Abstract
Stearidonic acid (SDA; 18:4, n-3) is the delta 15-desaturase product of gamma linolenic acid (GLA; 18:3, n-6) and delta 6-desaturase product of alpha linolenic acid (ALA; 18:3, n-3). Construction of engineered oleaginous microbes have been attracting significant interest in producing SDA because of its nutritional value and pharmaceutical applications. Mucor circinelloides is a GLA producing filamentous fungus, which can be a useful tool to produce SDA. This study has, therefore, overexpressed the delta-15 desaturase (D15D) gene from Mortierella alpina in this fungus to construct a SDA-producing cell factory. To produce SDA in M. circinelloides, the homologous overexpression of D15D gene was analyzed. When the gene was overexpressed in M. circinelloides CBS 277.49, up to 5.0% SDA was accumulated in this strain. According to current knowledge, this is the first study describing the construction of a SDA-producing cell factory by overexpression of D15D gene in oleaginous fungus M. circinelloides. A new scope for further research has been established by this work to improve SDA production in this fungus, specifically in its high lipid-producing strain, WJ11.
1. Introduction
Polyunsaturated fatty acids (PUFAs) have significant functions in maintaining human health []. They are very important nutritionally because mammals, including humans, can synthesize saturated fatty acids (SAFAs) and mono-unsaturated fatty acids (MUFAs) but cannot produce linoleic acid (LA; 18:2, n-6) and α-linolenic acid (ALA; 18:3, n-3) that are known as de novo omega-6 or omega-3 polyunsaturated fatty acids (PUFAs), respectively. Hence, these fatty acids are called essential fatty acids (EFAs) and must be obtained from external sources [,]. LA and ALA are metabolized to arachidonic acid (ARA; 20:4, n-6) and eicosapentaenoic acid (EPA; 20:5, n-3), respectively, in a series of reactions catalyzed by the same sets of enzymes []. However, the metabolism of EFAs can be altered in several diseases, such as hypertension, diabetes mellitus, obesity, atherosclerosis, Alzheimer’s disease, schizophrenia, cancer, etc. Therefore, the physiological role of EFAs and their products is very important as they are involved in various biological reactions []. Hence, PUFAs are attracting significant research interest in recent times [].
Stearidonic acid (SDA; 18:4, n-3) is produced from gamma linolenic acid (GLA; 18:3, n-6) and ALA (18:3, n-3) catalyzed by the enzymes delta 15-desaturase and delta 6-desaturase, respectively (Figure 1) []. SDA can be elongated to a longer chain omega-3 PUFA with related biological properties, such as EPA and docosahexaenoic acid (DHA) []. However, conversion of SDA from ALA is very inefficient in humans because delta 6-desaturase is rate-limiting, therefore it is difficult to change tissue levels of EPA (20:5, n-3) by the consumption of ALA. Consequently, the oral administration of SDA has been suggested as an alternative approach of increasing the amount of EPA in body tissues [,]. Again, in comparison to EPA and DHA, SDA is less unsaturated, more stable, less prone to oxidation, and therefore more amenable to a wide variety of food and beverage applications [].
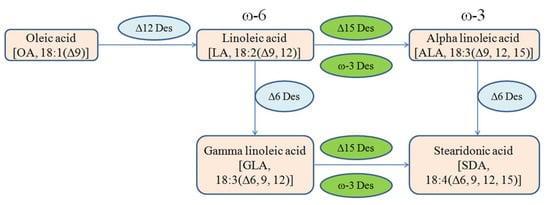
Figure 1.
Metabolic pathway of linoleic acid (LA) flux into n-6 or n-3 PUFAs to produce stearidonic acid (SDA).
SDA occurs naturally in seafood, fish and a few species of seaweed as a minor n-3 PUFA []. Among the plant sources, oils derived from blackcurrant seed and members of the Boraginaceae family contain SDA []. By weight, Echium oil is comparatively a better plant source of SDA and contains both ω-3 PUFA and ω-6 PUFA []. However, due to safety concerns, limited shelf-life, palatability issues and the risk of over-fishing, fish and fish oil are difficult to use as commercial source of (ω-3) LCPUFA [,]. Plant sources also need to be improved for large scale production due to their low yield. Therefore, there is a requirement to identify alternative sources of SDA that can be used commercially [].
Fungi, microalgae and bacteria that can accumulate lipids up to 20% of their dry cell weight are known as oleaginous microorganisms. Mucor circinelloides has been extensively investigated for GLA production since the 1980s []. It has been considered as an important classical organism for microbial lipid analysis because of its ability to produce oil that is rich in GLA as well as the availability of genome data and genetic tools []. In previous studies this fungus was used for improved GLA production [], increased lipid accumulation [], 13C-metabolic flux analysis of lipid accumulation [], investigation of the effects of 20 standard amino acids on growth, GLA and total fatty acids production [], the role of pentose phosphate pathway in lipid accumulation [], etc. Mortierella alpina, another filamentous fungus, can accumulate lipids up to 50% of its dry weight [,] and mostly composed of triacylglycerol with a high amount of arachidonic acid (AA; 20:4, n-6) [,], however at a low temperature it can also produce EPA []. In this experiment, the delta-15 desaturase (D15D) gene from M. alpina was cloned and recombined in M. circinelloides to make a SDA-producing cell factory.
2. Results
2.1. Generation of D15D-Overexpressing Strains of M. circinelloides by Genetic Engineering
According to the genomic data of M. alpina, this study found only one gene encoding for D15D (Genebank accession number KF433065), which is 1212 bp long. For overexpression of the target gene, the coding region of D15D was cloned into the M. circinelloides expression vector pMAT1552, which contained the strong zrt1 promoter and flanking sequence of carRP locus to allow integration of the whole over-expressing construction by homologous recombination []. Integration in carRP locus produced white colonies, which were easily distinguishable from yellow transformants that did not integrate the D15D gene. The target gene-overexpressing plasmids, pMAT1552-D15D, and the empty plasmids pMAT1552 were transformed into the uridine auxotrophic strain, pleu-MU402, and selection of the colonies was performed as reported by Rodríguez-Frómeta []. Three overexpressing and one control transformants named as Mc-D15D, Mc-D15D-1, Mc-D15D-2 and Mc-1552, respectively, were selected. Additional screening was carried out (data not shown) and only one strain (Mc-D15D) that produced a maximum amount of lipid and SDA was selected for further experiments.
PCR analysis was used to confirm the integration of the target gene in the genome of overexpressing transformants. A primer pair 1552-F/R (Table S1) was used to amplify the target gene and the 557 bp sequences of the plasmid pMAT1552. As expected, bands of 1796 bp of PCR products were seen on gels for transformants with the target gene but only 557 bp fragment was observed for Mc-1552 control strain (Figure 2). Thus, PCR amplification results confirmed the integration of the target gene in the genome of recombinant fungi.
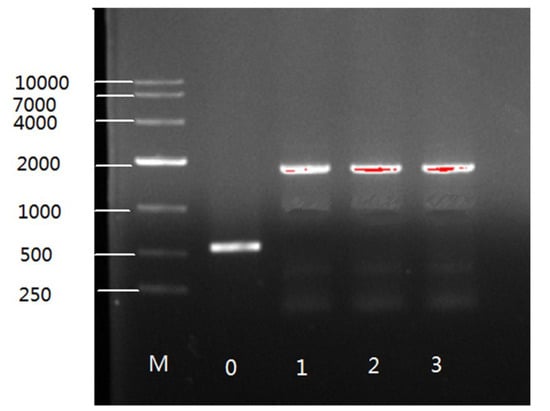
Figure 2.
PCR amplification of genome of control and recombinant strains. Band 0 representing the control strain and 1,2,3 showing the recombinant strains.
2.2. Expression Levels of D15D Genes in the Recombinant Strains
Real-time quantitative PCR were carried out to analyze the mRNA level of D15D in the recombinant strains at 3, 24, 48 and 72 h of growth in two liter fermenter with K & R medium (Figure 3). The mRNA expression level of Mc-D15D was considered as 1 at 3 h and by comparing with this value, the expression level increased in that strain by 5.10, 3.50 and 2.55 fold at 24, 48 and 72 h, respectively. Although it increased quickly from 3 to 24 h, there was a decreasing trend with the incubation time after 24 h. The fact that D15D mRNA was maintained at elevated levels throughout the culture time confirmed that it was overexpressed in the recombinant strains.
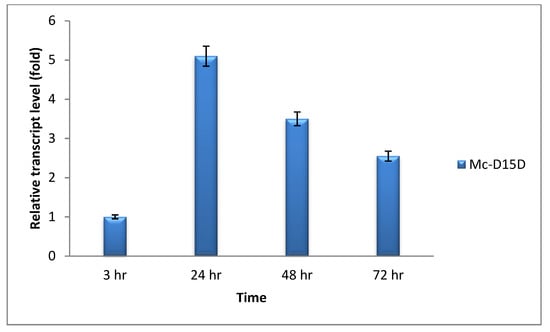
Figure 3.
Determination of expression levels of delta-15 desaturase (D15D) genes by RT-qPCR in the recombinant strains.
2.3. Effect of D15D Over-Expression in Cell Growth and Lipid Accumulation
The effect of the D15D over-expression in growth and lipid accumulation was analyzed in the strain Mc-D15D, as shown in Figure 4. The concentrations of ammonium and glucose in the culture medium are also highlighted in Figure 4. Adequate concentrations of glucose remained during the entire bioprocess (Figure 4a), however at approximately 24 h the ammonium was used up by Mc-D15D (Figure 4b). After 12 h of cultivation, cell dry weight (CDW) increased rapidly but slowed down after nitrogen depletion (Figure 4c). The fungus started to accumulate lipids immediately after nitrogen exhaustion from the culture medium. The total fatty acids (TFAs) content increased rapidly from 24 h, reaching its peak at 60 h and then slowed gradually. The maximum content of TFAs was found to be 15.53% in Mc-D15D (Figure 4d).
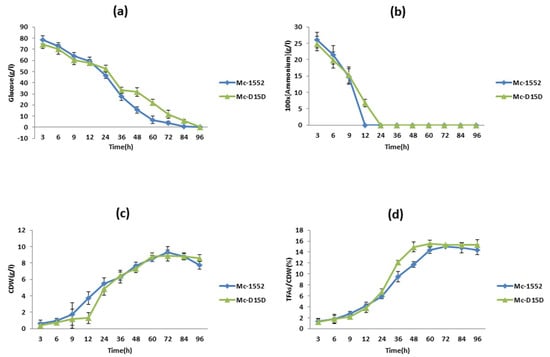
Figure 4.
Cell growth and lipid accumulation of D15D overexpressing strains. Recombinant Mc-D15D and control strain Mc-1552 cultures were grown in 1.5 L modified K&R medium and (a) glucose concentration, (b) ammonium concentration, (c) cell dry weight (CDW), and (d) lipid content were measured. Samples from the fermenter were taken at the indicated times. The values were mean of three biological replicates. Error bars represent the standard error of the mean.
2.4. SDA Accumulation in D15D Overexpressing Strains
The fatty acid composition in D15D genes overexpressing strains are presented in Table 1. In the transformants, SDA production started at 24 h, and at 48 h its content reached its peak (5.09%). From 36 to 72 h the SDA content remained elevated over 4%. In these recombinant fungi the other fatty acid contents were almost similar as found in the control strains.

Table 1.
The fatty acid composition in D15D overexpressing strains.
3. Discussion
The accumulation of very-long-chain polyunsaturated fatty acid (VLCPUFA) in recombinant microorganisms is a process which can further improve the accumulation of desired products []. The oleaginous fungus, M. circinelloides, is an attractive oleaginous fungus for researchers because of its ability to produce oil that is rich in gamma linolenic acid. On the other hand, M. alpina produces both ARA and EPA and possess both ω-6 and ω-3 biosynthetic pathways for the production of fatty acids. However, in all other features, both fungi share similar characteristics and phenotype (filamentous) []. Delta-15 desaturase (ω-3 desaturase) of M. alpina ATCC 32221, is reported to convert GLA to SDA and LA to ALA. ALA is converted to SDA by delta-6 desaturase, which is already present in M. circinelloides []. Therefore, SDA can be produced using both pathways that were constructed in this fungus by overexpressing D15D gene (Figure 1). Besides the production of SDA, a significant amount of ALA was also produced in this process. The RT-qPCR results revealed that D15D mRNA remained elevated during the entire culture time of Mc-D15D and fatty acid analysis revealed the presence of SDA. These results confirmed that D15D was overexpressed in the recombinant strains.
Deficiency of nitrogen (N) is a general approach to start the storage of lipid in most oleaginous microorganisms. Investigation of lipid accumulation is frequently carried out by comparing the N rich phase to the N deficiency phase in the whole culture time, and the quantity of lipid enhances under N depletion [,,]. The maximum concentration of TFAs is 15% in CBS 277.49 []. Similar trends and results were observed in the experiments of this study. Recombination and expression of D15D gene showed no significant effect on growth or total lipid content of this fungus.
In M. circinelloides the main fatty acids are 16:0, 18:0, 18:1, 18:2 (LA) and 18:3 (GLA) [,]. After overexpressing D15D gene, the same fatty acids were found as the major lipids and its expression did not affect the major lipid profile of this organism. SDA normally occurs only as a metabolic intermediate in the biosynthetic pathway of ω-3 fatty acids in higher plants or fungi and algae; it is not significantly accumulated in any organism []. In the recombinant Mc-D15D in this study, SDA is the end product of this pathway. Since M. circinelloides has been investigated extensively for GLA production since the 1980s [], it can also be used for SDA production industrially.
Previous research has confirmed that during the last 50 years the decreased consumption of ω-3 fatty acids is thought to be responsible for the increased occurrence of some diseases, such as hypertension, obesity, atherosclerosis, CHD, metabolic syndrome X, cancer, etc. The precursor of the prostaglandins of series 3 and the leukotrienes of series 5 are also produced from EPA []. Fish and sea food products are the major sources of these ω-3 PUFA, however due to their poor stability and chemical safety these are not frequently used in food processing []. Current plant sources also need to be improved for large scale production []. Scientists are trying to construct engineered SDA-producing microbes because of nutritional value and increased demand. Kimura et al. constructed genetically modified Saccharomyces cerevisiae which can produce up to 13% SDA when the fungus is supplied with increased histidine [,]. The recombinant M. circinelloides in this study can produce almost 5% SDA without any supplement and according to our knowledge, this is the first study describing the overexpression of D15D gene in M. circinelloides to construct SDA producing cell factory. However, more research is required to use this fungus for the industrial production of SDA.
4. Materials and Methods
4.1. Plasmids, Strains and Conditions of Culture Media
Mortierella alpina ATCC 32222 was used as the source of delta-15 desaturase (D15D) gene. The uracil auxotroph strain, pleu-MU402 of M. circinelloides CB277.49 [] was used as the recipient strain for recombination and overexpression of the desaturase gene. The expression vector pMAT1552 [] was used for gene cloning. Escherichia coli Top 10 was used to maintain and propagate recombinant plasmids, which was grown in lysogeny broth at 37 °C. Mortierella alpina ATCC 32222 was grown in a 1-L flask containing Potato Dextrose Water (PDW) for 72 h for mycelia collection. The initial cultivation of the recombinant strains Mc-D15D (D15D overexpresssion strains) and Mc-1552 (control strains carrying the vector pMAT1552) were carried out in 1-L flasks containing 150 mL K&R medium containing 30 g/L glucose, 3.3 g/L ammonium tartrate, 7.0 g/L KH2PO4, 2.0 g/L Na2HPO4, 1.5 g/L MgSO4·7H2O, 1.5 g/L yeast extract, 0.1 g/L CaCl2·2H2O, 8 mg/L FeCl3·6H2O, 1 mg/L ZnSO4·7H2O, 0.1 mg/L CuSO4·5H2O, 0.1 mg/L Co(NO3)2·6H2O and 0.1 mg/L MnSO4·5H2O, pH 6.0 [] for 24 h at 28 °C with shaking at 150 rpm. This initial culture was inoculated at 10% (v/v) into a 2-L fermenter (BioFlo/CelliGen115, New Brunswick Scientific, Edison, NJ, USA) where a modified K&R medium containing 80 g glucose/L was used as culture medium. The volume of medium was 1.5 L and fermenters were held at 28 °C, stirred at 700 rpm with aeration of 0.5 vvm. Automatic addition of 2 M NaOH was used to maintain the pH at 6.0.
4.2. Plasmids Construction
D15D-overexpressing plasmid for pleu-MU402 was constructed by using the expression vector pMAT1552, which contains the pyrG gene of M. circinelloides to encode orotidine 5′-phosphate decarboxylase to produce uridine as a selectable marker. It is surrounded up-stream and down-stream by 1 kb of carB-carRP sequences to allow its chromosomal integration by homologous recombination. PCR amplification was carried out to isolate the D15D gene from the genome of M. alpina ATCC 32222 using the primers D15D-F/R (Table S1). The 30 bp homologous sequences of both sides of XhoI restriction site in pMAT1552 were also included in these primers. After digestion with XhoI restriction endonuclease, the PCR fragment was ligated to generate plasmid pMAT1552-D15D (Figure 5) for mutant pleu-MU402. The recombinant plasmid was transformed into E. coli Top 10 competent cells for its propagation. The extracted plasmids from these bacteria were checked by PCR analysis and DNA sequencing was carried out to confirm the gene sequence.
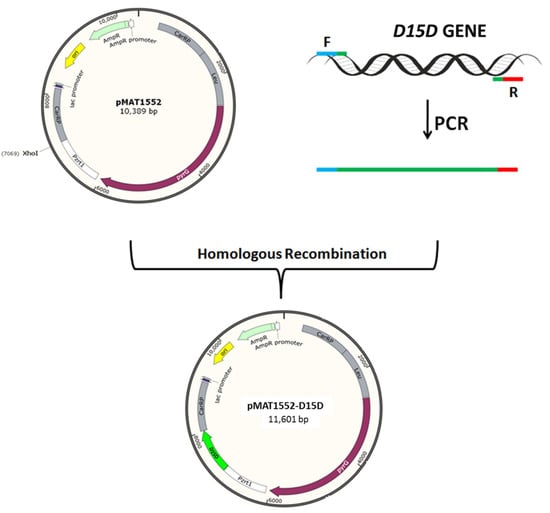
Figure 5.
The structure of plasmids pMAT1552 and pMAT1552-D15D. The D15D gene was isolated by PCR amplification with appropriate primers. The PCR fragment was ligated into XhoI restriction site to generate plasmid as pMAT1552-D15D.
4.3. Gene Expression and RT-qPCR Analysis
M. circenioides was cultivated in a 2-L fermenter using K&R medium and mycelium was collected at 3, 24, 48, and 72 h. Total RNA of M. circinelloides was extracted using Trizol after grinding mycelia under liquid N2 and reverse-transcribed by using the Prime ScriptRT reagent kit (Takara) as described by manufacturer’s instructions. LightCycler 96 Instrument (Roche Diagnostics GmbH, Switzerland) with FastStart Universal SYBR Green Master (ROX) Supermix (Roche) was used to perform RT-qPCR by using primers D15DqPCR-F/R (Table S1). The expression level of mRNA was standardized to levels of 18S rRNA mRNA, and the results were analyzed as relative expression levels. The method of 2−ΔΔC was used to quantify the obtained data.
4.4. Calculation of Glucose and Nitrogen Concentration from Culture Medium
The concentration glucose in the culture medium was measured by a glucose oxidase Perid-test kit according to the protocol supplied by the manufacturer (Shanghai Rongsheng Biotech Co., Ltd.). Ammonium concentration was determined by the indophenol method [].
4.5. Determination of Cell Dry Weight, Total Fatty Acid and Fatty Acid Analysis
Biomass samples were collected on a weighed filter paper by filtration through a Buchner funnel under reduced pressure. These were washed three times with distilled water, frozen at −80 °C overnight and then dried by freeze dryer. The weight of the empty fatty acid extraction tube was taken. The extraction of fatty acids was carried out using 20 mg dry mycelia. Chloroform/methanol (2:1, v/v) was used to extract total fatty acids. Finally, the final weight of tube with fatty acids was taken. Total fatty acid was determined using following equation:
Here, T1 = weight of tube with fatty acid; T0 = weight of empty tube; Wm = weight of mycelia.
For fatty acid analysis 10 mg of dry mycellia was taken. Fatty acid was extracted using the same method and methylated with 10% (v/v) methanolic HCl at 60 °C for 3 h. Pentadecanoic acid (15:0) (Sigma-Aldrich) was added into the freeze-dried cells as an internal standard and n-hexane were used to extract the fatty acid methyl esters, which were analyzed by GCFID equipped with a 30 m × 0.32 mm DB-Waxetr column with 0.25 μm film thickness. The program was as follows: 120 °C for 3 min, ramp to 200 °C at 5 °C per min, ramp to 220 °C at 4 °C per min, hold 2 min.
4.6. Statistical Analysis
SPSS 16.0 for Windows (SPSS Inc. Chicago, IL) software was used to complete the statistical analysis. Three independent experiments were carried out and the data obtained from these experiments were used to calculate the mean values and standard error of the mean. The differences between means of the test were calculated by Student’s t test, and p < 0.05 was considered as significantly different.
4.7. Ethics Approval and Consent to Participate
Any experiments with human or animal participants were not performed by any of the authors.
5. Conclusions
This study constructed a SDA-producing M. circinelloides CBS 277.49 by D15D gene overexpression. According to our knowledge, this is the first study about the construction of a SDA-producing strain by gene cloning and recombination in Mucor. This work establishes a new scope for further research for improved production of SDA in M. circinelloides, specifically in its high lipid-producing strain, WJ11.
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/20/7/1683/s1, Table S1. The sequences of primers used in this work.
Author Contributions
Data curation, M.A.K.K., J.Y. and S.A.H.; formal analysis, M.A.K.K. and J.Y.; investigation, H.Z.; methodology, V.G. and Y.S.; writing–original draft, M.A.K.K.; writing–review and editing, V.G. and Y.S.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31670064), the Natural Science Foundation of Shandong province (ZR2018BC058), the Science and Technology Project of Shandong College (No. J16LE20 and J17KA133), the Taishan Industry Leading Talent Project, and a starting grant from Shandong University of Technology.
Acknowledgments
We are grateful to Muhammad Hafiy Bin Mohd Halim and Mohamed Yusuf Bin Mahamed Nazir for their assistance to draw pictures and data analysis.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| SDA | Stearidonic acid |
| LA | Linoleic acid |
| ALA | Alpha linolenic acid |
| GLA | Gamma linolenic acid |
| ARA | Arachidonic acid |
| EPA | Eicosapentaenoic acid |
| DHA | Docosahexaenoic acid |
| PUFA | Polyunsaturated fatty acid |
| GC | Gas chromatography |
| VLCPUFA | Very-long-chain polyunsaturated fatty acid |
| CDW | Cell dry weight |
| SAFA | Saturated fatty acids |
| MUFA | Mono-unsaturated fatty acids |
| TFA | Total fatty acids |
References
- Yazawa, H.; Iwahashi, H.; Kamisaka, Y.; Kimura, K.; Aki, T.; Ono, K.; Uemura, H. Heterologous Production of Dihomo-gamma-Linolenic Acid in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2007, 73, 6965–6971. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Simopoulos, A.P. Essential fatty acids in health and chronic disease. Am. J. Clin. Nutr. 1999, 70, 560S–569S. [Google Scholar] [CrossRef]
- Elrazak, A.A.; Ward, A.C.; Glassey, J. Response surface methodology for optimising the culture conditions for eicosapentaenoic acid production by marine bacteria. J. Ind. Microbiol. Biotechnol. 2013, 40, 477. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Chen, J.; Zhang, H.; Song, Y. Molecular switch that controls the flux of linolenic acid into n-6 or n-3 polyunsaturated fatty acids in microorganisms. Am. J. Biochem. Biotechnol. 2014, 10, 105–115. [Google Scholar] [CrossRef]
- Das, U.N. Essential fatty acids: Biochemistry, physiology and pathology. Biotechnol. J. 2006, 1, 420–439. [Google Scholar] [CrossRef]
- Harris, W.S. Stearidonic Acid–Enhanced Soybean Oil: A Plant-Based Source of (n-3) Fatty Acids for Foods. J. Nutr. 2012, 142, 600S–604S. [Google Scholar] [CrossRef]
- Baik, J.Y.; Kim, N.H.; Oh, S.; Kim, I. Preparation of Highly Purified Stearidonic Acid from Echium Oil via an Enzymatic Method Combined with Preparative High Performance Liquid Chromatography. J. Oleo Sci. 2015, 64, 729–736. [Google Scholar] [CrossRef] [PubMed]
- Greg, C. Vegetarian Omegas: The Importance of SDA. 2015. Available online: http://www.ahiflower.com/wp-content/uploads/2015/06/NPI-Veg-omega-3s-SDA-Sep-2015.pdf (accessed on 28 December 2018).
- Abeywardena, M.Y.; Adams, M.; Dallimore, J.; Kitessa, S.M. Rise in DPA Following SDA-Rich Dietary Echium Oil Less Effective in Affording Anti-Arrhythmic Actions Compared to High DHA Levels Achieved with Fish Oil in Sprague-Dawley Rats. Nutrients 2016, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Petrik, M.B.H.; McEntee, M.F.; Johnson, B.T.; Obukowicz, M.G.; Whelan, J. Highly unsaturated (n-3) fatty acids, but not α-linolenic, conjugated linoleic or γ-linolenic acids, reduce tumorigenesis in ApcMin/+ mice. J. Nutr. 2000, 130, 2434–2443. [Google Scholar] [CrossRef] [PubMed]
- Bilgiç, S.; Yeşilçubuk, N.Ş. Lipase-catalyzed acidolysis of olive oil with echium oil stearidonic acid: Optimization by response surface methodology. J. Am. Oil Chem. Soc. 2012, 89, 1971–1980. [Google Scholar] [CrossRef]
- McIntyre, P.B.; Jones, L.E.; Flecker, A.S.; Vanni, M.J. Fish extinctions alter nutrient recycling in tropical freshwaters. Proc. Natl. Acad. Sci. USA 2007, 104, 4461–4466. [Google Scholar] [CrossRef]
- Taylor, B.W.; Flecker, A.S.; Hall, R.O. Loss of a harvested fish species disrupts carbon flow in a diverse tropical river. Science 2006, 313, 833–836. [Google Scholar] [CrossRef]
- Whelan, J. Dietary Stearidonic Acid Is a Long Chain (n-3) Polyunsaturated Fatty Acid with Potential Health Benefits. J. Nutr. 2009, 139, 5–10. [Google Scholar] [CrossRef]
- Zhang, Y.; Luan, X.; Zhang, H.; Garre, V.; Song, Y.; Ratledge, C. Improved γ-linolenic acid production in Mucor circinelloides by homologous overexpressing of delta-12 and delta-6 desaturases. Microb. Cell Fact. 2017, 16, 113. [Google Scholar] [CrossRef]
- Tang, X.; Zan, X.; Zhao, L.; Chen, H.; Chen, Y.Q.; Chen, W.; Song, Y.; Ratledge, C. Proteomics analysis of high lipid-producing strain Mucor circinelloides WJ11: An explanation for the mechanism of lipid accumulation at the proteomic level. Microb. Cell Fact. 2016, 15, 35. [Google Scholar] [CrossRef]
- Yang, J.; Li, S.; Khan, M.A.K.; Garre, V.; Vongsangnak, W.; Song, Y. Increased Lipid Accumulation in Mucor circinelloides by Overexpression of Mitochondrial Citrate Transporter Genes. Ind. Eng. Chem. Res. 2019, 58, 2125–2134. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, H.; Wang, L.; Chen, H.; Chen, Y.Q.; Chen, W.; Song, Y. 13C-metabolic flux analysis of lipid accumulation in the oleaginous fungus Mucor circinelloides. Bioresour. Technol. 2015, 197, 23–29. [Google Scholar] [CrossRef]
- Tang, X.; Zhang, H.; Chen, H.; Chen, Y.Q.; Chen, W.; Song, Y. Effects of 20 Standard Amino Acids on the Growth, Total Fatty Acids Production, and c-Linolenic Acid Yield in Mucor circinelloides. Curr. Microbiol. 2014, 69, 899–908. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Tang, X.; Luan, X.; Chen, H.; Chen, Y.Q.; Chen, W.; Song, Y.; Ratledge, C. Role of pentose phosphate pathway in lipid accumulation of oleaginous fungus Mucor circinelloides. RSC Adv. 2015, 5, 97658. [Google Scholar] [CrossRef]
- Bajpai, P.K.; Bajpai, P.; Ward, O.P. Arachidonic Acid Production by Fungi. Appl. Environ. Microbiol. 1991, 57, 1255–1258. [Google Scholar]
- Ratledge, C.; Wynn, J.P. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv. Appl. Microbiol. 2002, 51, 1–51. [Google Scholar]
- Jang, H.D.; Lin, Y.Y.; Yang, S.S. Effect of culture media and conditions on polyunsaturated fatty acids production by Mortierella alpina. Bioresour. Technol. 2005, 96, 1633–1644. [Google Scholar] [CrossRef]
- Ratledge, C. Fatty acid biosynthesis in microorganisms being used for Single Cell Oil production. Biochimie 2004, 86, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Michaelson, L.V.; Lazarus, C.M.; Griffiths, G.; Napieri, J.A.; Stobart, A.K. Isolation of a Δ5-Fatty Acid Desaturase Gene from Mortierella alpina. J. Biol. Chem. 1998, 273, 19055–19059. [Google Scholar] [CrossRef]
- Zhao, L.; Cánovas-Márquez, J.T.; Tang, X.; Chen, H.; Chen, Y.Q.; Chen, W.; Garre, V.; Song, Y.; Ratledge, C. Role of malate transporter in lipid accumulation of oleaginous fungus Mucor circinelloides. Appl. Microbiol. Biotechnol. 2016, 100, 1297–1305. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Frómeta, R.A.; Gutiérrez, A.; Torres-Martínez, S.; Garre, V. Malic enzyme activity is not the only bottleneck for lipid accumulation in the oleaginous fungus Mucor circinelloides. Appl. Microbiol. Biotechnol. 2013, 97, 3063–3072. [Google Scholar] [CrossRef] [PubMed]
- Tavares, S.; Grotkjaer, T.; Obsen, T.; Haslam, R.P.; Napier, J.A.; Gunnarsson, N. Metabolic Engineering of Saccharomyces cerevisiae for Production of Eicosapentanoic Acid, Using a Novel Δ5-Desaturase from Paramecium tetraurelia. Appl. Environ. Microbiol. 2011, 77, 1854–1861. [Google Scholar] [CrossRef] [PubMed]
- Sakuradani, E.; Murata, S.; Kanamaru, H.; Shimizu, S. Functional analysis of a fatty acid elongase from arachidonic acid-producing Mortierella alpina 1S-4. Appl. Microbiol. Biotechnol. 2008, 81, 497–503. [Google Scholar] [CrossRef]
- Wynn, J.P.; bin Abdul Hamid, A.; Ratledge, C. The role of malic enzyme in the regulation of lipid accumulation in filamentous fungi. Microbiology 1999, 145, 1911–1917. [Google Scholar] [CrossRef]
- Yang, Z.K.; Ma, Y.H.; Zheng, J.W.; Yang, W.D.; Liu, J.S.; Li, H.Y. Proteomics to reveal metabolic network shifts towards lipid accumulation following nitrogen deprivation in the diatom Phaeodactylum tricornutum. J. Appl. Phycol. 2014, 26, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Hao, G.; Wang, L.; Wang, H.; Gu, Z.; Liu, L.; Zhang, H.; Chen, W.; Chen, Y.Q. Identification of a critical determinant that enables efficient fatty acid synthesis in oleaginous fungi. Sci. Rep. 2015, 5, 11247. [Google Scholar] [CrossRef]
- Hussain, S.A.; Hameed, A.; Khan, M.A.K.; Zhang, Y.; Zhang, H.; Garre, V.; Song, Y. Engineering of fatty acid synthases (FASs) to boost the production of medium-chain fatty acids (MCFAs) in Mucor circinelloides. Int. J. Mol. Sci. 2019, 20, 786. [Google Scholar] [CrossRef]
- Das, U.N. Essential fatty acids and their metabolites could function as endogenous HMG-CoA reductase and ACE enzyme inhibitors, anti-arrhythmic, anti-hypertensive, anti-atherosclerotic, anti-inflammatory, cytoprotective, and cardioprotective molecules. Lipids Health Dis. 2008, 7, 37. [Google Scholar] [CrossRef]
- Ursin, V.M. Modification of plant lipids for human health: Development of functional land-based omega-3 fatty acids. J. Nutr. 2003, 133, 4271–4274. [Google Scholar] [CrossRef] [PubMed]
- Kimura, K.; Kamisaka, Y.; Uemura, H.; Yamaoka, M. Increase in stearidonic acid by increasing the supply of histidine to oleaginous Saccharomyces cerevisiae. J. Biosci. Bioeng. 2014, 117, 53–56. [Google Scholar] [CrossRef]
- Kimura, K.; Tomita, N.; Uemura, H.; Aki, T.; Ono, K.; Kamisaka, Y. Improvement of Stearidonic Acid Production in Oleaginous Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 2009, 73, 1447–1449. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, F.E.; de Haro, J.P.; Torres-Martinez, S.; Ruiz-Vazquez, R.M. Mutants defective in a Mucor circinelloides dicer-like gene are not compromised in siRNA silencing but display developmental defects. Fungal Genet. Biol. 2007, 44, 504–516. [Google Scholar] [CrossRef] [PubMed]
- Roncero, M.I.; Jepsen, L.P.; Stroman, P.; van Heeswijck, R. Characterization of a leuA gene and an ARS element from Mucor circinelloides. Gene 1989, 84, 335–343. [Google Scholar] [CrossRef]
- Kendrick, A.; Ratledge, C. Desaturation of polyunsaturated fatty acids in Mucor circinelloides and the involvement of a novel membrane-bound malic enzyme. Eur. J. Biochem. 1992, 209, 667–673. [Google Scholar] [CrossRef]
- Chaney, A.L.; Marbach, E.P. Modified reagents for determination of urea and ammonia. Clin. Chem. 1962, 8, 130–132. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).