Exploring the Biochemical Origin of DNA Sequence Variation in Barley Plants Regenerated via in Vitro Anther Culture
Abstract
1. Introduction
2. Results
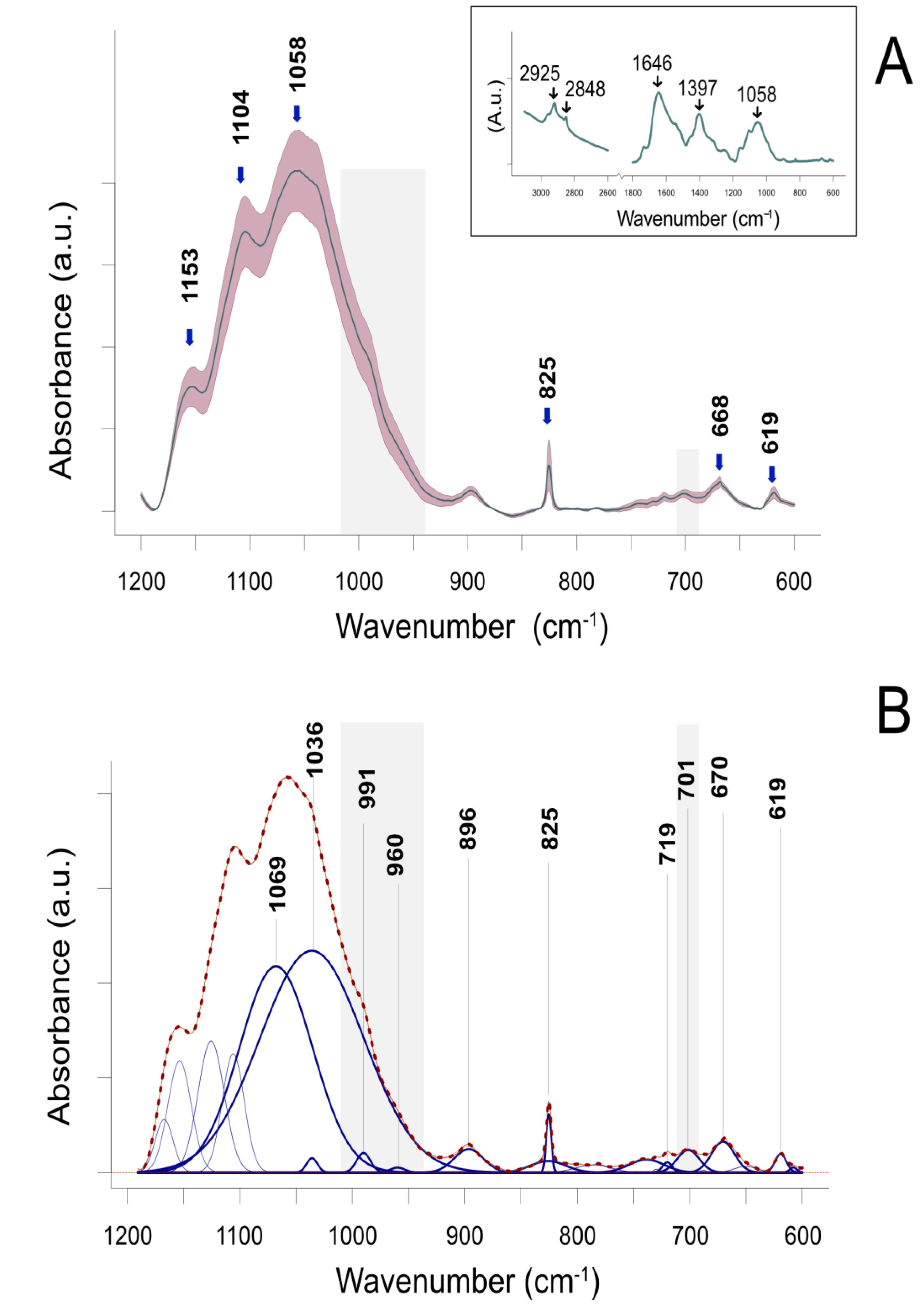
2.1. ATR-FTIR Spectroscopy
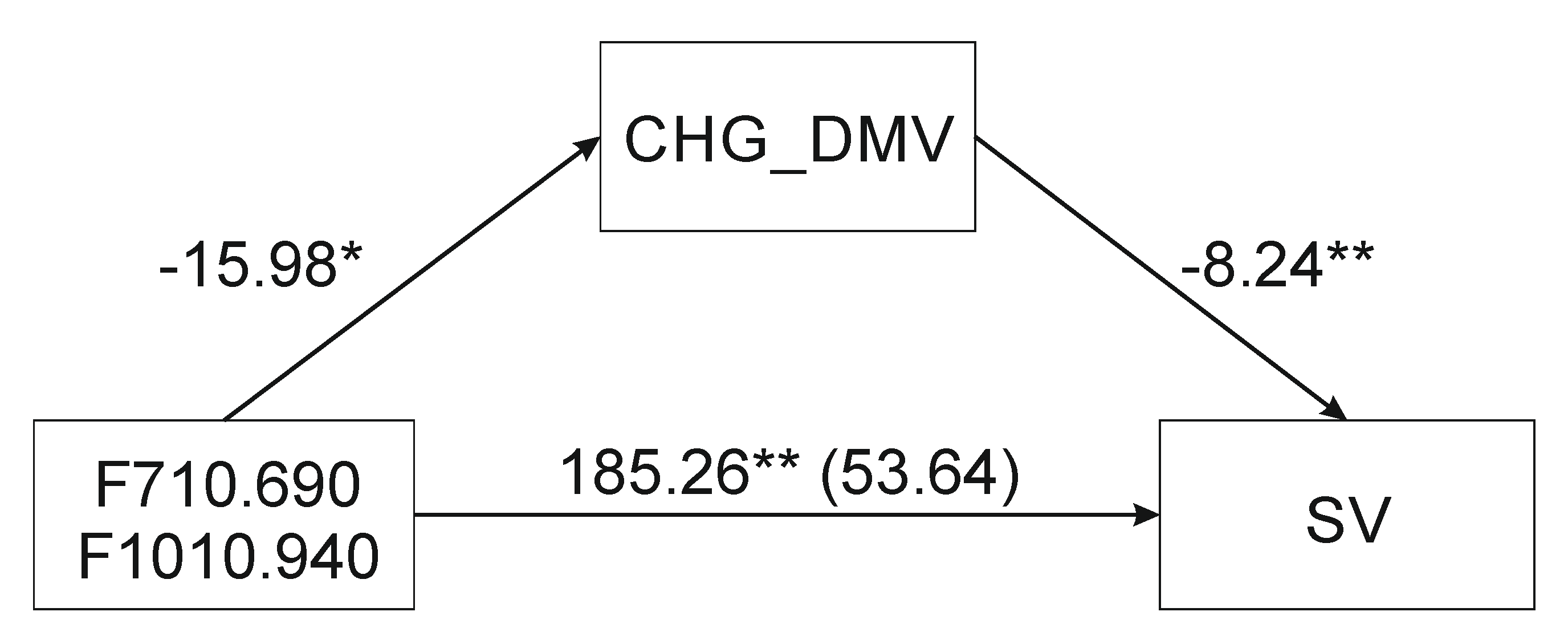
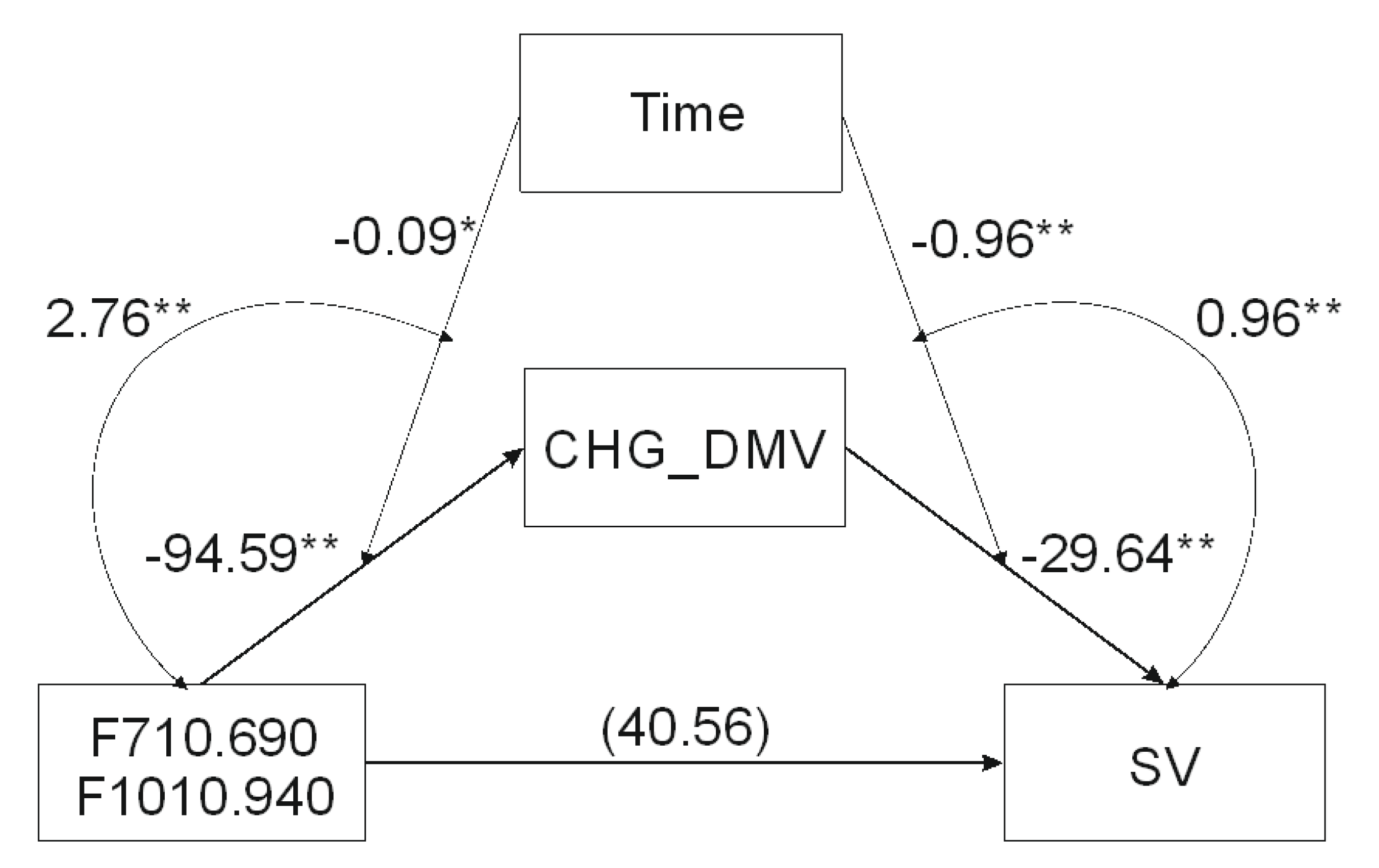
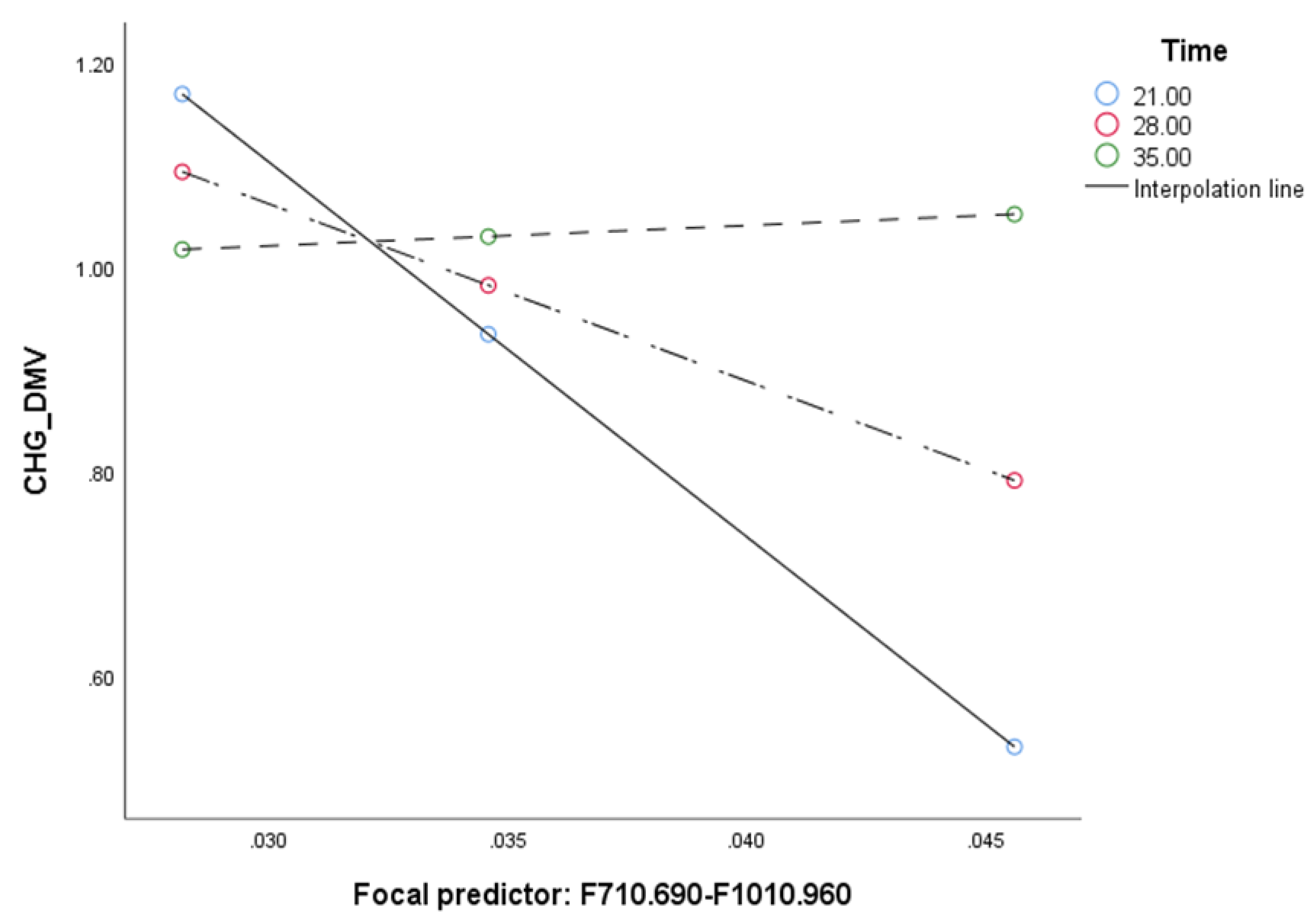
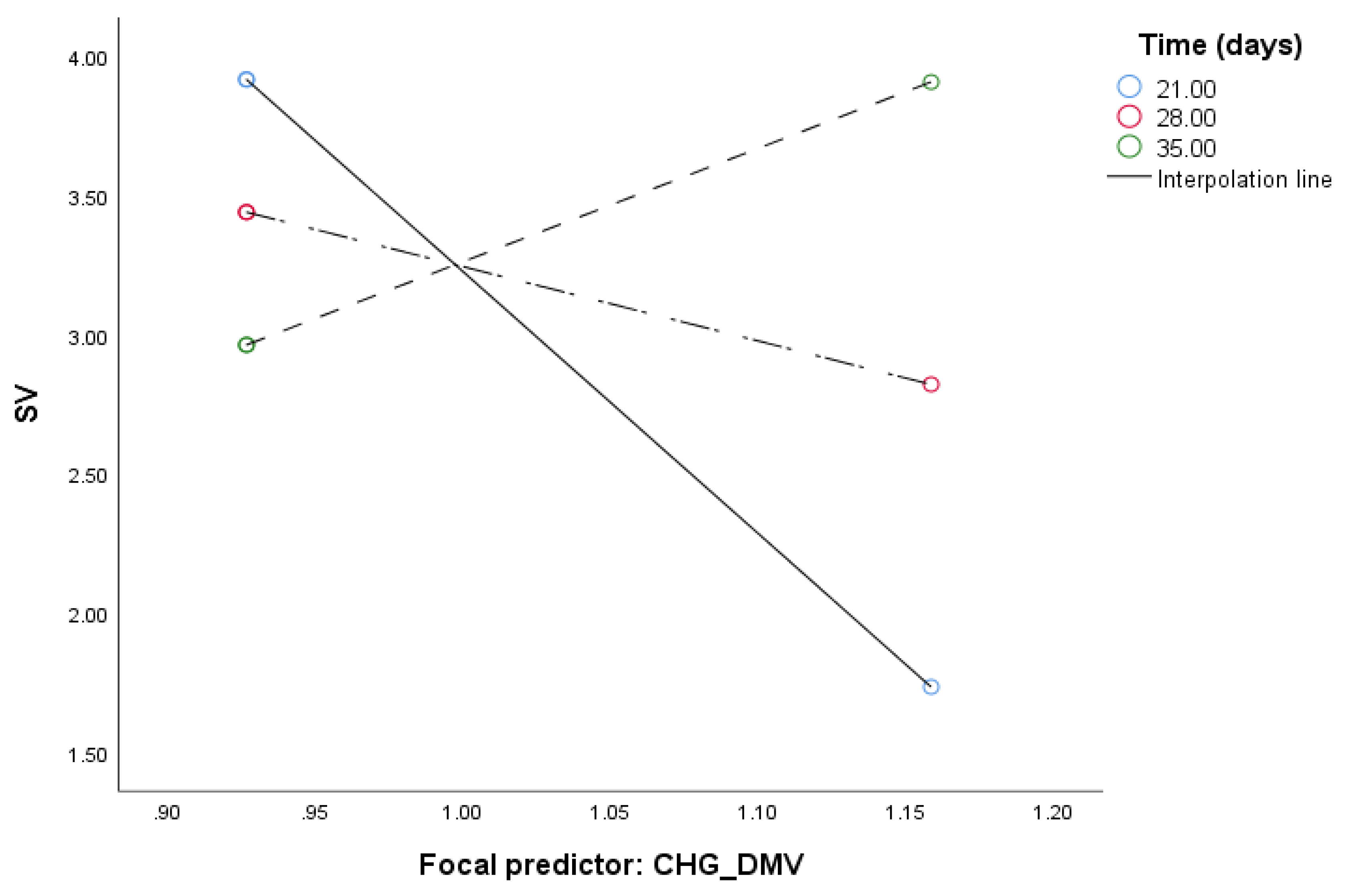
2.2. Mediation Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. DNA Isolation and Analysis
4.3. Mediation Analysis
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 5 hmC | 5-hydroxymethylcytosine |
| 5 mC | 5-methylcytosine |
| ATR-FTIR | Attenuated Total Reflectance Fourier Transform Infrared |
| CG_DMV | demethylation of the CG contexts |
| CHG_DMV | demethylation of the CHG contexts |
| dcAdoMet | decarboxylated form of SAM |
| DMV | demethylation |
| IDM | increased DNA methylation complex |
| IF | indirect effect |
| metAFLP | methylation-sensitive Amplified Fragment Length Polymorphism |
| MMR | mismatch repair |
| NER | nucleotide excision repair |
| RdDM | RNA-directed DNA methylation |
| ROS1 | REPRESSOR OF SILENCING 1 |
| SAM | S-adenosyl-L-methionine, |
| SV | sequence variation |
| TCA | tricarboxylic acid cycle |
| TE | total effect |
| VAF | variance accounted for |
References
- Naito, K.; Zhang, F.; Tsukiyama, T.; Saito, H.; Hancock, C.N.; Richardson, A.O.; Okumoto, Y.; Tanisaka, T.; Wessler, S.R. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 2009, 461, 1130. [Google Scholar] [CrossRef] [PubMed]
- Tanurdzic, M.; Vaughn, M.W.; Jiang, H.; Lee, T.-J.; Slotkin, R.K.; Sosinski, B.; Thompson, W.F.; Doerge, R.W.; Martienssen, R.A. Epigenomic consequences of immortalized plant cell suspension culture. PLoS Biol. 2008, 6, e302. [Google Scholar] [CrossRef] [PubMed]
- Birnbaum, K.D.; Roudier, F. Epigenetic memory and cell fate reprogramming in plants. Regeneration 2017, 4, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Maki, H. Origins of Spontaneous Mutations: Specificity and Directionality of Base-Substitution, Frameshift, and Sequence-Substitution Mutageneses. Annu. Rev. Genet. 2002, 36, 279–303. [Google Scholar] [CrossRef]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [CrossRef]
- Fujii, S.; Akiyama, M.; Aoki, K.; Sugaya, Y.; Higuchi, K.; Hiraoka, M.; Miki, Y.; Saitoh, N.; Yoshiyama, K.; Ihara, K.; et al. DNA Replication Errors Produced by the Replicative Apparatus of Escherichia coli. J. Mol. Biol. 1999, 289, 835–850. [Google Scholar] [CrossRef]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204. [Google Scholar] [CrossRef]
- Zhang, H.; Lang, Z.; Zhu, J.-K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef]
- Slotkin, R.K.; Vaughn, M.; Borges, F.; Tanurdzic, M.; Becker, J.D.; Feijo, J.A.; Martienssen, R.A. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 2009, 136, 461–472. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.K. Active DNA demethylation in plants and animals. Cold Spring Harb. Symp. Quant. Biol. 2012, 77, 161–173. [Google Scholar] [CrossRef]
- Ooi, S.K.; Bestor, T.H. The colorful history of active DNA demethylation. Cell 2008, 133, 1145–1148. [Google Scholar] [CrossRef]
- Li, Y.; Kumar, S.; Qian, W. Active DNA demethylation: Mechanism and role in plant development. Plant Cell Rep. 2018, 37, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Galisteo, A.P.; Morales-Ruiz, T.; Ariza, R.R.; Roldán-Arjona, T. Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol. Biol. 2008, 67, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Gehring, M.; Bubb, K.L.; Henikoff, S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 2009, 324, 1447–1451. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.F.; Ibarra, C.A.; Silva, P.; Zemach, A.; Eshed-Williams, L.; Fischer, R.L.; Zilberman, D. Genome-wide demethylation of Arabidopsis endosperm. Science 2009, 324, 1451–1454. [Google Scholar] [CrossRef]
- Ausin, I.; Greenberg, M.V.C.; Simanshu, D.K.; Hale, C.J.; Vashisht, A.A.; Simon, S.A.; Lee, T.-F.; Feng, S.; Española, S.D.; Meyers, B.C.; et al. INVOLVED IN DE NOVO 2-containing complex involved in RNA-directed DNA methylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 8374–8381. [Google Scholar] [CrossRef]
- He, X.-J.; Chen, T.; Zhu, J.-K. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011, 21, 442–465. [Google Scholar] [CrossRef]
- Globisch, D.; Münzel, M.; Müller, M.; Michalakis, S.; Wagner, M.; Koch, S.; Brückl, T.; Biel, M.; Carell, T. Tissue Distribution of 5-Hydroxymethylcytosine and Search for Active Demethylation Intermediates. PLoS ONE 2010, 5, e15367. [Google Scholar] [CrossRef]
- Nabel, C.S.; Manning, S.A.; Kohli, R.M. The Curious Chemical Biology of Cytosine: Deamination, Methylation, and Oxidation as Modulators of Genomic Potential. ACS Chem. Biol. 2012, 7, 20–30. [Google Scholar] [CrossRef]
- Wibowo, A.; Becker, C.; Marconi, G.; Durr, J.; Price, J.; Hagmann, J.; Papareddy, R.; Putra, H.; Kageyama, J.; Becker, J.; et al. Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity. Elife 2016, 5, e13546. [Google Scholar] [CrossRef]
- Yong-Villalobos, L.; González-Morales, S.I.; Wrobel, K.; Gutiérrez-Alanis, D.; Cervantes-Peréz, S.A.; Hayano-Kanashiro, C.; Oropeza-Aburto, A.; Cruz-Ramírez, A.; Martínez, O.; Herrera-Estrella, L. Methylome analysis reveals an important role for epigenetic changes in the regulation of the Arabidopsis response to phosphate starvation. Proc. Natl. Acad. Sci. USA 2015, 112, E7293–E7302. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Gaubert, H.; Bucher, E.; Mirouze, M.; Vaillant, I.; Paszkowski, J. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 2011, 472, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Coronel, C.J.; González, A.I.; Ruiz, M.L.; Polanco, C. Analysis of somaclonal variation in transgenic and regenerated plants of Arabidopsis thaliana using methylation related metAFLP and TMD markers. Plant Cell Rep. 2018, 37, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Machczyńska, J.; Zimny, J.; Bednarek, P. Tissue culture-induced genetic and epigenetic variation in triticale (× Triticosecale spp. Wittmack ex A. Camus 1927) regenerants. Plant Mol. Biol. 2015, 89, 279–292. [Google Scholar] [CrossRef]
- Orłowska, R.; Bednarek, P.T. Precise evaluation of tissue culture-induced variation during optimisation of in vitro regeneration regime in barley. Plant Mol. Biol. 2020. [Google Scholar] [CrossRef]
- Polanco, C.; Ruiz, M.L. AFLP analysis of somaclonal variation in Arabidopsis thaliana regenerated plants. Plant Sci. 2002, 162, 817–824. [Google Scholar] [CrossRef]
- Bednarek, P.T.; Orłowska, R. Plant tissue culture environment as a switch-key of (epi) genetic changes. Plant Cell Tissue Organ Cult. (PCTOC) 2020, 140, 245–257. [Google Scholar] [CrossRef]
- Machczyńska, J.; Orłowska, R.; Zimny, J.; Bednarek, P.T. Extended metAFLP approach in studies of the tissue culture induced variation (TCIV) in case of triticale. Mol. Breed. 2014, 34, 845–854. [Google Scholar] [CrossRef]
- Bednarek, P.T.; Orłowska, R.; Koebner, R.M.D.; Zimny, J. Quantification of the tissue-culture induced variation in barley (Hordeum vulgare L.). BMC Plant Biol. 2007, 7, 10. [Google Scholar] [CrossRef][Green Version]
- Kazarian, S.G.; Chan, K.L.A. ATR-FTIR spectroscopic imaging: Recent advances and applications to biological systems. Analyst 2013, 138, 1940–1951. [Google Scholar] [CrossRef]
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A.; et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 2014, 9, 1771–1791. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Lahlali, R.; Liu, X.; Karunakaran, C. Infrared spectroscopy combined with imaging: A new developing analytical tool in health and plant science. Appl. Spectrosc. Rev. 2016, 51, 466–483. [Google Scholar] [CrossRef]
- Gorgulu, S.T.; Dogan, M.; Severcan, F. The Characterization and Differentiation of Higher Plants by Fourier Transform Infrared Spectroscopy. Appl. Spectrosc. 2007, 61, 300–308. [Google Scholar] [CrossRef]
- Lu, H.-F.; Shen, J.-B.; Lin, X.-Y.; Fu, J.-L. Relevance of Fourier transform infrared spectroscopy and leaf anatomy for species classification in Camellia (Theaceae). TAXON 2008, 57, 1274–1278. [Google Scholar] [CrossRef]
- Alonso-Simón, A.; García-Angulo, P.; Mélida, H.; Encina, A.; Álvarez, J.M.; Acebes, J.L. The use of FTIR spectroscopy to monitor modifications in plant cell wall architecture caused by cellulose biosynthesis inhibitors. Plant Signal. Behav. 2011, 6, 1104–1110. [Google Scholar] [CrossRef]
- Westworth, S.; Ashwath, N.; Cozzolino, D. Application of FTIR-ATR spectroscopy to detect salinity response in Beauty Leaf Tree (Calophyllum inophyllum L). Energy Procedia 2019, 160, 761–768. [Google Scholar] [CrossRef]
- Willick, I.R.; Lahlali, R.; Vijayan, P.; Muir, D.; Karunakaran, C.; Tanino, K.K. Wheat flag leaf epicuticular wax morphology and composition in response to moderate drought stress are revealed by SEM, FTIR-ATR and synchrotron X-ray spectroscopy. Physiol. Plant. 2018, 162, 316–332. [Google Scholar] [CrossRef]
- Bednarek, P.T.; Orłowska, R. CG Demethylation Leads to Sequence Mutations in an Anther Culture of Barley Due to the Presence of Cu, Ag Ions in the Medium and Culture Time. Int. J. Mol. Sci. 2020, 21, 4401. [Google Scholar] [CrossRef]
- Meyer, C.-L.; Juraniec, M.; Huguet, S.; Chaves-Rodriguez, E.; Salis, P.; Isaure, M.-P.; Goormaghtigh, E.; Verbruggen, N. Intraspecific variability of cadmium tolerance and accumulation, and cadmium-induced cell wall modifications in the metal hyperaccumulator Arabidopsis halleri. J. Exp. Bot. 2015, 66, 3215–3227. [Google Scholar] [CrossRef]
- Stuart, B.H. Biological Applications of Infrared Spectroscopy; John WIley & Sons, Ltd.: Chichester, UK, 1997. [Google Scholar]
- Fan, M.; Dai, D.; Huang, B. Fourier Transform Infrared Spectroscopy for Natural Fibres. In Fourier Transform—Materials Analysis; Salih, S., Ed.; InTech: Rijeka, Croatia, 2012; pp. 45–68. [Google Scholar]
- Kacuráková, M.; Capek, P.; Sasinková, V.; Wellner, N.; Ebringerová, A. FT-IR study of plant cell wall model compounds: Pectic polysaccharides and hemicelluloses. Carbohydr. Polym. 2000, 43, 195–203. [Google Scholar] [CrossRef]
- Mathlouthi, M.; Koenig, J.L. Vibrational Spectra of Carbohydrates. In Advances in Carbohydrate Chemistry and Biochemistry; Tipson, R.S., Horton, D., Eds.; Academic Press: Cambridge, MA, USA, 1987; Volume 44, pp. 7–89. [Google Scholar]
- Šandula, J.; Kogan, G.; Kačuráková, M.; Machová, E. Microbial (1→3)-β-d-glucans, their preparation, physico-chemical characterization and immunomodulatory activity. Carbohydr. Polym. 1999, 38, 247–253. [Google Scholar] [CrossRef]
- Dovbeshko, G.I.; Gridina, N.Y.; Kruglova, E.B.; Pashchuk, O.P. FTIR spectroscopy studies of nucleic acid damage. Talanta 1997, 53, 233–246. [Google Scholar] [CrossRef]
- Stuart, B.H. Infrared Spectroscopy: Fundamentals and Applications; John Wiley & Sons: Chichester, UK, 2004. [Google Scholar]
- Ilharco, L.M.; Garcia, A.R.; Lopes da Silva, J.; Vieira Ferreira, L.F. Infrared Approach to the Study of Adsorption on Cellulose: Influence of Cellulose Crystallinity on the Adsorption of Benzophenone. Langmuir 1997, 13, 4126–4132. [Google Scholar] [CrossRef]
- Kondo, T.; Sawatari, C. A Fourier transform infra-red spectroscopic analysis of the character of hydrogen bonds in amorphous cellulose. Polymer 1996, 37, 393–399. [Google Scholar] [CrossRef]
- Cardoso, P.B.; Machado, T.O.; Feuser, P.E.; Sayer, C.; Meier, M.A.R.; Araújo, P.H.H. Biocompatible Polymeric Nanoparticles From Castor Oil Derivatives via Thiol-Ene Miniemulsion Polymerization. Eur. J. Lipid Sci. Technol. 2018, 120, 1700212. [Google Scholar] [CrossRef]
- Hostetler, M.J.; Stokes, J.J.; Murray, R.W. Infrared Spectroscopy of Three-Dimensional Self-Assembled Monolayers: N-Alkanethiolate Monolayers on Gold Cluster Compounds. Langmuir 1996, 12, 3604–3612. [Google Scholar] [CrossRef]
- Nyquist, R.A. Thiols, Sulfides and Disulfides, Alkanethiols, and Alkanedithiols (S-H stretching). In Interpreting Infrared, Raman, and Nuclear Magnetic Resonance Spectra; Academic Press: Cambridge, MA, USA, 2001; pp. 65–83. [Google Scholar]
- MacKinnon, D.P.; Fairchild, A.J.; Fritz, M.S. Mediation Analysis. Annu. Rev. Psychol. 2007, 58, 593–614. [Google Scholar] [CrossRef]
- Sauter, M.; Moffatt, B.; Saechao, M.C.; Hell, R.; Wirtz, M. Methionine salvage and S-adenosylmethionine: Essential links between sulfur, ethylene and polyamine biosynthesis. Biochem. J. 2013, 451, 145–154. [Google Scholar] [CrossRef]
- Chiang, P.K.; Gordon, R.K.; Tai, J.; Zeng, G.C.; Doctor, B.P.; Pardhasaradhi, K.; McCann, P.P. S-Adenosylmethionine and methylation. FASEB J. 1996, 10, 471–480. [Google Scholar] [CrossRef]
- Cui, X.-H.; Murthy, H.N.; Wu, C.-H.; Paek, K.-Y. Sucrose-induced osmotic stress affects biomass, metabolite, and antioxidant levels in root suspension cultures of Hypericum perforatum L. Plant Cell Tissue Organ Cult. (PCTOC) 2010, 103, 7–14. [Google Scholar] [CrossRef]
- Carone, S.B.; Santa-Catarina, C.; Silveira, V.; Floh, E.I.S. Polyamine patterns in haploid and diploid tobacco tissues and in vitro cultures. Braz. Arch. Biol. Technol. 2010, 53, 409–417. [Google Scholar] [CrossRef]
- Bishimbayeva, N.; Murtazina, A.; McDougall, G. Influence of Phytohormones on Monosaccharide Composition of Polysaccharides from Wheat Suspension Culture. Eurasian Chem. Technol. J. 2017, 19, 237–321. [Google Scholar] [CrossRef]
- Bednarek, P.T.; Orłowska, R. Time of in vitro anther culture may moderate action of copper and silver ions that affect the relationship between DNA methylation change and the yield of barley green regenerants. Plants 2020, 9. unpublished work. [Google Scholar]
- Ganeshan, S.; Chodaparambil, S.V.; Båga, M.; Fowler, D.B.; Hucl, P.; Rossnagel, B.G.; Chibbar, R.N. In vitro regeneration of cereals based on multiple shoot induction from mature embryos in response to thidiazuron. Plant Cell Tissue Organ Cult. 2006, 85, 63–73. [Google Scholar] [CrossRef]
- Miroshnichenko, D.; Chaban, I.; Chernobrovkina, M.; Dolgov, S. Protocol for efficient regulation of in vitro morphogenesis in einkorn (Triticum monococcum L.), a recalcitrant diploid wheat species. PLoS ONE 2017, 12, e0173533. [Google Scholar] [CrossRef]
- Biesaga-Kościelniak, J.; Kościelniak, J.; Janeczko, A. The impact of zearalenone and thidiazuron on indirect plant regeneration of oilseed rape and wheat. Acta Physiol. Plant. 2010, 32, 1047–1053. [Google Scholar] [CrossRef]
- Burton, R.A.; Fincher, G.B. (1,3;1,4)-β-D-Glucans in Cell Walls of the Poaceae, Lower Plants, and Fungi: A Tale of Two Linkages. Mol. Plant 2009, 2, 873–882. [Google Scholar] [CrossRef]
- Bilska-Kos, A.; Mytych, J.; Suski, S.; Magoń, J.; Ochodzki, P.; Zebrowski, J. Sucrose phosphate synthase (SPS), sucrose synthase (SUS) and their products in the leaves of Miscanthus × giganteus and Zea mays at low temperature. Planta 2020, 252, 23. [Google Scholar] [CrossRef]
- Hofmann, N.R. A Functional Link between Mitochondria and the Cell Wall in Stress Responses. Plant Cell 2016, 28, 1996. [Google Scholar] [CrossRef]
- Huang, S.; Van Aken, O.; Schwarzländer, M.; Belt, K.; Millar, A.H. The Roles of Mitochondrial Reactive Oxygen Species in Cellular Signaling and Stress Response in Plants. Plant Physiol. 2016, 171, 1551–1559. [Google Scholar] [CrossRef]
- Bulone, V.; Schwerdt, J.G.; Fincher, G.B. Co-evolution of Enzymes Involved in Plant Cell Wall Metabolism in the Grasses. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Roulin, S.; Buchala, A.J.; Fincher, G.B. Induction of (1→3,1→4)-β-D-glucan hydrolases in leaves of dark-incubated barley seedlings. Planta 2002, 215, 51–59. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hrmova, M.; Fincher, G.B. Dissecting the catalytic mechanism of a plant β-d-glucan glucohydrolase through structural biology using inhibitors and substrate analogues. Carbohydr. Res. 2007, 342, 1613–1623. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Z.; Liu, H.; Xu, C.; Shanklin, J. Sugar Potentiation of Fatty Acid and Triacylglycerol Accumulation. Plant Physiol. 2017, 175, 696–707. [Google Scholar] [CrossRef] [PubMed]
- Funck, D.; Clauß, K.; Frommer, W.; Hellmann, H. The Arabidopsis CstF64-Like RSR1/ESP1 Protein Participates in Glucose Signaling and Flowering Time Control. Front. Plant Sci. 2012, 3, 80. [Google Scholar] [CrossRef]
- Price, J.; Laxmi, A.; St Martin, S.K.; Jang, J.-C. Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 2004, 16, 2128–2150. [Google Scholar] [CrossRef]
- Jang, J.C.; Sheen, J. Sugar sensing in higher plants. Plant Cell 1994, 6, 1665–1679. [Google Scholar] [CrossRef]
- Pego, J.V.; Kortstee, A.J.; Huijser, C.; Smeekens, S.C.M. Photosynthesis, sugars and the regulation of gene expression. J. Exp. Bot. 2000, 51, 407–416. [Google Scholar] [CrossRef]
- Kesten, C.; Menna, A.; Sánchez-Rodríguez, C. Regulation of cellulose synthesis in response to stress. Curr. Opin. Plant Biol. 2017, 40, 106–113. [Google Scholar] [CrossRef]
- Ding, W.; Smulan, L.J.; Hou, N.S.; Taubert, S.; Watts, J.L.; Walker, A.K. s-Adenosylmethionine Levels Govern Innate Immunity through Distinct Methylation-Dependent Pathways. Cell Metab. 2015, 22, 633–645. [Google Scholar] [CrossRef]
- Roje, S. S-Adenosyl-L-methionine: Beyond the universal methyl group donor. Phytochemistry 2006, 67, 1686–1698. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.L.-C.; Li, H.; Ecker, J.R. Ethylene Biosynthesis and Signaling Networks. Plant Cell 2002, 14, S131–S151. [Google Scholar] [CrossRef] [PubMed]
- Zuo, J.; Wang, Y.; Zhu, B.; Luo, Y.; Wang, Q.; Gao, L. Comparative Analysis of DNA Methylation Reveals Specific Regulations on Ethylene Pathway in Tomato Fruit. Genes 2018, 9, 266. [Google Scholar] [CrossRef] [PubMed]
- Yu, J. Obesity, Fatty Liver and Liver Cancer; Yu, J., Ed.; Springer: Singapore, 2018; p. 157. [Google Scholar]
- Lu, S.C.; Mato, J.M. S-adenosylmethionine in Liver Health, Injury, and Cancer. Physiol. Rev. 2012, 92, 1515–1542. [Google Scholar] [CrossRef] [PubMed]
- Modrich, P. DNA MISMATCH CORRECTION. Annu. Rev. Biochem. 1987, 56, 435–466. [Google Scholar] [CrossRef] [PubMed]
- Booker, S.J.; Grove, T.L. Mechanistic and functional versatility of radical SAM enzymes. F1000 Biol. Rep. 2010, 2, 52. [Google Scholar] [CrossRef]
- Zhang, B.; Arcinas, A.J.; Radle, M.I.; Silakov, A.; Booker, S.J.; Krebs, C. First Step in Catalysis of the Radical S-Adenosylmethionine Methylthiotransferase MiaB Yields an Intermediate with a [3Fe-4S]0-Like Auxiliary Cluster. J. Am. Chem. Soc. 2020, 142, 1911–1924. [Google Scholar] [CrossRef]
- Broderick, J.B.; Duffus, B.R.; Duschene, K.S.; Shepard, E.M. Radical S-Adenosylmethionine Enzymes. Chem. Rev. 2014, 114, 4229–4317. [Google Scholar] [CrossRef]
- Manova, V.; Gruszka, D. DNA damage and repair in plants—from models to crops. Front. Plant Sci. 2015, 6, 885. [Google Scholar] [CrossRef]
- Harkey, A.F.; Yoon, G.M.; Seo, D.H.; DeLong, A.; Muday, G.K. Light Modulates Ethylene Synthesis, Signaling, and Downstream Transcriptional Networks to Control Plant Development. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Hu, Z.; Vanderhaeghen, R.; Cools, T.; Wang, Y.; De Clercq, I.; Leroux, O.; Nguyen, L.; Belt, K.; Millar, A.H.; Audenaert, D.; et al. Mitochondrial Defects Confer Tolerance against Cellulose Deficiency. Plant Cell 2016, 28, 2276–2290. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Li, L.; De Clercq, I.; Narsai, R.; Xu, Y.; Hartmann, A.; Lozano Claros, D.; Custovic, E.; Lewsey, M.G.; Whelan, J.; et al. ANAC017 coordinates organellar functions and stress responses by reprogramming retrograde signaling. Plant Physiol. 2019. [Google Scholar] [CrossRef]
- Lozoya, O.A.; Martinez-Reyes, I.; Wang, T.; Grenet, D.; Bushel, P.; Li, J.; Chandel, N.; Woychik, R.P.; Santos, J.H. Mitochondrial nicotinamide adenine dinucleotide reduced (NADH) oxidation links the tricarboxylic acid (TCA) cycle with methionine metabolism and nuclear DNA methylation. PLoS Biol. 2018, 16, e2005707. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Gao, C.; Cueto, R.; Liu, L.; Fu, H.; Shao, Y.; Yang, W.Y.; Fang, P.; Choi, E.T.; Wu, Q.; et al. Homocysteine-methionine cycle is a metabolic sensor system controlling methylation-regulated pathological signaling. Redox Biol. 2020, 28, 101322. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Hématy, K.; Höfte, H. Growth Control and Cell Wall Signaling in Plants. Annu. Rev. Plant Biol. 2012, 63, 381–407. [Google Scholar] [CrossRef] [PubMed]
- Parrilla-Doblas, J.T.; Roldán-Arjona, T.; Ariza, R.R.; Córdoba-Cañero, D. Active DNA Demethylation in Plants. Int. J. Mol. Sci. 2019, 20, 4683. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Lang, Z. The mechanism and function of active DNA demethylation in plants. J. Integr. Plant Biol. 2020, 62, 148–159. [Google Scholar] [CrossRef]
- Bartels, A.; Han, Q.; Nair, P.; Stacey, L.; Gaynier, H.; Mosley, M.; Huang, Q.Q.; Pearson, J.K.; Hsieh, T.-F.; An, Y.-Q.C.; et al. Dynamic DNA Methylation in Plant Growth and Development. Int. J. Mol. Sci. 2018, 19, 2144. [Google Scholar] [CrossRef]
- Chu, C.C. The N6 medium and its applications to anther culture of cereal crops. Proc. Symp. Plant Tissue Cult. Sci. Press Pekin. 1978, 43–50. [Google Scholar]
- Kumlehn, J.; Serazetdinova, L.; Hensel, G.; Becker, D.; Loerz, H. Genetic transformation of barley (Hordeum vulgare L.) via infection of androgenetic pollen cultures with Agrobacterium tumefaciens. Plant Biotechnol. J. 2006, 4, 251–261. [Google Scholar] [CrossRef]
- Hanson, B.A. ChemoSpec: Exploratory Chemometrics for Spectroscopy. R Package Version 4.4.97. 2017. Available online: https://CRAN.R-project.org/package=ChemoSpec (accessed on 12 December 2017).
- Beleites, C.; Sergo, V. hyperspec: A Package to Handle Hyperspectral Data Sets in R. R Package Version 0.98-20161118. 2016. Available online: http://hyperspec.R-Forge.R-project.org/ (accessed on 19 November 2016).
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. 2018. Available online: http://www.R-project.org/ (accessed on 20 December 2018).
- Wojdyr, M. Fityk: A general-purpose peak fitting program. J. Appl. Crystallogr. 2010, 43, 1126–1128. [Google Scholar] [CrossRef]
- Hayes, A.F. Introduction to Mediation, Moderation, and Conditional Process Analysis. In A Regression Bases Approach; A Division of Guilford Publications, Inc.: New York, NY, USA, 2018; p. 507. [Google Scholar]
- Nitzl, C.; Roldan Jose, L.; Cepeda, G. Mediation analysis in partial least squares path modeling: Helping researchers discuss more sophisticated models. Ind. Manag. Data Syst. 2016, 116, 1849–1864. [Google Scholar] [CrossRef]
- Preacher, K.J.; Kelley, K. Effect size measures for mediation models: Quantitative strategies for communicating indirect effects. Psychol. Methods 2011, 16, 93–115. [Google Scholar] [CrossRef] [PubMed]
- Preacher, K.J.; Leonardelli, G.J. Calculation for the Sobel Test: An Interactive Calculation Tool for Mediation Tests. 2001. Available online: http://quantpsy.org/sobel/sobel.htm (accessed on 20 January 2009).
- Faul, F.; Erdfelder, E.; Lang, A.-G.; Buchner, A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 2007, 39, 175–191. [Google Scholar] [CrossRef] [PubMed]
| Sample No. | Trial | Independent Variable 1 | Mediator 1 | Dependent Variable 1 | Moderator | ||||
|---|---|---|---|---|---|---|---|---|---|
| F710.690 | F1010.940 | F710.690-F1010.940 | DMV | CG_DMV | CHG_DMV | SV | Time (Days) | ||
| 1 | M1 | 0.0012 | 0.0443 | 0.0455 | 1.6210 | 0.4636 | 1.1583 | 3.7058 | 21 |
| 2 | M1 | 0.0021 | 0.0267 | 0.0288 | 1.3900 | 0.2325 | 1.1588 | 2.5488 | 21 |
| 3 | M1 | 0.0018 | 0.0371 | 0.0389 | 1.3900 | 0.2315 | 1.1584 | 2.5479 | 21 |
| 4 | M1 | 0.0014 | 0.0317 | 0.0332 | 1.3900 | 0.2315 | 1.1584 | 2.7794 | 21 |
| 5 | M1 | 0.0011 | 0.0388 | 0.0399 | 1.6210 | 0.4630 | 1.1583 | 3.2423 | 21 |
| 6 | M2 | 0.0010 | 0.0283 | 0.0293 | 1.6220 | 0.4637 | 1.1585 | 2.5481 | 28 |
| 7 | M2 | 0.0015 | 0.0287 | 0.0302 | 1.6220 | 0.4650 | 1.1584 | 2.3171 | 28 |
| 8 | M2 | 0.0018 | 0.0322 | 0.0339 | 1.3900 | 0.4640 | 0.9268 | 2.5482 | 28 |
| 9 | M3 | 0.0014 | 0.0274 | 0.0288 | 1.3900 | 0.4640 | 0.9268 | 2.5482 | 35 |
| 10 | M3 | 0.0014 | 0.0264 | 0.0279 | 1.6210 | 0.4636 | 1.1583 | 3.7058 | 35 |
| 11 | M3 | 0.0008 | 0.0454 | 0.0461 | 1.6210 | 0.4637 | 1.1584 | 3.9377 | 35 |
| 12 | M3 | 0.0018 | 0.0263 | 0.0281 | 1.3900 | 0.4640 | 0.9268 | 2.5482 | 35 |
| 13 | M3 | 0.0019 | 0.0255 | 0.0275 | 1.3830 | 0.4616 | 0.9224 | 4.3802 | 35 |
| 14 | M4 | 0.0011 | 0.0282 | 0.0293 | 1.3900 | 0.4640 | 0.9268 | 2.5482 | 28 |
| 15 | M4 | 0.0013 | 0.0325 | 0.0338 | 1.3900 | 0.4640 | 0.9268 | 2.3166 | 28 |
| 16 | M5 | 0.0010 | 0.0329 | 0.0339 | 1.3900 | 0.4637 | 0.9268 | 2.5482 | 35 |
| 17 | M5 | 0.0009 | 0.0484 | 0.0493 | 1.3900 | 0.4630 | 0.9266 | 3.7055 | 35 |
| 18 | M5 | 0.0009 | 0.0483 | 0.0491 | 1.6220 | 0.6952 | 0.9271 | 3.4748 | 35 |
| 19 | M5 | 0.0018 | 0.0294 | 0.0312 | 1.8530 | 0.6954 | 1.1582 | 3.7057 | 35 |
| 20 | M6 | 0.0011 | 0.0353 | 0.0364 | 0.9270 | 0.0000 | 0.9271 | 2.5487 | 21 |
| 21 | M6 | 0.0013 | 0.0343 | 0.0356 | 1.3900 | 0.4640 | 0.9268 | 2.5482 | 21 |
| 22 | M6 | 0.0016 | 0.0238 | 0.0254 | 0.9270 | 0.0000 | 0.9268 | 2.5484 | 21 |
| 23 | M6 | 0.0018 | 0.0311 | 0.0329 | 1.1580 | 0.2323 | 0.9268 | 2.5483 | 21 |
| 24 | M6 | 0.0015 | 0.0331 | 0.0346 | 1.1580 | 0.2315 | 0.9271 | 2.3167 | 21 |
| 25 | M7 | 0.0014 | 0.0268 | 0.0283 | 1.8530 | 0.6944 | 1.1581 | 4.6315 | 35 |
| 26 | M7 | 0.0009 | 0.0482 | 0.0492 | 1.6210 | 0.4630 | 1.1584 | 2.5478 | 35 |
| 27 | M7 | 0.0010 | 0.0373 | 0.0383 | 1.3900 | 0.4630 | 0.9266 | 3.9370 | 35 |
| 28 | M8 | 0.0009 | 0.0440 | 0.0449 | 0.0000 | 0.0000 | 0.0000 | 13.5169 | 21 |
| 29 | M8 | 0.0004 | 0.0533 | 0.0537 | 0.0000 | 0.0000 | 0.0000 | 13.5170 | 21 |
| 30 | M8 | 0.0014 | 0.0426 | 0.0440 | 0.0000 | 0.0000 | 0.0000 | 14.0089 | 21 |
| 31 | M9 | 0.0013 | 0.0441 | 0.0454 | 2.0840 | 0.9267 | 1.1582 | 4.4006 | 28 |
| 32 | M9 | 0.0015 | 0.0361 | 0.0377 | 1.1580 | 0.2320 | 0.9268 | 2.7798 | 28 |
| 33 | M9 | 0.0013 | 0.0388 | 0.0400 | 1.1580 | 0.2320 | 0.9268 | 2.7798 | 28 |
| 34 | M9 | 0.0016 | 0.0335 | 0.0351 | 1.1580 | 0.2320 | 0.9268 | 2.5486 | 28 |
| 35 | M9 | 0.0019 | 0.0223 | 0.0243 | 2.0 940 | 0.9320 | 1.1651 | 3.4931 | 28 |
| Model | Effects | 95% CI | ||||||
|---|---|---|---|---|---|---|---|---|
| R2 | c’ | a | b | c | IE | L | U | |
| F710.690- > CHG_DMV- > SV | 0.79 | −436.37 | 274.82 * | −8.59 *** | −2797.49 * | −0.299 † | −0.56 † | 0.06 † |
| F1010.940- > CHG_DMV- > SV | 0.80 | 51.35 | −15.57 * | −8.24 *** | 179.63 ** | 128.28 | 0.18 | 286.98 |
| F710.690-F1010.940- > CHG_DMV- > SV | 0.80 | 53.64 | −15.98 * | −8.24 *** | 185.26 *** | 131.62 | 0.18 | 298.27 |
| Model: Moderated Mediation | B | SE | 95% CI L | 95% CI U |
|---|---|---|---|---|
| c’ | 40.5613 | 25.167 | −10.8377 | 91.9603 |
| a | −94.5994 *** | 25.8835 | −147.3903 | −41.8085 |
| Time (moderator) | −0.0887 * | 0.0335 | −0.1571 | −0.0204 |
| Moderation: F710.690-F1010.940 × Time | 2.7594 ** | 0.8776 | 0.9696 | 4.5493 |
| b | −29.6424 **** | 4.0666 | −37.9477 | −21.3371 |
| Time (moderator) | −0.9608 **** | 0.1848 | −1.3382 | −0.5834 |
| Moderation: CHG_DMV × Time | 0.9633 **** | 0.181 | 0.5936 | 1.333 |
| c | 40.5613 | 25.167 | −10.8377 | 91.9603 |
| Conditional indirect effect of F710.690-F1010.940 on SV (F710.690-F1010.940 --> CHG_DMV --> SV) | ||||
| Time | B | SE | L | U |
| 21 | 344.9937 | 154.151 | 9.9749 | 633.6907 |
| 28 | 46.2799 | 114.4689 | −7.6518 | 113.7094 |
| 35 | 8.0692 | 53.3305 | −43.0736 | 72.1826 |
| Trial | Cu2+ (µM) | Ag+ (µM) | Time (Days) |
|---|---|---|---|
| M1 | 0.1 | 0 | 21 |
| M2 | 0.1 | 10 | 28 |
| M3 | 0.1 | 60 | 35 |
| M4 | 5 | 60 | 28 |
| M5 | 5 | 0 | 35 |
| M6 | 5 | 10 | 21 |
| M7 | 10 | 10 | 35 |
| M8 | 10 | 60 | 21 |
| M9 | 10 | 0 | 28 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bednarek, P.T.; Zebrowski, J.; Orłowska, R. Exploring the Biochemical Origin of DNA Sequence Variation in Barley Plants Regenerated via in Vitro Anther Culture. Int. J. Mol. Sci. 2020, 21, 5770. https://doi.org/10.3390/ijms21165770
Bednarek PT, Zebrowski J, Orłowska R. Exploring the Biochemical Origin of DNA Sequence Variation in Barley Plants Regenerated via in Vitro Anther Culture. International Journal of Molecular Sciences. 2020; 21(16):5770. https://doi.org/10.3390/ijms21165770
Chicago/Turabian StyleBednarek, Piotr T., Jacek Zebrowski, and Renata Orłowska. 2020. "Exploring the Biochemical Origin of DNA Sequence Variation in Barley Plants Regenerated via in Vitro Anther Culture" International Journal of Molecular Sciences 21, no. 16: 5770. https://doi.org/10.3390/ijms21165770
APA StyleBednarek, P. T., Zebrowski, J., & Orłowska, R. (2020). Exploring the Biochemical Origin of DNA Sequence Variation in Barley Plants Regenerated via in Vitro Anther Culture. International Journal of Molecular Sciences, 21(16), 5770. https://doi.org/10.3390/ijms21165770





