Abstract
Photosynthesis is not only a primary generator of reactive oxygen species (ROS) but also a component of plant defence. To determine the relationships among photosynthesis, ROS, and defence responses to powdery mildew in wheat, we compared the responses of the Pm40-expressing wheat line L658 and its susceptible sister line L958 at 0, 6, 12, 24, 48, and 72 h post-inoculation (hpi) with powdery mildew via analyses of transcriptomes, cytology, antioxidant activities, photosynthesis, and chlorophyll fluorescence parameters. The results showed that H2O2 accumulation in L658 was significantly greater than that in L958 at 6 and 48 hpi, and the enzymes activity and transcripts expression of peroxidase and catalase were suppressed in L658 compared with L958. In addition, the inhibition of photosynthesis in L658 paralleled the global downregulation of photosynthesis-related genes. Furthermore, the expression of the salicylic acid-related genes non-expressor of pathogenesis related genes 1 (NPR1), pathogenesis-related 1 (PR1), and pathogenesis-related 5 (PR5) was upregulated, while the expression of jasmonic acid- and ethylene-related genes was inhibited in L658 compared with L958. In conclusion, the downregulation of photosynthesis-related genes likely led to a decline in photosynthesis, which may be combined with the inhibition of peroxidase (POD) and catalase (CAT) to generate two stages of H2O2 accumulation. The high level of H2O2, salicylic acid and PR1 and PR5 in L658 possible initiated the hypersensitive response.
1. Introduction
Plants are challenged by various biotic stresses, including viral, bacterial, and fungal pathogenic stresses, which have substantial economic and ecological impacts [1,2]. Plants depend on their innate immune system to perceive and respond to biotic stimuli [3]. The first line of the immune response is called pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI), which is triggered by various PAMPs [4]. During this period, the physiological and biochemical statuses of plants correspondingly change, which include phenomena such as cytoskeletal reorganization, cell wall reinforcement, reactive oxygen species (ROS) bursts, and stomatal closure [5,6,7]. To defend against pathogens further, resistance (R) proteins of plants recognize specific effectors generated by pathogens in the infection process to trigger the second line of defence referred to as effector-triggered immunity (ETI) [4]. These defence responses include expression of pathogenesis-related (PR) genes and initiation of the hypersensitive response (HR) to restrict biotrophic pathogen growth [4,8]. ETI may also trigger systemic acquired resistance (SAR) responses in distal uninfected tissues [9,10]. Although PTI and ETI responses are triggered by different pathogen-derived molecules, they share several downstream signals, including ROS and phytohormones [11]. The signaling overlap between PTI and ETI suggests that they are complementary, however, the differences in the strength or timing of signals lead to differences in defence strength [12]. In this process, chloroplasts, as the main production sites of ROS and phytohormones, are considered to be an important battleground for the interaction between hosts and pathogens [13,14].
In general, ROS are composed mainly of singlet oxygen (1O2), superoxide anion radical (O2•−), hydrogen peroxide (H2O2), and hydroxyl radical (•OH) [15,16]. In plants, ROS are produced mainly by photosystem I (PSI) and photosystem II (PSII) during photosynthesis [17]. On the one hand, ground-state oxygen (3O2) is continuously excited to produce 1O2 in the PSII reaction centre [18,19]. On the other hand, O2 is reduced to O2•- directly by electron transport and electron leakage from the acceptor side (QA and QB) of PSI and PSII, respectively [20,21]. Finally, O2•-, which presents high reactivity and short life, is easily dismutated to H2O2 by superoxide dismutase (SOD) [22]. Traditionally, ROS are considered inevitably harmful by-products during aerobic metabolism. In fact, ROS, especially H2O2, not only have basic defence functions but also act as second messengers to activate the expression of defence genes [23]. Various studies have demonstrated that H2O2 could be both a signal molecule that induces defence responses at low/moderate concentrations and a defensive weapon to resist pathogen infection directly at high concentrations [16,23]. The concentration of H2O2 can be rapidly and precisely regulated by various antioxidant enzymes, mainly SOD, catalase (CAT), peroxidase (POD), and ascorbate peroxidase (APX), with CAT acting as a key H2O2-scavenging enzyme and playing a particularly crucial role in regulating H2O2 concentrations [16]. Overall, whether H2O2 acts as a signaling molecule or as a defensive weapon relies on the precise regulation between H2O2 production and scavenging, and the different interplay between H2O2-producing and H2O2-scavenging pathways during stress determines the different kinds of defence responses put forth by plants and their tolerance to stress [16,24].
In addition to H2O2, phytohormones such as salicylic acid (SA), jasmonic acid (JA), and ethylene (ET), which are mainly synthesized in chloroplasts, also play important roles in the regulation of different defence responses [13]. In host plants, the biosynthesis of ET includes the following three steps: firstly, s-adenosine Met (SAM) synthetase converts methionine (Met) into SAM; secondly, SAM is converted into 1-aminocyclopropane-1-carboxylic acid (ACC) by ACC synthase; finally, ACC oxidase (ACO) catalyses ACC to ET. The precursor for ET biosynthesis, methionine, is produced in the chloroplast [19]. In addition, linolenic acid released from chloroplast membranes is the substrate of JA biosynthesis [13]. Moreover, the isochorismate synthase (ICS) pathway, which is one of the major contributors to SA synthesis from chorismate, is localized in the chloroplast as well [25]. It is well known that there is a mutually antagonistic interaction between SA- and JA/ET-dependent signaling against distinct pathogens that have different lifestyles [4]. For example, biotrophic pathogens are inhibited mainly by SA-mediated defences, whereas defences against necrotrophic pathogens are regulated by JA/ET [26]. Furthermore, some studies have suggested that there is an interplay between plant hormones and both photosynthesis and H2O2 [24,27,28]. For instance, the expression of photosynthesis-related genes is universally downregulated under biotic stress while the expression of genes involved in the synthesis of JA, SA, and ET is upregulated, which indicates that the downregulated expression of photosynthesis-related genes is part of the defence response [29]. Furthermore, SA can inhibit H2O2-scavenging enzymes, such as CAT and APX [30,31], and in turn, H2O2 accumulation in the chloroplast can result in elevated SA levels and enhanced SA responses [32].
In addition to indirectly participating in plant defence, some photosynthesis-related proteins can directly interact with pathogens to regulate defence responses. For example, Arabidopsis PsbP, a PSII subunit, interacts with the coat protein of Alfalfa mosaic virus to inhibit viral replication [33]. The electron acceptor protein ferredoxin-I (Fd-I) is associated with the production of ROS and the HR in defence against Pseudomonas syringae in sweet pepper [34], and the overexpression of the Fd-I gene also confers enhanced resistance to virulent bacteria in other plants [35,36,37,38]. In fact, light-harvesting chlorophyll a/b-binding proteins (LHCBs) also partly modulate ROS homeostasis to affect abscisic acid (ABA) signaling [39]. Furthermore, some pathogen effectors, such as P. syringae HopI1 and HopN1, interact with the oxygen-evolving complex PsbQ of PSII and target chloroplast-localized heat shock protein Hsp70, respectively, to supress immunity reactions, including the suppression of SA accumulation and the reduction of ROS production and cell death [40,41]. Furthermore, some photorespiratory genes are involved in plant defence response as well [42,43]. For instance, silencing of photorespiration-related gene glycolate oxidase (GOX) in Nicotiana benthamiana increased susceptibility to P. syringae and Xanthomonas campestris. Furthermore, Arabidopsis gox mutants exhibited a reduction in H2O2 accumulation after P. syringae infection [44]. In tomato, GOX2 and another two photorespiratory genes serine glyoxylate aminotransferase (SGT) and serine hydroxyl methyltransferase (SHMT1) positively regulated the defence response against P. syringae. H2O2 accumulation plays critical role in GOX2-regulated but not SGT-regulated and SHMT1-regulated SA signaling [45]. These studies indicate that photosynthesis-related proteins combined with both ROS and phytohormones play a central role in plant defence.
Wheat powdery mildew, caused by the obligate biotrophic fungus Blumeria graminis f. sp. tritici (Bgt), is a destructive wheat disease worldwide [46,47]. The wheat powdery mildew resistance gene Pm40 originally derived from Elytrigia intermedia was mapped onto chromosome arm 7BS. This gene is associated with strong resistance against various Bgt strains and has been widely used in Chinese wheat breeding programmes [48,49,50]. Our pfrevious studies suggested that components associated with photosynthesis, especially the PSII reaction centre, were important sites for Pm40-mediated resistance, and some photosynthesis-related proteins, early ROS accumulation, papilla formation, and defence-related gene expression are involved in resistance [51,52,53].
RNA sequencing (RNA-seq) is an effective and powerful method for investigating pathogen infection, host defence and plant-pathogen interaction mechanisms. In our previous study, we compared the transcriptome of Bgt during interactions with the Pm40-expressing wheat line L658 and the Pm40-deficient sister line L958 to determine the pathogenesis of Bgt [54]. In this study, we further analysed the sequencing data with a focus on determining the resistance responses of the host from the other standpoint. In addition, differences between the two genotypes were revealed via light microscopy and antioxidant enzyme activities, and photosynthesis and chlorophyll fluorescence parameters were measured at various time points to further elucidate the relationships among photosynthesis, ROS, and plant hormones in wheat during the response to Bgt.
2. Results
2.1. Transcriptome Analysis of L658 and L958 during Bgt Infection
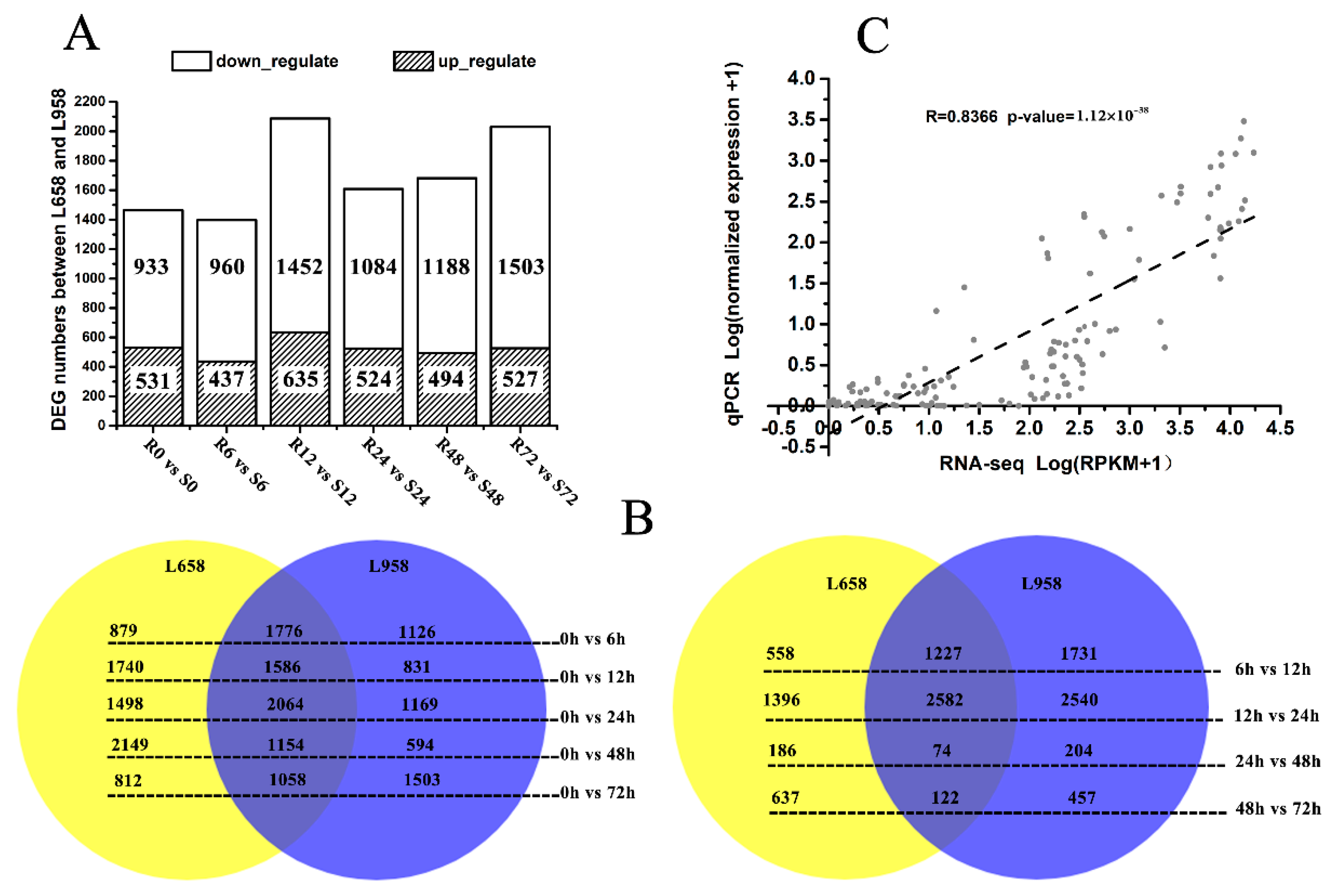
In our previous study, differences in the infection mechanism of Bgt during the interactions with L658 and L958 were determined [54]. To further clarify the difference in the molecular response mechanisms between L658 and L958, we continued to analyse the RNA-seq data from the plant side. Briefly, the percentage of reads mapped to the wheat reference genome was high for all samples and the smallest percentage of mapped reads reached 84.69%, with 21 out of 36 percentages higher than 90% (Table S1). Furthermore, we screened the differentially expressed genes (DEGs) between the two genotypes at the same time point and the maximal DEG number was 2087 at 12 h post-inoculation (hpi) (Figure 1A). In addition, we also screened the DEGs in the same genotype at each inoculation time point compared with the uninoculated time point (0 h), and between two adjacent time points (Figure 1B). Interestingly, a large portion of these DEGs were the same in both L658 and L958 (shown as overlaps), and the expression trend of nearly all of them was the same (Figure 1B), which could be explained by their similar genetic background. To validate the reliability of the transcriptome data, 12 randomly selected genes were analysed via quantitative real-time PCR (qRT-PCR) (Table S2). The correlation between normalized RNA-seq reads per kilobase per million (RPKM) values and normalized expression values from qRT-PCR was high (R = 0.8366, p-value = 1.12 × 10−38), which confirmed the reliability of RNA-seq data (Figure 1C).
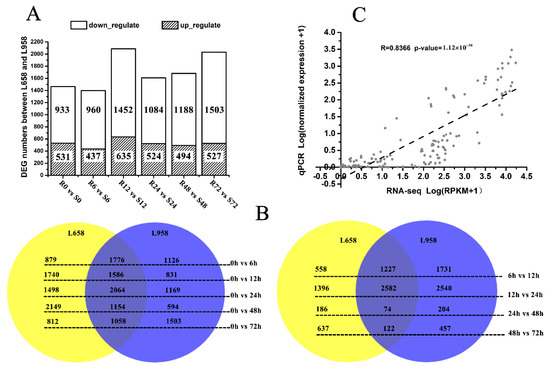
Figure 1.
Number of differentially expressed genes (DEGs) in different comparisons and reliability of RNA-seq data as demonstrated by quantitative real-time PCR (qRT-PCR). (A) Number of DEGs between L658 (R) and L958 (S) at different inoculation time points; (B) Venn diagram showing the number of DEGs shared between (overlap) and specific to L658 (yellow) and L958 (blue) at various inoculation time points compared with 0 h (left) and between two adjacent time points (right); and (C) correlations between normalized RNA-seq reads per kilobase per million (RPKM) values and normalized qRT-PCR expression values. The scatterplot shows the log10 of RPKM values +1 and the log10 of qRT-PCR expression values +1; a trend line is shown as the dotted line.
2.2. Difference in H2O2 Accumulation and Related DEG Expression between L658 and L958
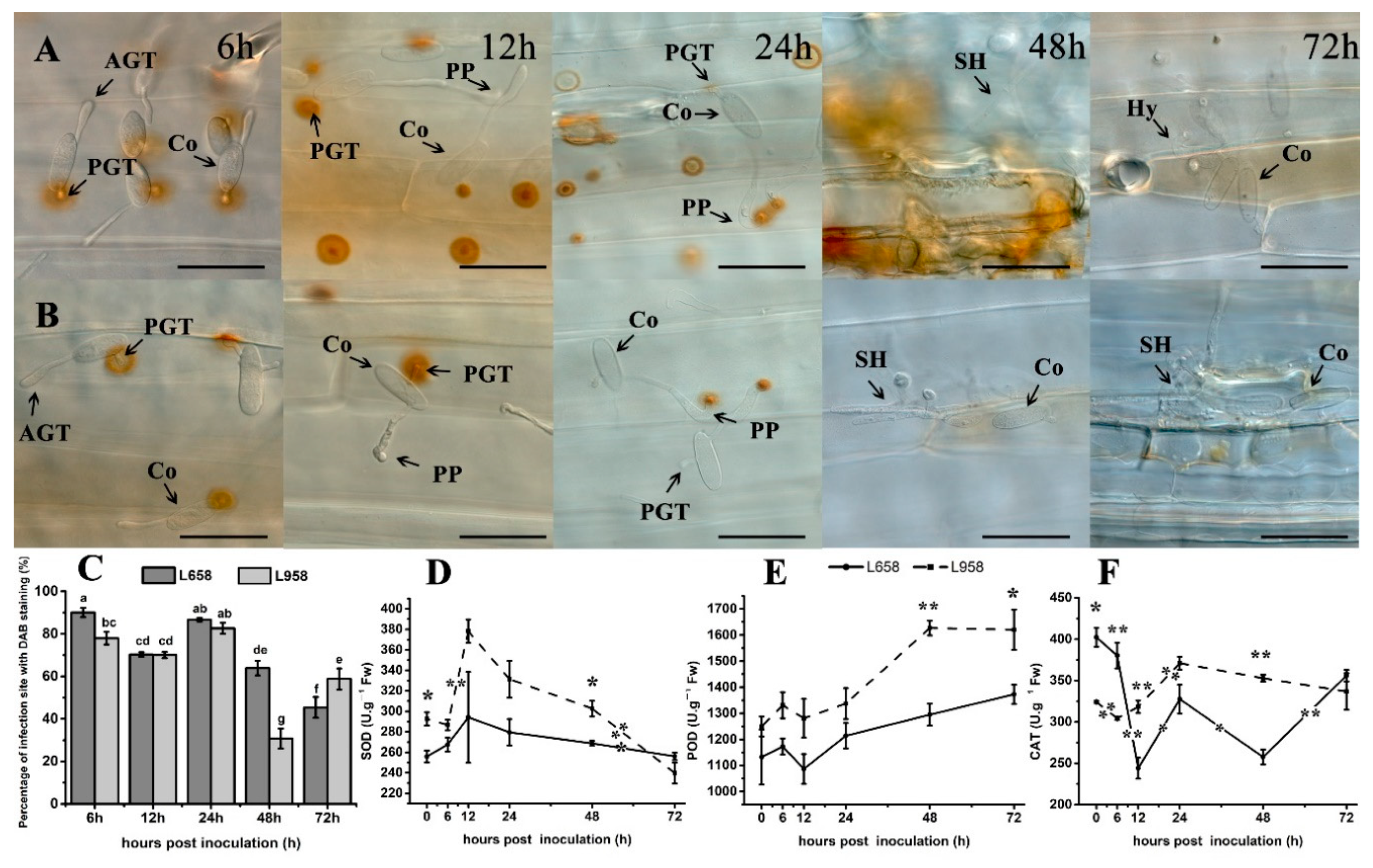
H2O2 accumulation is a typical host defence response to biotrophic fungi [4]. In this study, the results of 3′-diaminobenzidine (DAB) staining showed that H2O2 similarly accumulated under primary germ tube (PGT) at 6 and 12 hpi and then accumulated under penetration peg (PP) at 24 hpi in both L658 and L958 (Figure 2A,B). Subsequently, a stronger H2O2 response was generated in the whole infected and adjacent cells in L658 at 48 hpi, while H2O2 faded gradually in L958 at 48 and 72 hpi (Figure 2A,B). In addition, the percentage of interaction sites stained by DAB had no significant difference between L658 and L958 at 12 and 24 hpi, while that in L658 was significantly (p < 0.05) higher at both 6 and 48 hpi and lower at 72 hpi than that in L958 (Figure 2C).
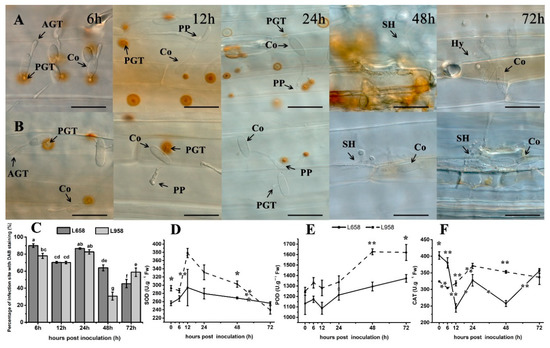
Figure 2.
H2O2 accumulation revealed by 3’-diaminobenzidine (DAB) staining at interaction sites in the resistant wheat line L658 and susceptible wheat line L958 after inoculation with Bgt and activity of antioxidant enzymes at various time points. H2O2 accumulation (reddish-brown) staining by DAB at interaction sites in L658 (A) and L958 (B) at various time points. AGT: appressorium germ tube; PGT, primary germ tube; PP: penetration peg; SH: secondary hyphae; Hy, hyphae; Co: conidia. The dark bar indicates 50 μm. (C) Dark grey and light grey bars represent the percentage of infection sites exhibiting H2O2 accumulation in L658 and L958, respectively, after inoculation with Bgt at various time points. Each point represents at least 100 infection sites of each of three leaf pieces, and the lowercase letter at the top of the bar chart represents statistically significant differences at p < 0.05. Activities of SOD (D), POD (E), and CAT (F) at different inoculation time points in the two genotypes; the solid and dotted lines represent L658 and L958, respectively. The vertical bars represent the means ± SEs. The asterisks represent significant differences as follows: ** p < 0.01 and * p < 0.05. The asterisk at the top of the line chart represents the difference between L658 and L958 at each time point. The asterisk on the trend line represents the difference between two adjacent time points for the same genotype.
To further determine the difference of H2O2 at the transcript level, we identified some DEGs related to H2O2 generation and H2O2 scavenging (Table 1). In L658, the expression of H2O2 generation-related DEGs, such as oxalate oxidase (OXO) was upregulated, especially at 12 hpi, while the expression of many H2O2-scavenging DEGs, such as POD and CAT isozymes, was downregulated in L658 compared with L958 (Table 1). In L958, the expression of other H2O2 generation-related DEGs, including respiratory burst oxidase homologue protein (RBOH), polyamine oxidase (POX), and amine oxidase (AOX), were usually upregulated, while DEGs encoding POD and CAT were commonly upregulated in L958 compared with L658 (Table 1). This result indicated that although different DEGs regulated H2O2 generation in L658 and L958, the capacity to scavenge H2O2 was more seriously inhibited in L658 than L958.

Table 1.
List of DEGs involved in H2O2 production and scavenging between L658 and L958 at each time point.
The antioxidant enzyme activities further confirmed the transcriptome data. The results showed that although the activity of SOD and POD had no significant change in L658 (Figure 2D,E), CAT activity was sharply decreased from 6 to 12 hpi (p < 0.01) and from 24 to 48 hpi (p < 0.05) (Figure 2F). In L958, the activity of SOD significantly increased from 6 to 12 hpi while the CAT activity also significantly increased from 12 to 24 hpi (Figure 2F). In contrast, the CAT activity in L958 was significantly higher (p < 0.01) than that in L658 at 12 and 48 hpi (Figure 2F). Therefore, the inhibition of H2O2 scavenging-related genes and enzyme activities in L658 likely caused a more abundant accumulation of H2O2 than in L958.
2.3. Changes in Photosynthesis and Photosynthesis-Related Genes in Response to Bgt
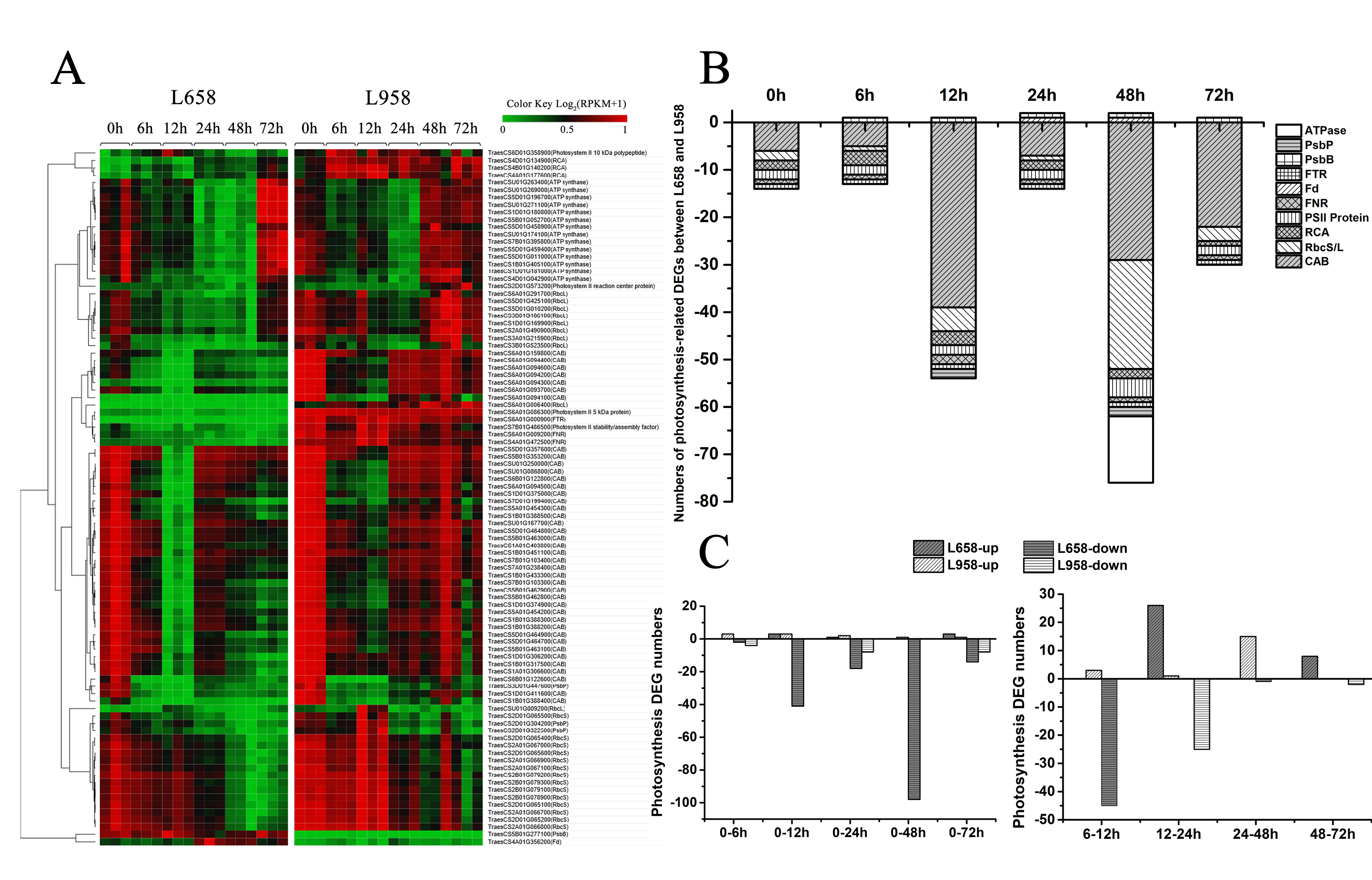
Photosynthesis is not only the main generator of H2O2, but also regarded as a plant defence component [13]. To investigate the influence of Bgt in photosynthesis, we identified a large number of photosynthesis-related DEGs, most of which encoded chlorophyll a/b-binding proteins (Cab), proteins in reaction centres, ATP synthase (ATPase), ribulose bisphosphate carboxylase small chain/large chain (RbcS/L) and Rubisco activase (RCA) (Figure 3A). Interestingly, we found that almost all of the photosynthesis-related DEGs were downregulated in L658 compared with L958, and 54 (98%) out of 55 and 75 (96%) out of 78 photosynthesis-related DEGs were downregulated in L658 at 12 and 48 hpi, respectively (Figure 3B, Table S3). Furthermore, 41 and 98 DEGs were also downregulated in L658 at 12 hpi and 48 hpi compared with 0 hpi, respectively, and 45 DEGs were also downregulated at 12 hpi compared with 6 hpi in L658 (Figure 3C, Table S4). On the contrary, only eight photosynthesis-related DEGs were downregulated in L958 at both 24 and 72 hpi compared with 0 hpi, and 25 DEGs were downregulated at 24 hpi compared with 12 hpi (Figure 3C, Table S4). This result indicated that the expression of photosynthesis-related genes in L658 was more seriously inhibited compared with that in L958, which could lead to the disruption of photosynthesis.
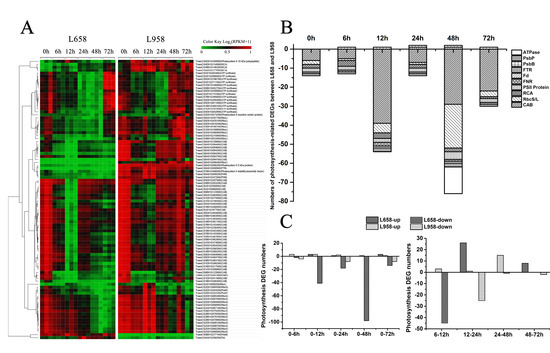
Figure 3.
Number of DEGs and expression patterns of photosynthesis-related genes. (A) Different expression patterns of photosynthesis-related DEGs in L658 and L958 at various time points on the basis of log2 of RPKM values +1; the green colour represents low expression levels, and the red colour represents high expression levels. (B) Number of different photosynthesis-related DEGs between L658 and L958 at various time points. The negative numbers and positive numbers represent downregulated and upregulated DEGs in L658 compared with L958, respectively. ATPase: ATP synthase; FTR: ferredoxin-thioredoxin reductase; Fd: ferredoxin; FNR: ferredoxin: NADP+ oxidoreductase; RCA: Rubisco activase; RbcS/L: ribulose bisphosphate carboxylase small chain/large chain; Cab: chlorophyll a/b-binding protein. (C) Number of photosynthesis-related DEGs in L658 and L958 at various inoculation time points compared with 0 h (left) and between two adjacent time points (right).
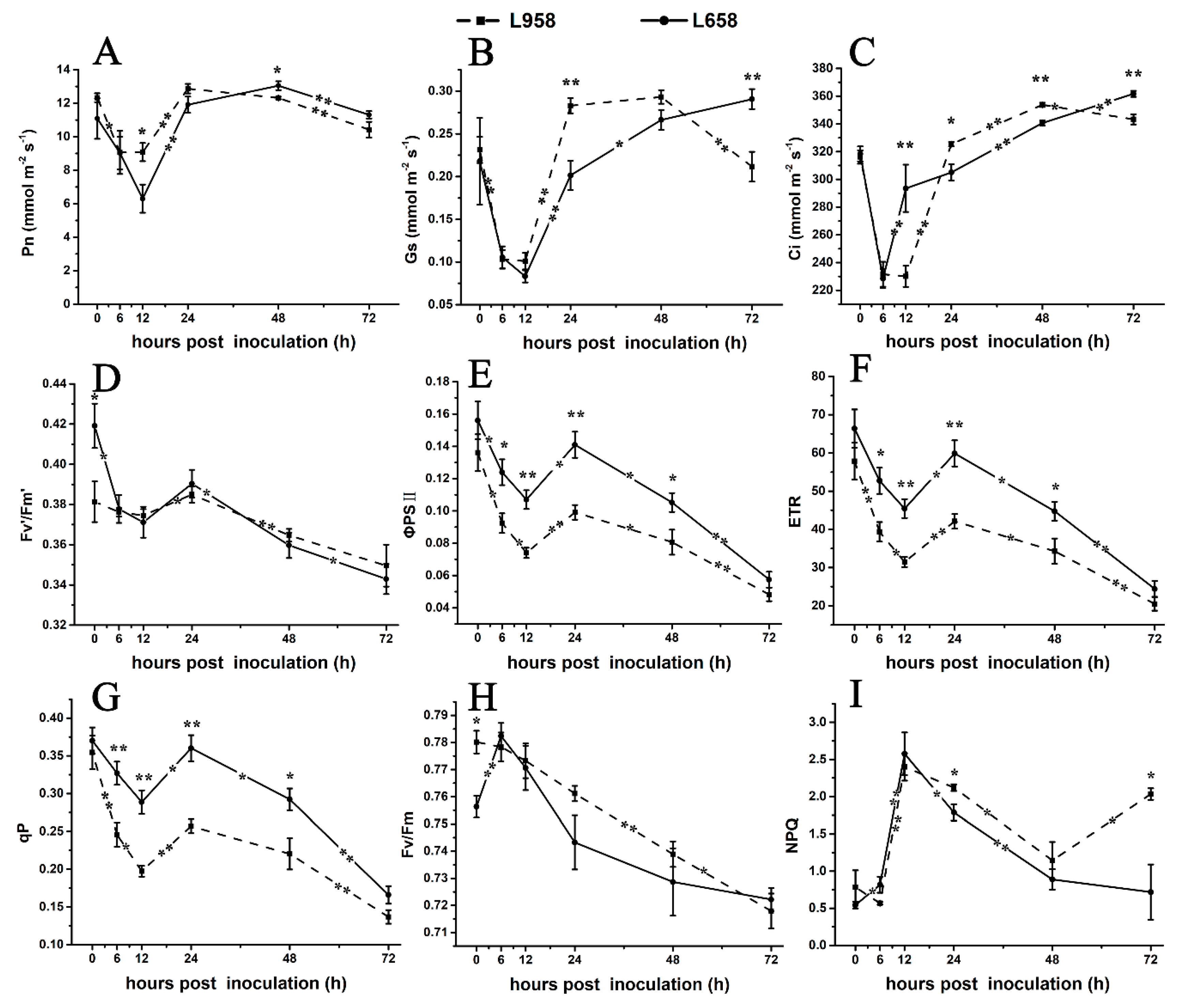
To further confirm the photosynthesis-related transcriptome data, we detected the photosynthetic parameters. As shown in Figure 4, the net photosynthetic rate (Pn) of L658 decreased from 0 hour (without inoculation) to 12 hpi and from 48 to 72 hpi (Figure 4A) and stomatal conductance (Gs) exhibited a reduction from 0 to 12 hpi and an increase from 12 to 72 hpi (Figure 4B). Compared with Pn, the intercellular CO2 concentration (Ci) of L658 increased significantly (p < 0.01) from 6 to 12 hpi and from 24 to 72 hpi (Figure 4C). The opposite change trend of Pn and Ci in L658 indicated that non-stomatal limitation, such as the inhibition of photosynthesis-related gene expression, was the main reason for the decline of Pn. However, the Pn, Gs, and Ci of L958 showed similar change trends, with a decrease from 0 to 12 hpi and from 48 to 72 hpi (Figure 4A–C), which indicated that the Pn of L958 decreased under stomatal limitation.
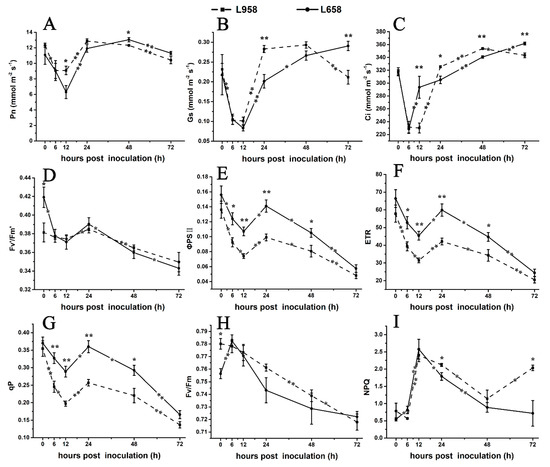
Figure 4.
Changes in photosynthesis parameters and chlorophyll florescence parameters in L658 and L958. The solid and dotted lines represent different trends in the Pn (A), Gs (B), Ci (C), Fv’/Fm’ (D), ΦPSII (E), ETR (F), qP (G), Fv/Fm (H), and NPQ (I) in L658 and L958. The meanings of the symbols are the same as those in Figure 1. The asterisks represent significant differences as follows: ** p < 0.01 and * p < 0.05. The asterisk at the top of the line chart represents the difference between L658 and L958 at each time point. The asterisk on the trend line represents the difference between two adjacent time points for the same genotype.
Furthermore, we used chlorophyll fluorescence to evaluate changes in PSII. In L658, the efficiency of excitation capture by open PSII reaction centres (Fv’/Fm’), actual photochemical efficiency of PSII (ΦPSII), electron transport rate (ETR), and coefficient of photochemical chlorophyll fluorescence quenching (qP) decreased from 0 to 12 hpi and from 24 to 72 hpi (Figure 4D–G), and the maximal photochemical efficiency of PSII in dark-adapted leaves (Fv/Fm) increased from 0 to 6 hpi and then decreased to 72 hpi (Figure 4H). Although the change tendency of Fv’/Fm’, ΦPSII, ETR and qP of L958 was similar to that of L658, the ΦPSII, ETR and qP of L958 were inhibited more strongly than those of L658 (Figure 4D–G). Interestingly, non-photochemical quenching (NPQ) significantly (p < 0.05) increased from 0 to 12 hpi and decreased from 12 to 72 hpi in L658, while it decreased from 0 to 6 hpi and increased from 48 to 72 hpi in L958 (Figure 4I). These data indicated that although the electron transport chain was blocked in both L658 and L958, the different changes of NPQ from 48 to 72 hpi may cause the differences in H2O2 accumulation at 48 and 72 hpi between L658 and L958.
2.4. Identification of DEGs Related to Phytohormones and PR Proteins
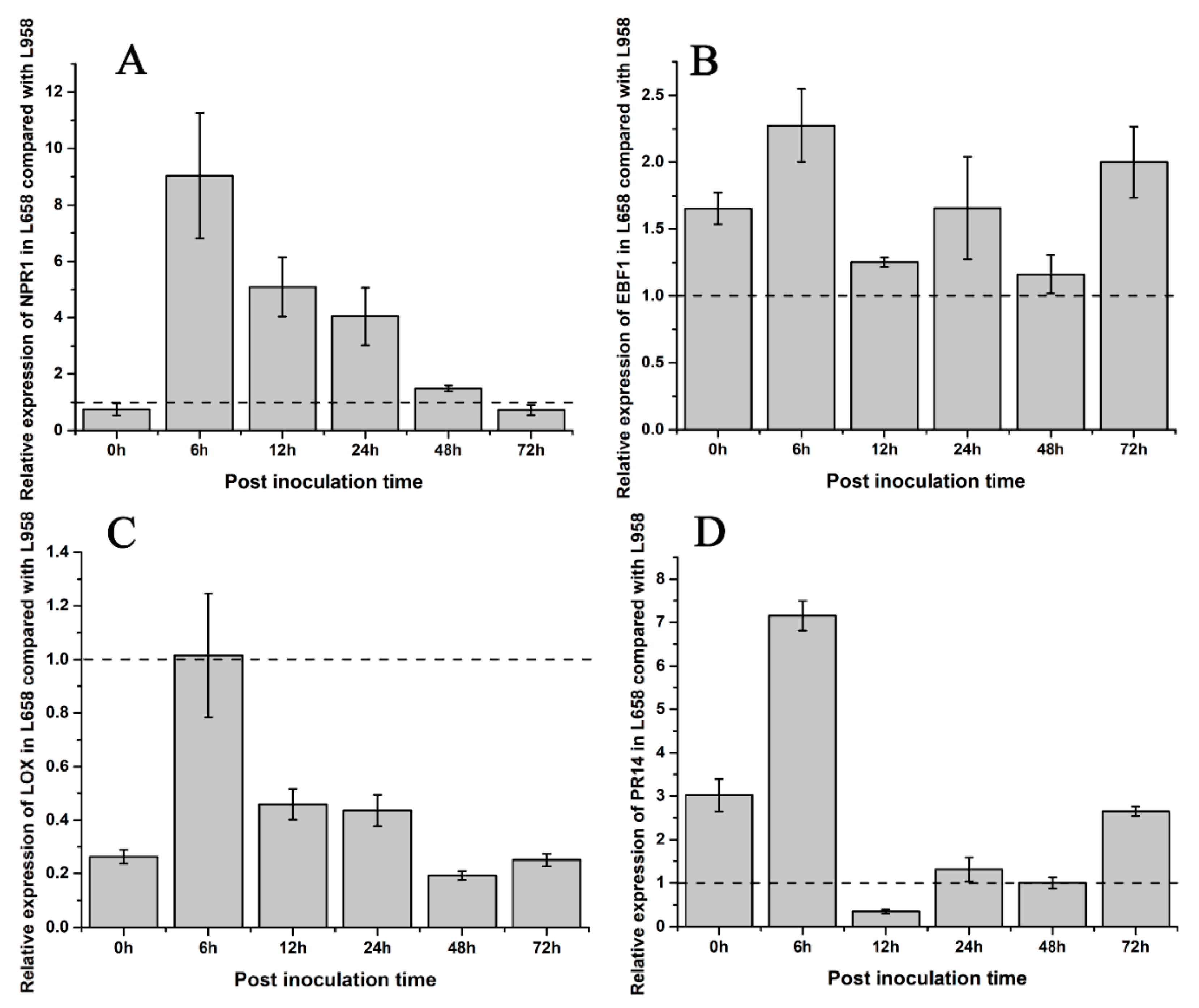
To understand the difference in hormone regulation between L658 and L958, we identified some phytohormone DEGs. The DEGs involved in SA synthesis, such as salicylic acid-binding protein (SABP) and isochorismate synthase (ICS), were downregulated in L658. However, NPR1, which is a downstream key regulator of SA, was upregulated in L658 at all time points (Table 2, Figure S1). In contrast, the DEGs involved in JA/ET pathways, such as the JA biosynthesis-related DEGs encoding lipoxygenase (LOX) and allene oxide synthase (AOS), and nearly all ET-related DEGs, including ethylene-responsive transcription factor (ERF), ethylene insensitive (EIN), 1-aminocyclopropane-1-carboxylate synthase (ACS), and 1-aminocyclopropane-1-carboxylate oxidase (ACO), were globally downregulated in L658, especially at 72 hpi (Table 2, Figure S1). On the contrary, the expression of NPR1 was inhibited in L958, whereas JA/ET-related genes were highly expressed (Table 2, Figure S1). The results of the qRT-PCR analysis further confirmed that the expression of NPR1 and EBF1 in L658 was greater than that in L958 (Figure 5A,B), and the expression of LOX was lower in L658 than L958 (Figure 5C). These results showed that the SA pathway was activated and the JA/ET pathways were supressed in L658, while the JA pathway was overactivated in L958.

Table 2.
List of DEGs involved in hormone pathways between L658 and L958 at each time point.
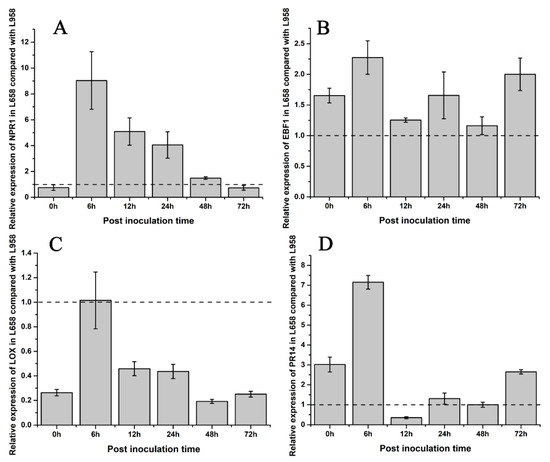
Figure 5.
Fold change in the relative expression of defence-related genes detected by qRT-PCR in L658 compared with L958. The light grey bars represent the fold change in the relative expression of defence genes in L658 compared with L958. These genes included the following: (A) SA-related gene non-expressor of pathogenesis related genes 1, NPR1; (B) ethylene-related gene EIN3-binding F-box protein 1, EBF1; (C) JA-related gene lipoxygenase, LOX; and (D) pathogenesis-related protein 14, PR14. The dotted line means that the relative expression in L958 was set as 1.
The dynamic expression patterns revealed that most of the DEGs encoding pathogenesis-related 1 (PR1) and pathogenesis-related 5 (PR5) were expressed at greater levels in L658, especially at 24 and 48 hpi, while nearly all DEGs encoding pathogenesis-related 10 (PR10) and pathogenesis-related 14 (PR14) were downregulated at all times in L658 (Table 3, Figure S2). However, both PR10 and PR14 exhibited opposite expression tendencies, with higher expression in L958 compared with L658 (Table 3, Figure S2). Furthermore, the results of the qRT-PCR analysis also confirmed that the relative expression of PR14 was lower in L658 than in L958 at 12 and 48 hpi (Figure 5D). Overall, the high expression of NPR1 in L658 activated the expression of PR1 and PR5, which further induced host cell death (Figure S3). However, the high expression of JA/ET-related genes and PR10 and PR14 in L958 may have suppressed the formation of HR (Figure S3).

Table 3.
List of DEGs encoding pathogenesis-related genes between L658 and L958 at each time point.
3. Discussion
Powdery mildew is a leaf disease that causes substantial injuries to host photosynthesis, which results in the reduction of photosynthesis-related protein abundance and gene expression [52,55,56]. Previously, plant defence and photosynthesis were studied separately. However, a growing number of studies suggest that photosynthesis is highly related to plant immunity to pathogens [13,14,19,42,43]. Therefore, it is meaningful to investigate the cross-talk between photosynthesis and plant immunity. Dual RNA-seq is widely used to detect the gene expression in various pathosystems, thus providing an effective way to explain the molecular mechanism of plant-fungal interactions [57]. In addition, the measurement of photosynthesis-related parameters, including the net photosynthetic rate and chlorophyll fluorescence, is a very powerful method to monitor and quantify changes in photosynthesis [58,59]. In the present study, we characterized the expression profile of photosynthesis-related genes in resistant and susceptible wheat lines by comparing the wheat transcriptome after inoculation with Bgt, at the same time we measured the photosynthesis-related parameters. In addition, we screened some H2O2- and defence-related genes and determined cytological reactions to reveal the relationship between photosynthesis and immunity response.
3.1. Initial Stronger H2O2 Burst and Secondary Lasting H2O2 Burst Were Regulated by Antioxidant Enzyme Activities and H2O2-Related Genes and Occurred in Incompatible Reactions
ROS burst is one of the earliest immunity responses after the successful recognition of a pathogen [60]. In general, there are two distinct peaks of ROS burst induced during incompatible reactions: the first phase is often a low-amplitude and transient peak that is followed by a sustained burst of comparatively greater intensity that correlates with disease resistance; however, only the first transient burst of ROS is induced in compatible reactions [60,61,62,63]. In the present study, the first localized H2O2 burst was observed at 6 hpi, and the strength was stronger in L658 than L958 (Figure 2A–C). Subsequently, a stronger H2O2 accumulation generated around the whole infected and adjacent cells at 48 hpi in L658 while H2O2 faded gradually in L958 (Figure 2A–C). This result indicated that both the first stronger H2O2 burst and the second lasting H2O2 burst in L658 had an important role in the establishment of resistance.
To reveal the difference in H2O2 accumulation at the molecular level, we identified H2O2-related DEGs. Interestingly, the expression of the H2O2-generating DEG OXO was upregulated in L658 at 12 hpi (Table 1). In previous studies, OXO catalyses the degradation of oxalic acid secreted by fungi into H2O2 and is involved in H2O2 accumulation in the apoplasm during different cereal plant and fungal interactions [16,64,65]. However, powdery mildew does not secrete oxalic acid, thus, the primary role of OXO in cereals would seem to be the production of H2O2 to lignify cell walls against further fungal invasion [65,66]. Hence, DEGs encoding OXO in L658 may be involved in the first H2O2 burst to reinforce the cell wall or transmit defence signals. In addition to OXO, the expression of H2O2-scavenging DEGs in L658, including those encoding POD and CAT isozymes, emerged as being universally downregulated (Table 1), which corresponded with the inhibition of the POD and CAT enzyme activities in L658 (Figure 2E,F). The suppression of CAT enzyme activity promotes the increase of H2O2 concentration [29], and transgenic lines that overexpress CAT are more sensitive to pathogens because the capacity to remove H2O2 is over activated [67]. Furthermore, although the correlation between POD genes and plant defence is equivocal, PODs are generally considered to be H2O2-detoxifying enzymes that may indirectly modulate resistance by regulating H2O2 concentrations [68]. Taken together, the downregulation of POD and CAT gene expression in L658 was likely related to the suppression of enzyme activity, which further led to the increase of H2O2 accumulation. Moreover, the first H2O2 accumulation during the stage from 6 to 12 hpi was possibly involved in reinforcing the cell wall while the second lasting and stronger H2O2 accumulation from 48 to 72 hpi further induced the HR (Figure S3).
As for L958, three DEGs encoding RBOH were upregulated (Table 1). The RBOH family is reported to mediate the production of apoplastic ROS during the defence responses, however, various RBOH proteins might have different functions in disease resistance [69,70]. For example, silencing Rboh of N. benthamiana led to a suppression of HR and increased the susceptibility to Phytophthora infestans [71]. However, the Arabidopsis atrbohF mutant exhibited more resistance to Peronospora parasitica and increased the HR response [72]. Therefore, whether RBOH is involved in positive or negative resistance to Bgt in L958 needs to be further investigated, but DEGs encoding POD and CAT were clearly upregulated in L958 compared with L658, and the enzyme activities of SOD, POD, and CAT were also significantly higher in L958 compared with L658 (Table 1, Figure 2E,F). Therefore, although other DEGs likely regulated H2O2 generation, the capacity to scavenge H2O2 was enhanced in L958 compared with L658, which caused the first weaker H2O2 burst and failure of the second H2O2 burst.
3.2. Global Downregulation of Photosynthesis-Related Genes Likely Disrupted Photosynthesis and Promoted the Generation of H2O2
Generally, virulent or avirulent pathogen invasion often results in a decrease in photosynthesis to different extents, and the reduction in photosynthesis capability may represent a “hidden cost” of defence [29,73]. The plant transcriptome revealed that many photosynthesis-related genes are downregulated under biotic stress [29]. For example, photosynthesis and the expression of the photosynthesis-related transcripts, such as Cab and RbcS, were reduced in barley-Blumeria graminis f. sp. hordei (Bgh) incompatible interaction [74], and the downregulation of Rubisco and ATPase also occurred under other biotic stresses [10,29,75]. However, in some cases, such as in the incompatible interaction between Arabidopsis and P. syringae, the repression of photosynthesis was not correlated with the downregulation of RbcS and Cab transcripts [76], which may be related to the lack of participation of a majority of plant cells in the defence reaction, thus, the repression of RbcS and Cab cannot be detected in the whole leaves’ RNA. In the present study, the downregulation of Cab, RbcS, and ATPase transcripts paralleled the decline in photosynthetic rate in L658, especially from 6 to 12 hpi and from 48 to 72 hpi (Figure 3A and Figure 4A). Meanwhile, the reduction of Pn in L658 was accompanied by the decrease of Gs and the increase of Ci (Figure 4B,C), which indicated that non-stomatal limitation led to the decrease of Pn in L658 [77,78], which was consistent with the above results. Taken together, the downregulation of photosynthesis-related DEGs in L658 likely switched off photosynthesis activity to strengthen the defence reaction. Inversely, only a small part of photosynthesis-related genes in L958 were downregulated (Figure 3C). Furthermore, the Pn, Gs, and Ci decreased consistently in L958 at 12 and 72 hpi (Figure 4A–C), which suggested that the decrease of Pn in L958 was primally controlled by stomatal limitation rather than to regulate immune response.
In addition, the expression of a DEG encoding Fd (TraesCS4A01G356200) was upregulated in L658 compared with L958 at 24 and 48 hpi (Figure 3A,B). In photosynthesis, Fd is a major element of photosynthesis-associated proteins that accept electrons from PSI and reduce NADP+ by ferredoxin: NADP+ oxidoreductase (FNR). In some plant-pathogen interactions, Fd has been suggested to participate in pathogen defence directly or to induce the production of ROS to mount the HR [29,34,38]. Therefore, it would be reasonable to assume that the upregulation of Fd in L658 may be involved in plant defence.
To further reveal the defence mechanism of photosynthesis, we monitored the status of PSII by detecting chlorophyll fluorescence indexes. In a previous study, Bgh infection led to a reduction of ΦPSII, an increase of NPQ, and a reduction of Fv/Fm in barley [79]. In general, the increase of NPQ is a typical host response in plant–fungus interactions to protect plants when light energy absorption exceeds the capacity for light utilization, but the decrease in NPQ predisposes chloroplasts to the production of ROS, which might be a priming mechanism for chloroplast ROS signaling during later immune responses [14,80,81]. In our study, Fv’/Fm’, Fv/Fm, ΦPSII, ETR, and qP significantly decreased in L658 at 12 and 72 hpi (Figure 4D–H). This response implied that photosynthetic electron transport was blocked, which could result in electron leakage in L658 for the generation of H2O2. Furthermore, the increase in NPQ in L658 at 12 hpi and the continuous decrease after 12 hpi may indicate that L658 needed photoprotection before 12 hpi but tended to generate ROS at the late stage to induce the HR (Figure 4I). In L958, the ΦPSII, ETR, and qP were supressed more seriously than in L658 (Figure 4E–G), which may be due to damage to the chloroplasts after Bgt invasion. Interestingly, the NPQ of L958 increased at 72 hpi (Figure 4I), indicating that L958 did not activate the second ROS burst and thus was insufficient to induce the HR.
3.3. Mutually Antagonistic Interactions between SA and JA/ET Are Involved in the Regulation of Defence Against Bgt by Regulating Different PR Genes Expression
The difference of H2O2 accumulation and photosynthesis might influence the different regulation of phytohormones, including SA, JA, and ET, after inoculation with Bgt. Generally, SA and JA interact antagonistically under different pathogen attacks and the SA bone fide receptor NPR1 acts as an important regulator to mediate cross-talk between SA and JA [82]. In addition, ET has positive effects on JA but may have positive or negative impacts on SA depending on the pathosystem [83,84]. Regarding the defences against biotrophic fungi, the SA signaling pathway is commonly activated and then directly regulates the conformation of NPR1 to induce the formation of SAR, the activation of PR1 and PR5 and to suppress the JA/ET signaling pathways [82,85,86]. In previous studies, the overexpression of NPR1 in tobacco and apple enhanced resistance to Botrytis cinereal and apple powdery mildew, respectively [87,88]. In a recent study, the repression of NPR1 in Brassica napus L. by RNAi significantly reduced resistance to Sclerotinia sclerotiorum, and silencing NPR1 inhibited SA defence response but enhanced the JA/ET defence response [89]. In the present study, although ICS expression was downregulated at 12 hpi, the transcript TraesCS7A01G021800 encoding NPR1 was upregulated in L658 (Table 2, Figure S1). Compared with SA, the expression of JA biosynthesis genes, including LOX and AOS, ET biosynthesis genes, including ACS and ACO, and ET-mediated signaling genes, such as ERF and EIN, were downregulated in L658, especially at 72 hpi (Table 2, Figure S1). Furthermore, the expression of PR1 and PR5 was upregulated in L658 (Table 3, Figure S2). Taken together, the downregulated expression of JA/ET-related genes indicated that JA/ET levels were also decreased, which could be attributed to increased SA levels in L658. The increased SA and the upregulated expression of NPR1, PR1, and PR5 likely induced the HR in L658.
However, in L958, JA/ET-related genes were strongly expressed (Table 2, Figure S1), which suggested that JA/ET levels increased in L958. Interestingly, the expression of the other two clusters of genes encoding PR10 and PR14 was largely upregulated in L958 as well (Table 3, Figure S2). It is reported that the expression of the PR10 gene is induced by JA and ABA [90]. Furthermore, silencing of PR10-like proteins in Medicago truncatula resulted in the reduced colonization of the oomycete Aphanomyces euteiches and induced the expression of PR5 [91]. In another research, a wheat orthologue of LTP, which is a member of the PR14 family, acted as a negative regulator to resist Puccinia striiformis f. sp. tritici (Pst), and the knockdown of this gene increased wheat resistance to Pst, which was accompanied by the accumulation of H2O2 and SA [92,93]. Therefore, the increase in JA/ET level and the high expression of PR10 and PR14 in L958 may act as negative regulators in the defence response to promote the establishment of susceptibility.
4. Materials and Methods
4.1. Plant and Pathogen Materials
The resistant wheat line L658 carrying Pm40 and its susceptible sister line L958 without Pm40 were used for comparative analyses by observations of cytological changes and determinations of physiological and biochemical indices. In addition, RNA-seq was performed for wheat plants after they were inoculated with Bgt, as described previously [54]. The fungus was a single-spore isolate collected from a field at Wenjiang Farm, Chengdu, Sichuan Province, and propagated weekly on the highly susceptible wheat line CY20. The wheat plants were cultivated in a growth chamber (Microclima MC1750E, Snijders Scientific, Tilburg, Holland) at 18 °C and 80% relative humidity under a 16-h light/8-h dark photoperiod. Seven-day-old seedlings of L658 and L958 were inoculated with fresh Bgt spores at a density of 100–200 conidia/mm2 by shaking infected leaf segments.
4.2. Cytological Observations of H2O2 and Cell Death
H2O2 accumulation was detected by the DAB (Sigma-Aldrich, Shanghai, China) staining method as described by Wang et al. [94]. The first leaves of L658 and L958 were harvested at 6, 12, 24, 48, and 72 hpi. The inoculated leaves were cut off and the end of cut leaves were immersed in 0.1% (w/v) DAB solution (pH 3.8) for 12 h to take up and react with the DAB fully. Cell death was detected by the trypan blue (Sigma-Aldrich, Shanghai, China) [53]. Leaf segments were immersed in 1% (w/v) TPB solution and then boiled for 2 min to stain the necrotic cells. Afterward, the leaf segments were decoloured in an ethanol-acetic acid (1:1, v/v) mixture until the chlorophyll was removed and then cleared in saturated chloral hydrate. The leaf segments were subsequently stored in 50% glycerol solution for examination and image collection via bright-field microscopy (Nikon Eclipse 80i, Nikon Corporation, Tokyo, Japan).
4.3. Determination of Antioxidant Enzyme Activity
A total of 0.2 g of fresh leaves was harvested at 6, 12, 24, 48, and 72 hpi for preparation of the crude enzyme, and samples at 0 h (without inoculation) were used as controls. Each sample was detected with three biological replicates. The fresh leaves were immediately frozen in liquid nitrogen and then homogenized in 2 mL of 50 mM pre-chilled phosphate buffer (pH 7.8) containing 1% (w/v) polyvinylpyrrolidone. The homogenate was then centrifuged at 12,000× g for 20 min at 4 °C, after which the supernatant was used for determination of enzyme activities. The activities of SOD, CAT, and POD were measured at 560, 240, and 470 nm, respectively, as described previously [51].
4.4. Determination of Photosynthesis Indices
The photosynthesis index data, including the Pn, Gs, and Ci, were collected with a portable photosynthesis system (LI-6400-02B, LI-COR, Lincoln, NE, USA) as described previously [51], and the measurement conditions included a vapour pressure deficit (VPD) of 0.6 kPa under an actinic light intensity of 1000 μmol/m2/s and an air temperature of 22 °C. The first measurements were performed before inoculation, and it was set as the controls (0 h). Subsequently, wheat leaves were inoculated with 100–200 conidia/mm2 fresh spores at 9:00 am. After 6 h (15:00 p.m.), 12 h (21:00 p.m.), 24 h (9:00 a.m.), 48 h (9:00 a.m.), and 72 h (9:00 a.m.) post inoculation, the photosynthesis indices were collected. The measurements at six time points were performed under the light period of the growth chamber. The data of six independent replications of each genotype were collected by measuring the centre of the first leaf of the wheat plants. The means of these six replications were then analysed statistically.
4.5. Measurements of Chlorophyll Fluorescence and Quantum Yields of PSII
Chlorophyll fluorescence was measured with a portable photosynthesis system (LI-COR 6400XT, Lincoln, NE, USA) equipped with a leaf chamber fluorometer (LI-6400-40; LI-COR, Lincoln, NE, USA) as described previously [51]. Measurements were taken from the centre of the first leaves of L658 and L958 at 0 (without inoculation), 6, 12, 24, 48, and 72 hpi. Six plants of each genotype were checked independently at the different timepoints, and the average represented the phenotypic value for subsequent statistical analysis. The data of the qP, maximum fluorescence in the light (Fm’), variable chlorophyll fluorescence yield in the light (Fv’), and ΦPSII were collected in the light. Furthermore, the maximum fluorescence (Fm) and variable chlorophyll fluorescence yield (Fv) were measured with a fluorescence meter after the plants were dark adapted for 30 min.
4.6. RNA Extraction, cDNA Library Construction, and RNA-Seq
First leaves were randomly collected from L658 and L958 plants at 0, 6, 12, 24, 48, and 72 hpi, with each sample comprising three biological replicates. A total of 36 leaf samples were frozen in liquid nitrogen and then sent to a service company (Beijing Biomarker Technology Co, Ltd., Beijing, China) for RNA-seq. Briefly, the total RNA in wheat was extracted according to the TRIzol reagent method (Invitrogen Life Technologies, CA, USA). After the integrity and purity of the RNA was checked via an Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA) and a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, DE, USA), a cDNA library was constructed by the use of a NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, USA). Last, RNA-seq was performed on an Illumina HiSeq™ 2500 platform in accordance with the 150-bp paired-end sequencing strategy. All the raw sequencing data have been submitted to the NCBI Sequence Read Archive (SRA), and the accession number is SRP117269 [54].
4.7. Alignment of RNA-Seq Reads and Differential Gene Expression Analysis
Reads that contained an adapter or a poly-N tail or those that were of low quality were removed from the raw reads. The filtered high-quality reads were then mapped to the IWGSC RefSeq v1.0 wheat genome reference (https://wheat-urgi.versailles.inra.fr/) by HISAT2 [95], the process of which allowed up to 2 base mismatches. All identified genes were functionally annotated via BLASTX of the non-redundant (NR) database and the Swiss-Prot protein database [96]. The expression level of the transcripts was subsequently calculated and normalized to an RPKM value by StringTie [97]. Furthermore, EdgeR [98] was applied to detect DEGs in pairwise comparisons with a threshold of a false discovery rate (FDR) ≤ 0.001 and |log2(fold change) | ≥ 2.
4.8. Reliable Analysis by qRT-PCR
To confirm the reliability of the RNA-seq data, we randomly chose 12 expressed genes for qRT-PCR experiments. Transcript One-Step gDNA Removal and a cDNA Synthesis Supermix Kit (Transgen Biotech, Beijing, China) were used to synthesize cDNA for qRT-PCR. The primer sequences used were designed by NCBI Primer-BLAST and are listed in Additional Table S2. The reactions were conducted on a Thermal Cycler CFX96 Real-Time System (Bio-Rad Laboratories, Hercules, CA, USA) together with SYBR Green qPCR Master Mix (Omega, Beijing, China). The relative expression of the abovementioned 12 genes was calculated according to the 2−ΔΔCt method [99], with tubulin [100] and GAPDH [101] used as reference genes. The experiments were performed for three biological replicates, and each biological replicate involved three technical repeats.
4.9. Statistical Analysis
Significant differences in the mean physiological parameters, photosynthesis parameters and relative gene expression were determined by independent sample t-tests or Duncan’s multiple range test via IBM Statistical Package for Social Science (SPSS) 19.0 software version 19.0 (SPSS Inc., Chicago, IL, USA). The data are presented as the mean values ± standard error (SEs).
5. Conclusions
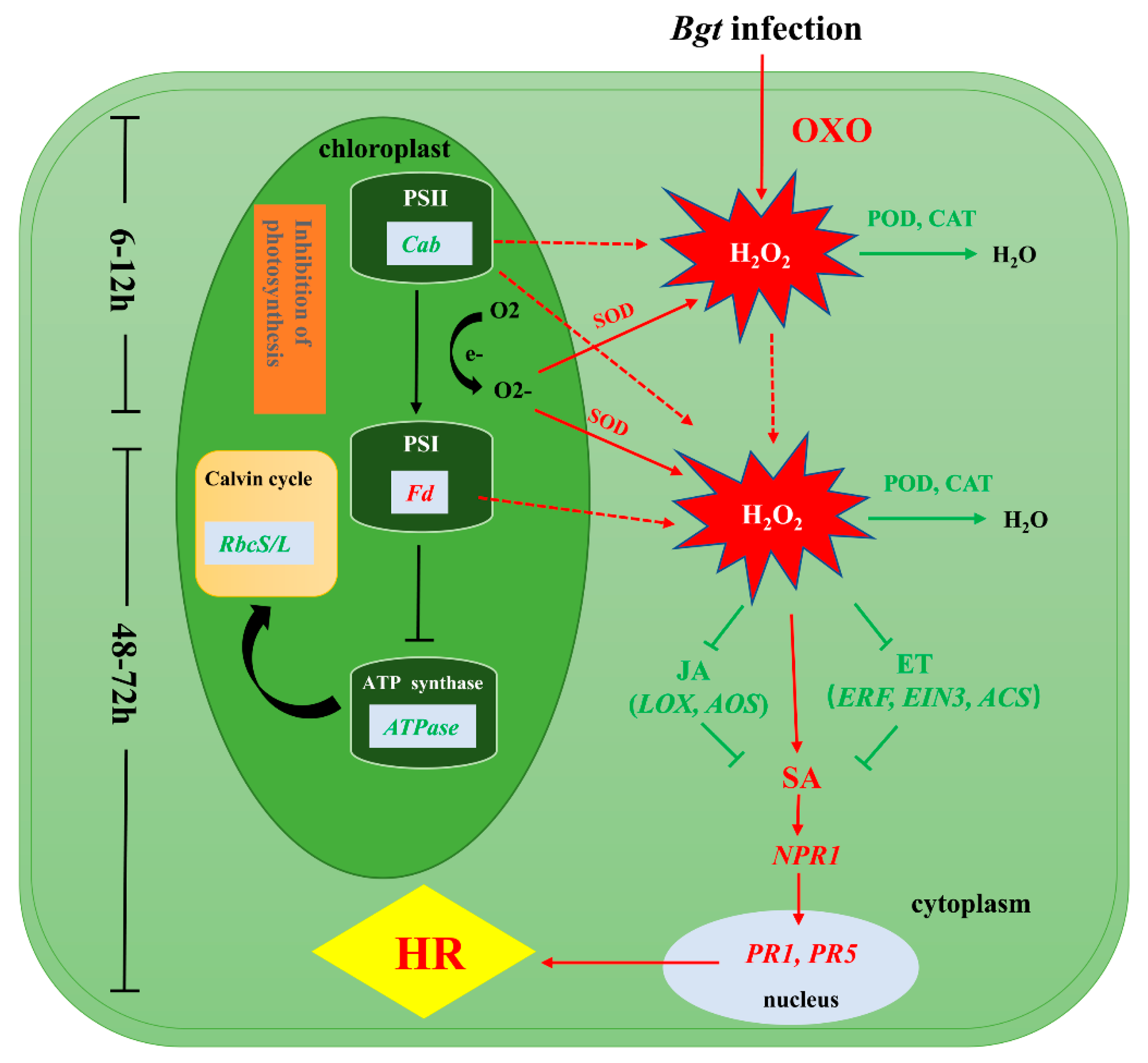
We investigated the differences in the physiological and biochemical status and in gene expression between L658 and L958 to reveal the mechanisms of Pm40-regulated resistance against Bgt, as shown in Figure 6. During the pre-penetration stage, the expression of genes encoding OXO was upregulated in L658 to generate H2O2. At the same time, the inhibition of photosynthesis caused by the downregulated expression of genes encoding Cab, ATPase, and RbcS/L generated O2-, which was dismutated into H2O2 by SOD. The suppression of POD and CAT further promoted the accumulation of H2O2 in L658. The first stage of H2O2 accumulation was at 6–12 hpi, and the H2O2 generated during this stage likely acted as a signaling molecule to provoke the second and relatively more intense burst of H2O2 at 48–72 hpi. The second H2O2 accumulation possibly enhanced SA levels by directly promoting the expression of SA-related genes or by indirectly inhibiting the expression of JA-/ET-related genes. Subsequently, SA likely activated the expression of NPR1, PR1, and PR5 and ultimately induced HR in L658.
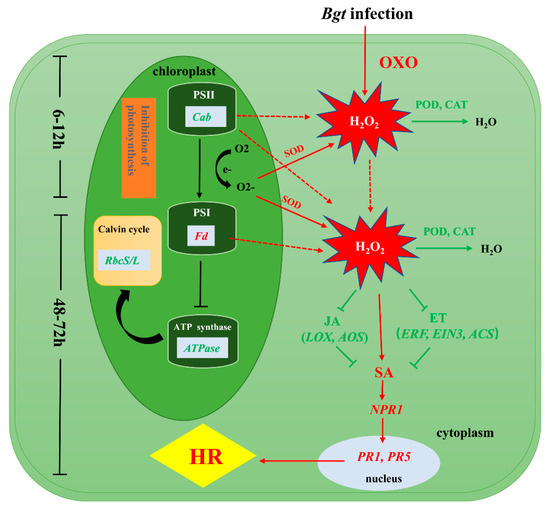
Figure 6.
Presumptive schematic of the resistance mechanism synergistically regulated by photosynthesis, ROS and plant hormones in the Pm40-expressing wheat line L658. The green colour represents low expression levels, and the red colour represents high expression levels.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/16/5767/s1. Figure S1. Expression patterns of DEGs involved in the SA, JA, and ET pathways in L658 and L958. The green colour represents low expression levels, and the red colour represents high expression levels. Figure S2. Expression patterns of DEGs encoding different PR proteins in L658 and L958. The green colour represents low expression levels, and the red colour represents high expression levels. Figure S3. Localization of hypersensitive cell death at interaction sites in L658 and L958 after inoculation with Bgt and the changes in main photosynthesis parameters in the two genotypes. Necrosis (dark blue) staining by trypan blue at interaction sites in L658 (A) and L958 (B) at various time points. Table S1. Summary of RNA-seq data and read mapping. Table S2. List of forward and reverse primers designed for use in qRT-PCR. Table S3. Number of different photosynthesis-related DEGs between L658 and L958 at various time points. Table S4. Number of photosynthesis-related DEGs in L658 and L958 at various inoculation time points compared with 0 h and between two adjacent time points.
Author Contributions
M.Z., P.L., G.G., and X.C. designed the research; Y.H., Y.L., S.Z., X.Q., L.L., and H.Y. performed the research; Y.H., S.Z., F.T., and P.L. analysed the data; Y.H. and P.L. wrote the paper; M.Z. and P.L. acquired the financial support for the project leading to this publication. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key R&D Program of China (No. 2018YFD0200500).
Acknowledgments
We are grateful to Biomarker Technology Co., Ltd. (Beijing, China) for the technical assistance.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Abbreviations
| ABA | Abscisic Acid |
| ACO | 1-aminocyclopropane-1-carboxylate oxidase |
| ACS | 1-aminocyclopropane-1-carboxylate synthase |
| AOS | Allene Oxide Synthase |
| AOX | Amine Oxidase |
| APX | Peroxidase |
| ATPase | ATP synthase |
| Bgt | Blumeria graminis f. sp. tritici |
| Cabs | Chlorophyll a/b-binding proteins |
| CAT | Catalase |
| EIN | Ethylene insensitive |
| ERF | Ethylene-responsive transcription factor |
| ET | Ethylene |
| Fd-I | Ferredoxin-I |
| FNR | Ferredoxin: NADP+ oxidoreductase |
| ICS | Isochorismate Synthase |
| JA | Jasmonic Acid |
| LOX | Lipoxygenase |
| NPR1 | Non-expressor of Pathogenesis Related genes 1 |
| OXO | Oxalate Oxidase |
| PGT | Primary Germ Tube |
| POD | Peroxidase |
| POX | Polyamine Oxidase |
| PP | Penetration Peg |
| PR1 | Pathogenesis-related 1 |
| PR5 | Pathogenesis-related 5 |
| PR10 | Pathogenesis-related 10 |
| PR14 | Pathogenesis-related 14 |
| RbcS/L | Ribulose bisphosphate carboxylase small chain/large chain |
| RBOH | Respiratory Burst Oxidase Homologue protein |
| RCA | Rubisco Activase |
| ROS | Reactive Oxygen Species |
| SA | Salicylic Acid |
| SABP | Salicylic Acid-binding protein |
| SOD | Superoxide dismutase |
References
- Pimentel, D. Diversification of biological control strategies in agriculture. Crop. Prot. 1991, 10, 243–253. [Google Scholar] [CrossRef]
- Pimentel, D. (Ed.) Biological Invasions: Economic and Environmental Costs of Alien Plant Animal and Microbe Species; CRC Press: Boca Raton, FL, USA, 2002; p. 384. [Google Scholar]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.A. ROS in biotic interactions. Physiol. Plantarum. 2010, 138, 414–429. [Google Scholar] [CrossRef]
- Melotto, M.; Underwood, W.; He, S.Y. Role of stomata in plant innate immunity and foliar bacterial diseases. Annu. Rev. Phytopathol. 2008, 46, 101–122. [Google Scholar] [CrossRef]
- Hardham, A.R.; Jones, D.A.; Takemoto, D. Cytoskeleton and cell wall function in penetration resistance. Curr. Opin. Plant Biol. 2007, 10, 342–348. [Google Scholar] [CrossRef]
- Coll, N.S.; Epple, P.; Dangl, J.L. Programmed cell death in the plant immune system. Cell Death Differ. 2011, 18, 1247–1256. [Google Scholar] [CrossRef]
- Fu, Z.Q.; Dong, X. Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 2013, 64, 839–863. [Google Scholar] [CrossRef]
- Li, X.; Zhong, S.F.; Chen, W.Q.; Fatima, S.A.; Huang, Q.L.; Li, Q.; Tan, F.Q.; Luo, P.G. Transcriptome analysis identifies a 140 kb region of chromosome 3B containing genes specific to Fusarium Head Blight resistance in wheat. Int. J. Mol. Sci. 2018, 19, 852. [Google Scholar] [CrossRef]
- Herrera-Vásquez, A.; Salinas, P.; Holuigue, L. Salicylic acid and reactive oxygen species interplay in the transcriptional control of defense genes expression. Front. Plant Sci. 2015, 6, 171. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, F.; Kenichi, T. Understanding the plant immune system. Mol. Plant Microbe Interact. 2010, 23, 1531–1536. [Google Scholar] [CrossRef] [PubMed]
- Kretschmer, M.; Damoo, D.; Djamei, A.; James, K. Chloroplasts and Plant Immunity: Where Are the Fungal Effectors? Pathogens 2020, 9, 19. [Google Scholar] [CrossRef]
- Sowden, R.G.; Watson, S.J.; Jarvis, P. The role of chloroplasts in plant pathology. Essays Biochem. 2017, 62, 21–39. [Google Scholar]
- Saxena, I.; Srikanth, S.; Chen, Z. Cross talk between H2O2 and interacting signal molecules under plant stress response. Front. Plant Sci. 2016, 7, 570. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive oxygen species oxidative damage and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef]
- Asada, K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006, 141, 391–396. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism oxidative stress and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Lu, Y.; Yao, J. Chloroplasts at the crossroad of photosynthesis pathogen infection and plant defense. Int. J. Mol. Sci. 2018, 19, 3900. [Google Scholar] [CrossRef]
- Asada, K. The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Biol. 1999, 50, 601–639. [Google Scholar] [CrossRef]
- Badger, M.R.; von Caemmerer, S.; Ruuska, S.; Nakano, H. Electron flow to oxygen in higher plants and algae: Rates and control of direct photoreduction (Mehler reaction) and rubisco oxygenase. Philos. T. Roy. Soc. B 2000, 355, 1433–1446. [Google Scholar] [CrossRef]
- Sabater, B.; Martín, M. Hypothesis: Increase of the ratio singlet oxygen plus superoxide radical to hydrogen peroxide changes stress defense response to programmed leaf death. Front. Plant Sci. 2013, 4, 479. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Suzuki, N.; Miller, G.A.D.; Tognetti, V.B.; Vandepoele, K.; Gollery, M.; Shulaev, V.; Breusegeme, F.V. ROS signaling: The new wave? Trends Plant Sci. 2011, 16, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.; Coutu, J.; Shulaev, V.; Mittler, R. Reactive oxygen signaling in plants. Annu. Plant Rev. Online 2018, 33, 189–201. [Google Scholar]
- Rekhter, D.; Lüdke, D.; Ding, Y.; Feussner, K.; Zienkiewicz, K.; Lipka, V.; Wiermer, M.; Zhang, Y.L.; Feussner, I. Isochorismate-derived biosynthesis of the plant stress hormone salicylic acid. Science 2019, 365, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C. Jasmonates: An update on biosynthesis signal transduction and action in plant stress response growth and development. Annu. Bot. 2007, 100, 681–697. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis perception signal transduction and action in plant stress response growth and development. An update to the 2007 review in Annals of Botany. Annu. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Bilgin, D.D.; Zavala, J.A.; Zhu, J.; Clough, S.J.; Ort, D.R.; DeLucia, E.H. Biotic stress globally downregulates photosynthesis genes. Plant Cell Environ. 2010, 33, 1597–1613. [Google Scholar] [CrossRef]
- Chen, Z.; Silva, H.; Klessig, D.F. Active oxygen species in the induction of plant systemic acquired resistance by salicylic acid. Science 1993, 262, 1883–1886. [Google Scholar] [CrossRef]
- Durner, J.; Klessig, D.F. Inhibition of ascorbate peroxidase by salicylic acid and 2 6-dichloroisonicotinic acid two inducers of plant defense responses. Proc. Natl Acad Sci. USA 1995, 92, 11312–11316. [Google Scholar] [CrossRef]
- Maruta, T.; Noshi, M.; Tanouchi, A.; Tamoi, M.; Yabuta, Y.; Yoshimura, K.; Takahiro Ishikawa, T.; Shigeoka, S. H2O2-triggered retrograde signaling from chloroplasts to nucleus plays specific role in response to stress. J. Biol. Chem. 2012, 287, 11717–11729. [Google Scholar] [CrossRef] [PubMed]
- Balasubramaniam, M.; Kim, B.S.; Hutchens-Williams, H.M.; Loesch-Fries, L.S. The photosystem II oxygen-evolving complex protein PsbP interacts with the coat protein of Alfalfa mosaic virus and inhibits virus replication. Mol. Plant Microbe Interact. 2014, 27, 1107–1118. [Google Scholar] [CrossRef] [PubMed]
- Dayakar, B.V.; Lin, H.J.; Chen, C.H.; Ger, M.J.; Lee, B.H.; Pai, C.H.; Chow, D.; Huang, H.E.; Hwang, S.Y.; Chung, M.C.; et al. Ferredoxin from sweet pepper (Capsicum annuum L.) intensifying harpinpss-mediated hypersensitive response shows an enhanced production of active oxygen species (AOS). Plant Mol. Biol. 2003, 51, 913–924. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.E.; Ger, M.J.; Yip, M.K.; Chen, C.Y.; Pandey, A.K.; Feng, T.Y. A hypersensitive response was induced by virulent bacteria in transgenic tobacco plants overexpressing a plant ferredoxin-like protein (PFLP). Physiol. Mol. Plant Pathol. 2004, 64, 103–110. [Google Scholar] [CrossRef]
- Liau, C.H.; Lu, J.C.; Prasad, V.; Hsiao, H.H.; You, S.J.; Lee, J.T.; Yang, N.S.; Huang, H.E.; Feng, T.Y.; Chen, W.H.; et al. The sweet pepper ferredoxin-like protein (pflp) conferred resistance against soft rot disease in Oncidium Orchid. Transgenic Res. 2003, 12, 329–336. [Google Scholar] [CrossRef]
- Tang, K.; Sun, X.; Hu, Q.; Wu, A.; Lin, C.H.; Lin, H.J.; Twyman, R.M.; Christou, P.; Feng, T.Y. Transgenic rice plants expressing the ferredoxin-like protein (AP1) from sweet pepper show enhanced resistance to Xanthomonas oryzae pv. oryzae. Plant Sci. 2001, 160, 1035–1042. [Google Scholar] [CrossRef]
- Huang, H.E.; Ger, M.J.; Chen, C.Y.; Pandey, A.K.; Yip, M.K.; Chou, H.W.; Feng, T.Y. Disease resistance to bacterial pathogens affected by the amount of ferredoxin-I protein in plants. Mol. Plant Pathol. 2007, 8, 129–137. [Google Scholar] [CrossRef]
- Xu, Y.H.; Liu, R.; Yan, L.; Liu, Z.Q.; Jiang, S.C.; Shen, Y.Y.; Wang, X.F.; Zhang, D.P. Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis. J. Exp. Bot. 2011, 63, 1095–1106. [Google Scholar] [CrossRef]
- Jelenska, J.; Yao, N.; Vinatzer, B.A.; Wright, C.M.; Brodsky, J.L.; Greenberg, J.T. A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Curr. Biol. 2007, 17, 499–508. [Google Scholar] [CrossRef]
- Rodríguez-Herva, J.J.; González-Melendi, P.; Cuartas-Lanza, R.; Antúnez-Lamas, M.; Río-Alvarez, I.; Li, Z.; López-Torrejón, G.; Díaz, I.; Del Pozo, J.C.; Chakravarthy, S.; et al. bacterial cysteine protease effector protein interferes with photosynthesis to suppress plant innate immune responses. Cell Microbiol. 2012, 14, 669–681. [Google Scholar] [CrossRef]
- Kangasjärvi, S.; Neukermans, J.; Li, S.; Aro, E.M.; Noctor, G. Photosynthesis, photorespiration, and light signaling in defence responses. J. Exp. Bot. 2012, 63, 1619–1636. [Google Scholar] [CrossRef] [PubMed]
- Rojas, C.M.; Senthil-Kumar, M.; Tzin, V.; Mysore, K. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 2014, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Rojas, C.M.; Senthil-Kumar, M.; Wang, K.; Ryu, C.M.; Kaundal, A.; Mysore, K.S. Glycolate oxidase modulates reactive oxygen species-mediated signal transduction during nonhost resistance in Nicotiana benthamiana and Arabidopsis. Plant Cell 2012, 24, 336–352. [Google Scholar] [CrossRef] [PubMed]
- Ahammed, G.J.; Li, X.; Zhang, G.; Zhang, H.; Shi, J.; Pan, C.; Yu, J.; Shi, K. Tomato photorespiratory glycolate-oxidase-derived H2O2 production contributes to basal defence against Pseudomonas syringae. Plant Cell Environ. 2018, 41, 1126–1138.87. [Google Scholar] [CrossRef] [PubMed]
- Bruinsma, J. By how much do land water and crop yields need to increase by 2050. In Proceedings of the FAO Expert Meeting on How to Feed the World in 2050, Rome, Italy, 24–26 June 2009. [Google Scholar]
- Conner, R.L.; Kuzyk, A.D.; Su, H. Impact of powdery mildew on the yield of soft white spring wheat cultivars. Can. J. Plant Sci. 2003, 83, 725–728. [Google Scholar] [CrossRef]
- Luo, P.G.; Luo, H.Y.; Chang, Z.J.; Zhang, H.Y.; Zhang, M.; Ren, Z.L. Characterization and chromosomal location of Pm40 in common wheat: A new gene for resistance to powdery mildew derived from Elytrigia intermedium. Theor. Appl. Genet. 2009, 118, 1059–1064. [Google Scholar] [CrossRef]
- Liu, Z.H.; Xu, M.; Xiang, Z.P.; Li, X.; Chen, W.Q.; Luo, P.G. Registration of the novel wheat lines L658 L693 L696 and L699 with resistance to Fusarium Head blight stripe rust and powdery mildew. J. Plant Regist. 2015, 9, 121–124. [Google Scholar] [CrossRef]
- Shen, X.K.; Ma, L.X.; Zhong, S.F.; Liu, N.; Zhang, M.; Chen, W.Q.; Zhou, Y.L.; Li, H.J.; Chang, Z.J.; Li, X.; et al. Identification and genetic mapping of the putative Thinopyrum intermedium-derived dominant powdery mildew resistance gene PmL962 on wheat chromosome arm 2BS. Theor. Appl. Genet. 2015, 128, 517–528. [Google Scholar] [CrossRef]
- Ma, L.X.; Zhong, S.F.; Liu, N.; Chen, W.Q.; Liu, T.G.; Li, X.; Zhang, M.; Ren, Z.L.; Yang, J.Z.; Luo, P.G. Gene expression profile and physiological and biochemical characterization of hexaploid wheat inoculated with Blumeria graminis f. sp. tritici. Physiol. Mol. Plant Pathol. 2015, 90, 39–48. [Google Scholar] [CrossRef]
- Liang, Y.P.; Xia, Y.; Chang, X.L.; Gong, G.S.; Yang, J.Z.; Hu, Y.T.; Cahill, M.; Luo, L.Y.; Li, T.; He, L.; et al. Comparative proteomic analysis of wheat carrying Pm40 response to Blumeria graminis f. sp. tritici using two-dimensional electrophoresis. Int. J. Mol. Sci. 2019, 20, 933. [Google Scholar] [CrossRef]
- Chang, X.L.; Luo, L.Y.; Liang, Y.P.; Hu, Y.T.; Luo, P.G.; Gong, G.S.; Chen, H.B.; Khaskheli, M.I.; Liu, T.G.; Chen, W.Q.; et al. Papilla formation defense gene expression and HR contribute to the powdery mildew resistance of the novel wheat line L699 carrying Pm40 gene. Physiol. Mol. Plant Pathol. 2019, 106, 208–216. [Google Scholar] [CrossRef]
- Hu, Y.T.; Liang, Y.P.; Zhang, M.; Tan, F.Q.; Zhong, S.F.; Li, X.; Gong, G.S.; Chang, X.L.; Shang, J.; Tang, S.W.; et al. Comparative transcriptome profiling of Blumeria graminis f. sp. tritici during compatible and incompatible interactions with sister wheat lines carrying and lacking Pm40. PLoS ONE 2018, 13, e0198891. [Google Scholar] [CrossRef]
- Li, J.; Yang, X.; Liu, X.; Yu, H.; Du, C.; Li, M.; He, D. Proteomic analysis of the compatible interaction of wheat and powdery mildew (Blumeria graminis f. sp. tritici). Plant Physiol. Bioch. 2017, 111, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guo, G.; Zhou, L.; Chen, Y.; Zong, Y.; Huang, J.; Liu, C. Transcriptome analysis identifies candidate genes and functional pathways controlling the response of two contrasting barley varieties to powdery mildew infection. Int. J. Mol. Sci. 2020, 21, 151. [Google Scholar] [CrossRef]
- Westermann, A.J.; Lars, B.; Joerg, V. Resolving host–pathogen interactions by dual RNA-seq. PLoS Pathog. 2017, 13, e1006033. [Google Scholar] [CrossRef]
- Kuckenberg, J.; Tartachnyk, I.; Noga, G. Temporal and spatial changes of chlorophyll fluorescence as a basis for early and precise detection of leaf rust and powdery mildew infections in wheat leaves. Precis. Agric. 2009, 10, 34–44. [Google Scholar] [CrossRef]
- Ajigboye, O.O.; Bousquet, L.; Murchie, E.H.; Ray, R.V. Chlorophyll fluorescence parameters allow the rapid detection and differentiation of plant responses in three different wheat pathosystems. Funct. Plant Boil. 2016, 43, 356–369. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.A.; Jones, J.D.; Dangl, J.L. Reactive oxygen species signaling in response to pathogens. Plant Physiol. 2006, 141, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Lamb, C.; Dixon, R.A. The oxidative burst in plant disease resistance. Annu. Rev. Plant Biol. 1997, 48, 251–275. [Google Scholar] [CrossRef]
- Nishimura, M.T.; Dangl, J.L. Arabidopsis and the plant immune system. Plant J. 2010, 61, 1053–1066. [Google Scholar] [CrossRef]
- Shetty, N.P.; Jørgensen, H.J.L.; Jensen, J.D.; Collinge, D.B.; Shetty, H.S. Roles of reactive oxygen species in interactions between plants and pathogens. Eur. J. Plant Pathol. 2008, 121, 267–280. [Google Scholar] [CrossRef]
- Hu, X.; Bidney, D.L.; Yalpani, N.; Duvick, J.P.; Crasta, O.; Folkerts, O.; Lu, G.H. Overexpression of a gene encoding hydrogen peroxide-generating oxalate oxidase evokes defense responses in sunflower. Plant Physiol. 2003, 133, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Lane, B.G. Oxalate germins and higher-plant pathogens. IUBMB Life 2002, 53, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Schulze-Lefert, P.; Vogel, J. Closing the ranks to attack by powdery mildew. Trends Plant Sci. 2000, 5, 343–348. [Google Scholar] [CrossRef]
- Polidoros, A.N.; Mylona, P.V.; Scandalios, J.G. Transgenic tobacco plants expressing the maize Cat2 gene have altered catalase levels that affect plant-pathogen interactions and resistance to oxidative stress. Transgenic Res. 2001, 10, 555–569. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Sheng, X.; Greenshields, D.L.; Ogieglo, A.; Kaminskyj, S.; Selvaraj, G.; Wei, Y.D. Profiling of wheat class III peroxidase genes derived from powdery mildew-attacked epidermis reveals distinct sequence-associated expression patterns. Mol. Plant Microbe Interact. 2005, 18, 730–741. [Google Scholar] [CrossRef]
- Torres, M.A.; Dangl, J.L. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 2005, 8, 397–403. [Google Scholar] [CrossRef]
- Torres, M.A.; Jones, J.D.; Dangl, J.L. Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 2005, 37, 1130–1134. [Google Scholar] [CrossRef]
- Yoshioka, H.; Numata, N.; Nakajima, K.; Katou, S.; Kawakita, K.; Rowland, O.; Jones, J.D.G.; Doke, N. Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 2003, 15, 706–718. [Google Scholar] [CrossRef]
- Torres, M.A.; Dangl, J.L.; Jones, J.D. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 2002, 99, 517–522. [Google Scholar] [CrossRef]
- Scharte, J.; Schon, H.; Weis, E. Photosynthesis and carbohydrate metabolism in tobacco leaves during an incompatible interaction with Phytophthora nicotianae. Plant Cell Environ. 2005, 28, 1421–1435. [Google Scholar] [CrossRef]
- Swarbrick, P.J.; Schulze-Lefert, P.; Scholes, J.D. Metabolic consequences of susceptibility and resistance (race-specific and broad-spectrum) in barley leaves challenged with powdery mildew. Plant Cell Environ. 2006, 29, 1061–1076. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Vivancos, P.; Clemente-Moreno, M.J.; Rubio, M.; Olmos, E.; García, J.A.; Martínez-Gómez, P.; Hernández, J.A. Alteration in the chloroplastic metabolism leads to ROS accumulation in pea plants in response to plum pox virus. J. Exp. Bot. 2008, 59, 2147–2160. [Google Scholar] [CrossRef] [PubMed]
- Bonfig, K.B.; Schreiber, U.; Gabler, A.; Roitsch, T.; Berger, S. Infection with virulent and avirulent P. syringae strains differentially affects photosynthesis and sink metabolism in Arabidopsis leaves. Planta 2006, 225, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sharkey, T.D.; Bernacchi, C.J.; Farquhar, G.D.; Singsaas, E.L. Fitting photosynthetic carbon dioxide response curves for C3 leaves. Plant Cell Environ. 2007, 30, 1035–1040. [Google Scholar] [CrossRef]
- Gerganova, M.; Popova, A.V.; Stanoeva, D.; Velitchkova, M. Tomato plants acclimate better to elevated temperature and high light than to treatment with each factor separately. Plant Physiol. Bioch. 2016, 104, 234–241. [Google Scholar] [CrossRef]
- Brugger, A.; Kuska, M.T.; Mahlein, A.K. Impact of compatible and incompatible barley-Blumeria graminis f. sp. hordei interactions on chlorophyll fluorescence parameters. J. Plant Dis. Protect. 2018, 125, 177–186. [Google Scholar]
- Göhre, V.; Jones, A.M.; Sklenář, J.; Robatzek, S.; Weber, A.P. Molecular crosstalk between PAMP-triggered immunity and photosynthesis. Mol. Plant Microbe Interact. 2012, 25, 1083–1092. [Google Scholar] [CrossRef]
- Rolfe, S.A.; Scholes, J.D. Chlorophyll fluorescence imaging of plant–pathogen interactions. Protoplasma 2010, 247, 163–175. [Google Scholar] [CrossRef]
- Backer, R.; Naidoo, S.; van den Berg, N. The nonexpressor of pathogenesis-related genes 1 (NPR1) and related Family: Mechanistic insights in plant disease resistance. Front. Plant Sci. 2019, 10, 102. [Google Scholar] [CrossRef]
- Broekgaarden, C.; Caarls, L.; Vos, I.A.; Pieterse, C.M.; Van Wees, S.C. Ethylene: Traffic controller on hormonal crossroads to defense. Plant Physiol. 2015, 169, 2371–2379. [Google Scholar] [CrossRef] [PubMed]
- Van der Ent, S.; Van Wees, S.C.; Pieterse, C.M. Jasmonate signaling in plant interactions with resistance-inducing beneficial microbes. Phytochemistry 2009, 70, 1581–1588. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Glazebrook, J.; Clarke, J.D.; Volko, S.; Dong, X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 1997, 88, 57–63. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, D.; Chu, J.Y.; Boyle, P.; Wang, Y.; Brindle, I.D.; Luca, V.D.; Després, C. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 2012, 1, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Qiao, Y.S.; Lv, D.; Gao, Z.H.; Qu, S.C.; Zhang, Z. Malus hupehensis NPR1 induces pathogenesis-related protein gene expression in transgenic tobacco. Plant Biol. 2012, 14, 46–56. [Google Scholar] [CrossRef]
- Chen, X.K.; Zhang, J.Y.; Zhang, Z.; Du, X.L.; Du, B.B.; Qu, S.C. Overexpressing MhNPR1 in transgenic Fuji apples enhances resistance to apple powdery mildew. Mol. Boil. Rep. 2012, 39, 8083–8089. [Google Scholar] [CrossRef]
- Wang, Z.; Ma, L.Y.; Li, X.; Zhao, F.Y.; Sarwar, R.; Cao, J.; Li, Y.L.; Ding, L.N.; Zhu, K.M.; Yang, Y.H.; et al. Genome-wide identification of the NPR1-like gene family in Brassica napus and functional characterization of BnaNPR1 in resistance to Sclerotinia sclerotiorum. Plant Cell Rep. 2020, 39, 709–722. [Google Scholar] [CrossRef]
- Wang, C.S.; Huang, J.C.; Hu, J.H. Characterization of two subclasses of PR-10 transcripts in lily anthers and induction of their genes through separate signal transduction pathways. Plant Mol. Biol. 1999, 40, 807–814. [Google Scholar] [CrossRef]
- Colditz, F.; Niehaus, K.; Krajinski, F. Silencing of PR-10-like proteins in Medicago truncatula results in an antagonistic induction of other PR proteins and in an increased tolerance upon infection with the oomycete Aphanomyces euteiches. Planta 2007, 226, 57–71. [Google Scholar] [CrossRef]
- Sudisha, J.; Sharathchandra, R.G.; Amruthesh, K.N.; Kumar, A.; Shetty, H.S. Pathogenesis related proteins in plant defense response. In Plant Defence: Biological Control. Progress in Biological Control; Mérillon, J., Ramawat, K., Eds.; Springer: Dordrecht, Germany, 2012; Volume 12, pp. 379–403. [Google Scholar]
- Ahmed, S.M.; Liu, P.; Xue, Q.; Ji, C.; Qi, T.; Guo, J.; Guo, J.; Kang, Z.S. TaDIR1-2 a wheat ortholog of lipid transfer protein AtDIR1 contributes to negative regulation of wheat resistance against Puccinia striiformis f. sp. tritici. Front. Plant Sci. 2017, 8, 521. [Google Scholar] [CrossRef]
- Wang, C.F.; Huang, L.L.; Buchenauer, H.; Han, Q.M.; Zhang, H.C.; Kang, Z.S. Histochemical studies on the accumulation of reactive oxygen species (O2− and H2O2) in the incompatible and compatible interaction of wheat-Puccinia striiformis f. sp. tritici. Physiol. Mol. Plant Pathol. 2007, 71, 230–239. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast-spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.Z.; Lopez, R.; Magrane, M.; et al. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2004, 32, D115–D119. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Faheem, M.; Li, Y.B.; Arshad, M.; Cheng, J.Y.; Jia, Z.; Wang, Z.K.; Xiao, J.; Wang, H.Y.; Cao, A.Z.; Xing, L.P.; et al. A disulphide isomerase gene (PDI-V) from Haynaldia villosa contributes to powdery mildew resistance in common wheat. Sci. Rep. 2016, 6, 24227. [Google Scholar] [CrossRef]
- Scholtz, J.J.; Visser, B. Reference gene selection for qPCR gene expression analysis of rust-infected wheat. Physiol. Mol. Plant Pathol. 2013, 81, 22–25. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).