Isolation and Functional Characterization of MsFTa, a FLOWERING LOCUS T Homolog from Alfalfa (Medicago sativa)
Abstract
1. Introduction
2. Results
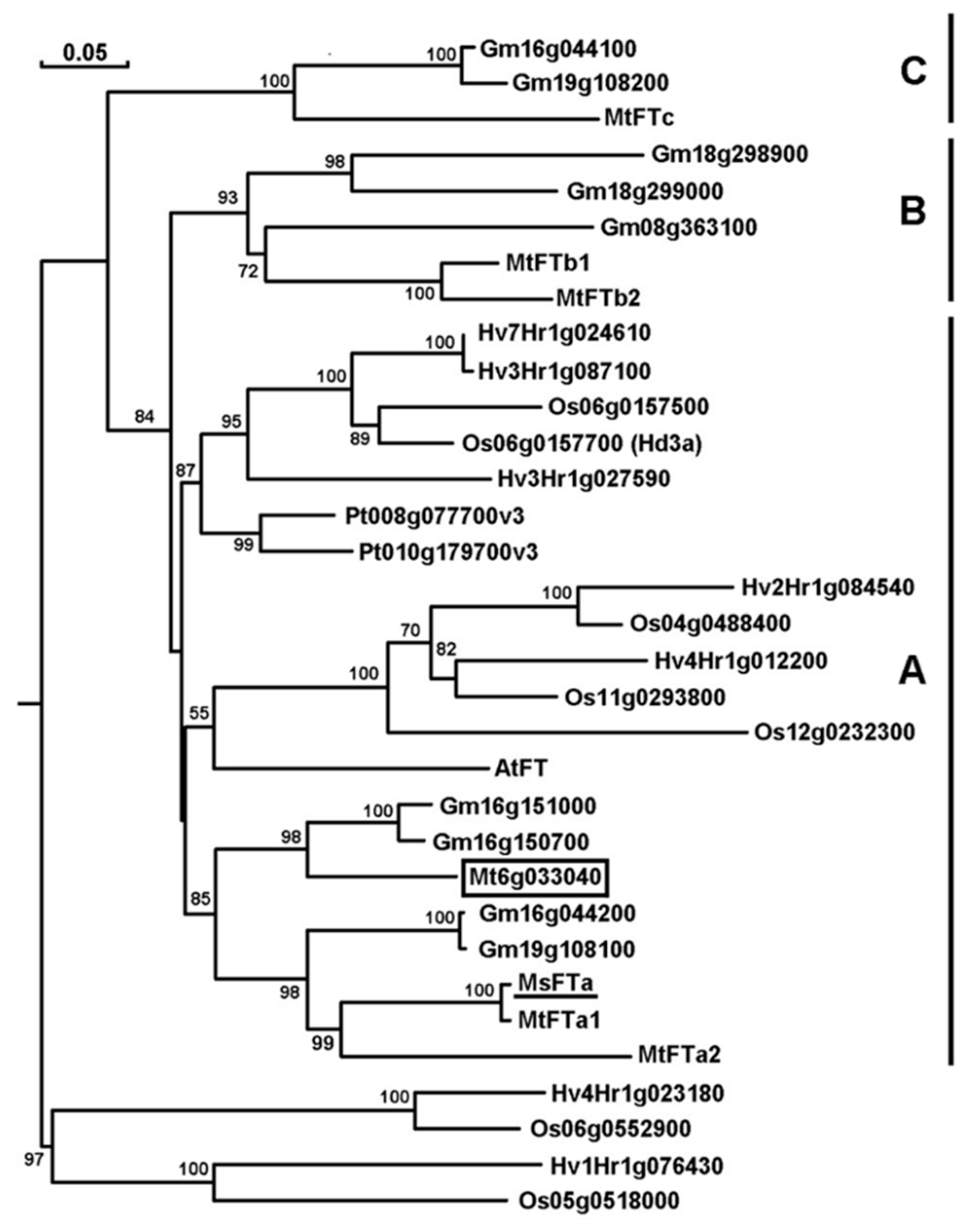
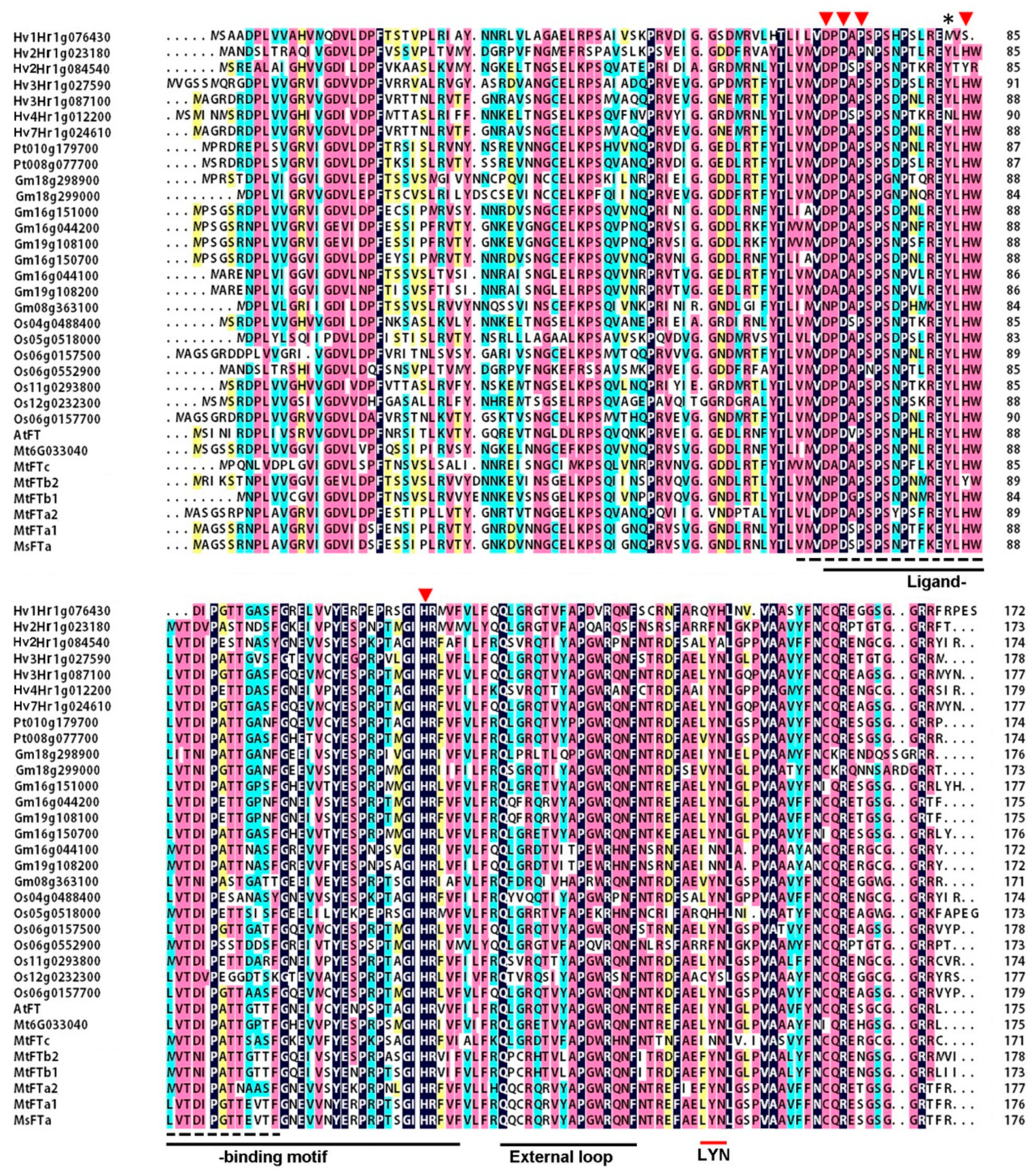
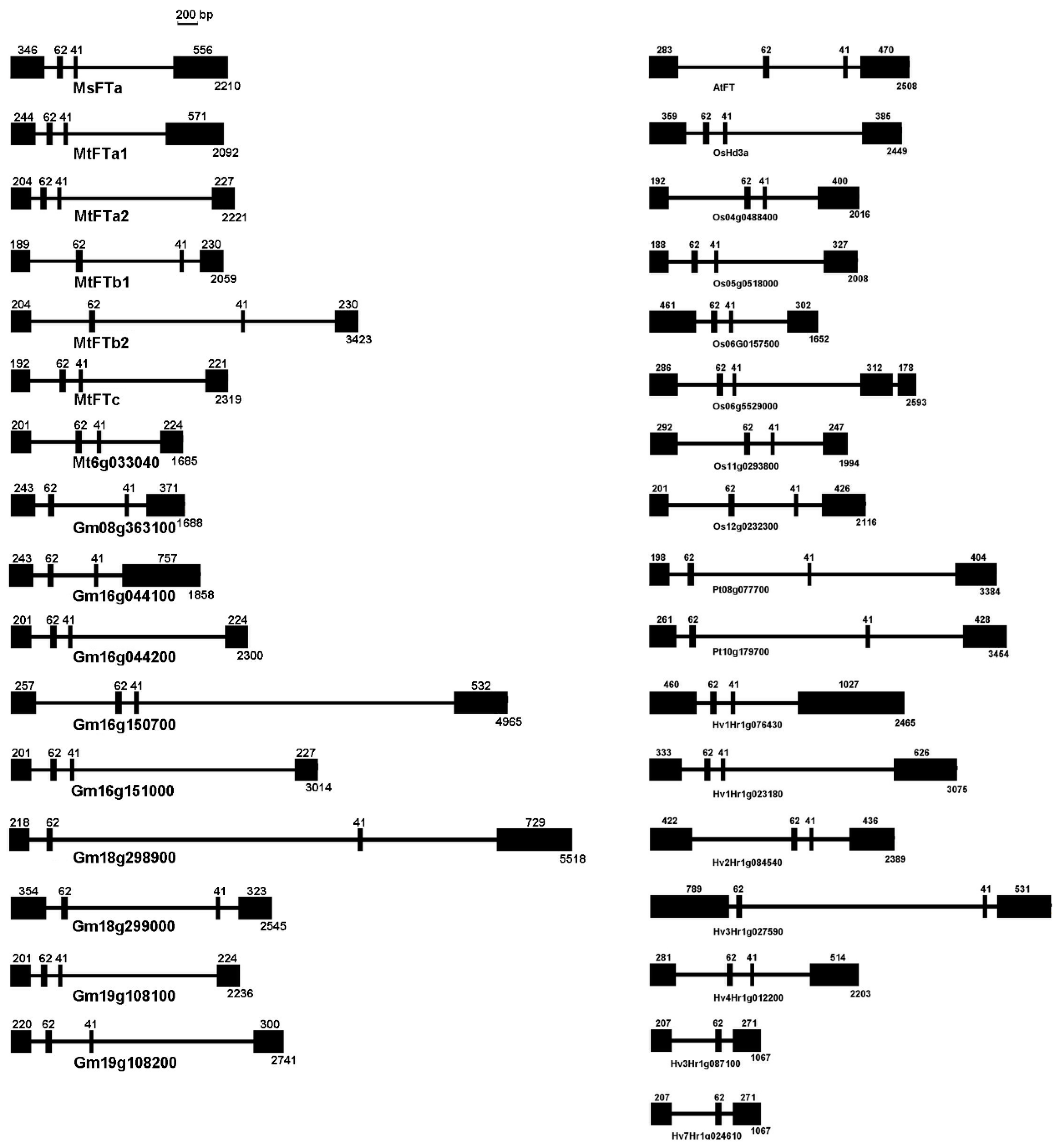
2.1. MsFTa Sequence Shared the Common Features of FT-like Genes
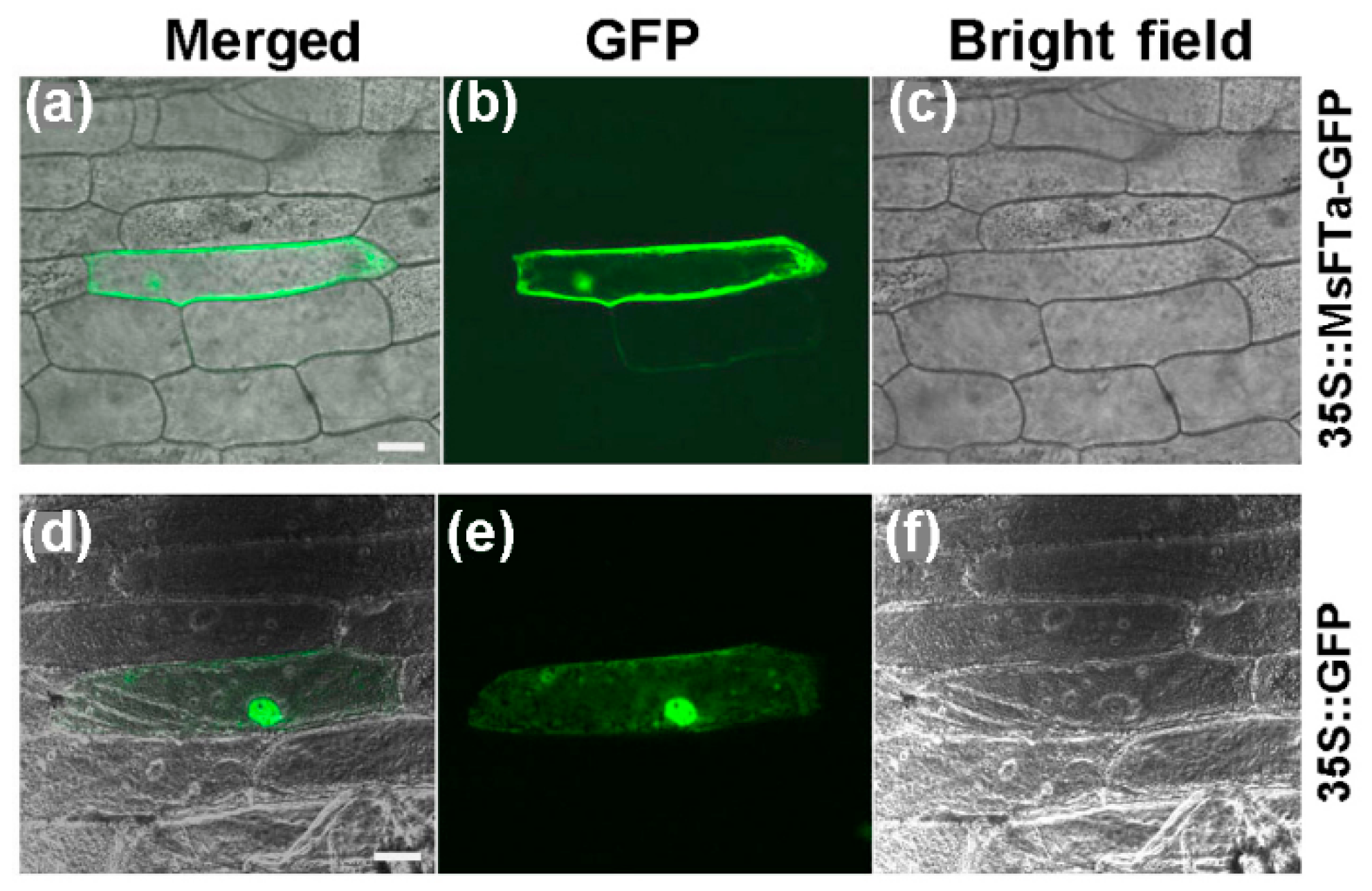
2.2. The MsFTa-GFP Recombinant Protein Resided in Both the Cytoplasm and Nucleus
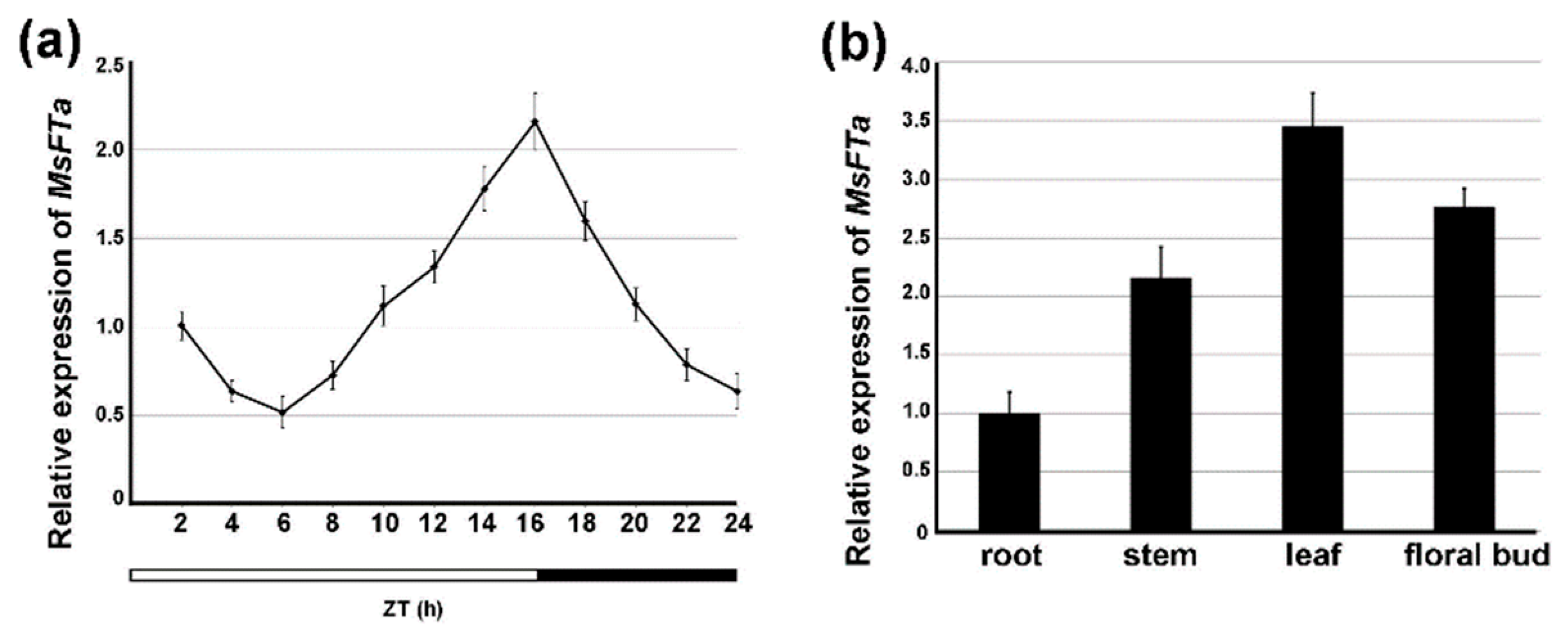
2.3. Diurnal and Tissue-Specific Expression Pattern of MsFTa
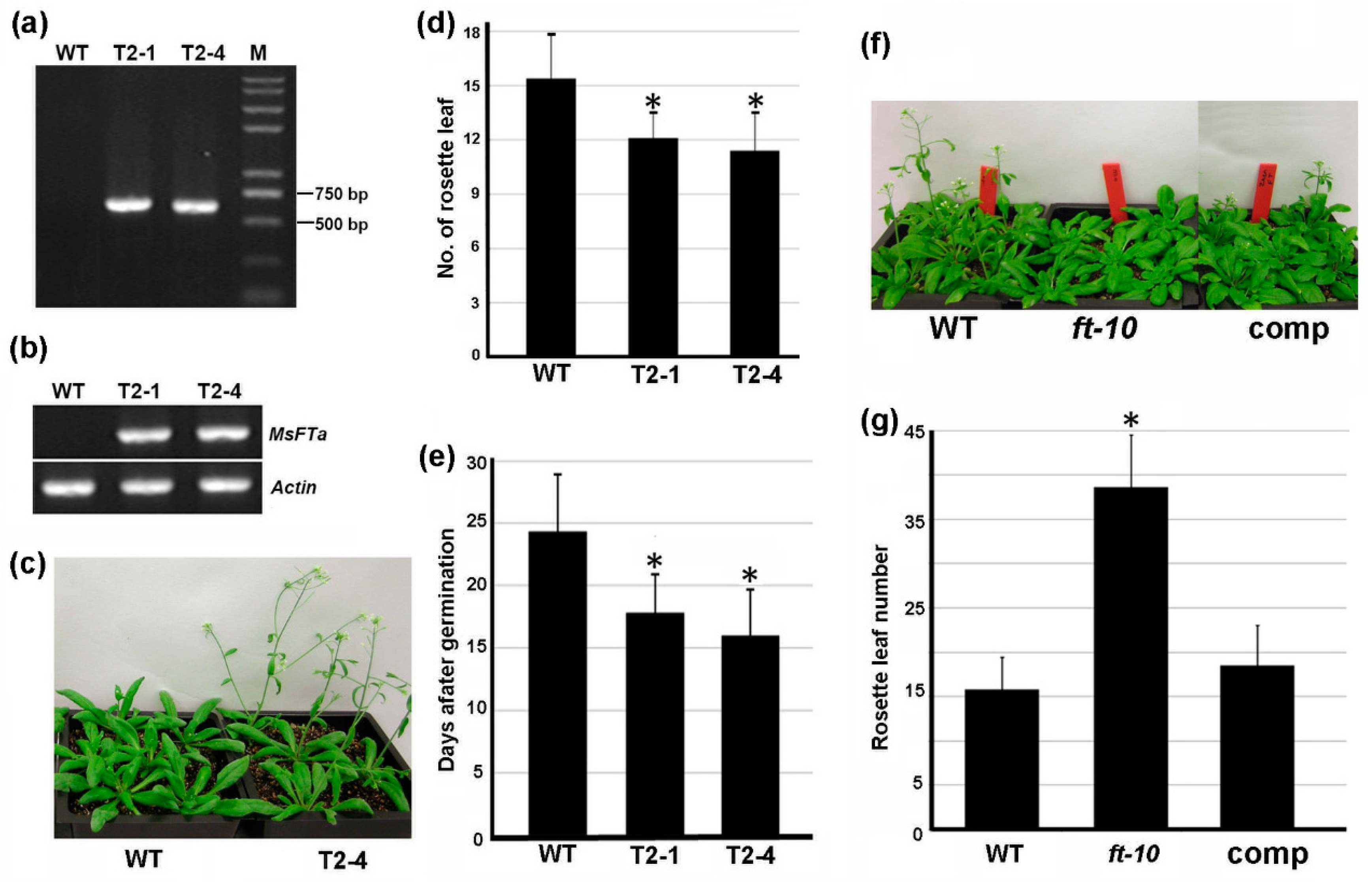
2.4. Ectopic Expression of MsFTa in Arabidopsis Strongly Accelerated Flowering
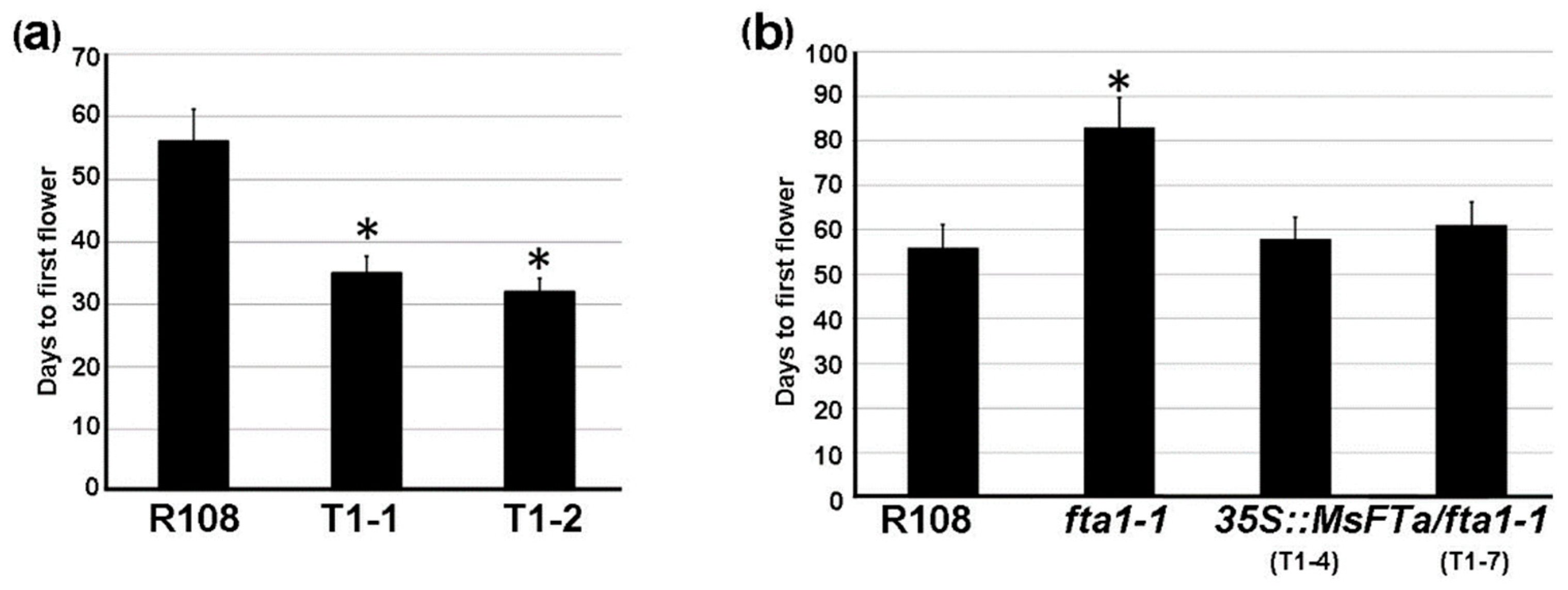
2.5. Ectopic Expression of MsFTa Promoted Flowering in Transgenic M. truncatula Plants
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Cloning of MsFTa from Alfalfa
4.3. Bioinformatic Analysis
4.4. Expression Analysis by RT-PCR
4.5. Subcellular Localization Analysis
4.6. Plasmid construct and plant transformation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Blazquez, M.A.; Green, R.; Nilsson, O.; Sussman, M.R.; Weigel, D. Gibberellins promote flowering of arabidopsis by activating the LEAFY promoter. Plant Cell 1998, 10, 791–800. [Google Scholar] [CrossRef]
- Huq, E.; Tepperman, J.M.; Quail, P.H. GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2000, 97, 9789–9794. [Google Scholar] [CrossRef] [PubMed]
- Okamuro, J.K.; Szeto, W.; Lotys-Prass, C.; Jofuku, K.D. Photo and hormonal control of meristem identity in the Arabidopsis flower mutants apetala2 and apetala1. Plant Cell 1997, 9, 37–47. [Google Scholar] [CrossRef]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Bohlenius, H.; Eriksson, S.; Parcy, F.; Nilsson, O. The mRNA of the Arabidopsis gene FT moves from leaf to shoot apex and induces flowering. Science 2005, 309, 1694–1696. [Google Scholar] [CrossRef] [PubMed]
- Putterill, J.; Robson, F.; Lee, K.; Simon, R.; Coupland, G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 1995, 80, 847–857. [Google Scholar] [CrossRef]
- Samach, A.; Onouchi, H.; Gold, S.E.; Ditta, G.S.; Schwarz-Sommer, Z.; Yanofsky, M.F.; Coupland, G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 2000, 288, 1613–1616. [Google Scholar] [CrossRef] [PubMed]
- Hecht, V.; Foucher, F.; Ferrandiz, C.; Macknight, R.; Navarro, C.; Morin, J.; Vardy, M.E.; Ellis, N.; Beltran, J.P.; Rameau, C.; et al. Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 2005, 137, 1420–1434. [Google Scholar] [CrossRef]
- Ruelens, P.; de Maagd, R.A.; Proost, S.; Theissen, G.; Geuten, K.; Kaufmann, K. FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nat. Commun. 2013, 4, 2280. [Google Scholar] [CrossRef]
- Wong, A.C.; Hecht, V.F.; Picard, K.; Diwadkar, P.; Laurie, R.E.; Wen, J.; Mysore, K.; Macknight, R.C.; Weller, J.L. Isolation and functional analysis of CONSTANS-LIKE genes suggests that a central role for CONSTANS in flowering time control is not evolutionarily conserved in Medicago truncatula. Front. Plant Sci. 2014, 5, 486. [Google Scholar] [CrossRef]
- Laurie, R.E.; Diwadkar, P.; Jaudal, M.; Zhang, L.; Hecht, V.; Wen, J.; Tadege, M.; Mysore, K.S.; Putterill, J.; Weller, J.L.; et al. The Medicago FLOWERING LOCUS T homolog, MtFTa1, is a key regulator of flowering time. Plant Physiol. 2011, 156, 2207–2224. [Google Scholar] [CrossRef] [PubMed]
- Tamaki, S.; Matsuo, S.; Wong, H.L.; Yokoi, S.; Shimamoto, K. Hd3a protein is a mobile flowering signal in rice. Science 2007, 316, 1033–1036. [Google Scholar] [CrossRef]
- Wang, Z.; Zhou, Z.; Liu, Y.; Liu, T.; Li, Q.; Ji, Y.; Li, C.; Fang, C.; Wang, M.; Wu, M.; et al. Functional evolution of phosphatidylethanolamine binding proteins in soybean and Arabidopsis. Plant Cell 2015, 27, 323–336. [Google Scholar] [CrossRef] [PubMed]
- Komiya, R.; Yokoi, S.; Shimamoto, K. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 2009, 136, 3443–3450. [Google Scholar] [CrossRef]
- Navarro, C.; Abelenda, J.A.; Cruz-Oro, E.; Cuellar, C.A.; Tamaki, S.; Silva, J.; Shimamoto, K.; Prat, S. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 2011, 478, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Niu, L.; Fu, C.; Lin, H.; Wolabu, T.W.; Wu, Y.; Wang, Z.Y.; Tadege, M. Control of floral transition in the bioenergy crop switchgrass. Plant Cell Environ. 2016, 39, 2158–2171. [Google Scholar] [CrossRef] [PubMed]
- Skot, L.; Sanderson, R.; Thomas, A.; Skot, K.; Thorogood, D.; Latypova, G.; Asp, T.; Armstead, I. Allelic variation in the perennial ryegrass FLOWERING LOCUS T gene is associated with changes in flowering time across a range of populations. Plant Physiol. 2011, 155, 1013–1022. [Google Scholar] [CrossRef]
- Hsu, C.Y.; Adams, J.P.; Kim, H.; No, K.; Ma, C.; Strauss, S.H.; Drnevich, J.; Vandervelde, L.; Ellis, J.D.; Rice, B.M.; et al. FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc. Natl. Acad. Sci. USA 2011, 108, 10756–10761. [Google Scholar] [CrossRef]
- Song, G.Q.; Walworth, A.; Zhao, D.; Jiang, N.; Hancock, J.F. The Vaccinium corymbosum FLOWERING LOCUS T-like gene (VcFT): A flowering activator reverses photoperiodic and chilling requirements in blueberry. Plant Cell Rep. 2013, 32, 1759–1769. [Google Scholar] [CrossRef]
- Trankner, C.; Lehmann, S.; Hoenicka, H.; Hanke, M.V.; Fladung, M.; Lenhardt, D.; Dunemann, F.; Gau, A.; Schlangen, K.; Malnoy, M.; et al. Over-expression of an FT-homologous gene of apple induces early flowering in annual and perennial plants. Planta 2010, 232, 1309–1324. [Google Scholar] [CrossRef]
- Hao, X.; Chao, W.; Yang, Y.; Horvath, D. Coordinated Expression of FLOWERING LOCUS T and DORMANCY ASSOCIATED MADS-BOX-Like Genes in Leafy Spurge. PLoS ONE 2015, 10, e0126030. [Google Scholar] [CrossRef]
- Yarur, A.; Soto, E.; Leon, G.; Almeida, A.M. The sweet cherry (Prunus avium) FLOWERING LOCUS T gene is expressed during floral bud determination and can promote flowering in a winter-annual Arabidopsis accession. Plant Reprod. 2016, 29, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Varkonyi-Gasic, E.; Moss, S.M.; Voogd, C.; Wang, T.; Putterill, J.; Hellens, R.P. Homologs of FT, CEN and FD respond to developmental and environmental signals affecting growth and flowering in the perennial vine kiwifruit. New Phytol. 2013, 198, 732–746. [Google Scholar] [CrossRef]
- Ahn, J.H.; Miller, D.; Winter, V.J.; Banfield, M.J.; Lee, J.H.; Yoo, S.Y.; Henz, S.R.; Brady, R.L.; Weigel, D. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 2006, 25, 605–614. [Google Scholar] [CrossRef]
- Hanzawa, Y.; Money, T.; Bradley, D. A single amino acid converts a repressor to an activator of flowering. Proc. Natl. Acad. Sci. USA 2005, 102, 7748–7753. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Luo, L.; Fu, Q.; Niu, L.; Xu, Z.F. Isolation and functional characterization of JcFT, a FLOWERING LOCUS T (FT) homologous gene from the biofuel plant Jatropha curcas. BMC Plant Biol. 2014, 14, 125. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.; Su, S.; Ma, L.; Wen, Y.; Wang, X. Molecular cloning and functional characterization of CoFT1, a homolog of FLOWERING LOCUS T (FT) from Camellia oleifera. Gene 2017, 626, 215–226. [Google Scholar] [CrossRef]
- Nelson, M.N.; Ksiazkiewicz, M.; Rychel, S.; Besharat, N.; Taylor, C.M.; Wyrwa, K.; Jost, R.; Erskine, W.; Cowling, W.A.; Berger, J.D.; et al. The loss of vernalization requirement in narrow-leafed lupin is associated with a deletion in the promoter and de-repressed expression of a Flowering Locus T (FT) homologue. New Phytol. 2017, 213, 220–232. [Google Scholar] [CrossRef]
- Li, Z.; Long, R.; Zhang, T.; Wang, Z.; Zhang, F.; Yang, Q.; Kang, J.; Sun, Y. Molecular cloning and functional analysis of the drought tolerance gene MsHSP70 from alfalfa (Medicago sativa L.). J. Plant Res. 2017, 130, 387–396. [Google Scholar] [CrossRef]
- Lifschitz, E.; Eshed, Y. Universal florigenic signals triggered by FT homologues regulate growth and flowering cycles in perennial day-neutral tomato. J. Exp. Bot. 2006, 57, 3405–3414. [Google Scholar] [CrossRef]
- Andres, F.; Coupland, G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012, 13, 627–639. [Google Scholar] [CrossRef]
- Ho, W.W.; Weigel, D. Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. Plant Cell 2014, 26, 552–564. [Google Scholar] [CrossRef]
- Abe, M.; Kobayashi, Y.; Yamamoto, S.; Daimon, Y.; Yamaguchi, A.; Ikeda, Y.; Ichinoki, H.; Notaguchi, M.; Goto, K.; Araki, T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 2005, 309, 1052–1056. [Google Scholar] [CrossRef] [PubMed]
- Kaneko-Suzuki, M.; Kurihara-Ishikawa, R.; Okushita-Terakawa, C.; Kojima, C.; Nagano-Fujiwara, M.; Ohki, I.; Tsuji, H.; Shimamoto, K.; Taoka, K.I. TFL1-Like Proteins in Rice Antagonize Rice FT-Like Protein in Inflorescence Development by Competition for Complex Formation with 14-3-3 and FD. Plant Cell Physiol. 2018, 59, 458–468. [Google Scholar] [CrossRef]
- Taoka, K.; Ohki, I.; Tsuji, H.; Furuita, K.; Hayashi, K.; Yanase, T.; Yamaguchi, M.; Nakashima, C.; Purwestri, Y.A.; Tamaki, S.; et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 2011, 476, 332–335. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, R.; Devisetty, U.K.; Maloof, J.N.; Zuo, Y.; Li, J.; Shen, Y.; Zhao, J.; Bao, M.; Ning, G. The Divergence of Flowering Time Modulated by FT/TFL1 Is Independent to Their Interaction and Binding Activities. Front. Plant Sci. 2017, 8, 697. [Google Scholar] [CrossRef]
- Pin, P.A.; Nilsson, O. The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ. 2012, 35, 1742–1755. [Google Scholar] [CrossRef]
- Voogd, C.; Brian, L.A.; Wang, T.; Allan, A.C.; Varkonyi-Gasic, E. Three FT and multiple CEN and BFT genes regulate maturity, flowering, and vegetative phenology in kiwifruit. J. Exp. Bot. 2017, 68, 1539–1553. [Google Scholar] [CrossRef] [PubMed]
- Wolabu, T.W.; Zhang, F.; Niu, L.; Kalve, S.; Bhatnagar-Mathur, P.; Muszynski, M.G.; Tadege, M. Three FLOWERING LOCUS T-like genes function as potential florigens and mediate photoperiod response in sorghum. New Phytol. 2016, 210, 946–959. [Google Scholar] [CrossRef] [PubMed]
- Jaudal, M.; Yeoh, C.C.; Zhang, L.; Stockum, C.; Mysore, K.S.; Ratet, P.; Putterill, J. Retroelement insertions at the Medicago FTa1 locus in spring mutants eliminate vernalisation but not long-day requirements for early flowering. Plant J. 2013, 76, 580–591. [Google Scholar] [CrossRef]
- Yeoh, C.C.; Balcerowicz, M.; Zhang, L.; Jaudal, M.; Brocard, L.; Ratet, P.; Putterill, J. Fine mapping links the FTa1 flowering time regulator to the dominant spring1 locus in Medicago. PLoS ONE 2013, 8, e53467. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lifschitz, E.; Eviatar, T.; Rozman, A.; Shalit, A.; Goldshmidt, A.; Amsellem, Z.; Alvarez, J.P.; Eshed, Y. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 2006, 103, 6398–6403. [Google Scholar] [CrossRef]
- Rinne, P.L.; Welling, A.; Vahala, J.; Ripel, L.; Ruonala, R.; Kangasjarvi, J.; van der Schoot, C. Chilling of dormant buds hyperinduces FLOWERING LOCUS T and recruits GA-inducible 1,3-beta-glucanases to reopen signal conduits and release dormancy in Populus. Plant Cell 2011, 23, 130–146. [Google Scholar] [CrossRef] [PubMed]
- Reig, C.; Gil-Munoz, F.; Vera-Sirera, F.; Garcia-Lorca, A.; Martinez-Fuentes, A.; Mesejo, C.; Perez-Amador, M.A.; Agusti, M. Bud sprouting and floral induction and expression of FT in loquat [Eriobotrya japonica (Thunb.) Lindl.]. Planta 2017, 246, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Cosson, V.; Durand, P.; d’Erfurth, I.; Kondorosi, A.; Ratet, P. Medicago truncatula transformation using leaf explants. Methods Mol. Biol. 2006, 343, 115–127. [Google Scholar] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, J.; Zhang, T.; Guo, T.; Ding, W.; Long, R.; Yang, Q.; Wang, Z. Isolation and Functional Characterization of MsFTa, a FLOWERING LOCUS T Homolog from Alfalfa (Medicago sativa). Int. J. Mol. Sci. 2019, 20, 1968. https://doi.org/10.3390/ijms20081968
Kang J, Zhang T, Guo T, Ding W, Long R, Yang Q, Wang Z. Isolation and Functional Characterization of MsFTa, a FLOWERING LOCUS T Homolog from Alfalfa (Medicago sativa). International Journal of Molecular Sciences. 2019; 20(8):1968. https://doi.org/10.3390/ijms20081968
Chicago/Turabian StyleKang, Junmei, Tiejun Zhang, Tao Guo, Wang Ding, Ruicai Long, Qingchuan Yang, and Zhen Wang. 2019. "Isolation and Functional Characterization of MsFTa, a FLOWERING LOCUS T Homolog from Alfalfa (Medicago sativa)" International Journal of Molecular Sciences 20, no. 8: 1968. https://doi.org/10.3390/ijms20081968
APA StyleKang, J., Zhang, T., Guo, T., Ding, W., Long, R., Yang, Q., & Wang, Z. (2019). Isolation and Functional Characterization of MsFTa, a FLOWERING LOCUS T Homolog from Alfalfa (Medicago sativa). International Journal of Molecular Sciences, 20(8), 1968. https://doi.org/10.3390/ijms20081968





