Abstract
Birch pollen allergy is among the most prevalent pollen allergies in Northern and Central Europe. This IgE-mediated disease can be treated with allergen immunotherapy (AIT), which typically gives rise to IgG antibodies inducing tolerance. Although the main mechanisms of allergen immunotherapy (AIT) are known, questions regarding possible Fc-mediated effects of IgG antibodies remain unanswered. This can mainly be attributed to the unavailability of appropriate tools, i.e., well-characterised recombinant antibodies (rAbs). We hereby aimed at providing human rAbs of several classes for mechanistic studies and as possible candidates for passive immunotherapy. We engineered IgE, IgG1, and IgG4 sharing the same variable region against the major birch pollen allergen Bet v 1 using Polymerase Incomplete Primer Extension (PIPE) cloning. We tested IgE functionality and IgG blocking capabilities using appropriate model cell lines. In vitro studies showed IgE engagement with FcεRI and CD23 and Bet v 1-dependent degranulation. Overall, we hereby present fully functional, human IgE, IgG1, and IgG4 sharing the same variable region against Bet v 1 and showcase possible applications in first mechanistic studies. Furthermore, our IgG antibodies might be useful candidates for passive immunotherapy of birch pollen allergy.
Keywords:
allergy; IgE; IgG1; IgG4; FcεRI; CD23; antibodies; blocking antibodies; in vitro; allergen immunotherapy 1. Introduction
Birch pollen allergy is among the most common allergies in Central and Northern Europe, with a prevalence of up to 16% [1]. More than 90% of individuals allergic to birch pollen are sensitised to a single major allergen, Bet v 1 [1,2]. Therefore, Bet v 1 has been extensively studied for more than 30 years [3,4,5].
The molecular key player in any allergy is IgE, which interacts with IgE receptors FcεRI and CD23 on the surface of effector cells [6]. Binding to the high-affinity receptor FcεRI on mast cells and basophils is of major importance for the immediate allergic reaction: Upon allergen binding to IgE, FcεRs are cross-linked and cells release mediators that are responsible for typical allergy symptoms [7]. An allergic individual typically has a polyclonal IgE response, which allows receptor cross-linking by monomeric allergens. However, a cross-link of FcεRI loaded with monoclonal IgE is possible according to the concept of allergen-associated molecular patterns (AAMPs), e.g., by allergens with repetitive identical epitopes—such as tropomyosin—or the formation of aggregates of monovalent allergens [8,9]. Bet v 1 is known to non-covalently form dimers in a concentration-dependent manner [8,10]. Although protein aggregation is thought to increase immunogenicity [11], studies with Bet v 1 have shown both an increase and decrease of its allergenicity with multimer formation [10,12].
The low-affinity IgE receptor CD23 is mainly expressed on B cells, but its expression can also be found or stimulated on various other cell types, such as monocytes, macrophages, follicular dendritic cells, and epithelial cells [13,14]. In contrast to FcεRI, CD23 does not belong to the family of Ig receptors. It is a C-type lectin that binds several ligands besides IgE, such as CD21 and various integrins, and can directly interact with major histocompatibility complexes (MHC) [13,14,15]. CD23 has many functions depending on its ligand, form (soluble vs. membrane-bound), and subtype (CD23a vs. CD23b). With regards to allergy, two important functions have the capacity to regulate IgE synthesis in B cells and mediate allergen internalisation, processing, and presentation to T cells, also known as facilitated antigen presentation (FAP) [6].
Currently, the only effective treatment of birch pollen allergy—besides symptomatic relief medication—is active allergen immunotherapy (AIT) [16]. In this approach, increasing doses of allergen are administered to the patient and the immune response is shifted from allergy to tolerance. The development of tolerance is dependent on IgG4 antibodies to the allergen [17] and raised IgG1 and especially IgG4 antibody serum levels are considered to be useful biomarkers for the success of AIT [18,19]. Based on these observations, the first therapeutic IgG targeting an allergen for passive immunotherapy was recently developed [20].
Although advances have helped understand the mechanisms of AIT [18], questions regarding the full functional characteristics of IgGs remain unanswered. The capability of these antibodies to block allergen recognition by IgE is undisputed; however, their Fc-mediated functions are insufficiently explored [21]. Furthermore, current studies focus on IgG4 and its many unusual properties [22] rather than the IgG1 isotype, despite observed increases in the levels of both subclasses in individuals undergoing AIT. In addition, few mechanistic studies directly compare IgE, IgG1, and IgG4, likely due to limited availability of well-characterised recombinant antibodies produced in sufficient amounts.
Polymerase Incomplete Primer Extension (PIPE) cloning can be employed for rapid combination of any known variable antibody regions with the constant region sequences of any isotype to generate functional antibodies [23]. This has been applied in oncology to study antibodies of the most commonly used therapeutic agent isotypes IgG1 and IgG4 [24]. Furthermore, it has led to the recent emergence of IgE as a novel antibody isotype in cancer treatment in the AllergoOncology field [25,26].
We now applied PIPE cloning with the objective of deriving IgG1 and IgG4 isotypes from a known IgE antibody against the major birch pollen allergen Bet v 1 [27] and elucidating their target antigen engagement. Our main aim was to provide variable region-matched antibodies for mechanistic studies. These antibodies could help to understand the role of Bet v 1 aggregation on a cellular level and shed light on the Fc-mediated role of IgG antibodies in AIT. They furthermore might prove useful as candidates for passive immunotherapy.
2. Results
2.1. Biochemical Characterisation of PIPE-Cloned Anti-Bet v 1 Antibodies
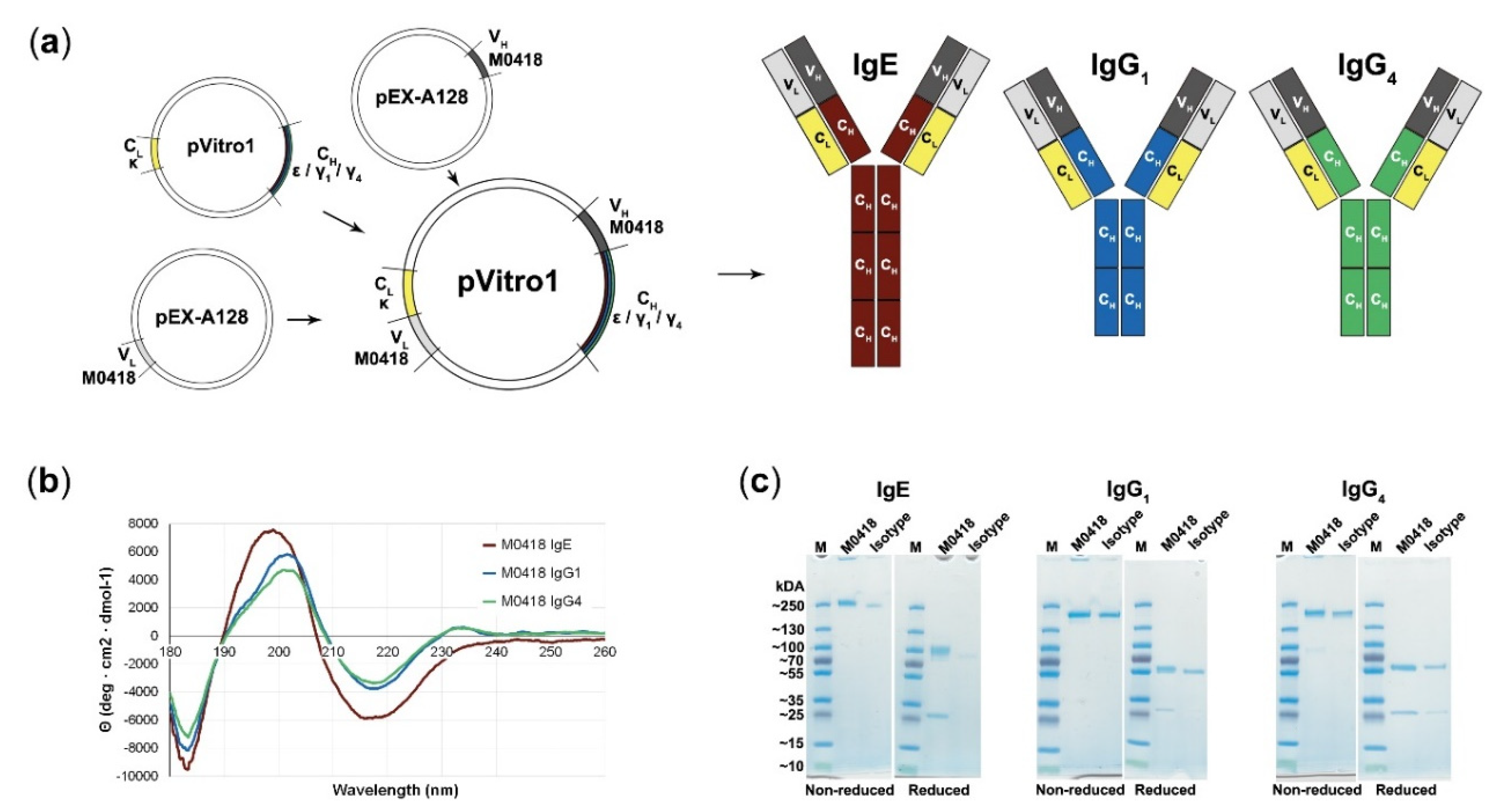
“M0418” anti-Bet v 1 variable heavy and light chain sequences [27] were successfully recombined with κ light chains and ε, γ1, or γ4 heavy chain sequences by PIPE cloning (see method schematic in Figure 1a) and expressed in the human Expi293F system.
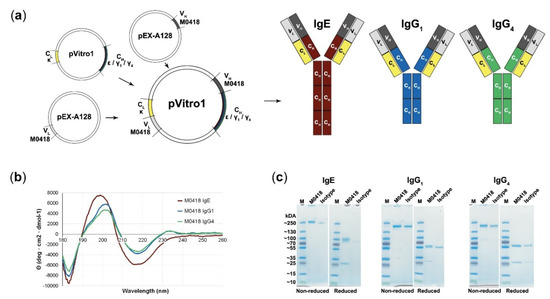
Figure 1.
Polymerase Incomplete Primer Extension (PIPE) cloning of several antibody classes and confirmation of their integrity. (a) Schematic representation of the PIPE cloning method and cloned antibodies. Variable regions (M0418) are recombined with different constant regions to create constructs for IgE, IgG1, and IgG4, respectively. (b) Circular dichroism spectra of M0418 IgE, IgG1, and IgG4 confirmed correct antibody assembly. (c) SDS-PAGE analysing purity and size of antibodies, as compared to the respective isotype controls.
CD spectroscopy was carried out in order to confirm antibody integrity (Figure 1b). Spectra showed a strong maximum at 199 (IgE)–201 (IgGs) nm and two minima at 183 and 218 nm (all isotypes). M0418 IgG antibodies furthermore showed a second slight maximum at 230 nm.
SDS-PAGE indicated high purity of our antibodies, showed migration patterns similar to a commercial isotype control and gave a first indication of the molecular weight of the antibodies, which was around 250 kDa for IgE and between 130–250 kDa for the IgGs (Figure 1c; see Figures S1–S4 for uncropped gel images).
SEC-MALS analysis confirmed this observation and yielded a molecular weight of 226 (IgE), 168 (IgG1), and 161 (IgG4) kDa (Table 1). Therefore, the measured molecular weight was higher than the expected theoretical molecular weight determined from the amino acid sequence.

Table 1.
Biochemical characteristics of recombinant M0418 antibodies.
2.2. Specificity Determination
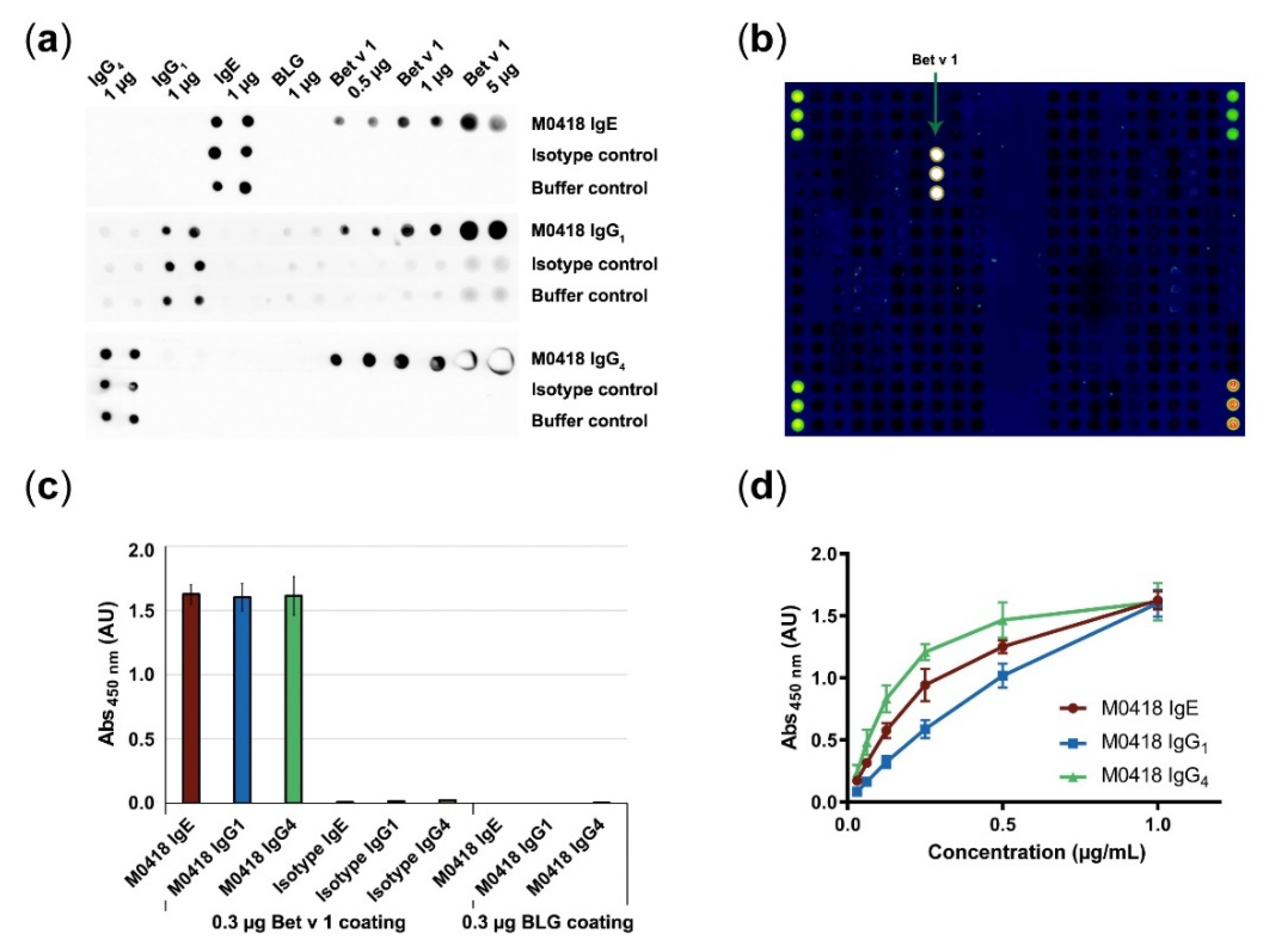
We confirmed antibody specificity to immobilised Bet v 1 by dot blot, ImmunoCAP ISAC112 (Thermo Fisher Scientific, Waltham, MA, USA) microarray and in-house ELISA (Figure 2).
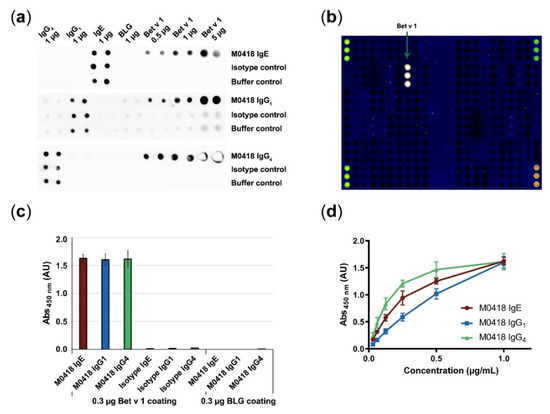
Figure 2.
Confirmation of antibody specificity. (a) Specificity dot blot confirmed binding of M0418 antibodies to membrane-bound Bet v 1, but not to a control allergen (BLG); spotted antibodies act as an integrity control for the assay. (b) ImmunoCAP ISAC112 microarray confirmed binding of M0418 IgE to Bet v 1 but no other of the spotted 112 allergens. (c) ELISA confirmed specific binding of M0418 antibodies to Bet v 1, but not to the control allergen BLG; respective isotype controls were included as an assay integrity control. (d) ELISA showed concentration-dependent binding of M0418 antibodies to immobilised Bet v 1. Results in (c,d) are represented as means ± SD of three independent experiments.
All M0418 isotypes recognised membrane-bound Bet v 1—but not the negative control allergen β-lactoglobulin (BLG)—in a dot blot (Figure 2a). The IgG1 control strips gave a faint background signal that can be attributed to unspecific binding of the secondary antibody. Therefore, another HRP-labelled antibody was used for IgG1 detection in all further experiments (i.e., ELISA).
M0418 IgE binding to Bet v 1 was furthermore confirmed in the ImmunoCAP ISAC112 microarray (Thermo Fisher Scientific, Waltham, MA, USA), where binding appeared to be exclusive to Bet v 1, i.e., it did not bind any of the other 111 allergens that were immobilised on the chip (Figure 2b).
Specificity was furthermore confirmed in an ELISA (Figure 2c), where M0418 antibodies bound Bet v 1, but not BLG. In addition, only M0418 antibodies—and not the respective isotype control antibodies—recognised Bet v 1.
Finally, we confirmed concentration-dependence of Bet v 1 binding in an ELISA for all M0418 isotypes (Figure 2d). Differences in the shape of the curves can be attributed to the different detection antibodies used.
2.3. M0418 IgG Inhibits Serum IgE Binding in a Quantitative ELISA
Next, we were interested in possible inhibitory functions of our IgG antibodies as well as utilising our IgE for Bet v 1—specific IgE quantification from human plasma.
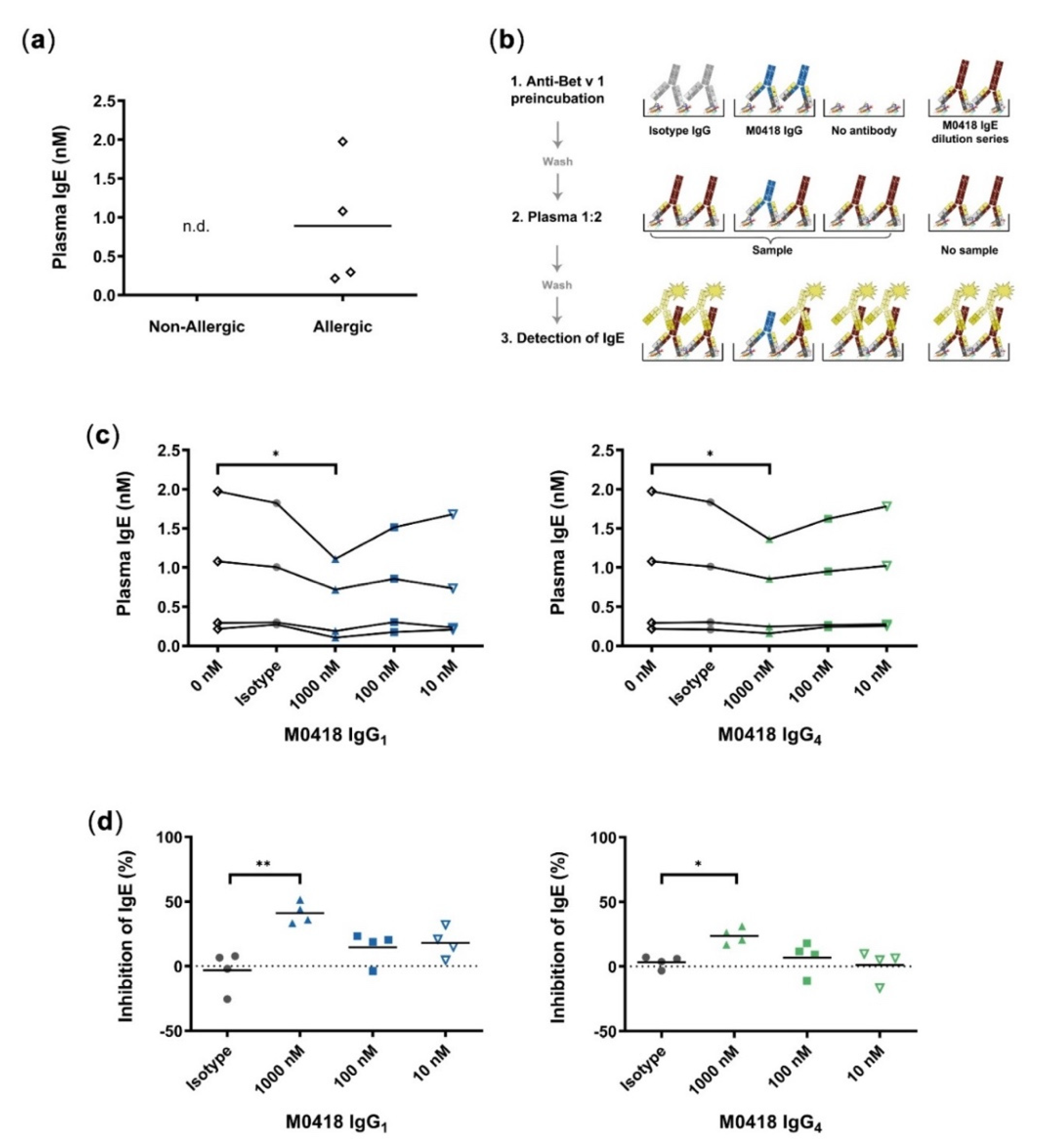
Therefore, we created a quantitative blocking ELISA, in which we used our recombinant M0418 IgE to set up a standard curve and determine the IgE plasma levels of four birch-pollen-allergic and non-allergic donors, respectively. The levels of IgE that were measured in the patients’ blood samples were between 0.2 and 2 nM, while those in serum samples from non-allergic donors were below the limit of detection in this assay (Figure 3a). In addition, we determined the capabilities of our IgG antibodies to block IgE binding to Bet v 1 in these allergic donor samples (see Figure 3b for a scheme describing the method). We chose relatively high maximum IgG concentrations to cover all possible IgE/IgG ratios that were reported in the literature [29].
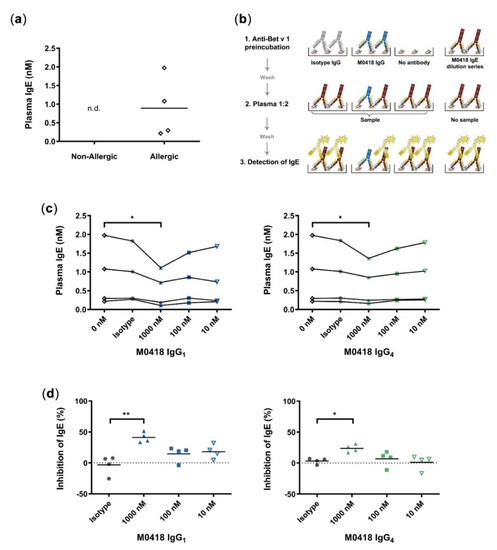
Figure 3.
Quantitative inhibition ELISA. (a) Quantification of Bet v 1-specific IgE from plasma of allergic and non-allergic individuals; n.d., not detectable (below limit of detection). (b) Schematic of quantitative blocking ELISA. 1, M0418 IgG antibodies (blue), or isotype controls (grey) were added for binding to Bet v 1 coated on ELISA plates. 2, Plates were incubated with plasma from birch pollen allergic individuals and 3, bound IgE (red) was detected by HRP-labelled Anti-IgE antibodies (yellow). The IgE standard curve from a M0418 IgE dilution series allowed quantification of plasma IgE levels. This blocking ELISA scheme was used in (c) for quantification of plasma IgE levels of Bet v 1-allergic individuals. 1000 nM M0418 IgG antibodies significantly decreased the amount of IgE-binding in contrast to the respective isotype controls. (d) Normalisation of IgE inhibition per individual shows that the M0418 IgG antibodies significantly decreased binding of IgE to Bet v 1 by 20–50%. Each data point represents the mean value of one allergic individual, as determined in three separate experiments, hence n = 4. * p < 0.05, ** p < 0.01.
M0418 IgG1 and IgG4—but not their respective non-specific isotype controls—could decrease the relative amount of plasma IgE binding to immobilised Bet v 1 at 1000 nM (Figure 3c). The normalisation of measurements per donor showed that antibodies significantly decreased binding of plasma IgEs to Bet v 1 by 20–50% (Figure 3d). M0418 IgG1 inhibited plasma IgE binding more effectively than M0418 IgG4.
2.4. M0418 IgE Upregulates FcεRI in Human LAD2 Mast Cells
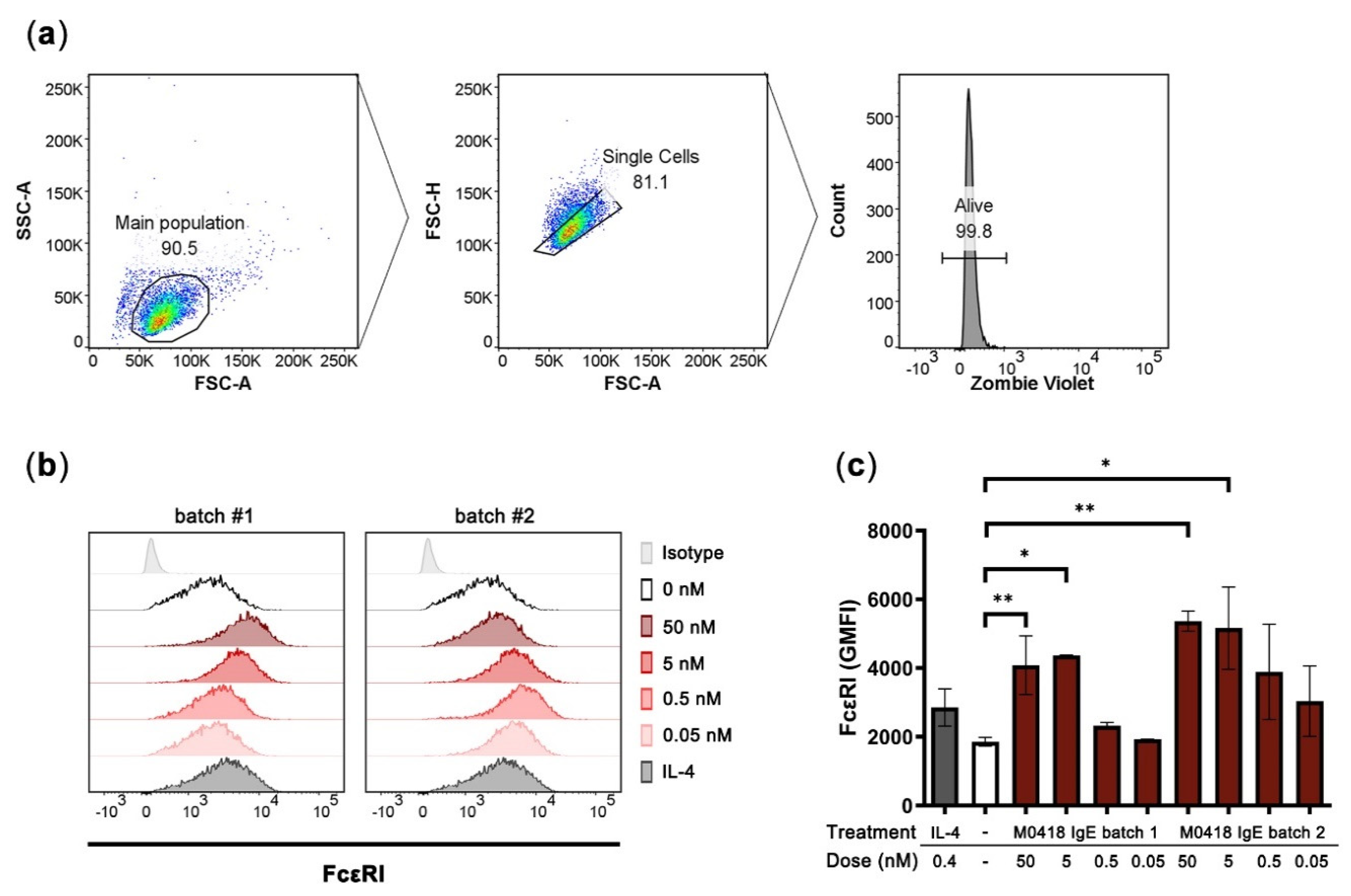
We then investigated the Fc-mediated functionality of M0418 IgE in vitro by investigating interaction with the high-affinity IgE receptor FcεRI. Because IgE can upregulate FcεRI expression in mast cells [7,30], we studied FcεRI expression in the human mast cell line LAD2 upon M0418 IgE treatment [31].
After four days of incubation with M0418 IgE, LAD2 cells showed a significant increase in FcεRI expression (Figure 4). The upregulation was dependent on the IgE concentration and more pronounced than upregulation triggered by IL-4 stimulation (positive control [32]). A maximum was reached at 5 nM of IgE. Furthermore, there was no significant difference between two batches of M0418 IgE with regard to receptor upregulation, indicating uniformity of our production system (Figure 4c).
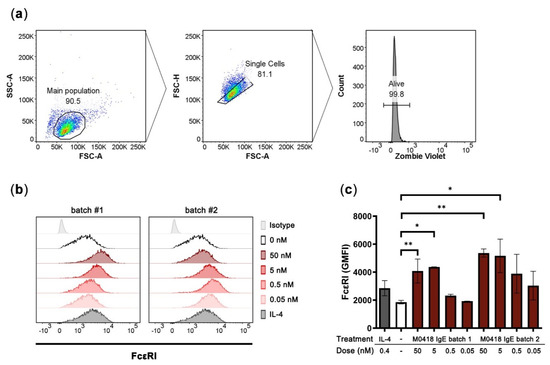
Figure 4.
Upregulation of FcεRI in human LAD2 mast cells upon stimulation with M0418 IgE. (a) Gating strategy. (b) Representative histograms showing FcεRI expression of LAD2 cells. Two different batches of M0418 IgE were tested, IL-4 was included as a positive control. (c) FcεRI was upregulated in LAD2 cells after a 4-day-incubation with two production batches of recombinant M0418 IgE. SSC-A, side scatter area; FSC-A, forward scatter area; FSC-H, forward scatter height; Zombie Violet, viability dye; GMFI, geometric mean fluorescence intensity. Data are represented as means ± SD of at least three independent experiments. * p < 0.05, ** p < 0.01.
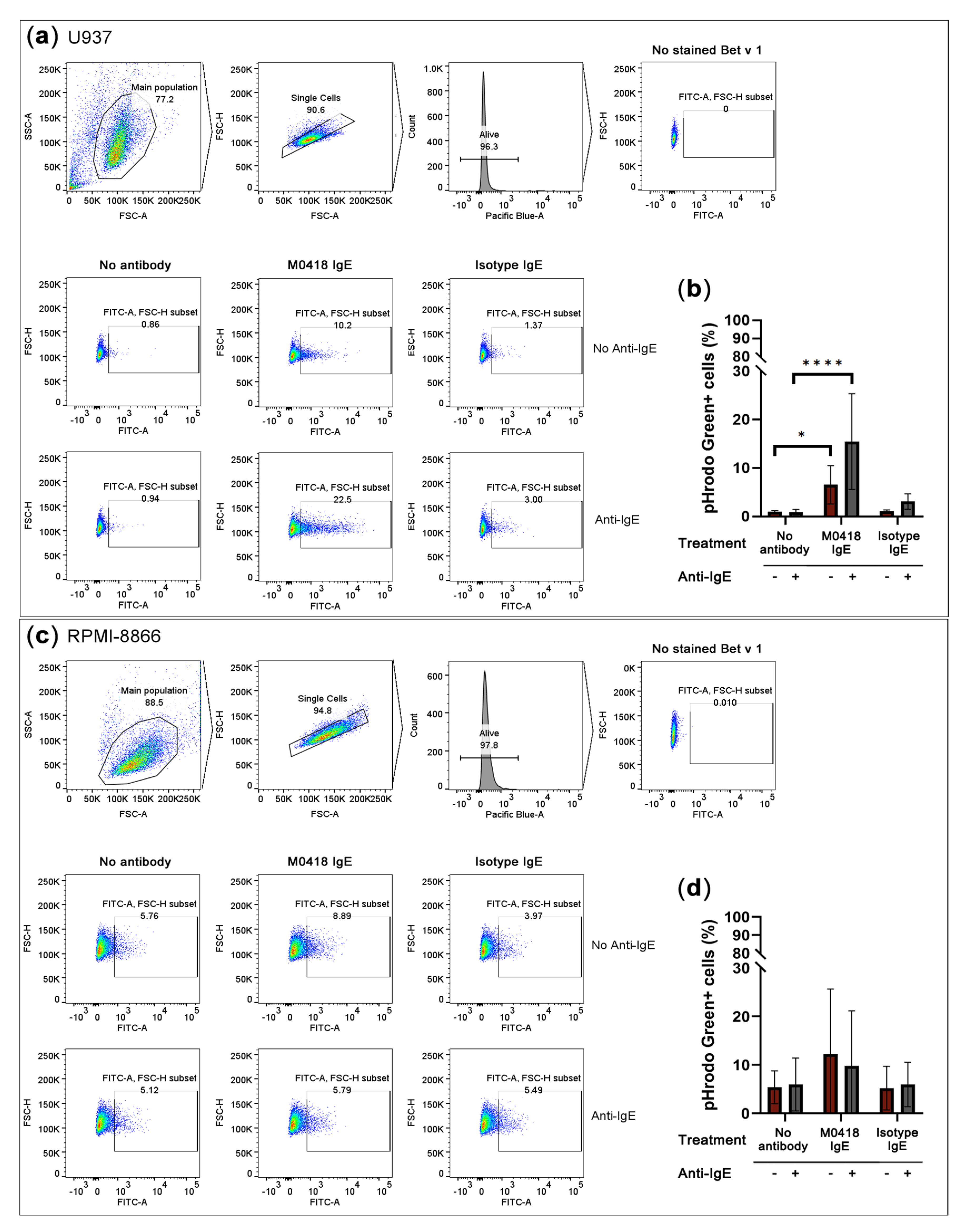
2.5. Bet v 1 Internalisation is Mediated by M0418 IgE in U937 Monocytes
Next, we sought to study Fc-mediated functionality via the low-affinity IgE receptor CD23. We labelled Bet v 1 with the pH-sensitive dye pHrodo Green and studied allergen internalisation via IgE immune complex formation in: (a) IL-4-primed human monocytic U937 cells that upregulate cell surface CD23 [33], and (b) the CD23hi human B-cell line RPMI-8866. Cells were either pre-treated with M0418 IgE or an isotype control to determine internalisation specificity. We furthermore added either Bet v 1 alone or Bet v 1 in combination with an anti-IgE antibody, to establish a positive control for Bet v 1 internalisation via artificial receptor crosslinking.
Both of the cell lines expressed CD23, but not FcεRI, as confirmed by flow cytometry (Figure S5). In U937 monocytic cells, Bet v 1 internalisation was dependent on allergen-specific IgE recognition (Figure 5a,b). Artificial crosslinking via anti-IgE antibody further enhanced internalisation.
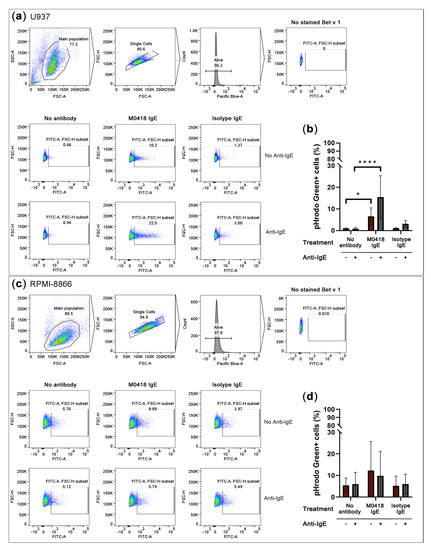
Figure 5.
Overnight internalisation of pHrodo Green-labelled Bet v 1 by IL-4-primed U937 monocytes and the CD23hi B-cell line RPMI-8866. (a) Gating strategy and representative flow cytometry data of U937 internalisation. (b) U937 monocytes internalised Bet v 1 in an IgE-dependent manner. (c) Gating strategy and representative flow cytometry data of RPMI-8866 internalisation. (d) Bet v 1-internalisation by RPMI-8866 cells was IgE-independent. SSC-A, side scatter area, FSC-A, forward scatter area, FSC-H, forward scatter height. Pacific Blue-A, area of Pacific Blue channel signal which indicates DAPI viability staining. Data are represented as means ± SD of at least three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
RPMI-8866 cells showed a high level of unspecific internalisation and, thus, internalised Bet v 1 independently of IgE (Figure 5c,d).
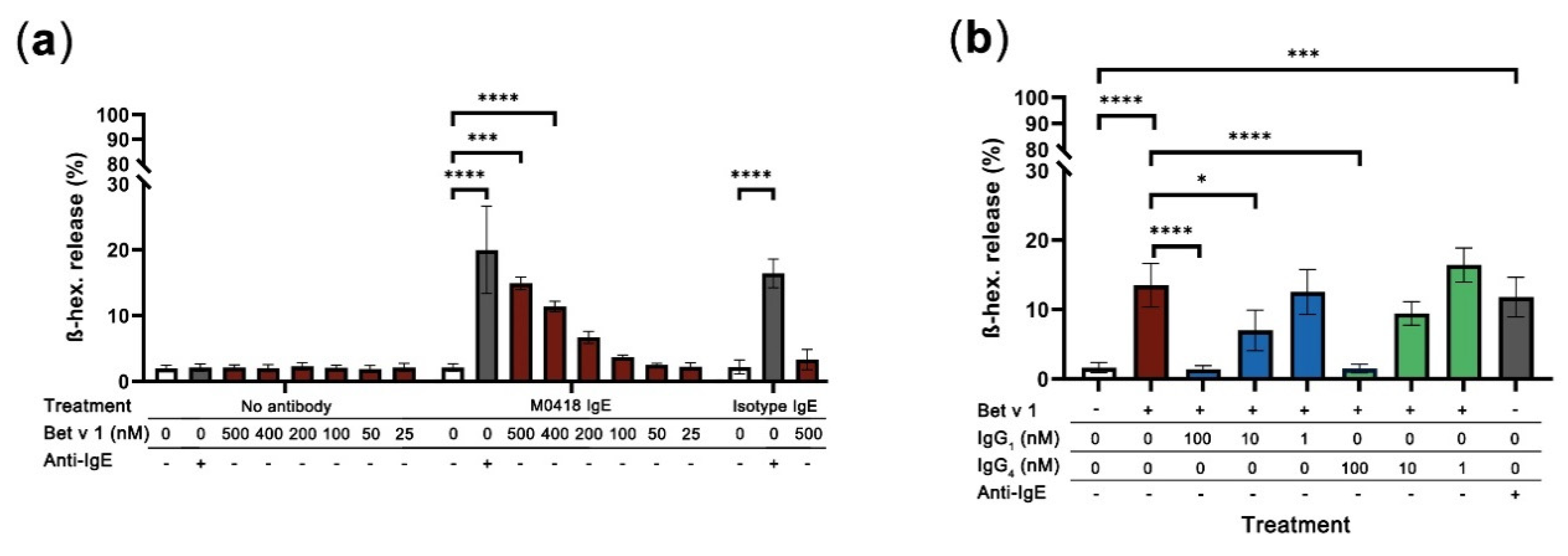
2.6. M0418 IgE Mediates Bet v 1-Degranulation in RBL-SX38 Basophils
Finally, we aimed to investigate the main effector function of our M0418 IgE, allergen-dependent degranulation. A well-established system for degranulation studies is the rat basophilic leukaemia cell line RBL-SX38, which is transfected with a fully functional human FcεRI [34]. We were curious as to whether we could trigger degranulation of cells that were sensitised with monoclonal antibody by addition of Bet v 1. To ensure the saturation of all Fcε Receptors on the cell surface, we used a sensitisation concentration of 5 nM IgE. This was furthermore in accordance with the values recommended in the literature [35].
Interestingly, Bet v 1 could crosslink FcεRI-bound allergen-specific IgE-but not non-specific isotype control IgE-and cause degranulation in RBL-SX38 cells (Figure 6a). Degranulation was concentration-dependent with regards to Bet v 1 and reached similar levels as those achieved with cross-linking RBL-SX38-bound IgE with polyclonal anti-IgE (positive control). Degranulation was significant with 400 and 500 nM of Bet v 1 and specific when compared to the isotype control.
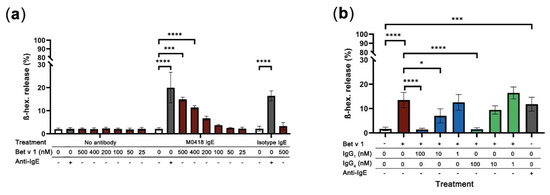
Figure 6.
RBL-SX38 degranulation assay. (a) Bet v 1 caused degranulation of RBL-SX38 basophils sensitised with M0418 IgE—but not with isotype control IgE—in a concentration-dependent manner. Degree of degranulation was comparable with the artificial crosslinker positive control (Anti-IgE). (b) Inhibition of Bet v 1-dependent degranulation of RBL-SX38 cells pre-treated with M0418 IgE by M0418 IgG antibodies was dependent on blocking antibody concentration. Data are represented as means ± SD of at least three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
When we tested whether our M0418 IgG antibodies could block epitopes on Bet v 1 and, thus, prevent degranulation of RBL-SX38 cells sensitised with M0418 IgE, both M0418 IgG1 and IgG4 blocked Bet v 1 degranulation in a concentration-dependent manner (Figure 6b). In both cases, 100 nM of IgG antibody were sufficient to completely inhibit degranulation caused by 400 nM Bet v 1.
3. Discussion
We successfully used PIPE cloning to produce fully human, recombinant anti-Bet v 1 IgE, IgG1, and IgG4 that share the same variable region. We confirmed antibody integrity and purity with CD spectroscopy, SDS-PAGE and SEC-MALS.
The shape of the presented CD spectra is typical for immunoglobulins—which consist mainly of β-sheets—and overall indicates correct assembly [36,37]. Deviations of the IgE spectrum from the IgG spectra can be attributed to the two additional constant domains in the Fc-region of IgE.
SDS-PAGE indicated a higher molecular weight than calculated from the sequence, which was confirmed by SEC-MALS. Such discrepancies between the theoretical and experimentally determined molecular weights may be attributed to antibody glycosylation. The fact that this effect was more pronounced for M0418 IgE than its IgG counterparts supports this notion, as IgE has substantially more glycosylation sites [38].
We confirmed specificity to Bet v 1 in a dot blot, allergen microarray and ELISA. ImmunoCAP ISAC112 data stringently confirmed the original observation that the M0418 variable region has very limited cross-reactivity with other structurally related PR-10 allergens [27].
After thorough characterisation, we approached functional studies and utilised all three M0418 antibody isotypes in a quantitative inhibition ELISA using allergic patient donor plasma. The Bet v 1-specific plasma IgE levels detected from allergic donors were consistent with those reported in the literature [29,39,40]. The partial blocking of Bet v 1 recognition by polyclonal patient-derived antibodies is likely due to the limited epitope coverage of our monoclonal antibodies and thus to incomplete blockade of multiple epitopes on Bet v 1 recognised by patient IgEs [41]. However, our overall results were in accordance with Bet v 1-inhibition ELISAs reported by others [42,43], see a comparison of relevant assay parameters in Appendix A in Table A1. Of note, M0418 IgG4 showed a weaker blocking capacity than its IgG1 counterpart, which can possibly be attributed to its unique property of Fab arm exchange with other IgG antibodies [22].
Furthermore, M0418 IgE was fully functional with regards to Fc-mediated effects in appropriate FcεRI and CD23 in vitro models.
It is well known that IgE levels positively correlate with the FcεRI expression on mast cells. Accordingly, in our model, culturing human LAD2 mast cells with M0418 IgE for several days resulted in an increase of of FcεRI on the cell surface, which are likely attributed to receptor upregulation [30]. Only few similar studies exist, mainly due to the lack of the necessary tools and we thus propose that M0418 IgE could be used as a practical reagent to improve mediator release of mast cells and other effector cells in in vitro. In fact, M0418 IgE incubation reached the plateau of receptor upregulation at similar concentrations as in previous reports [30].
The findings of our internalisation studies were rather surprising: Bet v 1 internalisation appeared to be strictly antibody-dependent in the case of U937 monocytes, and unspecific and antibody-independent in the case of RPMI-8866 B cells. Based on the nature of these cell types and previous findings in B cells showing their IgE-dependent allergen internalisation and presentation to T cells, we would have assumed opposite results [44,45,46]. To our knowledge, only one study exists, which investigates Bet v 1 internalisation by monocytes and B cells [47]. However, the analysis of these cell types is very brief, as the focus of the work lay on basophils. Furthermore, a comparison of our results with these findings is very difficult, as the authors analysed cellular subsets from PBMCs of allergic and non-allergic donors rather than cell lines treated with recombinant antibodies. In their findings, B cells of both allergic and non-allergic donors to a small, but similar extent, internalised Bet v 1. Monocytes, however, showed an overall strong and unspecific internalisation of Bet v 1, which is even more pronounced in an allergic setting. It is clear from the lack of available data, that these observations require additional studies. With our antibodies it should now be possible to investigate the antibody-dependence of Bet v 1 internalisation in detail. We suggest additional studies with several model cell lines as well as primary cells and investigating not only the role of IgE, but also IgG antibodies in internalisation or inhibition thereof.
In addition to these CD23-mediated internalisation studies, we were able to show allergen-dependent, IgE-mediated degranulation of RBL-SX38 basophils. M0418 IgE-mediated degranulation was caused by binding of increasing concentrations of Bet v 1. This finding appears to go against the textbook knowledge that a single-epitope allergen can only be crosslinked by polyclonal IgE. However, the formation of allergen multimers would not only theoretically allow crosslinking of monoclonal antibodies [8], but was recently shown in detail in a study with Timothy grass pollen allergen [48]. Recent degranulation studies with an engineered Bet v 1 trimer confirm this notion [44]. Additional studies of our group with monoclonal antibodies against the major cow’s milk allergen BLG confirm these findings [49]. It is furthermore well-known that Bet v 1 is prone to form dimers, as was shown previously [11,50].
Overall, we present, for the first time, a systematic characterisation of IgE, IgG1, and IgG4 sharing the same variable region targeting the major birch pollen allergen Bet v 1. Besides proving IgG blocking capabilities of our antibodies, we also confirmed full functionality of M0418 IgE in vitro.
Our findings may have raised further questions with regard to Bet v 1 aggregation and internalisation by B cells and monocytes but we now for the first time present the appropriate tools to address such mechanistic questions in vitro.
4. Materials and Methods
4.1. Cell Line Cultivation and Maintenance
Expi293F cells (cat# A14527, Thermo Fisher Scientific, Waltham, MA, USA) were cultivated in 30 or 125 mL of Expi293 Expression Medium (cat# A1435101, Gibco, Thermo Fisher Scientific, Waltham, MA, USA) in 125 mL or 500 mL baffled Erlenmeyer flasks with vented caps (Corning, New York, NY, USA), respectively, at 37 °C, 8% CO2, according to the manufacturer’s instructions. LAD2 [31] (kind gift of A. Kirshenbaum and D. Metcalfe, National Institute of Allergy and Infectious Diseases, Bethesda, MD, USA) were grown in StemPro-34 medium (cat# 61870044, Gibco, Thermo Fisher Scientific, Waltham, MA, USA) that was supplemented with 13 mL StemPro-34 Nutrient Supplement (cat# 10639011, Gibco, Thermo Fisher Scientific, Waltham, MA, USA) per 500 mL medium, L-Glutamine (2 mM), penicillin (100 U/mL)/streptomycin (100 µg/mL), and rhSCF (100 ng/mL; cat#11343327, Immunotools GmbH, Friesoythe, Germany) in T75 cell culture flasks (Corning, New York, NY, USA) at 37 °C, 5% CO2. RBL-SX38 [34], RPMI-8866 (ECACC 95041316), and U937 cells (ATCC® CRL-1593.2™) were cultivated in RPMI 1640 medium with GlutaMAXTM (cat# 618700, Thermo Fisher Scientific, Waltham, MA, USA), supplemented with 10% FBS (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), and maintained in T75 flasks (Corning, New York, NY, USA) at 37 °C, 5% CO2.
4.2. Allergens
Recombinant Bet v 1 (Bet v 1.0101) was produced in E. coli and quality was tested, as previously described [51,52]. Commercially available milk allergen β-lactoglobulin (BLG; cat# L01030, Sigma-Aldrich, St. Louis, MO, USA) was used as a negative control allergen in some of the experiments.
4.3. pHrodo Green-Labelling of Bet v 1
Bet v 1 was labelled with a pHrodoTM Green STP ester kit (cat# P35369, Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instruction. Briefly, 200 µg of Bet v 1 was labelled at a concentration of 1 mg/mL in 0.1 M bicarbonate buffer. For the labelling reaction, a molar dye: protein ratio of approximately 10:1 was used. The reaction mixture was incubated at room temperature for 1 h, protected from light. Tube was inverted every 15 min. to ensure mixing. After labelling, pHrodo Green-Bet v 1 was dialysed against phosphate-buffered saline (PBS) in order to remove excess dye using a Slide-A-Lyzer™ MINI Dialysis Device with a cut-off of 3.5 kDa (cat# 88401, Thermo Fisher Scientific, Waltham, MA, USA), overnight at 4 °C. Buffer was exchanged and the sample was dialysed for another 4.5 h at 4 °C. Labelled Bet v 1 was stored at 4 °C until used.
4.4. PIPE Cloning
M0418 variable region sequences were obtained from a previously published human IgE sequence [27]. If necessary, amino acid sequences were translated into nucleotide sequences and manually optimised for expression in human cells using various online tools included in the Sequence Manipulation Suite [53]. Optimised sequences were synthesised commercially and provided in pEX_A128 vectors (Eurofins Genomics AT GmbH, Vienna, Austria). PIPE cloning was carried out as previously described [23,54]. Briefly, light chain regions (M0418L, κ) and heavy chain regions (M0418H, ε/γ1/γ4) were amplified in four separate PIPE PCRs using Phusion Flash High-Fidelity PCR master mix (cat# F548S, Thermo Fisher Scientific, Waltham, MA, USA) and a MyCyclerTM thermal cycler (Bio-Rad Laboratories, Hercules, CA, USA). The respective template/primer combinations as well as PCR extension times are listed in Table 2. After confirmation of the expected size via agarose gel electrophoresis, the PCR products were digested for 1–2 h with DpnI (cat# R0176S, New England Biolabs, Ipswich, MA, USA) in the provided CutSmart® buffer at 37 °C, followed by 20 min. enzyme deactivation at 80 °C. Digested PCR products were then mixed in a 1:1:1:1 ratio and incubated at room temperature for at least 2 h up to overnight for ligation. NEB 10-beta Competent E. coli (cat# C3019, New England Biolabs, Ipswich, MA, USA) was transformed with the resulting pVitro1-hygro-mcs constructs according to the manufacturer’s instructions. Selection was carried out on LB-agar plates supplemented with 150 µg/mL hygromycin B (cat# 1287, Carl Roth GmbH & Co KG, Karlsruhe, Germany). Colonies were screened for constructs via colony PCR. The integrity of construct antibody sequences was finally confirmed by Cycle sequencing (Mix2seq overnight kit; Eurofins Genomics AT GmbH, Vienna, Austria). The resulting antibody sequences are summarised in Appendix A in Table A2 and heavy and light chain sequences are deposited at GenBank (accession numbers MT435130, MT435131, MT435132 and MT435133).

Table 2.
PIPE PCR parameters and primers for amplification of four fragments that then self-assemble into the vector pVitro1-hygro carrying M0418 IgE, IgG1 or IgG4.
4.5. Expression of Constructs and Antibody Purification
Expi293F cells were transfected with 30 µg of plasmid DNA according to the manufacturer’s instructions, with some modifications: 50 µg/mL of hygromycin B (cat# 1287, Carl Roth GmbH & Co KG, Karlsruhe, Germany) were added 72 h after transfection to further enhance production. Supernatant was harvested on day 7 after transfection by two consecutive centrifugation steps at low speed (280 x g, 7 min.), followed by a final centrifugation step at 3500 x g for 35 min. Supernatant was then filtered through a 0.22 µm PES membrane filter (cat# 124-0020, Thermo Fisher Scientific, Waltham, MA, USA) and diluted 1:2 with PBS prior to purification. The antibodies were purified via affinity chromatography using either a HiTrap KappaSelect column (IgE purification, 1 mL; GE Healthcare Life Sciences, Marlborough, MA, USA) or a HiTrap Protein A HP antibody purification column (IgG1 and IgG4 purification, 1 mL; GE Life Sciences) and an AEKTA chromatography system (GE Life Sciences/Amersham Biosciences, Marlborough, MA, USA). The antibodies were eluted from the column with 0.1 M glycine, pH 2.5, step elution (IgE) or 0.1 M citric acid, pH 3, gradient elution (IgG antibodies), respectively. Eluate was collected in 1 mL fractions and pH was immediately neutralised with 100 µL of Tris-HCl, pH 9.3. Antibody-Containing fractions were pooled and dialysed overnight at 4 °C against PBS using Tube-O-Dialyzer medi dialysis devices with a molecular cut-off of 15 kDa (cat# 786-618, GBiosciences, St. Louis, MO, USA). Concentration was then determined by measuring the UV absorption at 280 nm with a UV/VIS spectrophotometer (DeNovix DS-11 FX+, DeNovix Inc., Wilmington, DE, USA), using the respective extinction coefficients, as determined from the protein sequence. Antibody solutions were sterilised by passing them through a 0.2 µm PES filter (cat# 16532, Sartorius AG, Göttingen, Germany) and stored at 4 °C until use.
4.6. Determination of Antibody Integrity
4.6.1. Circular Dichroism Spectroscopy
The circular dichroism (CD) spectra of antibodies were measured in 75 mM phosphate buffer (pH 7.2) with a 0.1 mm quartz cuvette with on a Jasco J-1500 spectropolarimeter (Jasco International Co., Tokyo, Japan) at room temperature. Sample concentrations were between 0.2 and 0.6 mg/mL. Spectra were obtained from 260–180 nm, data pitch 0.2 nm, at a scan speed of 20 nm/min., with a response (D. I. T., digital integration time) of 8s. Data represent the average of 5 accumulated measurements and they are given as mean residue ellipticity.
4.6.2. SDS-PAGE
2–3µg of M0418 IgE, IgG1 and IgG4 or the respective commercially available isotype controls (human plasma IgE, human myeloma IgG1, and human myeloma IgG4, cat# 16-16-090705, 16-16-090707-1M, and 16-16-090707-4M, respectively, Athens Research & Technology, Athens, GA, USA) were mixed with Laemmli loading buffer with and without β-mercaptoethanol (reducing vs. non-reducing conditions), respectively, and denatured at 95 °C for 5 min. Samples were loaded into 50 µL pockets of a 4–15% gradient polyacrylamide gel (Mini-PROTEAN® TGX™ precast gel, cat# 4561084, Bio-Rad Laboratories, Hercules, CA, USA) and gel was run in SDS-Tris-glycine buffer at 200 V for 30–40 min. For size determination, PageRuler Plus prestained protein ladder (cat# 26619, Thermo Fisher Scientific, Waltham, MA, USA) was run on each gel. After electrophoresis, gel was stained with SimplyBlue safe stain (cat# LC6060, Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions.
4.6.3. SEC-MALS
The samples in PBS were injected onto a SEC column (Superdex 200 Increase 10/300, GE Healthcare Life Sciences, Marlborough, MA, USA) at a flow rate of 0.5 mL/min. at 7 °C, detected at 280 nm, and coupled to a multi angle light scattering detector (miniDAWN TREOS, Wyatt, Santa Barbara, CA, USA).
4.7. Specificity Testing
4.7.1. Dot Blot
Bet v 1 (0.5, 1, and 5 µg), BLG (1 µg), and isotype antibodies (1 µg; human plasma IgE, human myeloma IgG1 and human myeloma IgG4, cat# 16-16-090705, 16-16-090707-1M, and 16-16-090707-4M, respectively, Athens Research & Technology, Athens, GA, USA) were spotted onto a nitrocellulose membrane (0.2 µm nitrocellulose, cat# 10600001, GE Healthcare Life Sciences, Marlborough, MA, USA) in duplicate and dried overnight. Membrane was cut into strips and blocked with wash buffer (PBS with 0.1% Tween 20) supplemented with 5% skimmed milk powder (Heirler Genovis GmbH, Radolfzell am Bodensee) for 1 h. The membrane was washed with wash buffer three times and incubated either with antibody diluted with wash buffer to a final concentration of 1 µg/mL (i.e., M0418 IgE, IgG1, IgG4, or the respective isotype control) or wash buffer only for 1 h. Strips were briefly rinsed with wash buffer, three times, and then washed for 5 min. The strips were then incubated with the respective HRP-labelled detection antibodies diluted with wash buffer (anti-hu-IgE, 1:6000, cat# A18793, Invitrogen; anti-hu-IgG1, 1:6000, cat# 9054-05, Southern Biotech, Birmingham, AL, USA; anti-hu-IgG4, 1:10 000, cat# 9200-05, Southern Biotech, Birmingham, AL, USA, respectively) for another 1 h. Strips were again briefly rinsed, three times and then washed for another 10 min. For signal detection, Clarity Max Western ECL substrate (cat# 1705062, Bio-Rad Laboratories, Hercules, CA, USA) was prepared and then added to the membrane according to the manufacturer’s instructions. Membrane pictures were acquired using a ChemiDocTM Touch Imaging System (Bio-Rad Laboratories, Hercules, CA, USA). All incubation and wash steps were carried out at room temperature.
4.7.2. ImmunoCAP ISAC112
In addition to dot blot and ELISA, M0418 IgE specificity was tested using the ImmunoCAP ISAC112 microarray (Thermo Fisher Scientific, Waltham, MA, USA), strictly following the manufacturer’s instructions. A total of 1 µg M0418 IgE was used in the test, diluted with non-allergic serum.
4.7.3. ELISA
Maxisorp 96-well plates (Thermo Fisher Scientific, Waltham, MA, USA) were coated with 0.3 µg of Bet v 1 or BLG (100 µL/well; concentration of 3 µg/mL) in bicarbonate buffer, pH 9.6, overnight at 4 °C. Liquid was removed and the plate washed with wash buffer (Tris-Buffered saline, TBS with 0.05% Tween 20). Plate was then blocked with 200 µL/well wash buffer with 1% BSA for 2 h. Plate was then washed twice and sample or control antibodies (either M0418 IgE, IgG1, or IgG4 and the respective myeloma isotype controls, see Section 4.7.1 above) diluted in wash buffer were added: Either at a final concentration of 1 µg/mL (100 µL per well; added to Bet v 1 and BLG-coated plate wells) or in a 1:2 dilution series starting with 1 µg/mL to 0.03 µg/mL (M0418 antibodies only; Bet v 1-coated wells only). After 1 h of incubation, plate was washed with wash buffer three times and incubated with the respective HRP-labelled detection antibodies (anti-hu-IgE, 1:6000, cat# A18793, Invitrogen, Carlsbad, CA, USA; anti-hu-IgG1, 1:1000, cat# A10648, Invitrogen, Carlsbad, CA, USA; anti-hu-IgG4, 1:24,000, cat# 9200-05, Southern Biotech, Birmingham, AL, USA, respectively) for another 1 h. The plate was again washed three times and 100 µL TMB/well (cat# 00-4201-56, Invitrogen, Carlsbad, CA, USA) was added. The reaction was stopped with 100 µL/well 1 M H2SO4 and absorbance at 450 nm as well as 630 nm (reference wavelength) was measured using an Infinite 200M PRO plate reader (Tecan, Männerdorf, Switzerland). All of the incubation and wash steps were carried out at room temperature, unless stated otherwise. Each condition was tested in triplicate and each experiment was repeated at least three times. The values corrected by the reference wavelength were used for analysis. The limit of detection (LOD) was defined as the mean of the blank plus three times the corresponding standard deviation. Raw values below the LOD were excluded from the analysis. Measurements from each well were blank-corrected and the mean values of replicate wells of each experiment were calculated with Excel 365 (Microsoft Corporation, Albuquerque, NM, USA).
4.8. Quantitative Plasma IgE Blocking ELISA
Maxisorp 96-well plates (Thermo Fisher Scientific, Waltham, MA, USA) were coated with 0.17 µg Bet v 1/well in coating buffer (bicarbonate buffer, pH 9.6) overnight at 4 °C. The plates were washed with wash buffer (TBS with 0.05% Tween 20) once and then blocked with wash buffer supplemented with 1% BSA for 2 h at room temperature. Sample wells (as opposed to standard row wells, which were blocked further) were washed with wash buffer four times and incubated with 10, 100, or 1000 nM of M0418 IgG1 or IgG4, 1000 nM of the respective isotype control (human myeloma IgG1 and IgG4, cat# 16-16-090707-1M and 16-16-090707-4M, respectively, Athens Research & Technology, Athens, GA, USA; stock concentration was determined via UV/VIS spectroscopy prior to preparing the working solution) or wash buffer, respectively, for 1 h. The plate was washed four times and 100 µL of 1:2 diluted plasma samples from birch pollen allergic individuals was added to sample wells. A 1:2 dilution series of M0418 IgE was prepared in the standard row wells, ranging from 0.5 nM to 0.007 nM and 3 nM to 0.05 nM, respectively; matching the expected IgE levels in the plasma samples). After 1.5 h of incubation, the plate was washed four times and HRP-labelled detection antibody added (anti-hu-IgE, 1:6000, cat# A18793, Invitrogen, Carlsbad, CA, USA). After another 1 h of incubation wash step was repeated and 100 µL TMB/well (cat# 00-4201-56, Invitrogen, Carlsbad, CA, USA) were added. Reaction was stopped with 100 µL of 1 M H2SO4 and absorbance at 450 nm and 630 nm (reference wavelength) was measured using an Infinite 200M PRO plate reader (Tecan, Männerdorf, Switzerland). IgE in plasma of four non-allergic donors was quantified in the same manner, but without the addition of any blocking antibodies. All of the incubation and wash steps were carried out at room temperature, unless stated otherwise. Each condition was tested in duplicate and each experiment was repeated at least three times. Reference-Wavelength corrected values were used for analysis. Values below the LOD were excluded from the analysis. Measurements from each well were corrected by the blank and duplicate means were calculated with Excel 365 (Microsoft Corporation, Albuquerque, NM, USA). Further statistical analysis was carried out with GraphPad Prism version 8 (GraphPad Software LLC, San Diego, CA, USA).
4.9. In Vitro Experiments
4.9.1. FcεRI Upregulation of LAD2 Cells
A total of 1.5 x 105 LAD2 cells (500 µL of 3 x 105 cells/mL suspension) was incubated with 0.5, 5, or 50 nM of M0418 IgE, 6 ng/mL of IL-4 (positive control) or medium, respectively, in a 24-well plate. The cells were harvested after four days of incubation at 37 °C, 5% CO2 and washed with PBS twice. Cells were then stained with Zombie Violet (1:500 final; cat# 423114, Biolegend, San Diego, CA, USA) for 15 min., followed by co-incubation with APC-Cy7-labelled anti-FcεRI antibody (cat# 334632, clone# AER-37 (CRA-1), Biolegend, San Diego, CA, USA) or the respective isotype control antibody (IgG2b, kappa, mouse; cat# 400328, clone# MPC-11, Biolegend, San Diego, CA, USA) for another 15 min. The cells were then washed with Hank’s Balanced Salt Solution (HBSS; Gibco, Thermo Fisher Scientific, Waltham, MA, USA), centrifuged at 300 x g for 5 min., and then re-suspended in 200 µL of HBSS for flow cytometric analysis. 10,000 events were acquired using a FACSCanto II (BD Biosciences, Franklin Lakes, NJ, USA) and further analysis was carried out in FlowJoTM version 10 (FlowJo LCC, Ashland, OR, USA) and Graphpad Prism version 8 (GraphPad Software LLC, San Diego, CA, USA).
4.9.2. Internalisation of Labelled Bet v 1
U937 cells were primed with 10 ng/mL IL-4 for 48 h, twice, to upregulate CD23, as previously described [55]. Expression of CD23 and FcεRI was determined via flow cytometry while using the same staining protocol as in Section 4.9.1 but the following antibody panel: APC-Cy7-labelled anti-FcεRI antibody (cat# 334632, clone# AER-37 (CRA-1), Biolegend, San Diego, CA, USA), APC-labelled anti-CD23 antibody(cat# 338513, clone# EBVCS-5, Biolegend, San Diego, CA, USA), isotype control antibodies: APC-Cy7-labelled IgG2b, kappa, mouse (cat# 400328, clone# MPC-11, Biolegend, San Diego, CA, USA), and APC labelled IgG1, kappa, mouse (cat# 17-4714-42, clone#P3.6.2.8.1, eBioscience). Compensation was carried out with compensation beads (UltraComp eBeadsTM, cat# 01-2222-42, Thermo Fisher Scientific, Waltham, MA, USA). LAD2 cells were included as a FcεRI-positive, CD23-negative staining control. For the determination of Bet v 1 internalisation, U937 cells and RPMI-8866 cells were harvested and washed with assay buffer (HBSS supplemented with 1% FBS) once. 100 µL containing 1 x 105 cells were seeded into FACS tubes and 5 µg of M0418 IgE or isotype control IgE (NIP IgE, cloned in pVitro1, as previously described [23]) was added. One set of cells was left untreated. After 30 min. of incubation at 37 °C, cells were washed with assay buffer and 500 nM of labelled Bet v 1, diluted in assay buffer was added with or without additional crosslinker (polyclonal rabbit anti-IgE at a final concentration of 1.5 µg/sample; cat# A0094, Dako, Glostrup, Denmark). The cells were incubated with allergen for 1 h on ice, in the dark. Cells were again washed with assay buffer, re-suspended in 300 µL of medium supplemented with 2% FBS and incubated overnight at 37 °C, 5% CO2. After 16–18 h of incubation, the cells were washed and re-suspended in 200 µL of assay buffer with DAPI (final concentration: 3 µM; cat# 422801, Biolegend, San Diego, CA, USA) and subjected to flow cytometric analysis. 10,000 events were acquired using a FACSCanto II (BD Biosciences, Franklin Lakes, NJ, USA) and further analysis was carried out in FlowJoTM version 10 (FlowJo LLC, Ashland, OR, USA) and GraphPad Prism version 8 (GraphPad Software LLC, San Diego, CA, USA).
4.9.3. RBL-SX38 Degranulation Assay and Blocking of Degranulation
To determine cell degranulation, β-hexosaminidase release of RBL-SX38 cells was measured, as described previously [25]. Briefly, 1 × 104 RBL-SX38 cells (in 100 µL medium) were seeded into the wells of a flat-bottom 96 well cell culture plate (Thermo Fisher Scientific, Waltham, MA, USA, Waltham, MA, USA) and incubated overnight to attach to the plate. The cells were briefly washed with assay buffer (HBSS supplemented with 1% of FBS) and sensitised with 5 nM of M0418 IgE or isotype control IgE (NIP IgE, cloned in pVitro1 as previously described [23]) for 2–3 h. Cells were washed three times with 200 µL assay buffer per well and stimulated with 5–500 nM of Bet v 1, 1.5 µg/mL polyclonal rabbit Anti-IgE (positive control; cat# A0094, Dako, Glostrup, Denmark) or buffer only (negative control) in a volume of 100 µL. Furthermore, a Triton-X control was included as a reference (100% degranulation). Cells were stimulated for 45 min. and assay plate was then centrifuged at 400 x g for 5 min. at 4 °C. 25 µL supernatant were transferred to a black 96 well plate (FluoroNuncTM, cat# 437796, Thermo Fisher Scientific, Waltham, MA, USA, Waltham, MA, USA). The samples were diluted with 25 μL of assay buffer and 50 µL of fluorogenic substrate (1 mM 4-methylumbelliferyl N-acetyl-b-D-glucosaminide in 100 mM citrate buffer, pH 4.5, with 0.1% DMSO) were added. The plate was incubated at 37 °C for 1 h in the dark and the reaction quenched with 100 μL of 0.5 M Trizma® base (cat#6066, Sigma-Aldrich, St. Louis, MO, USA), pH 8.2, per well and fluorescence signal was measured using a FLUOstar Omega Microplate Reader (BMG Labtech, Ortenberg, Germany); excitation at 350 nM, emission at 450 nm. In the case of blocking experiments, stimulation was carried out with 400 nM of Bet v 1 with or without 1, 10, or 100 nM of M0418 IgG1 or IgG4, respectively. All of the measurements were made in quadruplicates and experiments were repeated at least three times. The signal gain was adjusted to 90% for the well giving the highest signal (Triton-X-lysed cells). The limit of detection (LOD) was defined as the mean of the respective blank (regular buffer vs. Triton-X) plus three times the corresponding standard deviation. Raw values that were below the limit of detection were eliminated from the analysis. All other values were then blank-corrected, normalised to the 100% degranulation control (Triton-X lysed cells) and their mean values were used for further statistical analysis. All of the incubation steps were carried out at 37 °C, 5% CO2, if not indicated otherwise. Basic calculations were carried out in Excel 365 (Microsoft Corporation, Albuquerque, NM, USA) and further statistical analysis was done in GraphPad Prism version 8 (GraphPad Software LLC, San Diego, CA, USA).
4.10. Patient Samples
Plasma samples from birch pollen-allergic individuals were obtained with their informed consent. Experiments with blood samples were in accordance with the Helsinki declaration of 1975 and they were approved by the institutional ethics committee of the Medical University of Vienna (EK number 2007/2016).
4.11. Statistical Analysis
For outlier identification, ROUT analysis (Q = 1%) was carried out. Data were then analysed via repeated measures one-way or two-way ANOVA or a corresponding mixed-effects model, as appropriate, followed by Dunnett’s multiple comparisons test. All of the statistical analyses were carried out using GraphPad Prism version 8 (GraphPad Software LLC, San Diego, CA, USA).
5. Conclusions
Overall, we present, for the first time, the rapid PIPE cloning of fully human, variable region-matched recombinant IgE, IgG1, and IgG4 antibodies specific to Bet v 1. Our IgE engaged with high and low affinity FcεRs and stimulated FcεRI upregulation in culture, while our IgG antibodies effectively inhibited IgE-mediated degranulation in vitro. Therefore, these basic tools may help to systematically investigate Fc-mediated effects of IgE versus IgG antibodies in allergy or tolerance at a cellular level. Future studies may prove our IgG1 and IgG4 to be useful candidates for passive immunotherapy of birch pollen allergy.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/16/5693/s1.
Author Contributions
Conceptualization, V.K.K. and E.J.-J.; methodology, V.K.K., S.C., J.F.-S., H.J.B., G.H., R.B., K.H.; validation, V.K.K. and C.L.P.; formal analysis, V.K.K.; investigation, V.K.K., C.L.P., G.H., R.B.; resources, E.J.-J., W.K., G.H., K.H., H.J.B., S.C., S.N.K.; data curation, V.K.K.; writing—original draft preparation, V.K.K. and E.J.-J.; writing—review and editing, V.K.K., S.C., J.F.-S., H.J.B., C.L.P., G.H., K.H., R.B., S.F., W.K., S.N.K., E.J.-J.; visualization, V.K.K.; supervision, E.J.-J., S.F., W.K., S.N.K.; project administration, E.J.-J.; funding acquisition, E.J.-J., S.N.K., S.F., W.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Austrian Science Fund (FWF) grants MCCA W1248-B30 and SFB F4606-B28 to E.J.-J., by grant SFB-F4604 to W.K., and by grant SFB-F4607 to S.F. V.K.K. received an EFIS-IL short-term research fellowship of the European Federation of Immunological Societies (EFIS) for a research visit at King’s College London. G.H. received financial support by the “Förderung Wissenschaftlicher Nachwuchs” program of the University of Graz. The research was supported by the National Institute for Health Research (NIHR) Biomedical Research Centre (BRC) based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London (IS-BRC-1215-20006). The authors acknowledge support by the Medical Research Council (MR/L023091/1); Breast Cancer Now (147; KCL-BCN-Q3), working in partnership with Walk the Walk; Cancer Research UK (C30122/A11527; C30122/A15774); Cancer Research UK King’s Health Partners Centre at King’s College London (C604/A25135); The Guy’s and St Thomas’s Foundation Trust Charity Melanoma Special Fund; CRUK/NIHR in England/DoH for Scotland, Wales and Northern Ireland Experimental Cancer Medicine Centre (C10355/A15587). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. Open Access Funding by the Austrian Science Fund (FWF).
Acknowledgments
We acknowledge the Austrian Science fund (FWF), European Federation of Immunological Societies (EFIS) and “Förderung Wissenschaftlicher Nachwuchs” program at the University of Graz for their financial support. We acknowledge the Biomedical Research Centre Immune Monitoring Core Facility team at Guy’s and St Thomas’ NHS Foundation Trust. We acknowledge Gerlinde Hofstetter and Mira Matz for their assistance with rBet v 1 production and antibody purification, respectively. We furthermore acknowledge Judith Wortmann, Mano Nakamura and Gabriel Osborn for practical assistance in the respective labs and fruitful discussions. Open Access Funding by the Austrian Science Fund (FWF).
Conflicts of Interest
E.J.-J. is founder and shareholder of Biomedical Int. R + D and business partner of Bencard Allergie, Germany as well as AllergyTherapeutics, UK, but has no COI in relation to the presented work. S.N.K. is founder and shareholder of Epsilogen Ltd. and holds patents on IgE antibodies for cancer therapy. H.J.B. is employed through a fund provided by Epsilogen Ltd. The other authors have no potential conflicts of interest to disclose. Furthermore, the funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Abbreviations
| AIT | Allergen immunotherapy |
| Bet v 1 | Major birch pollen allergen from Betula verrucosa |
| BLG | β-lactoglobulin |
| CD | circular dichroism |
| CD23 | Cluster of differentiation 23, the low-affinity Fc epsilon receptor II |
| D. I. T. | Digital integration time |
| DMSO | Dimethyl sulphoxide |
| ELISA | Enzyme-linked immunosorbent assay |
| FAP | Facilitated antigen presentation |
| FBS | Fetal bovine serum |
| FcεR | Fc epsilon receptor |
| FSC | Forward scatter |
| GMFI | Geometric mean fluorescence intensity |
| HBSS | Hank’s Balanced Salt Solution |
| Ig | Immunoglobulin |
| ISAC | Immuno-Solid phase Allergy Chip |
| kDa | Kilo Dalton |
| IL | Interleukin |
| LAD | Laboratory of allergic diseases |
| PCR | Polymerase chain reaction |
| PBS | Phosphate-buffered saline |
| PIPE | Polymerase incomplete primer extension |
| RBL | Rat basophilic leukaemia |
| rhSCF | Recombinant human stem cell factor |
| SDS-PAGE | Sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| SEC-MALS | Size exclusion chromatography with multi-angle light scattering |
| SSC | Side scatter |
| TBS | Tris-buffered saline |
Appendix A

Table A1.
Comparison of Key Parameters of Bet v 1 Inhibition ELISAs.
Table A1.
Comparison of Key Parameters of Bet v 1 Inhibition ELISAs.
| Blocking | Immobilised Bet v 1 | Amount of Blocking Protein | Serum | Inhibition |
|---|---|---|---|---|
| Protein | (pmol/well) | (pmol/well) | Dilution | (%) |
| M0418 IgG | 10 | 1–100 | 1:2 | 20–50 a |
| H3-1 ScFv [42] | 5.8 | 71.4 | 1:5 | 45 b |
| 5B4 IgG1 [43] | 14.7 | 0.007–130 c | 1:16 d | 20–50 c |
a Effective blocking with 100 pmol/well; IgG1 performed better than IgG4. b Mean of 30 sera; inhibition ranging from 0–62.9%. c Estimated from graph; effective blocking with ≥0.5 pmol/well. d Serum pool of 16 donors, diluted and co-incubated with blocking protein.

Table A2.
Nucleotide sequences of M0418 IgE, IgG1 and IgG4 light and heavy chains.
Table A2.
Nucleotide sequences of M0418 IgE, IgG1 and IgG4 light and heavy chains.
| Light chain sequence: | ATGTTGCCATCACAACTCATTGGGTTTCTGCTGCTCTGGGTTCCAGCTAGCCGCGGTAGCTACGAGCTGACCCAGCCTCCTAGCGCCAGCGGCACCCCTGGCCAGAGAGTGACCATCAGCTGCAGCGGCAGCAGCAGCAACATCGGCGGCAACACCGTGAACTGGTACCAGCAGGTGCCCGGCACCGCCCCTAGACTGCTGATCTACAAGAACAACCAGAGACCCAGCGGCGTGCCCGACAGATTCAGCGGCAGCAAGAGCGGCACCAGCGCCAGCCTGGCCATCAGCGGCCTGAGAAGCGAGGACGAGGCCGACTACTACTGCGAGGCCTGGGACGGCGGCCTGAGAGGCGGCGTGTTCGGCGGCGGCACCAAGCTGACCGTGCTGGGCAAGCGTACGGTGGCGGCGCCATCTGTCTTCATCTTCCCGCCATCTGATGAGCAGTTGAAATCTGGAACTGCCTCTGTTGTGTGCCTGCTGAATAACTTCTATCCCAGAGAGGCCAAAGTACAGTGGAAGGTGGATAACGCCCTCCAATCGGGTAACTCCCAGGAGAGTGTCACAGAGCAGGACAGCAAGGACAGCACCTACAGCCTCAGCAGCACCCTGACGCTGAGCAAAGCAGACTACGAGAAACACAAAGTCTACGCCTGCGAAGTCACCCATCAGGGCCTGAGCTCGCCCGTCACAAAGAGCTTCAACAGGGGAGAGTGTTAG |
| Italic: human leader, | |
| bold: M0418 VL sequence, | |
| underlined: hu κ sequence, | |
| bold italic: stop codon | |
| IgE heavy chain sequence: | ATGGACTGGACCTGGAGGATCCTCTTCTTGGTGGCGGCCGCCACAGGCGCGCACTCCCAGGTGCAGCTGGTGCAGAGCGGCGCCGAGGTGAAGAAGCCCGGCGAGAGCCTGAAGATCAGCTGCAAGGGCAGCGAGTACAGCTTCCCCAACTACTGGATCGCCTGGGTGAGACAGATGCCCGGCAAGGGCCTGGAGTGGATGGGCATGATCTACCCCGGCGACAGCGACACCAGATACAGCCCCAGCTTCCAGGGCCAGGTGAACATCAGCGCCGACAAGAGCAGCAGAACCGCCTTCCTGGAGTGGAGCAGCCTGAAGGCCAGCGACAGCGCCACCTACTTCTGCGCCAGACTGGGCGGCCAGCTGTGGAACAGCTACTACTACTACTACTACATGGACGTGTGGGGCAAGGGCACCACCGTGACCGTGAGCAGCGCTAGCACACAGAGCCCATCCGTCTTCCCCTTGACCCGCTGCTGCAAAAACATTCCCTCCAATGCCACCTCCGTGACTCTGGGCTGCCTGGCCACGGGCTACTTCCCGGAGCCGGTGATGGTGACCTGGGACACAGGCTCCCTCAACGGGACAACTATGACCTTACCAGCCACCACCCTCACGCTCTCTGGTCACTATGCCACCATCAGCTTGCTGACCGTCTCGGGTGCGTGGGCCAAGCAGATGTTCACCTGCCGTGTGGCACACACTCCATCGTCCACAGACTGGGTCGACAACAAAACCTTCAGCGTCTGCTCCAGGGACTTCACCCCGCCCACCGTGAAGATCTTACAGTCGTCCTGCGACGGCGGCGGGCACTTCCCCCCGACCATCCAGCTCCTGTGCCTCGTCTCTGGGTACACCCCAGGGACTATCAACATCACCTGGCTGGAGGACGGGCAGGTCATGGACGTGGACTTGTCCACCGCCTCTACCACGCAGGAGGGTGAGCTGGCCTCCACACAAAGCGAGCTCACCCTCAGCCAGAAGCACTGGCTGTCAGACCGCACCTACACCTGCCAGGTCACCTATCAAGGTCACACCTTTGAGGACAGCACCAAGAAGTGTGCAGATTCCAACCCGAGAGGGGTGAGCGCCTACCTAAGCCGGCCCAGCCCGTTCGACCTGTTCATCCGCAAGTCGCCCACGATCACCTGTCTGGTGGTGGACCTGGCACCCAGCAAGGGGACCGTGAACCTGACCTGGTCCCGGGCCAGTGGGAAGCCTGTGAACCACTCCACCAGAAAGGAGGAGAAGCAGCGCAATGGCACGTTAACCGTCACGTCCACCCTGCCGGTGGGCACCCGAGACTGGATCGAGGGGGAGACCTACCAGTGCAGGGTGACCCACCCCCACCTGCCCAGGGCCCTCATGCGGTCCACGACCAAGACCAGCGGCCCGCGTGCTGCCCCGGAAGTCTATGCGTTTGCGACGCCGGAGTGGCCGGGGAGCCGGGACAAGCGCACCCTCGCCTGCCTGATCCAGAACTTCATGCCTGAGGACATCTCGGTGCAGTGGCTGCACAACGAGGTGCAGCTCCCGGACGCCCGGCACAGCACGACGCAGCCCCGCAAGACCAAGGGCTCCGGCTTCTTCGTCTTCAGCCGCCTGGAGGTGACCAGGGCCGAATGGGAGCAGAAAGATGAGTTCATCTGCCGTGCAGTCCATGAGGCAGCGAGCCCCTCACAGACCGTCCAGCGAGCGGTGTCTGTAAATCCCGGTAAATGA |
| Italic: human leader, | |
| bold: M0418 VH sequence, | |
| underlined: hu ε sequence, | |
| bold italic: stop codon | |
| IgG1 heavy chain sequence: | ATGGACTGGACCTGGAGGATCCTCTTCTTGGTGGCGGCCGCCACAGGCGCGCACTCCCAGGTGCAGCTGGTGCAGAGCGGCGCCGAGGTGAAGAAGCCCGGCGAGAGCCTGAAGATCAGCTGCAAGGGCAGCGAGTACAGCTTCCCCAACTACTGGATCGCCTGGGTGAGACAGATGCCCGGCAAGGGCCTGGAGTGGATGGGCATGATCTACCCCGGCGACAGCGACACCAGATACAGCCCCAGCTTCCAGGGCCAGGTGAACATCAGCGCCGACAAGAGCAGCAGAACCGCCTTCCTGGAGTGGAGCAGCCTGAAGGCCAGCGACAGCGCCACCTACTTCTGCGCCAGACTGGGCGGCCAGCTGTGGAACAGCTACTACTACTACTACTACATGGACGTGTGGGGCAAGGGCACCACCGTGACCGTGAGCAGCGCTAGCACCAAGGGCCCATCGGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGGCACAGCGGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGCGGCGTGCACACCTTCCCGGCTGTCCTACAGTCCTCAGGACTCTACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGCAGCTTGGGCACCCAGACCTACATCTGCAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTGACAAAACTCACACATGCCCACCGTGCCCAGCACCTGAACTCCTGGGGGGACCGTCAGTCTTCCTCTTCCCCCCAAAACCCAAGGACACCCTCATGATCTCCCGGACCCCTGAGGTCACATGCGTGGTGGTGGACGTGAGCCACGAAGACCCTGAGGTCAAGTTCAACTGGTACGTGGACGGCGTGGAGGTGCATAATGCCAAGACAAAGCCGCGGGAGGAGCAGTACAACAGCACGTACCGTGTGGTCAGCGTCCTCACCGTCCTGCACCAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCAACAAAGCCCTCCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAGAACCACAGGTGTACACCCTGCCCCCATCCCGGGATGAGCTGACCAAGAACCAGGTCAGCCTGACCTGCCTGGTCAAAGGCTTCTATCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAACTACAAGACCACGCCTCCCGTGCTGGACTCCGACGGCTCCTTCTTCCTCTACAGCAAGCTCACCGTGGACAAGAGCAGGTGGCAGCAGGGGAACGTCTTCTCATGCTCCGTGATGCATGAGGCTCTGCACAACCACTACACGCAGAAGAGCCTCTCCCTGTCTCCGGGTAAATGA |
| Italic: human leader, | |
| bold: M0418 VH sequence, | |
| underlined: hu γ1 sequence, | |
| bold italic: stop codon | |
| IgG4 heavy chain sequence | ATGGACTGGACCTGGAGGATCCTCTTCTTGGTGGCGGCCGCCACAGGCGCGCACTCCCAGGTGCAGCTGGTGCAGAGCGGCGCCGAGGTGAAGAAGCCCGGCGAGAGCCTGAAGATCAGCTGCAAGGGCAGCGAGTACAGCTTCCCCAACTACTGGATCGCCTGGGTGAGACAGATGCCCGGCAAGGGCCTGGAGTGGATGGGCATGATCTACCCCGGCGACAGCGACACCAGATACAGCCCCAGCTTCCAGGGCCAGGTGAACATCAGCGCCGACAAGAGCAGCAGAACCGCCTTCCTGGAGTGGAGCAGCCTGAAGGCCAGCGACAGCGCCACCTACTTCTGCGCCAGACTGGGCGGCCAGCTGTGGAACAGCTACTACTACTACTACTACATGGACGTGTGGGGCAAGGGCACCACCGTGACCGTGAGCAGCGCTAGCACCAAGGGCCCATCCGTCTTCCCCCTGGCGCCCTGCTCCAGGAGCACCTCCGAGAGCACAGCCGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGCGGCGTGCACACCTTCCCGGCTGTCCTACAGTCCTCAGGACTCTACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGCAGCTTGGGCACGAAGACCTACACCTGCAACGTAGATCACAAGCCCAGCAACACCAAGGTGGACAAGAGAGTTGAGTCCAAATATGGTCCCCCATGCCCATCATGCCCAGCACCTGAGTTCCTGGGGGGACCATCAGTCTTCCTGTTCCCCCCAAAACCCAAGGACACTCTCATGATCTCCCGGACCCCTGAGGTCACGTGCGTGGTGGTGGACGTGAGCCAGGAAGACCCCGAGGTCCAGTTCAACTGGTACGTGGATGGCGTGGAGGTGCATAATGCCAAGACAAAGCCGCGGGAGGAGCAGTTCAACAGCACGTACCGTGTGGTCAGCGTCCTCACCGTCCTGCACCAGGACTGGCTGAACGGCAAGGAGTACAAGTGCAAGGTCTCCAACAAAGGCCTCCCGTCCTCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAGAGCCACAGGTGTACACCCTGCCCCCATCCCAGGAGGAGATGACCAAGAACCAGGTCAGCCTGACCTGCCTGGTCAAAGGCTTCTACCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAACTACAAGACCACGCCTCCCGTGCTGGACTCCGACGGCTCCTTCTTCCTCTACAGCAGGCTAACCGTGGACAAGAGCAGGTGGCAGGAGGGGAATGTCTTCTCATGCTCCGTGATGCATGAGGCTCTGCACAACCACTACACACAGAAGAGCCTCTCCCTGTCTCTGGGTAAATGA |
| Italic: human leader, | |
| bold: M0418 VH sequence, | |
| underlined: hu γ4 sequence, | |
| bold italic: stop codon |
References
- Biedermann, T.; Winther, L.; Till, S.J.; Panzner, P.; Knulst, A.; Valovirta, E. Birch pollen allergy in Europe. Allergy 2019, 74. [Google Scholar] [CrossRef] [PubMed]
- Jarolim, E.; Rumpold, H.; Endler, A.T.; Ebner, H.; Breitenbach, M.; Scheiner, O.; Kraft, D. IgE and IgG antibodies of patients with allergy to birch pollen as tools to define the allergen profile of Betula verrucose *. Allergy 1989, 44, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Breiteneder, H.; Pettenburger, K.; Bito, A.; Valenta, R.; Kraft, D.; Rumpold, H.; Scheiner, O.; Breitenbach, M. The gene coding for the major birch pollen allergen Bet v 1, is highly homologous to a pea disease resistance response gene. EMBO J. 1989, 8, 1935–1938. [Google Scholar] [CrossRef] [PubMed]
- Jensen-Jarolim, E. Happy 25th birthday, Bet v 1! World Allergy Organ. J. 2014, 7, 1–3. [Google Scholar] [CrossRef]
- Spangfort, M.D.; Larsen, J.N.; Gajhede, M. Crystallization and preliminary X-ray investigation at 2.0 A resolution of Bet v 1, a birch pollen protein causing IgE-mediated allergy. Proteins Struct. Funct. Genet. 1996, 26, 358–360. [Google Scholar] [CrossRef]
- Gould, H.J.; Sutton, B.J. IgE in allergy and asthma today. Nat. Rev. Immunol. 2008, 8, 205–217. [Google Scholar] [CrossRef]
- Galli, S.J.; Tsai, M. IgE and mast cells in allergic disease. Nat. Med. 2013, 18, 693–704. [Google Scholar] [CrossRef]
- Kofler, S.; Ackaert, C.; Samonig, M.; Asam, C.; Briza, P.; Horejs-Hoeck, J.; Cabrele, C.; Ferreira, F.; Duschl, A.; Huber, C.; et al. Stabilization of the dimeric birch pollen allergen Bet v 1 impacts its immunological properties. J. Biol. Chem. 2014, 289, 540–551. [Google Scholar] [CrossRef]
- Pali-Schöll, I.; Jensen-Jarolim, E. The concept of allergen-associated molecular patterns (AAMP). Curr. Opin. Immunol. 2016, 42, 113–118. [Google Scholar] [CrossRef]
- Vrtala, S.; Fohr, M.; Campana, R.; Baumgartner, C.; Valent, P.; Valenta, R. Genetic engineering of trimers of hypoallergenic fragments of the major birch pollen allergen, Bet v 1, for allergy vaccination. Vaccine 2011, 29, 2140–2148. [Google Scholar] [CrossRef][Green Version]
- Rosenberg, A.S. Effects of protein aggregates: An immunologic perspective. AAPS J. 2006, 8, E501–E507. [Google Scholar] [CrossRef] [PubMed]
- Schöll, I.; Kalkura, N.; Shedziankova, Y.; Bergmann, A.; Verdino, P.; Knittelfelder, R.; Kopp, T.; Hantusch, B.; Betzel, C.; Dierks, K.; et al. Dimerization of the Major Birch Pollen Allergen Bet v 1 Is Important for its In Vivo IgE-Cross-Linking Potential in Mice. J. Immunol. 2005, 175, 6645–6650. [Google Scholar] [CrossRef] [PubMed]
- Acharya, M.; Borland, G.; Edkins, A.L.; MacLellan, L.M.; Matheson, J.; Ozanne, B.W.; Cushley, W. CD23/FcεRII: Molecular multi-tasking. Clin. Exp. Immunol. 2010, 162, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Sutton, B.J.; Davies, A.M. Structure and dynamics of IgE-receptor interactions: FcεRI and CD23/FcεRII. Immunol. Rev. 2015, 268, 222–235. [Google Scholar] [CrossRef] [PubMed]
- Conrad, D.H.; Ford, J.W.; Sturgill, J.L.; Gibb, D.R. CD23: An overlooked regulator of allergic disease. Curr. Allergy Asthma Rep. 2007, 7, 331–337. [Google Scholar] [CrossRef]
- Wahn, U.; Bachert, C.; Heinrich, J.; Richter, H.; Zielen, S. Real-world benefits of allergen immunotherapy for birch pollen-associated allergic rhinitis and asthma. Allergy 2019, 74, 594–604. [Google Scholar] [CrossRef]
- James, L.K.; Shamji, M.H.; Walker, S.M.; Wilson, D.R.; Wachholz, P.A.; Francis, J.N.; Jacobson, M.R.; Kimber, I.; Till, S.J.; Durham, S.R. Long-term tolerance after allergen immunotherapy is accompanied by selective persistence of blocking antibodies. J. Allergy Clin. Immunol. 2011, 127, 509–516.e5. [Google Scholar] [CrossRef]
- Akdis, C.A.; Akdis, M. Mechanisms of allergen-specific immunotherapy and immune tolerance to allergens. World Allergy Organ. J. 2015, 8. [Google Scholar] [CrossRef]
- Shamji, M.H.; Kappen, J.H.; Akdis, M.; Jensen-Jarolim, E.; Knol, E.F.; Kleine-Tebbe, J.; Bohle, B.; Chaker, A.M.; Till, S.J.; Valenta, R.; et al. Biomarkers for monitoring clinical efficacy of allergen immunotherapy for allergic rhinoconjunctivitis and allergic asthma: An EAACI Position Paper. Allergy Eur. J. Allergy Clin. Immunol. 2017, 72, 1156–1173. [Google Scholar] [CrossRef]
- Orengo, J.M.; Radin, A.R.; Kamat, V.; Badithe, A.; Ben, L.H.; Bennett, B.L.; Zhong, S.; Birchard, D.; Limnander, A.; Rafique, A.; et al. Treating cat allergy with monoclonal IgG antibodies that bind allergen and prevent IgE engagement. Nat. Commun. 2018, 9, 1421. [Google Scholar] [CrossRef]
- Davies, A.M.; Sutton, B.J. Human IgG4: A structural perspective. Immunol. Rev. 2015, 268, 139–159. [Google Scholar] [CrossRef] [PubMed]
- Aalberse, R.C.; Stapel, S.O.; Schuurman, J.; Rispens, T. Immunoglobulin G4: An odd antibody. Clin. Exp. Allergy 2009, 39, 469–477. [Google Scholar] [CrossRef] [PubMed]
- Dodev, T.S.; Karagiannis, P.; Gilbert, A.E.; Josephs, D.H.; Bowen, H.; James, L.K.; Bax, H.J.; Beavil, R.; Pang, M.O.; Gould, H.J.; et al. A tool kit for rapid cloning and expression of recombinant antibodies. Sci. Rep. 2014, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kretschmer, A.; Schwanbeck, R.; Valerius, T.; Rösner, T. Antibody Isotypes for Tumor Immunotherapy. Transfus. Med. Hemother. 2017, 44, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Crescioli, S.; Chiaruttini, G.; Mele, S.; Ilieva, K.M.; Pellizzari, G.; Spencer, D.I.R.; Gardner, R.A.; Lacy, K.E.; Spicer, J.F.; Tutt, A.N.J.; et al. Engineering and stable production of recombinant IgE for cancer immunotherapy and AllergoOncology. J. Allergy Clin. Immunol. 2018, 141, 1519–1523.e9. [Google Scholar] [CrossRef] [PubMed]
- Fazekas-Singer, J.; Singer, J.; Ilieva, K.M.; Matz, M.; Herrmann, I.; Spillner, E.; Karagiannis, S.N.; Jensen-Jarolim, E. AllergoOncology: Generating a canine anticancer IgE against the epidermal growth factor receptor. J. Allergy Clin. Immunol. 2018, 142, 973–976.e11. [Google Scholar] [CrossRef] [PubMed]
- Levin, M.; Davies, A.M.; Liljekvist, M.; Carlsson, F.; Gould, H.J.; Sutton, B.J.; Ohlin, M. Human IgE against the major allergen Bet v 1 - defining an epitope with limited cross-reactivity between different PR-10 family proteins. Clin. Exp. Allergy 2014, 44, 288–299. [Google Scholar] [CrossRef]
- Swiss Institute of Bioinformatics ExPASy. ProtParam Tool. Available online: https://web.expasy.org/protparam/ (accessed on 11 September 2018).
- Subbarayal, B.; Schiller, D.; Möbs, C.; de Jong, N.W.; Ebner, C.; Reider, N.; Bartel, D.; Lidholm, J.; Pfützner, W.; Gerth van Wijk, R.; et al. Kinetics, cross-reactivity, and specificity of Bet v 1-specific IgG4 antibodies induced by immunotherapy with birch pollen. Allergy Eur. J. Allergy Clin. Immunol. 2013, 68, 1377–1386. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Lantz, C.S.; Oettgen, H.C.; Katona, I.M.; Fleming, T.; Miyajima, I.; Kinet, J.P.; Galli, S.J. IgE enhances mouse mast cell FcεRI expression in vitro and in vivo: Evidence for a novel amplification mechanism in IgE-dependent reactions. J. Exp. Med. 1997, 185, 663–672. [Google Scholar] [CrossRef]
- Kirshenbaum, A.S.; Akin, C.; Wu, Y.; Rottem, M.; Goff, J.P.; Beaven, M.A.; Rao, V.K.; Metcalfe, D.D. Characterization of novel stem cell factor responsive human mast cell lines LAD 1 and 2 established from a patient with mast cell sarcoma/leukemia; Activation following aggregation of FcεRI or FcγRI. Leuk. Res. 2003, 27, 677–682. [Google Scholar] [CrossRef]
- Toru, H.; Ra, C.; Nonoyama, S.; Suzuki, K.; Yata, J.-I.; Nakahata, T. Induction of the high-affinity IgE receptor (FcεRI) on human mast cells by IL-4. Int. Immunol. 1996, 8, 1367–1373. [Google Scholar] [PubMed]
- Boltz-Nitulescu, G.; Willheim, M.; Spittler, A.; Leutmezer, F.; Tempfer, C.; Winkler, S. Modulation of IgA, IgE, and IgG Fc receptor expression on human mononuclear phagocytes by 1α,25-dihydroxyvitamin D3 and cytokines. J. Leukoc. Biol. 1995, 58, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Dibbern, D.A.; Palmer, G.W.; Williams, P.B.; Bock, S.A.; Dreskin, S.C. RBL cells expressing human FcεRI are a sensitive tool for exploring functional IgE-allergen interactions: Studies with sera from peanut-sensitive patients. J. Immunol. Methods 2003, 274, 37–45. [Google Scholar] [CrossRef]
- Rådinger, M.; Jensen, B.M.; Swindle, E.; Gilfillan, A.M. Assay of mast cell mediators. In Mast Cells: Methods and Protocols; Humana Press: New York, NY, USA, 2014; pp. 307–323. ISBN 9781493915682. [Google Scholar]
- Dutta, U.; Cohenford, M.A.; Dain, J.A. Monitoring the effect of glucosamine and glyceraldehyde glycation on the secondary structure of human serum albumin and immunoglobulin G: An analysis based on circular dichroism, thermal melting profiles and UV-fluorescence spectroscopy. Anal. Chim. Acta 2006, 558, 187–194. [Google Scholar] [CrossRef]
- Doi, E.; Jirgensons, B. Circular Dichroism Studies on the Acid Denaturation of γ-Immunoglobulin G and Its Fragments. Biochemistry 1970, 9, 1066–1073. [Google Scholar] [CrossRef] [PubMed]
- Shade, K.-T.; Anthony, R. Antibody Glycosylation and Inflammation. Antibodies 2013, 2, 392–414. [Google Scholar] [CrossRef]
- Guhsl, E.E.; Hofstetter, G.; Lengger, N.; Hemmer, W.; Ebner, C.; Fröschl, R.; Bublin, M.; Lupinek, C.; Breiteneder, H.; Radauer, C. IgE, IgG4 and IgA specific to Bet v 1-related food allergens do not predict oral allergy syndrome. Allergy Eur. J. Allergy Clin. Immunol. 2015, 70, 59–66. [Google Scholar] [CrossRef]
- Huber, S.; Lang, R.; Steiner, M.; Aglas, L.; Ferreira, F.; Wallner, M.; Hawranek, T.; Gadermaier, G. Does clinical outcome of birch pollen immunotherapy relate to induction of blocking antibodies preventing IgE from allergen binding? A pilot study monitoring responses during first year of AIT. Clin. Transl. Allergy 2018, 8, 39. [Google Scholar] [CrossRef]
- Gieras, A.; Cejka, P.; Blatt, K.; Focke-Tejkl, M.; Linhart, B.; Flicker, S.; Stoecklinger, A.; Marth, K.; Drescher, A.; Thalhamer, J.; et al. Mapping of Conformational IgE Epitopes with Peptide-Specific Monoclonal Antibodies Reveals Simultaneous Binding of Different IgE Antibodies to a Surface Patch on the Major Birch Pollen Allergen, Bet v 1. J. Immunol. 2011, 186, 5333–5344. [Google Scholar] [CrossRef]
- Gadermaier, E.; Marth, K.; Lupinek, C.; Campana, R.; Hofer, G.; Blatt, K.; Smiljkovic, D.; Roder, U.; Focke-Tejkl, M.; Vrtala, S.; et al. Isolation of a high-affinity Bet v 1-specific IgG-derived ScFv from a subject vaccinated with hypoallergenic Bet v 1 fragments. Allergy Eur. J. Allergy Clin. Immunol. 2018, 73, 1425–1435. [Google Scholar] [CrossRef]
- Brier, S.; Le Mignon, M.; Jain, K.; Lebrun, C.; Peurois, F.; Kellenberger, C.; Bordas-Le Floch, V.; Mascarell, L.; Nony, E.; Moingeon, P. Characterization of epitope specificities of reference antibodies used for the quantification of the birch pollen allergen Bet v 1. Allergy Eur. J. Allergy Clin. Immunol. 2018, 73, 1032–1040. [Google Scholar] [CrossRef] [PubMed]
- Villazala-Merino, S.; Rodriguez-Dominguez, A.; Stanek, V.; Campion, N.J.; Gattinger, P.; Hofer, G.; Froeschl, R.; Fae, I.; Lupinek, C.; Vrtala, S.; et al. Allergen-specific IgE levels and the ability of IgE-allergen complexes to cross-link determine the extent of CD23-mediated T-cell activation. J. Allergy Clin. Immunol. 2020, 145, 958–967.e5. [Google Scholar] [CrossRef] [PubMed]
- Engeroff, P.; Fellmann, M.; Yerly, D.; Bachmann, M.F.; Vogel, M. A novel recycling mechanism of native IgE-antigen complexes in human B cells facilitates transfer of antigen to dendritic cells for antigen presentation. J. Allergy Clin. Immunol. 2018, 142, 557–568.e6. [Google Scholar] [CrossRef] [PubMed]
- Karagiannis, S.N.; Warrack, J.K.; Jennings, K.H.; Murdock, P.R.; Christie, G.; Moulder, K.; Sutton, B.J.; Gould, H.J. Endocytosis and recycling of the complex between CD23 and HLA-DR in human B cells. Immunology 2001, 103, 319–331. [Google Scholar] [CrossRef]
- Kitzmüller, C.; Nagl, B.; Deifl, S.; Walterskirchen, C.; Jahn-Schmid, B.; Zlabinger, G.J.; Bohle, B. Human blood basophils do not act as antigen-presenting cells for the major birch pollen allergen Bet v 1. Allergy Eur. J. Allergy Clin. Immunol. 2012, 67, 593–600. [Google Scholar] [CrossRef]
- Bucaite, G.; Kang-Pettinger, T.; Moreira, J.; Gould, H.J.; James, L.K.; Sutton, B.J.; McDonnell, J.M. Interplay between Affinity and Valency in Effector Cell Degranulation: A Model System with Polcalcin Allergens and Human Patient–Derived IgE Antibodies. J. Immunol. 2019, 203, 1693–1700. [Google Scholar] [CrossRef]
- Pranger, C.L.; Singer-Fazekas, J.; Köhler, V.K.; Pali-Schöll, I.; Fiocchi, A.; Karagiannis, S.N.; Zenarruzabeitia, O.; Borrego, F.; Jensen-Jarolim, E. PIPE-cloned human IgE, IgG1 and IgG4 antibodies: New tools for investigating cow’s milk allergy and tolerance. 2020. manuscript in preparation. [Google Scholar]
- Rouvinen, J.; Jänis, J.; Laukkanen, M.L.; Jylhä, S.; Niemi, M.; Päivinen, T.; Mäkinen-Kiljunen, S.; Haahtela, T.; Söderlund, H.; Takkinen, K. Transient dimers of allergens. PLoS ONE 2010, 5, e9037. [Google Scholar] [CrossRef]
- Hufnagl, K.; Moussa Afify, S.; Braun, N.; Wagner, S.; Wallner, M.; Hauser, M.; Wiederstein, M.; Gadermaier, G.; Wildner, S.; Redegeld, F.A.; et al. Retinoic acid-loading of the major birch pollen allergen Bet v 1 may improve specific allergen immunotherapy: In silico, in vitro and in vivo data in BALB/c mice. Allergy 2020. [Google Scholar] [CrossRef]
- Guhsl, E.E.; Hofstetter, G.; Hemmer, W.; Ebner, C.; Vieths, S.; Vogel, L.; Breiteneder, H.; Radauer, C. Vig r 6, the cytokinin-specific binding protein from mung bean (Vigna radiata) sprouts, cross-reacts with Bet v 1-related allergens and binds IgE from birch pollen allergic patients’ sera. Mol. Nutr. Food Res. 2014, 58, 625–634. [Google Scholar] [CrossRef]
- Stothard, P. The Sequence Manipulation Suite. Available online: https://www.bioinformatics.org/sms2/ (accessed on 9 November 2017).
- Ilieva, K.M.; Fazekas-Singer, J.; Achkova, D.Y.; Dodev, T.S.; Mele, S.; Crescioli, S.; Bax, H.J.; Cheung, A.; Karagiannis, P.; Correa, I.; et al. Functionally active Fc Mutant antibodies recognizing cancer antigens generated rapidly at high Yields. Front. Immunol. 2017, 8, 1112. [Google Scholar] [CrossRef] [PubMed]
- Josephs, D.H.; Bax, H.J.; Dodev, T.; Georgouli, M.; Nakamura, M.; Pellizzari, G.; Saul, L.; Karagiannis, P.; Cheung, A.; Herraiz, C.; et al. Anti-folate receptor-α IgE but not IgG recruits macrophages to attack tumors via TNFa/MCP-1 signaling. Cancer Res. 2017, 77, 1127–1141. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).