacting RNAs (piRNAs) and underlying molecular mechanisms. ExRNA 2019, 1. [Google Scholar] [CrossRef]
- Han, B.W.; Zamore, P.D. piRNAs. Curr. Biol. 2014, 24, R730–R733. [Google Scholar] [CrossRef]
- Lim, S.L.; Tsend-Ayush, E.; Kortschak, R.D.; Jacob, R.; Ricciardelli, C.; Oehler, M.K.; Grützner, F. Conservation and Expression of PIWI-Interacting RNA Pathway Genes in Male and Female Adult Gonad of Amniotes. Biol. Reprod. 2013, 89. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zimta, A.-A.; Sigurjonsson, O.E.; Gulei, D.; Tomuleasa, C. The Malignant Role of Exosomes as Nanocarriers of Rare RNA Species. Int. J. Mol. Sci. 2020, 21, 5866. https://doi.org/10.3390/ijms21165866
Zimta A-A, Sigurjonsson OE, Gulei D, Tomuleasa C. The Malignant Role of Exosomes as Nanocarriers of Rare RNA Species. International Journal of Molecular Sciences. 2020; 21(16):5866. https://doi.org/10.3390/ijms21165866
Chicago/Turabian StyleZimta, Alina-Andreea, Olafur Eysteinn Sigurjonsson, Diana Gulei, and Ciprian Tomuleasa. 2020. "The Malignant Role of Exosomes as Nanocarriers of Rare RNA Species" International Journal of Molecular Sciences 21, no. 16: 5866. https://doi.org/10.3390/ijms21165866
APA StyleZimta, A.-A., Sigurjonsson, O. E., Gulei, D., & Tomuleasa, C. (2020). The Malignant Role of Exosomes as Nanocarriers of Rare RNA Species. International Journal of Molecular Sciences, 21(16), 5866. https://doi.org/10.3390/ijms21165866






